Bridging Discovery and Treatment: Cancer Biomarker
Simple Summary
Abstract
1. Introduction
2. Current Cancer Biomarkers and Detection Modalities
2.1. Sample Type
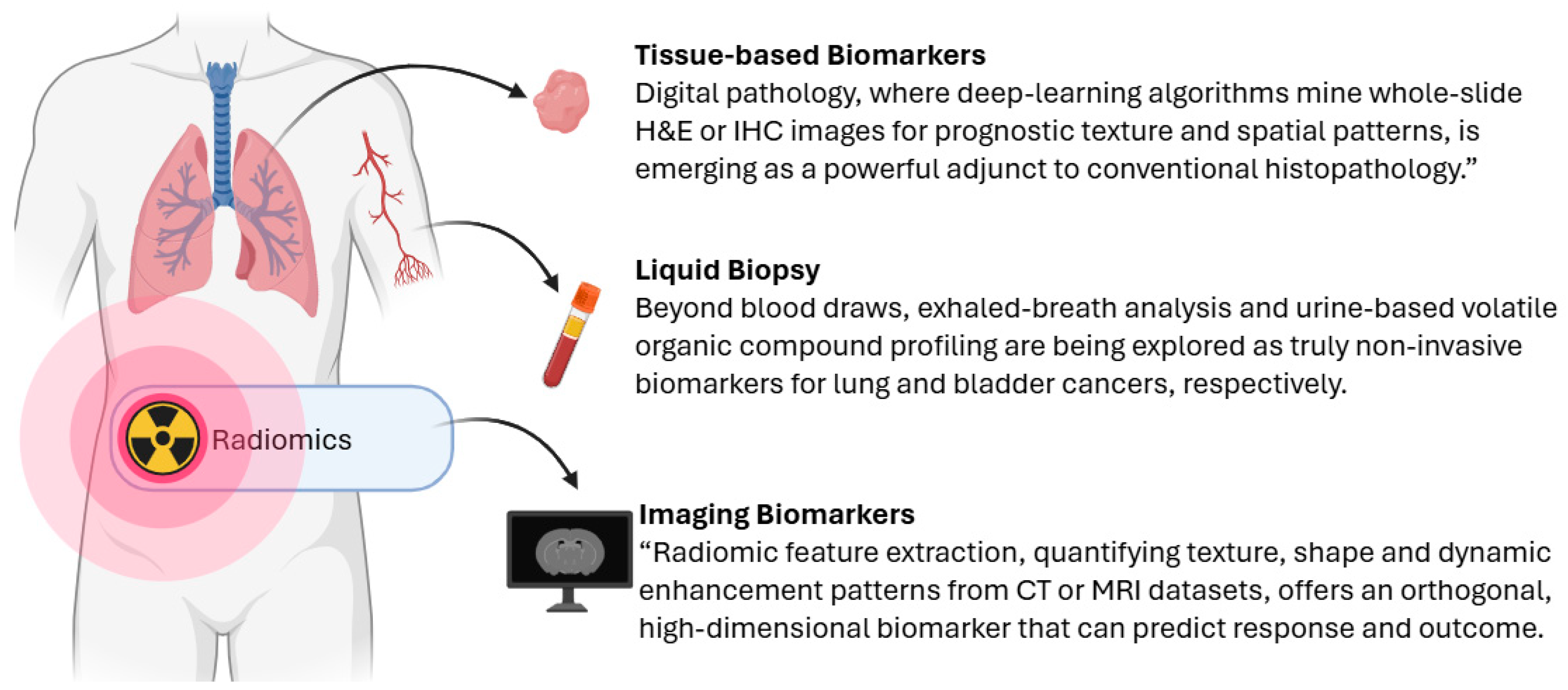
2.1.1. Tissue-Based Biomarkers
2.1.2. Liquid Biopsy
2.1.3. Imaging Biomarkers
2.1.4. Other Clinical Information
2.2. Detection “Dimensions” of Biomarkers
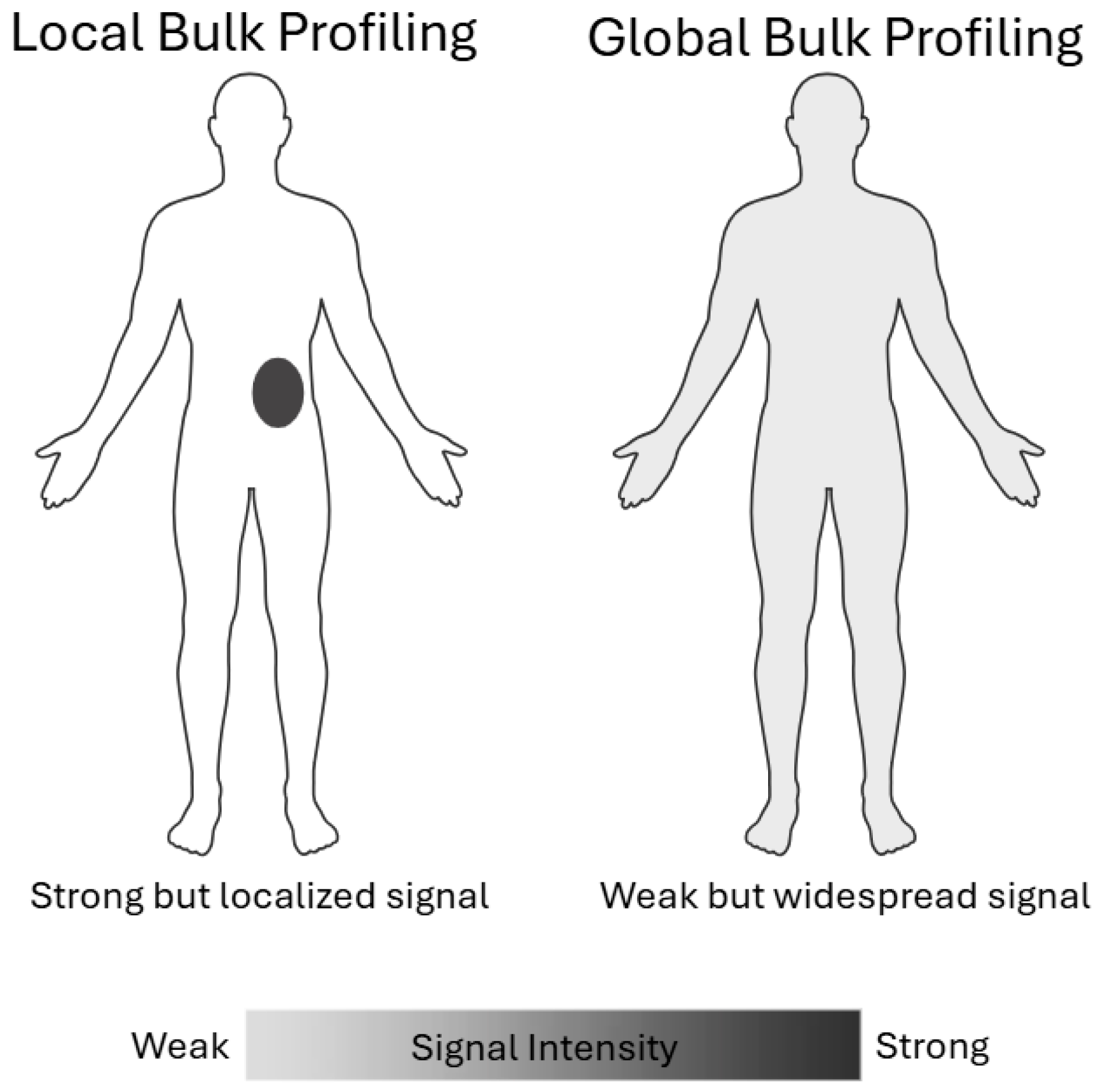
2.2.1. Local Bulk Profiling Versus Global Bulk Profiling
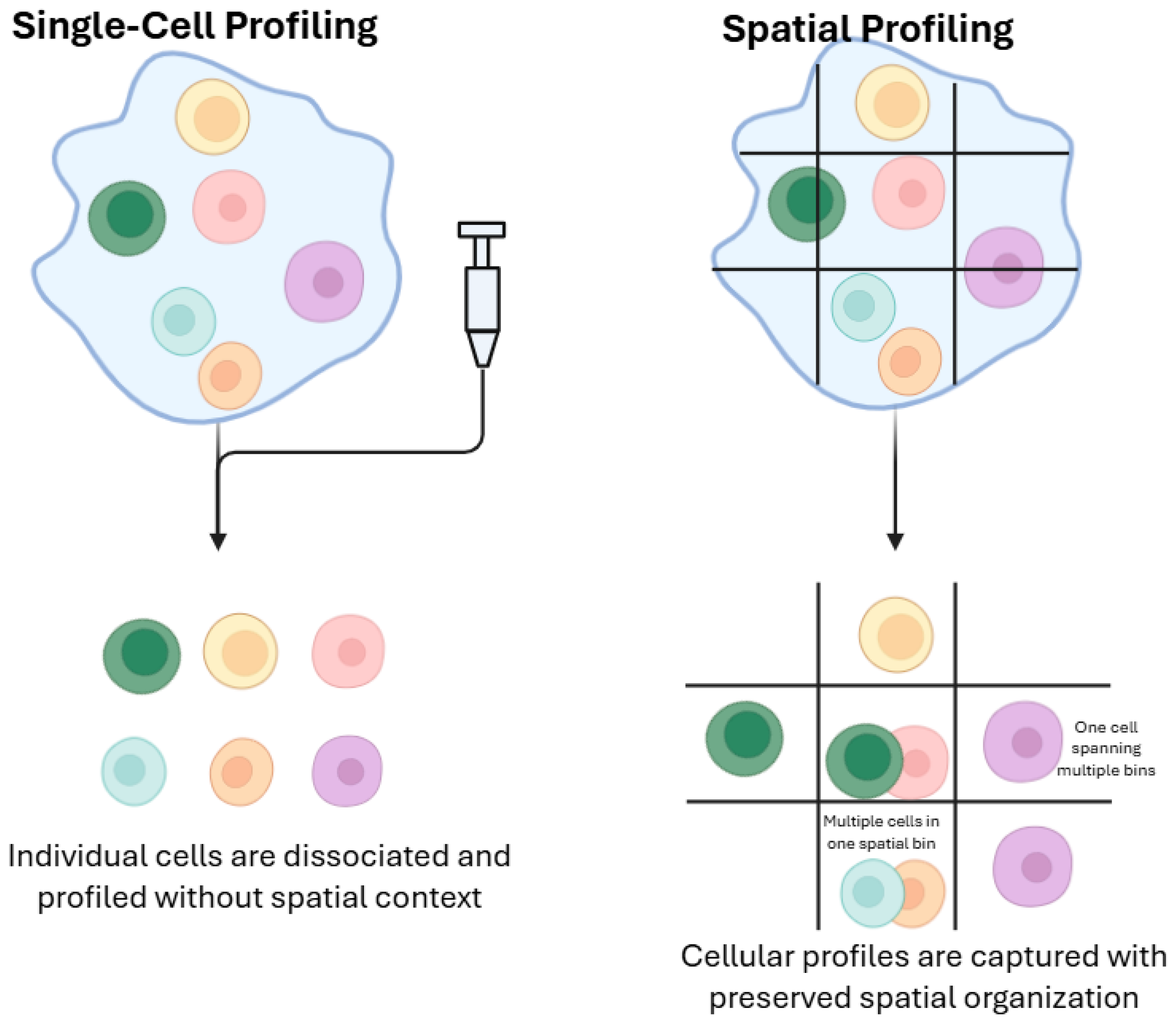
2.2.2. Single-Cell Profiling Versus Spatial Profiling
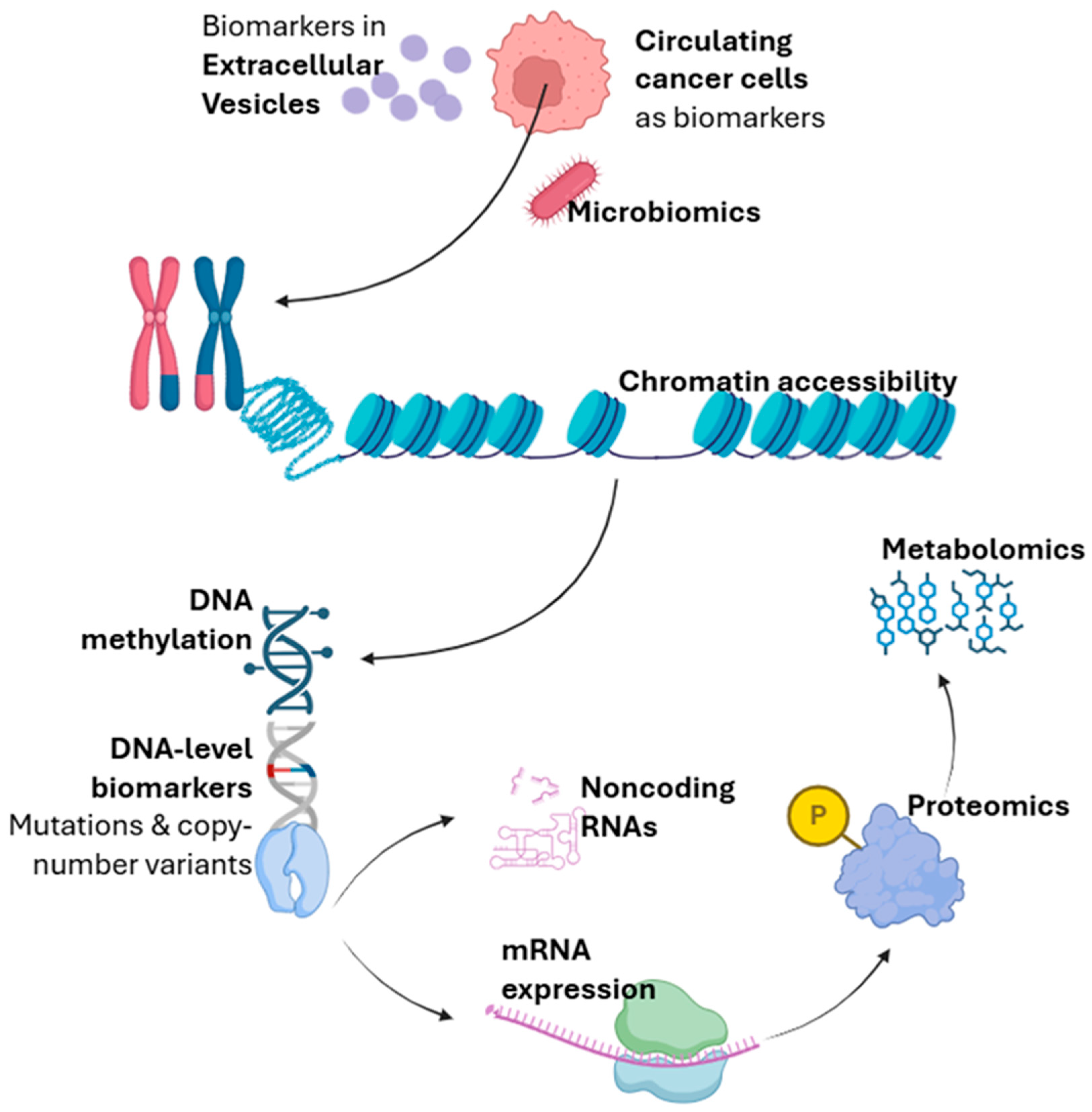
2.3. Detection Molecular Modality
2.3.1. DNA-Level Biomarkers
2.3.2. RNA-Level Biomarkers
2.3.3. Epigenetics
2.3.4. Proteomics
2.3.5. Metabolomics
2.3.6. Microbiomics
3. Clinical Applications of Biomarkers
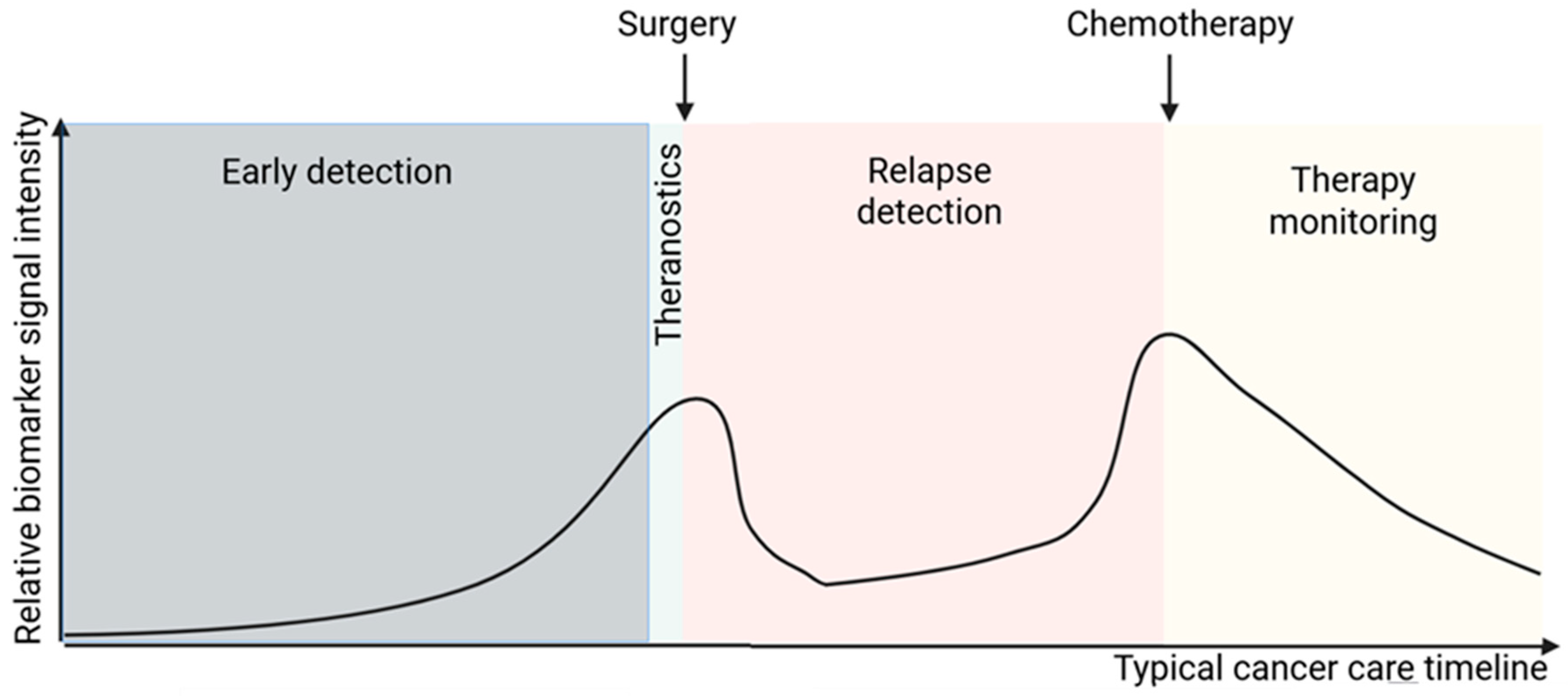
3.1. Early Detection
3.2. Theranostics
3.3. Monitoring and Surveillance
3.4. Prion-Based Detection Modalities
3.5. Summary of Biomarker Suitability Across Clinical Contexts
4. Impact of Biomarkers on Treatment Decisions
4.1. Biomarker-Driven Therapeutic Target Identification
4.2. Resistance Biomarkers
4.3. Early Intervention
4.4. Real-Time Intervention upon Relapse
5. Translational Challenges
5.1. Biomarker Validation and Clinical Trial Design
5.2. Regulatory Approval and IVD Registration
5.3. Standardization and Quality Control
5.4. Privacy and Ethical Issues
5.5. Controversies and Consensus
5.6. Leakage and the Reproducibility Crisis in Machine-Learning-Based Biomarkers
6. Personalized Precision Oncology Strategies
6.1. Intra- and Inter-Tumor Heterogeneity
6.2. Depth and Breadth of Detection: Multi-Omics Integration
6.3. Cost-Effectiveness and Clinical Accessibility
6.4. AI and Big-Data-Driven Personalization
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shin, S.H.; Bode, A.M.; Dong, Z. Addressing the challenges of applying precision oncology. Npj Precis. Oncol. 2017, 1, 28. [Google Scholar] [CrossRef]
- Azuaje, F. Artificial intelligence for precision oncology: Beyond patient stratification. Npj Precis. Oncol. 2019, 3, 6. [Google Scholar] [CrossRef]
- Cho, B.C.; Felip, E.; Hayashi, H.; Thomas, M.; Lu, S.; Besse, B.; Sun, T.; Martinez, M.; Sethi, S.N.; Shreeve, S.M.; et al. MARIPOSA: Phase 3 study of first-line amivantamab + lazertinib versus osimertinib in EGFR-mutant non-small-cell lung cancer. Future Oncol. 2022, 18, 639–647. [Google Scholar] [CrossRef]
- He, J.; Huang, Z.; Han, L.; Gong, Y.; Xie, C. Mechanisms and management of 3rd-generation EGFR-TKI resistance in advanced non-small cell lung cancer (Review). Int. J. Oncol. 2021, 59, 1–20. [Google Scholar] [CrossRef]
- Zou, J.; Wang, E. Cancer Biomarker Discovery for Precision Medicine: New Progress. Curr. Med. Chem. 2019, 26, 7655–7671. [Google Scholar] [CrossRef]
- Wu, L.; Qu, X. Cancer biomarker detection: Recent achievements and challenges. Chem. Soc. Rev. 2015, 44, 2963–2997. [Google Scholar] [CrossRef] [PubMed]
- Mäbert, K.; Cojoc, M.; Peitzsch, C.; Kurth, I.; Souchelnytskyi, S.; Dubrovska, A. Cancer biomarker discovery: Current status and future perspectives. Int. J. Radiat. Biol. 2014, 90, 659–677. [Google Scholar] [CrossRef]
- Pessoa, L.S.; Heringer, M.; Ferrer, V.P. ctDNA as a cancer biomarker: A broad overview. Crit. Rev. Oncol. Hematol. 2020, 155, 103109. [Google Scholar] [CrossRef]
- Dermani, F.K.; Samadi, P.; Rahmani, G.; Kohlan, A.K.; Najafi, R. PD-1/PD-L1 immune checkpoint: Potential target for cancer therapy. J. Cell Physiol. 2019, 234, 1313–1325. [Google Scholar] [CrossRef] [PubMed]
- Thunnissen, E.; de Langen, A.J.; Smit, E.F. PD-L1 IHC in NSCLC with a global and methodological perspective. Lung Cancer 2017, 113, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, E.; Feld, E.; Horn, L. Driven by Mutations: The Predictive Value of Mutation Subtype in EGFR-Mutated Non-Small Cell Lung Cancer. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2017, 12, 612–623. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Soda, M.; Togashi, Y.; Suzuki, R.; Sakata, S.; Hatano, S.; Asaka, R.; Hamanaka, W.; Ninomiya, H.; Uehara, H.; et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012, 18, 378–381. [Google Scholar] [CrossRef]
- Jiang, T.; Wang, G.; Liu, Y.; Feng, L.; Wang, M.; Liu, J.; Chen, Y.; Ouyang, L. Development of small-molecule tropomyosin receptor kinase (TRK) inhibitors for NTRK fusion cancers. Acta Pharm. Sin. B 2021, 11, 355–372. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharmacol. Res. 2019, 139, 395–411. [Google Scholar] [CrossRef]
- Kashima, K.; Kawauchi, H.; Tanimura, H.; Tachibana, Y.; Chiba, T.; Torizawa, T.; Sakamoto, H. CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation. Mol. Cancer Ther. 2020, 19, 2288–2297. [Google Scholar] [CrossRef]
- Kumaki, Y.; Oda, G.; Ikeda, S. Targeting MET Amplification: Opportunities and Obstacles in Therapeutic Approaches. Cancers 2023, 15, 4552. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, L. Updates on liquid biopsies in neuroblastoma for treatment response, relapse and recurrence assessment. Cancer Genet. 2024, 288–289, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Ling, S.; Zheng, S.; Xu, X. Liquid biopsy in hepatocellular carcinoma: Circulating tumor cells and circulating tumor DNA. Mol. Cancer 2019, 18, 114. [Google Scholar] [CrossRef]
- Wolf, J.; Rasmussen, D.K.; Sun, Y.J.; Vu, J.T.; Wang, E.; Espinosa, C.; Bigini, F.; Chang, R.T.; Montague, A.A.; Tang, P.H.; et al. Liquid-biopsy proteomics combined with AI identifies cellular drivers of eye aging and disease in vivo. Cell 2023, 186, 4868–4884.e12. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Rong, Z.; Wang, G.; Hou, Y.; Yang, F.; Qiu, M. Cancer metabolites: Promising biomarkers for cancer liquid biopsy. Biomark. Res. 2023, 11, 66. [Google Scholar] [CrossRef]
- Ren, F.; Fei, Q.; Qiu, K.; Zhang, Y.; Zhang, H.; Sun, L. Liquid biopsy techniques and lung cancer: Diagnosis, monitoring and evaluation. J. Exp. Clin. Cancer Res. 2024, 43, 96. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, S.; Wang, M.; Lopez-Beltran, A. Biological and clinical perspectives of TERT promoter mutation detection on bladder cancer diagnosis and management. Hum. Pathol. 2023, 133, 56–75. [Google Scholar] [CrossRef] [PubMed]
- Parihar, A.S.; Dehdashti, F.; Wahl, R.L. FDG PET/CT-based Response Assessment in Malignancies. Radiographics 2023, 43, e220122. [Google Scholar] [CrossRef]
- Kim, E.; Lee, E.; Plummer, C.; Gil, S.; Popel, A.S.; Pathak, A.P. Vasculature-specific MRI reveals differential anti-angiogenic effects of a biomimetic peptide in an orthotopic breast cancer model. Angiogenesis 2015, 18, 125–136. [Google Scholar] [CrossRef]
- Kong, Z.; Yan, C.; Zhu, R.; Wang, J.; Wang, Y.; Wang, Y.; Wang, R.; Feng, F.; Ma, W. Imaging biomarkers guided anti-angiogenic therapy for malignant gliomas. Neuroimage Clin. 2018, 20, 51–60. [Google Scholar] [CrossRef]
- Kim, Y.N.; Lee, H.Y.; Lee, K.S.; Seo, J.B.; Chung, M.J.; Ahn, M.J.; Park, K.; Kim, T.S.; Yi, C.A. Dual-energy CT in patients treated with anti-angiogenic agents for non-small cell lung cancer: New method of monitoring tumor response? Korean J. Radiol. 2012, 13, 702–710. [Google Scholar] [CrossRef]
- Templeton, A.J.; McNamara, M.G.; Šeruga, B.; Vera-Badillo, F.E.; Aneja, P.; Ocaña, A.; Leibowitz-Amit, R.; Sonpavde, G.; Knox, J.J.; Tran, B.; et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: A systematic review and meta-analysis. J. Natl. Cancer Inst. 2014, 106, dju124. [Google Scholar] [CrossRef]
- Ding, J.; Karp, J.E.; Emadi, A. Elevated lactate dehydrogenase (LDH) can be a marker of immune suppression in cancer: Interplay between hematologic and solid neoplastic clones and their microenvironments. Cancer Biomark. Sect. A Dis. Markers 2017, 19, 353–363. [Google Scholar] [CrossRef]
- Schorr, L.; Mathies, M.; Elinav, E.; Puschhof, J. Intracellular bacteria in cancer-prospects and debates. NPJ Biofilms Microbiomes 2023, 9, 76. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, J.Z.; Liu, Z.; Wei, J.F. The bacteria inside human cancer cells: Mainly as cancer promoters. Front. Oncol. 2022, 12, 897330. [Google Scholar] [CrossRef]
- Putzu, C.; Serra, R.; Campus, R.; Fadda, G.M.; Sini, C.; Marongiu, A.; Ginesu, G.C.; Fois, A.G.; Palmieri, G.; Zinellu, A.; et al. Complete Blood Count-Based Biomarkers as Predictors of Clinical Outcomes in Advanced Non-Small Cell Lung Cancer Patients with PD-L1 < 50% Treated with First-Line Chemoimmunotherapy. Curr. Oncol. 2024, 31, 4955–4967. [Google Scholar]
- Larroquette, C.A.; Hortobagyi, G.N.; Buzdar, A.U.; Holmes, F.A. Subclinical hepatic toxicity during combination chemotherapy for breast cancer. JAMA 1986, 256, 2988–2990. [Google Scholar] [CrossRef]
- Liu, H.; Li, Y.; Karsidag, M.; Tu, T.; Wang, P. Technical and Biological Biases in Bulk Transcriptomic Data Mining for Cancer Research. J. Cancer 2025, 16, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, A.; Liu, M.C.; Klein, E.A.; Venn, O.; Hubbell, E.; Beausang, J.F.; Gross, S.; Melton, C.; Fields, A.P.; Liu, Q.; et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell 2022, 40, 1537–1549.e12. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dong, A.; Rasteh, A.M.; Wang, P.; Weng, J. Identification of the novel exhausted T cell CD8 + markers in breast cancer. Sci. Rep. 2024, 14, 19142. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, W.C.; Budiarto, B.R.; Wang, Y.F.; Lin, C.Y.; Gwo, M.C.; So, D.K.; Tzeng, Y.S.; Chen, S.Y. Spatial multi-omics analyses of the tumor immune microenvironment. J. Biomed. Sci. 2022, 29, 96. [Google Scholar] [CrossRef]
- Huang, L.T.; Zhang, S.L.; Han, C.B.; Ma, J.T. Impact of EGFR exon 19 deletion subtypes on clinical outcomes in EGFR-TKI-Treated advanced non-small-cell lung cancer. Lung Cancer 2022, 166, 9–16. [Google Scholar] [CrossRef]
- Wang, X.; Qin, Z.; Qiu, W.; Xu, K.; Bai, Y.; Zeng, B.; Ma, Y.; Yang, S.; Shi, Y.; Fan, Y. Novel EGFR inhibitors against resistant L858R/T790M/C797S mutant for intervention of non-small cell lung cancer. Eur. J. Med. Chem. 2024, 277, 116711. [Google Scholar] [CrossRef]
- Zhao, Y.; He, Y.; Wang, W.; Cai, Q.; Ge, F.; Chen, Z.; Zheng, J.; Zhang, Y.; Deng, H.; Chen, Y.; et al. Efficacy and safety of immune checkpoint inhibitors for individuals with advanced EGFR-mutated non-small-cell lung cancer who progressed on EGFR tyrosine-kinase inhibitors: A systematic review, meta-analysis, and network meta-analysis. Lancet Oncol. 2024, 25, 1347–1356. [Google Scholar] [CrossRef]
- Prahallad, A.; Sun, C.; Huang, S.; Di Nicolantonio, F.; Salazar, R.; Zecchin, D.; Beijersbergen, R.L.; Bardelli, A.; Bernards, R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012, 483, 100–103. [Google Scholar] [CrossRef]
- Ueda, K.; Yamada, T.; Ohta, R.; Matsuda, A.; Sonoda, H.; Kuriyama, S.; Takahashi, G.; Iwai, T.; Takeda, K.; Miyasaka, T.; et al. BRAF V600E mutations in right-side colon cancer: Heterogeneity detected by liquid biopsy. Eur. J. Surg. Oncol. 2022, 48, 1375–1383. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, H.; Wang, D.; Hu, Y.; Yu, M.; Zhang, Q.; Qin, N.; Zhang, X.; Li, X.; Zhang, H.; et al. Dynamic cfDNA Analysis by NGS in EGFR T790M-Positive Advanced NSCLC Patients Failed to the First-Generation EGFR-TKIs. Front. Oncol. 2021, 11, 643199. [Google Scholar] [CrossRef]
- Su, K.Y.; Tseng, J.S.; Liao, K.M.; Yang, T.Y.; Chen, K.C.; Hsu, K.H.; Yang, P.C.; Yu, S.L.; Chang, G.C. Mutational monitoring of EGFR T790M in cfDNA for clinical outcome prediction in EGFR-mutant lung adenocarcinoma. PLoS ONE 2018, 13, e0207001. [Google Scholar] [CrossRef]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 131. [Google Scholar] [CrossRef]
- Oh, D.Y.; Bang, Y.J. HER2-targeted therapies—A role beyond breast cancer. Nat. Reviews Clin. Oncol. 2020, 17, 33–48. [Google Scholar] [CrossRef]
- Janjigian, Y.Y.; Kawazoe, A.; Yañez, P.; Li, N.; Lonardi, S.; Kolesnik, O.; Barajas, O.; Bai, Y.; Shen, L.; Tang, Y.; et al. The KEYNOTE-811 trial of dual PD-1 and HER2 blockade in HER2-positive gastric cancer. Nature 2021, 600, 727–730. [Google Scholar] [CrossRef]
- Okamoto, H.; Oitate, M.; Hagihara, K.; Shiozawa, H.; Furuta, Y.; Ogitani, Y.; Kuga, H. Pharmacokinetics of trastuzumab deruxtecan (T-DXd), a novel anti-HER2 antibody-drug conjugate, in HER2-positive tumour-bearing mice. Xenobiotica 2020, 50, 1242–1250. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, L.; Ma, B.; Liu, T.; Wang, Z.; Ye, Q.; Peng, Y.; Wang, B.; Chen, Y.; Xu, S.; et al. MYC induces CDK4/6 inhibitors resistance by promoting pRB1 degradation. Nat. Commun. 2024, 15, 1871. [Google Scholar] [CrossRef] [PubMed]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef]
- Thierry, A.R. Circulating DNA fragmentomics and cancer screening. Cell Genom. 2023, 3, 100242. [Google Scholar] [CrossRef]
- Qi, T.; Pan, M.; Shi, H.; Wang, L.; Bai, Y.; Ge, Q. Cell-Free DNA Fragmentomics: The Novel Promising Biomarker. Int. J. Mol. Sci. 2023, 24, 1503. [Google Scholar] [CrossRef]
- Jiang, P.; Sun, K.; Peng, W.; Cheng, S.H.; Ni, M.; Yeung, P.C.; Heung, M.M.S.; Xie, T.; Shang, H.; Zhou, Z.; et al. Plasma DNA End-Motif Profiling as a Fragmentomic Marker in Cancer, Pregnancy, and Transplantation. Cancer Discov. 2020, 10, 664–673. [Google Scholar] [CrossRef]
- Elzanowska, J.; Semira, C.; Costa-Silva, B. DNA in extracellular vesicles: Biological and clinical aspects. Mol. Oncol. 2021, 15, 1701–1714. [Google Scholar] [CrossRef]
- Sánchez-Herrero, E.; Campos-Silva, C.; Cáceres-Martell, Y.; Robado de Lope, L.; Sanz-Moreno, S.; Serna-Blasco, R.; Rodríguez-Festa, A.; Ares Trotta, D.; Martín-Acosta, P.; Patiño, C.; et al. ALK-Fusion Transcripts Can Be Detected in Extracellular Vesicles (EVs) from Nonsmall Cell Lung Cancer Cell Lines and Patient Plasma: Toward EV-Based Noninvasive Testing. Clin. Chem. 2022, 68, 668–679. [Google Scholar] [CrossRef]
- Dong, J.; Zhang, R.Y.; Sun, N.; Smalley, M.; Wu, Z.; Zhou, A.; Chou, S.J.; Jan, Y.J.; Yang, P.; Bao, L.; et al. Bio-Inspired NanoVilli Chips for Enhanced Capture of Tumor-Derived Extracellular Vesicles: Toward Non-Invasive Detection of Gene Alterations in Non-Small Cell Lung Cancer. ACS Appl. Mater. Interfaces 2019, 11, 13973–13983. [Google Scholar] [CrossRef]
- Claesson-Welsh, L.; Welsh, M. VEGFA and tumour angiogenesis. J. Intern. Med. 2013, 273, 114–127. [Google Scholar] [CrossRef]
- Yi, M.; Zheng, X.; Niu, M.; Zhu, S.; Ge, H.; Wu, K. Combination strategies with PD-1/PD-L1 blockade: Current advances and future directions. Mol. Cancer 2022, 21, 28. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, T.R.; Taube, J.M. PD-L1 and Emerging Biomarkers in Immune Checkpoint Blockade Therapy. Cancer J. 2018, 24, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, W.; Jiang, H.; Zhang, Y.; Ma, Z.; Wang, Z.; Xu, C.; Jiang, M.; Chen, J.; Cao, Z. MKI67 with arterial hypertension predict a poor survival for prostate cancer patients, a real-life investigation. Clin. Transl. Oncol. 2024, 26, 3037–3049. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Y.; Liao, P.; Yan, L.Y.; Zhao, Q.Y.; Xie, Z.Y.; Dong, J.; Sun, H.T. Correlation of MKI67 with prognosis, immune infiltration, and T cell exhaustion in hepatocellular carcinoma. BMC Gastroenterol. 2021, 21, 416. [Google Scholar] [CrossRef] [PubMed]
- Cardano, M.; Tribioli, C.; Prosperi, E. Targeting Proliferating Cell Nuclear Antigen (PCNA) as an Effective Strategy to Inhibit Tumor Cell Proliferation. Curr. Cancer Drug Targets 2020, 20, 240–252. [Google Scholar] [CrossRef]
- Bouchalova, K.; Kharaishvili, G.; Bouchal, J.; Vrbkova, J.; Megova, M.; Hlobilkova, A. Triple negative breast cancer—BCL2 in prognosis and prediction. Review. Curr. Drug Targets 2014, 15, 1166–1175. [Google Scholar] [CrossRef]
- Hakobyan, S.; Schmidt, M.; Binder, H.; Arakelyan, A. Topology-aware pathway analysis of spatial transcriptomics. PeerJ 2025, 13, e19729. [Google Scholar] [CrossRef]
- Ghorbani, A.; Hosseinie, F.; Khorshid Sokhangouy, S.; Islampanah, M.; Khojasteh-Leylakoohi, F.; Maftooh, M.; Nassiri, M.; Hassanian, S.M.; Ghayour-Mobarhan, M.; Ferns, G.A.; et al. The prognostic, diagnostic, and therapeutic impact of Long noncoding RNAs in gastric cancer. Cancer Genet. 2024, 282–283, 14–26. [Google Scholar] [CrossRef]
- Raju, G.S.R.; Pavitra, E.; Bandaru, S.S.; Varaprasad, G.L.; Nagaraju, G.P.; Malla, R.R.; Huh, Y.S.; Han, Y.K. HOTAIR: A potential metastatic, drug-resistant and prognostic regulator of breast cancer. Mol. Cancer 2023, 22, 65. [Google Scholar] [CrossRef]
- Tufail, M. HOTAIR in colorectal cancer: Structure, function, and therapeutic potential. Med. Oncol. 2023, 40, 259. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.W.; Jin, J.; Wu, X.Y.; Ren, Q.L.; Farzaneh, M. MALAT1-related signaling pathways in colorectal cancer. Cancer Cell Int. 2022, 22, 126. [Google Scholar] [CrossRef]
- Goyal, B.; Yadav, S.R.M.; Awasthee, N.; Gupta, S.; Kunnumakkara, A.B.; Gupta, S.C. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188502. [Google Scholar] [CrossRef] [PubMed]
- Arun, G.; Aggarwal, D.; Spector, D.L. MALAT1 Long Non-Coding RNA: Functional Implications. Noncoding RNA 2020, 6, 22. [Google Scholar] [CrossRef]
- Ren, J.; Zhang, X.; Cao, J.; Tian, J.; Luo, J.; Yu, Y.; Wang, F.; Zhao, Q. Radiosynthesis of a novel antisense imaging probe targeting LncRNA HOTAIR in malignant glioma. BMC Cancer 2022, 22, 79. [Google Scholar] [CrossRef]
- Miceli, R.T.; Chen, T.Y.; Nose, Y.; Tichkule, S.; Brown, B.; Fullard, J.F.; Saulsbury, M.D.; Heyliger, S.O.; Gnjatic, S.; Kyprianou, N.; et al. Extracellular vesicles, RNA sequencing, and bioinformatic analyses: Challenges, solutions, and recommendations. J. Extracell. Vesicles 2024, 13, e70005. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef]
- Ren, W.; Hou, J.; Yang, C.; Wang, H.; Wu, S.; Wu, Y.; Zhao, X.; Lu, C. Extracellular vesicles secreted by hypoxia pre-challenged mesenchymal stem cells promote non-small cell lung cancer cell growth and mobility as well as macrophage M2 polarization via miR-21-5p delivery. J. Exp. Clin. Cancer Res. CR 2019, 38, 62. [Google Scholar] [CrossRef]
- Wang, D.; Wang, X.; Song, Y.; Si, M.; Sun, Y.; Liu, X.; Cui, S.; Qu, X.; Yu, X. Exosomal miR-146a-5p and miR-155-5p promote CXCL12/CXCR7-induced metastasis of colorectal cancer by crosstalk with cancer-associated fibroblasts. Cell Death Dis. 2022, 13, 380. [Google Scholar] [CrossRef]
- Qiao, Z.; Zhang, Y.; Ge, M.; Liu, S.; Jiang, X.; Shang, Z.; Liu, H.; Cao, C.; Xiao, H. Cancer Cell Derived Small Extracellular Vesicles Contribute to Recipient Cell Metastasis Through Promoting HGF/c-Met Pathway. Mol. Cell Proteom. 2019, 18, 1619–1629. [Google Scholar] [CrossRef] [PubMed]
- Tey, S.K.; Wong, S.W.K.; Yeung, C.L.S.; Li, J.Y.K.; Mao, X.; Chung, C.Y.S.; Yam, J.W.P. Liver cancer cells with nuclear MET overexpression release translation regulatory protein-enriched extracellular vesicles exhibit metastasis promoting activity. J. Extracell. Biol. 2022, 1, e39. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, M.; Huang, D.; Ma, Y.; Ye, G.; Wen, Q.; Li, Y.; Deng, L.; Qi, Q.; Liu, T.; et al. Tumor perivascular cell-derived extracellular vesicles promote angiogenesis via the Gas6/Axl pathway. Cancer Lett. 2022, 524, 131–143. [Google Scholar] [CrossRef]
- Klutstein, M.; Nejman, D.; Greenfield, R.; Cedar, H. DNA Methylation in Cancer and Aging. Cancer Res. 2016, 76, 3446–3450. [Google Scholar] [CrossRef]
- Sprang, M.; Paret, C.; Faber, J. CpG-Islands as Markers for Liquid Biopsies of Cancer Patients. Cells 2020, 9, 1820. [Google Scholar] [CrossRef]
- Mansouri, A.; Hachem, L.D.; Mansouri, S.; Nassiri, F.; Laperriere, N.J.; Xia, D.; Lindeman, N.I.; Wen, P.Y.; Chakravarti, A.; Mehta, M.P.; et al. MGMT promoter methylation status testing to guide therapy for glioblastoma: Refining the approach based on emerging evidence and current challenges. Neuro Oncol. 2019, 21, 167–178. [Google Scholar] [CrossRef]
- Ryu, H.; Kim, J.H.; Kim, Y.J.; Jeon, H.; Kim, B.C.; Jeon, Y.; Kim, Y.; Bak, H.; Kang, Y.; Kim, C.; et al. Quantification method of ctDNA using cell-free DNA methylation profile for noninvasive screening and monitoring of colon cancer. Clin. Epigenetics 2024, 16, 95. [Google Scholar] [CrossRef]
- Heeke, S.; Gay, C.M.; Estecio, M.R.; Tran, H.; Morris, B.B.; Zhang, B.; Tang, X.; Raso, M.G.; Rocha, P.; Lai, S.; et al. Tumor- and circulating-free DNA methylation identifies clinically relevant small cell lung cancer subtypes. Cancer Cell 2024, 42, 225–237.e5. [Google Scholar] [CrossRef]
- Maurillo, L.; Spagnoli, A.; Genuardi, M.; Lunghi, M.; Di Renzo, N.; D’Arco, A.M.; Mele, G.; Levis, A.; Gozzini, A.; Petrini, M.; et al. 5-Azacytidine for the Treatment of Acute Myeloid Leukemia: A Retrospective, Multicenter Study of 55 Patients. Blood 2008, 112, 1947. [Google Scholar] [CrossRef]
- Cheng, A.P.; Cheng, M.P.; Loy, C.J.; Lenz, J.S.; Chen, K.; Smalling, S.; Burnham, P.; Timblin, K.M.; Orejas, J.L.; Silverman, E.; et al. Cell-free DNA profiling informs all major complications of hematopoietic cell transplantation. Proc. Natl. Acad. Sci. USA 2022, 119, e2113476118. [Google Scholar] [CrossRef] [PubMed]
- Corces, M.R.; Granja, J.M.; Shams, S.; Louie, B.H.; Seoane, J.A.; Zhou, W.; Silva, T.C.; Groeneveld, C.; Wong, C.K.; Cho, S.W.; et al. The chromatin accessibility landscape of primary human cancers. Science 2018, 362, eaav1898. [Google Scholar] [CrossRef] [PubMed]
- Mansisidor, A.R.; Risca, V.I. Chromatin accessibility: Methods, mechanisms, and biological insights. Nucleus 2022, 13, 236–276. [Google Scholar] [CrossRef]
- McClure, J.J.; Li, X.; Chou, C.J. Advances and Challenges of HDAC Inhibitors in Cancer Therapeutics. Adv. Cancer Res. 2018, 138, 183–211. [Google Scholar]
- Ali, I.; Choi, G.; Lee, K. BET Inhibitors as Anticancer Agents: A Patent Review. Recent Patents Anti-Cancer Drug Discov. 2017, 12, 340–364. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, W.; Kim, Y.; Lee, S.J.; Choi, W.; Lee, G.K.; Park, S.J.; Ju, S.; Kim, S.Y.; Lee, C.; et al. Proteogenomic characterization identifies clinical subgroups in EGFR and ALK wild-type never-smoker lung adenocarcinoma. Exp. Mol. Med. 2024, 56, 2082–2095. [Google Scholar] [CrossRef]
- Hayashi, N.; Iwamoto, T.; Gonzalez-Angulo, A.M.; Ferrer-Lozano, J.; Lluch, A.; Niikura, N.; Bartholomeusz, C.; Nakamura, S.; Hortobagyi, G.N.; Ueno, N.T. Prognostic impact of phosphorylated HER-2 in HER-2+ primary breast cancer. Oncologist 2011, 16, 956–965. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, G.; Zhang, Q.; Zhao, S.; Li, X.; Cao, W.; Luo, H.; Zhou, C. In-depth proteomic analysis identifies key gene signatures predicting therapeutic efficacy of anti-PD-1/PD-L1 monotherapy in non-small cell lung cancer. Transl. Lung Cancer Res. 2024, 13, 34–45. [Google Scholar] [CrossRef]
- Heil, L.R.; Remes, P.M.; Canterbury, J.D.; Yip, P.; Barshop, W.D.; Wu, C.C.; MacCoss, M.J. Dynamic Data-Independent Acquisition Mass Spectrometry with Real-Time Retrospective Alignment. Anal. Chem. 2023, 95, 11854–11858. [Google Scholar] [CrossRef]
- Vistain, L.F.; Tay, S. Single-Cell Proteomics. Trends Biochem. Sci. 2021, 46, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Suri, G.S.; Kaur, G.; Carbone, G.M.; Shinde, D. Metabolomics in oncology. Cancer Rep. 2023, 6, e1795. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Chen, C.X. Metabonomic analysis of tumor microenvironments: A mini-review. Front. Oncol. 2023, 13, 1164266. [Google Scholar] [CrossRef]
- Han, J.; Li, Q.; Chen, Y.; Yang, Y. Recent Metabolomics Analysis in Tumor Metabolism Reprogramming. Front. Mol. Biosci. 2021, 8, 763902. [Google Scholar] [CrossRef]
- Chou, F.J.; Liu, Y.; Lang, F.; Yang, C. D-2-Hydroxyglutarate in Glioma Biology. Cells 2021, 10, 2345. [Google Scholar] [CrossRef] [PubMed]
- Kadiyala, P.; Carney, S.V.; Gauss, J.C.; Garcia-Fabiani, M.B.; Haase, S.; Alghamri, M.S.; Núñez, F.J.; Liu, Y.; Yu, M.; Taher, A.; et al. Inhibition of 2-hydroxyglutarate elicits metabolic reprogramming and mutant IDH1 glioma immunity in mice. J. Clin. Investig. 2021, 131, e139542. [Google Scholar] [CrossRef]
- Fathi, A.T.; Nahed, B.V.; Wander, S.A.; Iafrate, A.J.; Borger, D.R.; Hu, R.; Thabet, A.; Cahill, D.P.; Perry, A.M.; Joseph, C.P.; et al. Elevation of Urinary 2-Hydroxyglutarate in IDH-Mutant Glioma. Oncologist 2016, 21, 214–219. [Google Scholar] [CrossRef]
- Riviere-Cazaux, C.; Suzuki, Y.; Kizilbash, Z.; Laxen, W.J.; Lacey, J.M.; Wipplinger, T.M.; Warrington, A.E.; Keough, M.B.; Kamga, L.F.; Andersen, K.M.; et al. Cerebrospinal fluid D-2-hydroxyglutarate for IDH-mutant glioma: Utility for detection versus monitoring. medRxiv 2025. [Google Scholar] [CrossRef]
- Li, X.; Zhao, Y.; Jiang, W.; Li, S.; Zhan, M.; Liu, H.; Zhang, C.; Liang, H.; Liu, H.; Lu, L.; et al. Ultralong circulating choline phosphate liposomal nanomedicines for cascaded chemo-radiotherapy. Biomater. Sci. 2019, 7, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Xu, R.; Shao, W.; Tan, J.; Wang, S.; Chen, S.; Zhuang, A.; Liu, X.; Jia, R. A Novel Nanozyme to Enhance Radiotherapy Effects by Lactic Acid Scavenging, ROS Generation, and Hypoxia Mitigation. Adv. Sci. 2024, 11, e2403107. [Google Scholar] [CrossRef] [PubMed]
- Matés, J.M.; Campos-Sandoval, J.A.; Márquez, J. Glutaminase isoenzymes in the metabolic therapy of cancer. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 158–164. [Google Scholar] [CrossRef]
- Du, D.; Liu, C.; Qin, M.; Zhang, X.; Xi, T.; Yuan, S.; Hao, H.; Xiong, J. Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma. Acta Pharm. Sin. B 2022, 12, 558–580. [Google Scholar] [CrossRef] [PubMed]
- Encarnación-Rosado, J.; Sohn, A.S.W.; Biancur, D.E.; Lin, E.Y.; Osorio-Vasquez, V.; Rodrick, T.; González-Baerga, D.; Zhao, E.; Yokoyama, Y.; Simeone, D.M.; et al. Targeting pancreatic cancer metabolic dependencies through glutamine antagonism. Nat. Cancer 2024, 5, 85–99. [Google Scholar] [CrossRef]
- Yang, W.H.; Qiu, Y.; Stamatatos, O.; Janowitz, T.; Lukey, M.J. Enhancing the Efficacy of Glutamine Metabolism Inhibitors in Cancer Therapy. Trends Cancer 2021, 7, 790–804. [Google Scholar] [CrossRef]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, eabc4552. [Google Scholar] [CrossRef]
- Cullin, N.; Azevedo Antunes, C.; Straussman, R.; Stein-Thoeringer, C.K.; Elinav, E. Microbiome and cancer. Cancer Cell 2021, 39, 1317–1341. [Google Scholar] [CrossRef]
- Fan, S.; Jiang, Z.; Zhang, Z.; Xing, J.; Wang, D.; Tang, D. Akkermansia muciniphila: A potential booster to improve the effectiveness of cancer immunotherapy. J. Cancer Res. Clin. Oncol. 2023, 149, 13477–13494. [Google Scholar] [CrossRef]
- Wang, G.; He, X.; Wang, Q. Intratumoral bacteria are an important “accomplice” in tumor development and metastasis. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188846. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Mussa, A.; Pagliardini, S.; Pagliardini, V.; Molinatto, C.; Baldassarre, G.; Corrias, A.; Silengo, M.C.; Ferrero, G.B. α-Fetoprotein assay on dried blood spot for hepatoblastoma screening in children with overgrowth-cancer predisposition syndromes. Pediatr. Res. 2014, 76, 544–548. [Google Scholar] [CrossRef]
- Hofman, M.S.; Hicks, R.J.; Maurer, T.; Eiber, M. Prostate-specific Membrane Antigen PET: Clinical Utility in Prostate Cancer, Normal Patterns, Pearls, and Pitfalls. Radiographics 2018, 38, 200–217. [Google Scholar] [CrossRef] [PubMed]
- Alberts, I.L.; Seifert, R.; Werner, R.A.; Rowe, S.P.; Afshar-Oromieh, A. Prostate-specific Membrane Antigen: Diagnostics. PET Clin. 2024, 19, 351–362. [Google Scholar] [CrossRef]
- Gubbi, S.; Koch, C.A.; Klubo-Gwiezdzinska, J. Peptide Receptor Radionuclide Therapy in Thyroid Cancer. Front. Endocrinol. 2022, 13, 896287. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Parghane, R.V.; Kamaldeep; Chakrabarty, S. Peptide Receptor Radionuclide Therapy of Neuroendocrine Tumors. Semin. Nucl. Med. 2020, 50, 447–464. [Google Scholar] [CrossRef]
- Hofland, J.; Brabander, T.; Verburg, F.A.; Feelders, R.A.; de Herder, W.W. Peptide Receptor Radionuclide Therapy. J. Clin. Endocrinol. Metab. 2022, 107, 3199–3208. [Google Scholar] [CrossRef]
- Moding, E.J.; Shahrokh Esfahani, M.; Jin, C.; Hui, A.B.; Nabet, B.Y.; Liu, Y.; Chabon, J.J.; Binkley, M.S.; Kurtz, D.M.; Hamilton, E.G.; et al. Integrating ctDNA Analysis and Radiomics for Dynamic Risk Assessment in Localized Lung Cancer. Cancer Discov. 2025, 15, 1609–1629. [Google Scholar] [CrossRef]
- Li, Q.; Liu, H.; Jin, J.; Zhang, Y.; Li, S.; Wang, L.; Zhao, S.; Wu, C.; Fang, Z.; Jin, J. Traditional Chinese Medicine Properties and Microcalorimetry: BioScience Evaluation. Med. Res. 2025, 1, 103–121. [Google Scholar] [CrossRef]
- Ehdaie, B.; Vertosick, E.; Spaliviero, M.; Giallo-Uvino, A.; Taur, Y.; O’Sullivan, M.; Livingston, J.; Sogani, P.; Eastham, J.; Scardino, P. The impact of repeat biopsies on infectious complications in men with prostate cancer on active surveillance. J. Urol. 2014, 191, 660–664. [Google Scholar] [CrossRef]
- Jin, Z.; Zhang, W.; Liu, H.; Ding, A.; Lin, Y.; Wu, S.X.; Lin, J. Potential Therapeutic Application of Local Anesthetics in Cancer Treatment. Recent Patents Anti-Cancer Drug Discov. 2022, 17, 326–342. [Google Scholar] [CrossRef]
- Li, R.; Mukherjee, M.B.; Jin, Z.; Liu, H.; Lin, K.; Liu, Q.; Dilger, J.P.; Lin, J. The Potential Effect of General Anesthetics in Cancer Surgery: Meta-Analysis of Postoperative Metastasis and Inflammatory Cytokines. Cancers 2023, 15, 2759. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Cho, H.Y.; Jeon, J.; Kim, K.A.; Han, Y.D.; Ahn, J.B.; Wortzel, I.; Lyden, D.; Kim, H.S. Detection of circulating KRAS mutant DNA in extracellular vesicles using droplet digital PCR in patients with colon cancer. Front. Oncol. 2022, 12, 1067210. [Google Scholar] [CrossRef] [PubMed]
- Bádon, E.S.; Mokánszki, A.; Mónus, A.; András, C.; Méhes, G. Clonal diversity in KRAS mutant colorectal adenocarcinoma under treatment: Monitoring of cfDNA using reverse hybridization and DNA sequencing platforms. Mol. Cell Probes 2023, 67, 101891. [Google Scholar] [CrossRef]
- Dos Reis, M.B.; Dos Santos, W.; de Carvalho, A.C.; Lima, A.B.; Reis, M.T.; Santos, F.; Reis, R.M.; Guimarães, D.P. Plasma mutation profile of precursor lesions and colorectal cancer using the Oncomine Colon cfDNA Assay. BMC Cancer 2024, 24, 1547. [Google Scholar] [CrossRef] [PubMed]
- Perrier, A.; Gligorov, J.; Lefèvre, G.; Boissan, M. The extracellular domain of Her2 in serum as a biomarker of breast cancer. Lab. Investig. A J. Tech. Methods Pathol. 2018, 98, 696–707. [Google Scholar] [CrossRef]
- Lee, S.B.; Lee, J.W.; Yu, J.H.; Ko, B.S.; Kim, H.J.; Son, B.H.; Gong, G.; Lee, H.J.; Kim, S.B.; Jung, K.H.; et al. Preoperative serum HER2 extracellular domain levels in primary invasive breast cancer. BMC Cancer 2014, 14, 929. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Heider, K.; Gale, D.; Murphy, S.; Fisher, E.; Mouliere, F.; Ruiz-Valdepenas, A.; Santonja, A.; Morris, J.; Chandrananda, D.; et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci. Transl. Med. 2020, 12, eaaz8084. [Google Scholar] [CrossRef]
- Grant, C.; Nagasaka, M. Neoadjuvant EGFR-TKI therapy in Non-Small cell lung cancer. Cancer Treat. Rev. 2024, 126, 102724. [Google Scholar] [CrossRef]
- Wu, Y.L.; Zhou, Q. Combination Therapy for EGFR-Mutated Lung Cancer. N. Engl. J. Med. 2023, 389, 2005–2007. [Google Scholar] [CrossRef]
- Bai, X.; Flaherty, K.T. Targeted and immunotherapies in BRAF mutant melanoma: Where we stand and what to expect. Br. J. Dermatol. 2021, 185, 253–262. [Google Scholar] [CrossRef]
- Ferrucci, P.F.; Lens, M.; Cocorocchio, E. Combined BRAF-Targeted Therapy with Immunotherapy in BRAF-Mutated Advanced Melanoma Patients. Curr. Oncol. Rep. 2021, 23, 138. [Google Scholar] [CrossRef] [PubMed]
- Doi, T.; Shitara, K.; Naito, Y.; Shimomura, A.; Fujiwara, Y.; Yonemori, K.; Shimizu, C.; Shimoi, T.; Kuboki, Y.; Matsubara, N.; et al. Safety, pharmacokinetics, and antitumour activity of trastuzumab deruxtecan (DS-8201), a HER2-targeting antibody-drug conjugate, in patients with advanced breast and gastric or gastro-oesophageal tumours: A phase 1 dose-escalation study. Lancet. Oncol. 2017, 18, 1512–1522. [Google Scholar] [CrossRef]
- Liu, H.; Tang, T. MAPK signaling pathway-based glioma subtypes, machine-learning risk model, and key hub proteins identification. Sci. Rep. 2023, 13, 19055. [Google Scholar] [CrossRef] [PubMed]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef]
- Lovly, C.M.; Iyengar, P.; Gainor, J.F. Managing Resistance to EFGR- and ALK-Targeted Therapies. Am. Soc. Clin. Oncol. Educ. Book. 2017, 37, 607–618. [Google Scholar] [CrossRef]
- Fuqua, S.A.; Gu, G.; Rechoum, Y. Estrogen receptor (ER) α mutations in breast cancer: Hidden in plain sight. Breast Cancer Res. Treat. 2014, 144, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Brett, J.O.; Spring, L.M.; Bardia, A.; Wander, S.A. ESR1 mutation as an emerging clinical biomarker in metastatic hormone receptor-positive breast cancer. Breast Cancer Res. 2021, 23, 85. [Google Scholar] [CrossRef]
- Liu, H.; Weng, J.; Huang, C.L.; Jackson, A.P. Is the voltage-gated sodium channel β3 subunit (SCN3B) a biomarker for glioma? Funct. Integr. Genom. 2024, 24, 162. [Google Scholar] [CrossRef]
- Liu, H. Expression and potential immune involvement of cuproptosis in kidney renal clear cell carcinoma. Cancer Genet. 2023, 274–275, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Weng, J. A Pan-Cancer Bioinformatic Analysis of RAD51 Regarding the Values for Diagnosis, Prognosis, and Therapeutic Prediction. Front. Oncol. 2022, 12, 858756. [Google Scholar] [CrossRef] [PubMed]
- Medina, J.E.; Dracopoli, N.C.; Bach, P.B.; Lau, A.; Scharpf, R.B.; Meijer, G.A.; Andersen, C.L.; Velculescu, V.E. Cell-free DNA approaches for cancer early detection and interception. J. Immunother. Cancer 2023, 11, e006013. [Google Scholar] [CrossRef]
- Zhao, F.; Bai, P.; Xu, J.; Li, Z.; Muhammad, S.; Li, D.; Zhang, Z.; Gao, Y.; Liu, Q. Efficacy of cell-free DNA methylation-based blood test for colorectal cancer screening in high-risk population: A prospective cohort study. Mol. Cancer 2023, 22, 157. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Mei, W.; Ma, K.; Zeng, C. Circulating Tumor DNA and Minimal Residual Disease (MRD) in Solid Tumors: Current Horizons and Future Perspectives. Front. Oncol. 2021, 11, 763790. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra392. [Google Scholar] [CrossRef]
- Tarazona, N.; Gimeno-Valiente, F.; Gambardella, V.; Zuñiga, S.; Rentero-Garrido, P.; Huerta, M.; Roselló, S.; Martinez-Ciarpaglini, C.; Carbonell-Asins, J.A.; Carrasco, F.; et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1804–1812. [Google Scholar] [CrossRef]
- Zook, J.M.; Chapman, B.; Wang, J.; Mittelman, D.; Hofmann, O.; Hide, W.; Salit, M. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol. 2014, 32, 246–251. [Google Scholar] [CrossRef]
- Pascual, J.; Attard, G.; Bidard, F.C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef]
- Kapoor, S.; Narayanan, A. Leakage and the reproducibility crisis in machine-learning-based science. Patterns 2023, 4, 100804. [Google Scholar] [CrossRef]
- Stanowicka-Grada, M.; Senkus, E. Anti-HER2 Drugs for the Treatment of Advanced HER2 Positive Breast Cancer. Curr. Treat. Options Oncol. 2023, 24, 1633–1650. [Google Scholar] [CrossRef] [PubMed]
| Aspect | Key Issue | Major Challenges | Current Mitigation Strategies |
|---|---|---|---|
| Biomarker Validation and Clinical Trial Design | Moving from discovery to clinical utility via analytical and clinical validation; biomarker-driven trial design (e.g., enrichment, adaptive trials) | Misclassification of biomarker-positive patients; complexity of trial design; limited generalizability | Enrichment and adaptive trial designs; clearly defined predictive value; appropriate control arms |
| Regulatory Approval and IVD Registration | In vitro diagnostic (IVD) registration requires demonstration of safety and effectiveness; companion diagnostics must co-develop with therapeutics | Regulatory heterogeneity across regions; burden of technical documentation; timeline mismatch with drug development | Early coordination of drug-CDx timelines; compliance with FDA/EMA/CE marking standards |
| Standardization and Quality Control | Ensuring reproducibility across sites; minimizing pre-analytical and analytical variability | Inter-lab variability in results; lack of harmonized materials and protocols | External quality assurance, proficiency testing, use of reference materials (e.g., NIST standards) |
| Privacy and Ethical Issues | Handling large-scale multi-omics data; protecting patient privacy and consent; ensuring equity | Data integration complexity; regulatory requirements for consent and access; disparities in test availability | Ethical frameworks for consent and data sharing; compliance with GDPR/HIPAA; policies to reduce access gaps |
| Controversies and Consensus | Discordance between cfDNA and tissue-based assays; inter-lab variant detection inconsistency | False positives/negatives due to methodological differences; biopsy type trade-offs | Standardization using spiked reference materials; dual-validation approach (cfDNA screening + tissue confirmation) per 2023 ESMO guidelines |
| Leakage and Reproducibility Crisis in ML-Based Biomarkers | Overfitting and data leakage in ML biomarker models lead to inflated performance | Use of test data in model training; lack of transparency in workflow reporting | Adoption of Model Information Sheets; workflow auditing; emphasis on independent validation and reproducibility checks |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Karsidag, I.; Golin, R.; Wu, G. Bridging Discovery and Treatment: Cancer Biomarker. Cancers 2025, 17, 3720. https://doi.org/10.3390/cancers17223720
Liu H, Karsidag I, Golin R, Wu G. Bridging Discovery and Treatment: Cancer Biomarker. Cancers. 2025; 17(22):3720. https://doi.org/10.3390/cancers17223720
Chicago/Turabian StyleLiu, Hengrui, Ilayda Karsidag, Rebecca Golin, and Guangzhen Wu. 2025. "Bridging Discovery and Treatment: Cancer Biomarker" Cancers 17, no. 22: 3720. https://doi.org/10.3390/cancers17223720
APA StyleLiu, H., Karsidag, I., Golin, R., & Wu, G. (2025). Bridging Discovery and Treatment: Cancer Biomarker. Cancers, 17(22), 3720. https://doi.org/10.3390/cancers17223720






