Integrated Multi-Omics Analysis Uncovers Immune–Metabolic Interplay in Hepatocellular Carcinoma Tumor Microenvironment
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Information in the Study Cohort
2.2. RNA Sequencing and Methylation Array
2.3. Consensus Matrix
2.4. Whole-Exome Sequencing
2.5. Differentially Expressed Genes (DEG) Analysis
2.6. Immune Gene Sets
2.7. Differentially Methylated Probes (DMP) Analysis
2.8. Biological Network Analysis and Functional Analysis
2.9. Differentially Methylated Regions (DMR) Analysis
2.10. Single-Cell Sequencing Analysis
2.11. Survival Analysis
2.12. Statistical Analysis
3. Results
3.1. Overall Workflow of TME-Driven Gene Identification with Hepatocellular Carcinoma Cohort
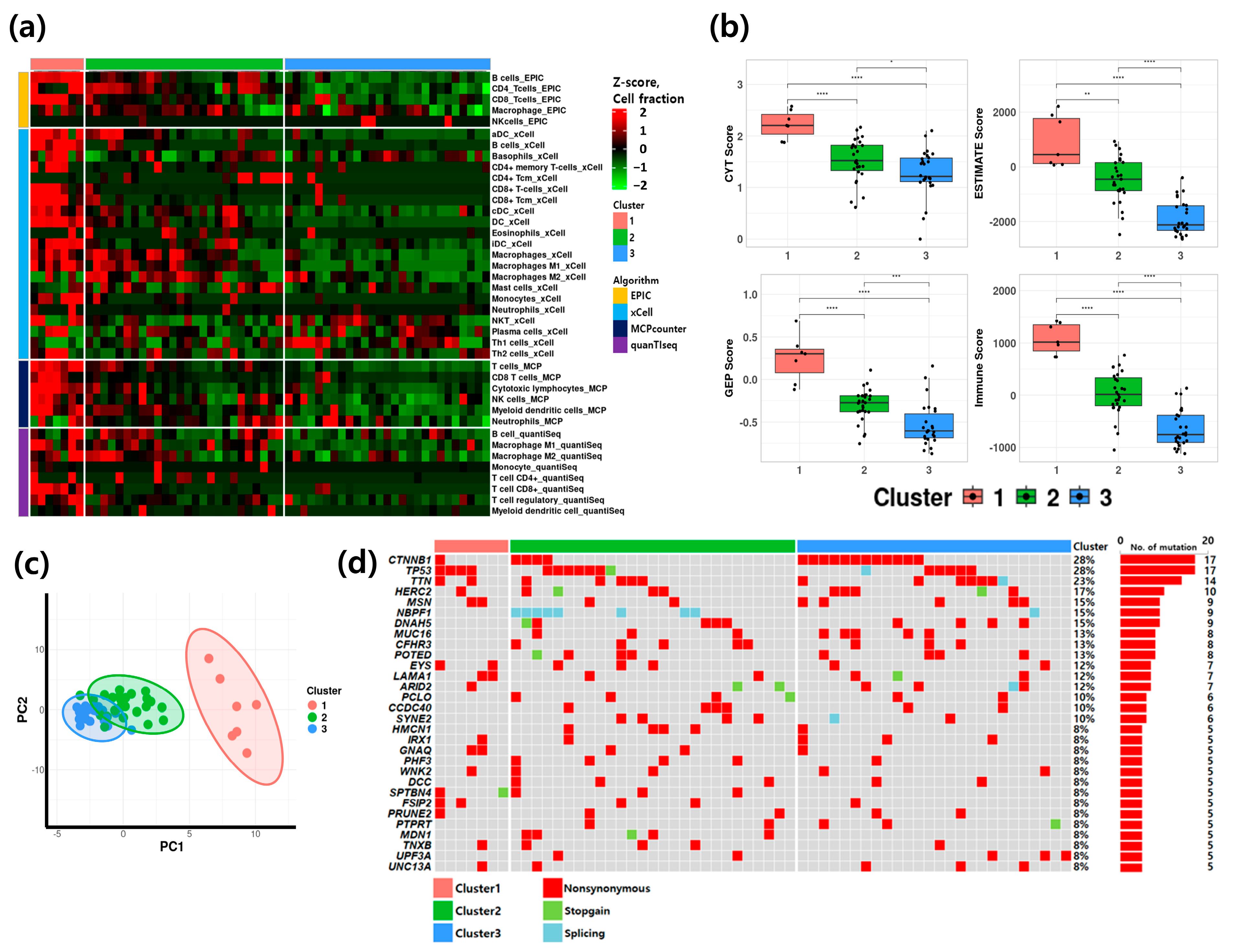
3.2. Classification and Characteristics of HCC Patients Based on TME
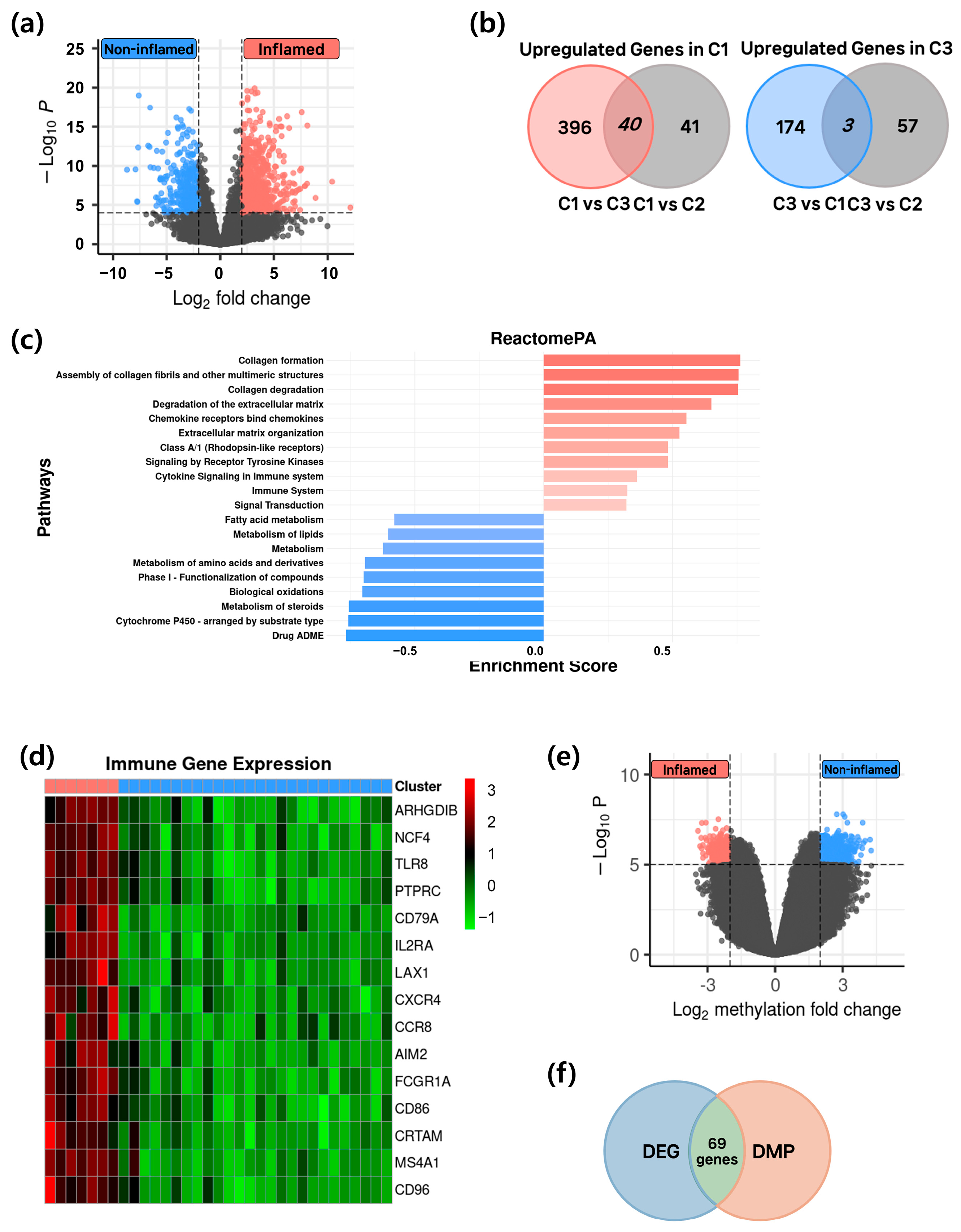
3.3. Identification of Epigenetic–Transcriptomic Regulated Genes
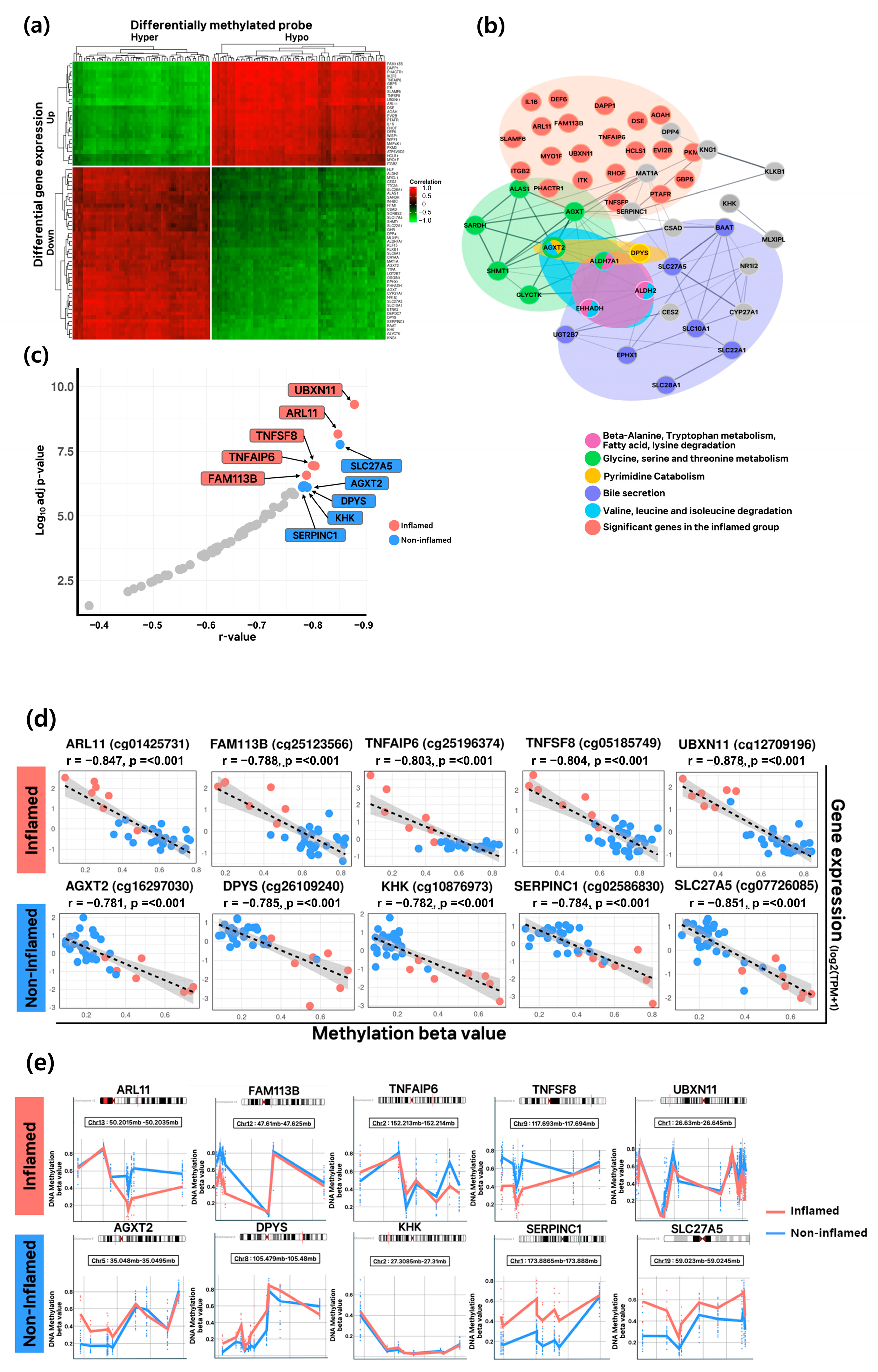
3.4. Epigenetic Regulation and Functional Characterization in Inflamed and Non-Inflamed HCC Groups
3.5. Cell Type Specificity and Prognostic Value of Key Epigenetic-Regulated Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HCC | Hepatocellular carcinoma |
| TME | Tumor microenvironment |
| DEG | Differentially expressed gene |
| DMP | Differentially methylated probe |
| DMR | Differentially methylated regions |
| GEP | Gene expression profile |
| CYT | Cytolytic activity |
| HCV | Hepatitis C virus |
| HBV | Hepatitis B virus |
| DFS | Disease-free survival |
| SUV | Standardized uptake value |
| NK | Natural killer |
| TAMs | Tumor-associated macrophages |
| MDSCs | Myeloid-derived suppressor cells |
| Tregs | Regulatory T cells |
| ECM | Extracellular matrix |
| PPI | Protein–protein interactions |
References
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 7. [Google Scholar] [CrossRef]
- Fu, S.; Deger, T.; Boers, R.G.; Boers, J.B.; Doukas, M.; Gribnau, J.; Wilting, S.M.; Debes, J.D.; Boonstra, A. Hypermethylation of DNA Methylation Markers in Non-Cirrhotic Hepatocellular Carcinoma. Cancers 2023, 15, 4784. [Google Scholar] [CrossRef]
- Johnson, P.J.; Kalyuzhnyy, A.; Boswell, E.; Toyoda, H. Progression of chronic liver disease to hepatocellular carcinoma: Implications for surveillance and management. BJC Rep. 2024, 2, 39. [Google Scholar] [CrossRef]
- Kanda, T.; Goto, T.; Hirotsu, Y.; Moriyama, M.; Omata, M. Molecular Mechanisms Driving Progression of Liver Cirrhosis towards Hepatocellular Carcinoma in Chronic Hepatitis B and C Infections: A Review. Int. J. Mol. Sci. 2019, 20, 1358. [Google Scholar] [CrossRef]
- Minami, T.; Tateishi, R.; Kondo, M.; Nakagomi, R.; Fujiwara, N.; Sato, M.; Uchino, K.; Enooku, K.; Nakagawa, H.; Asaoka, Y.; et al. Serum Alpha-Fetoprotein Has High Specificity for the Early Detection of Hepatocellular Carcinoma After Hepatitis C Virus Eradication in Patients. Medicine 2015, 94, e901. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Choi, J.Y.; Joung, J.G.; Joh, J.W.; Kim, J.M.; Hyun, S.H. Metabolism-Associated Gene Signatures for FDG Avidity on PET/CT and Prognostic Validation in Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 845900. [Google Scholar] [CrossRef]
- Li, S.; Lin, Y.; Gao, X.; Zeng, D.; Cen, W.; Su, Y.; Su, J.; Zeng, C.; Huang, Z.; Zeng, H.; et al. Integrative multi-omics analysis reveals a novel subtype of hepatocellular carcinoma with biological and clinical relevance. Front. Immunol. 2024, 15, 1517312. [Google Scholar] [CrossRef]
- Hansen, K.D.; Timp, W.; Bravo, H.C.; Sabunciyan, S.; Langmead, B.; McDonald, O.G.; Wen, B.; Wu, H.; Liu, Y.; Diep, D.; et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011, 43, 768–775. [Google Scholar] [CrossRef]
- Yu, M.; Du, G.; Xu, Q.; Huang, Z.; Huang, X.; Qin, Y.; Han, L.; Fan, Y.; Zhang, Y.; Han, X.; et al. Integrated analysis of DNA methylome and transcriptome identified CREB5 as a novel risk gene contributing to recurrent pregnancy loss. eBioMedicine 2018, 35, 334–344. [Google Scholar] [CrossRef]
- Goncalves, E.; Goncalves-Reis, M.; Pereira-Leal, J.B.; Cardoso, J. DNA methylation fingerprint of hepatocellular carcinoma from tissue and liquid biopsies. Sci. Rep. 2022, 12, 11512. [Google Scholar] [CrossRef]
- Zhao, Y.; Wan, K.; Wang, J.; Wang, S.; Chang, Y.; Du, Z.; Zhang, L.; Dong, L.; Zhou, D.; Zhang, W.; et al. DNA methylation and gene expression profiling reveal potential association of retinol metabolism related genes with hepatocellular carcinoma development. PeerJ 2024, 12, e17916. [Google Scholar] [CrossRef]
- Racle, J.; de Jonge, K.; Baumgaertner, P.; Speiser, D.E.; Gfeller, D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife 2017, 6, e26476. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautes-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [PubMed]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef]
- Argentiero, A.; Delvecchio, A.; Fasano, R.; Andriano, A.; Caradonna, I.C.; Memeo, R.; Desantis, V. The Complexity of the Tumor Microenvironment in Hepatocellular Carcinoma and Emerging Therapeutic Developments. J. Clin. Med. 2023, 12, 7469. [Google Scholar] [CrossRef]
- Zhen, Y.; Yang, J.; Song, J.; Xing, Z.; Zheng, J. Silencing ARL11 relieved atherosclerotic inflammation and lipid deposition via retraining JAK2/STAT1 pathway. Atherosclerosis 2024, 398, 118564. [Google Scholar] [CrossRef]
- Huang, H.; Yu, H.; Li, X.; Li, Y.; Zhu, G.; Su, L.; Li, M.; Chen, C.; Gao, M.; Wu, D.; et al. Genomic analysis of TNF-related genes with prognosis and characterization of the tumor immune microenvironment in lung adenocarcinoma. Front. Immunol. 2022, 13, 993890. [Google Scholar] [CrossRef]
- Lim, J.X.; McTaggart, T.; Jung, S.K.; Smith, K.J.; Hulme, G.; Laba, S.; Ng, Y.Q.; Williams, A.; Hussain, R.; Coxhead, J.; et al. PD-1 receptor deficiency enhances CD30(+) T(reg) cell function in melanoma. Nat. Immunol. 2025, 26, 1074–1086. [Google Scholar] [CrossRef]
- Fu, Z.; Cai, W.; Shao, J.; Xue, H.; Ge, Z.; Fan, H.; Dong, C.; Wang, C.; Zhang, J.; Shen, C.; et al. Genetic Variants in TNFSF4 and TNFSF8 Are Associated With the Risk of HCV Infection Among Chinese High-Risk Population. Front. Genet. 2021, 12, 630310. [Google Scholar] [CrossRef]
- Duan, K.; Fang, K.; Sui, C. TFAIP6 facilitates hepatocellular carcinoma cell glycolysis through upregulating c-myc/PKM2 axis. Heliyon 2024, 10, e30959. [Google Scholar] [CrossRef]
- Song, H.M.; Li, Z.W.; Huang, Q.; Wu, C.G.; Li, M.H.; Shen, J.K. A diagnostic signatures for intervertebral disc degeneration using TNFAIP6 and COL6A2 based on single-cell RNA-seq and bulk RNA-seq analyses. Ann. Med. 2025, 57, 2443568. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Yu, T.; Zheng, B.; Sun, W.; Li, Y.; Zhang, W.; Chen, Y.; Yuan, L.; Wang, X.J.; Wang, J.; et al. POSTN promotes the progression of NSCLC via regulating TNFAIP6 expression. Biochem. Biophys. Res. Commun. 2024, 736, 150891. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Yang, P.; Zhang, D.; Jin, C.; Peng, W.; Wang, T.; Sun, Q.; Chen, Z.; Feng, Y.; Sun, Y. KHK-A promotes fructose-dependent colorectal cancer liver metastasis by facilitating the phosphorylation and translocation of PKM2. Acta Pharm. Sin. B 2024, 14, 2959–2976. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xiang, L.; Zhang, W.; Xia, Z.; Chen, W. AGXT2 Suppresses the Proliferation and Dissemination of Hepatocellular Carcinoma Cells by Modulating Intracellular Lipid Metabolism. J. Hepatocell. Carcinoma 2024, 11, 1623–1639. [Google Scholar] [CrossRef]
- Xu, W.; Hu, M.; Lu, X.; Lao, Y.; Ma, N.; Wang, Y.; Li, J.; Chen, X.; Liu, S.; Liu, J.; et al. Inhibition of PCSK9 enhances the anti-hepatocellular carcinoma effects of TCR-T cells and anti-PD-1 immunotherapy. Int. J. Biol. Sci. 2024, 20, 3942–3955. [Google Scholar] [CrossRef]
- Sun, S.; Ma, J.; Zuo, T.; Shi, J.; Sun, L.; Meng, C.; Shu, W.; Yang, Z.; Yao, H.; Zhang, Z. Inhibition of PCSK9: A Promising Enhancer for Anti-PD-1/PD-L1 Immunotherapy. Research 2024, 7, 0488. [Google Scholar] [CrossRef]
- Tanianskii, D.A.; Jarzebska, N.; Birkenfeld, A.L.; O’Sullivan, J.F.; Rodionov, R.N. Beta-Aminoisobutyric Acid as a Novel Regulator of Carbohydrate and Lipid Metabolism. Nutrients 2019, 11, 524. [Google Scholar] [CrossRef]
- Yokoi, K.; Nakajima, Y.; Matsuoka, H.; Shinkai, Y.; Ishihara, T.; Maeda, Y.; Kato, T.; Katsuno, H.; Masumori, K.; Kawada, K.; et al. Impact of DPYD, DPYS, and UPB1 gene variations on severe drug-related toxicity in patients with cancer. Cancer Sci. 2020, 111, 3359–3366. [Google Scholar] [CrossRef]
- Le, Z.; Chen, S.; Feng, Y.; Lu, W.; Liu, M. SERPINC1, a new prognostic predictor of colon cancer, promote colon cancer progression through EMT. Cancer Rep. 2024, 7, e2079. [Google Scholar] [CrossRef]
- Xiao, X.; Mo, H.; Tu, K. CTNNB1 mutation suppresses infiltration of immune cells in hepatocellular carcinoma through miRNA-mediated regulation of chemokine expression. Int. Immunopharmacol. 2020, 89 Pt A, 107043. [Google Scholar] [CrossRef]
- Luke, J.J.; Bao, R.; Sweis, R.F.; Spranger, S.; Gajewski, T.F. WNT/β-catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin. Cancer Res. 2019, 25, 3074–3083. [Google Scholar] [CrossRef]
- Long, J.; Wang, A.; Bai, Y.; Lin, J.; Yang, X.; Wang, D.; Yang, X.; Jiang, Y.; Zhao, H. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. eBioMedicine 2019, 42, 363–374. [Google Scholar] [CrossRef]
- Vadakekolathu, J.; Lai, C.; Reeder, S.; Church, S.E.; Hood, T.; Lourdusamy, A.; Rettig, M.P.; Aldoss, I.; Advani, A.S.; Godwin, J.; et al. TP53 abnormalities correlate with immune infiltration and associate with response to flotetuzumab immunotherapy in AML. Blood Adv. 2020, 4, 5011–5024. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | Overall (n = 60) | Cluster 1 (n = 7) | Cluster 2 (n = 27) | Cluster 3 (n = 26) | p-Value |
|---|---|---|---|---|---|
| Age (range, years) | 58.1 (42–76) | 51.0 (42–61) | 60.0 (44–76) | 57.0 (43–73) | 0.0629 |
| Sex | 0.6951 | ||||
| Male | 49 (81.7%) | 5 (71.4%) | 23 (85.1%) | 21 (80.7%) | |
| Female | 11 (18.3%) | 2 (28.6%) | 4 (14.9%) | 5 (19.3%) | |
| Tumor size (cm) | 4.4 (±1.4) | 5.3 (±1.5) | 4.0 (±2.0) | 4.5 (±2.0) | 0.0767 |
| Tumor SUV 1 | 4.5 (±2.6) | 8.9 (±5.0) | 3.7 (±1.1) | 4.2 (±1.9) | 0.023 |
| Etiology | 0.5355 | ||||
| HBV | 46 (76.7%) | 6 (85.7%) | 22 (81.4%) | 18 (69.2%) | |
| HCV | 5 (8.3%) | 1 (14.3%) | 2 (7.4%) | 2 (7.7%) | |
| Alcohol and others | 9 (15%) | 0 (0.0%) | 3 (11.2%) | 6 (23.1%) | |
| Cirrhosis | 0.5923 | ||||
| Yes | 22 (36.7%) | 3 (42.8%) | 8 (29.6%) | 11 (42.3%) | |
| No | 38 (63.3%) | 4 (57.2%) | 19 (70.4%) | 15 (57.7%) | |
| Microvessel invasion | 0.0275 | ||||
| Yes | 30 (50.0%) | 6 (85.7%) | 9 (33.3%) | 15 (57.7%) | |
| No | 30 (50.0%) | 1 (14.3%) | 18 (66.7%) | 11 (42.3%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.-H.; Sim, D.W.; Kim, S.-Y.; Choi, J.Y.; Hyun, S.H.; Joung, J.-G. Integrated Multi-Omics Analysis Uncovers Immune–Metabolic Interplay in Hepatocellular Carcinoma Tumor Microenvironment. Cancers 2025, 17, 3565. https://doi.org/10.3390/cancers17213565
Park J-H, Sim DW, Kim S-Y, Choi JY, Hyun SH, Joung J-G. Integrated Multi-Omics Analysis Uncovers Immune–Metabolic Interplay in Hepatocellular Carcinoma Tumor Microenvironment. Cancers. 2025; 17(21):3565. https://doi.org/10.3390/cancers17213565
Chicago/Turabian StylePark, Jong-Heon, Dae Won Sim, Sook-Young Kim, Joon Young Choi, Seung Hyup Hyun, and Je-Gun Joung. 2025. "Integrated Multi-Omics Analysis Uncovers Immune–Metabolic Interplay in Hepatocellular Carcinoma Tumor Microenvironment" Cancers 17, no. 21: 3565. https://doi.org/10.3390/cancers17213565
APA StylePark, J.-H., Sim, D. W., Kim, S.-Y., Choi, J. Y., Hyun, S. H., & Joung, J.-G. (2025). Integrated Multi-Omics Analysis Uncovers Immune–Metabolic Interplay in Hepatocellular Carcinoma Tumor Microenvironment. Cancers, 17(21), 3565. https://doi.org/10.3390/cancers17213565





