TILDA-X: Transcriptome-Informed Lung Cancer Disparities via Explainable AI
Simple Summary
Abstract
1. Introduction
- We assume that the local interpretation of SHAP or the interpretation of each patient of AA and EA cohorts under different disease classes/labels, such as LUAD, LUSC, and HEALTHY, will help extract patient-specific significant genes reflecting patient-specific disparity related to those disease conditions.
- We also assume that the interpretation of a patient using the different combinations of disease classes (LUAD-LUSC-HEALTHY; LUAD-LUSC; LUAD-HEALTHY; LUSC-HEALTHY) would help derive a robust set of patient-specific genes. Each of the LUAD patients (irrespective of race and sex) was interrogated via three classification problems containing the LUAD cohort, namely LUAD-LUSC-HEALTHY, LUAD-LUSC, and LUAD-HEALTHY. In a similar way, each LUSC patient was interrogated via three classification problems containing the LUSC cohort (LUAD-LUSC-HEALTHY, LUAD-LUSC, and LUSC-HEALTHY).
- We explored SHAP in a bottom-up approach (going from patient-specific biomarker genes to cohort-specific biomarker genes) to decipher the disparity between any two cohorts of patients, including AAMs vs. AAFs, AAMs vs. EAMs, AAMs vs. EAFs, AAFs vs. EAMs, AAFs vs. EAFs, and EAMs vs. EAFs.
- Note that the classification problems are designed based on disease conditions (i.e., LUAD, LUSC, and HEALTHY) to avoid the race-specific imbalance in the dataset, which is innovative in discovering the health disparity. The data is highly imbalanced regarding race (AA:EA = 1:8). But input to the SHAP is a classification problem with disease conditions LUAD (n = 356), LUSC (n = 295), and HEALTHY (n = 313) as class labels, meaning the data is balanced (each class consists of ~300 samples). The race-specific high accuracies (AAM: 90% and EAM: 95%) for an imbalanced cohort ratio (AAM:EAM = 1:7) support our hypothesis.
2. Materials and Methods
2.1. Data Collection
2.2. Data Preparation
2.3. Study Flow Diagram
2.3.1. Multiple Classification Problems and Rationale
2.3.2. Local Feature Interpretation Using SHAP
2.3.3. Combined Patient-Specific Biomarker Genes
2.3.4. Race- and Sex-Cohort-Specific Disparity Information
2.3.5. Validation
3. Results
3.1. Selecting the Best Machine Learning Approach
3.2. Patient-Specific Disparity
3.3. Race- and Sex-Specific Correct Prediction
3.4. Recovery of Gene Set-Level Disparity Within LUAD and LUSC Cohorts
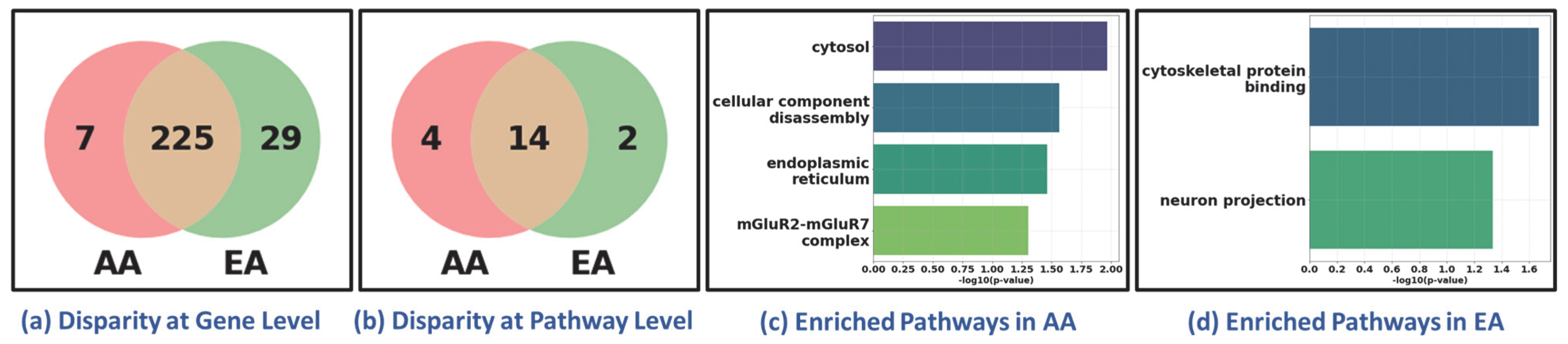
3.5. Recovery of Pathway-Level Disparity Within LUAD and LUSC Cohort
3.6. Generalizability
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Islami, F.; Goding Sauer, A.; Miller, K.D.; Siegel, R.L.; Fedewa, S.A.; Jacobs, E.J.; McCullough, M.L.; Patel, A.V.; Ma, J.; Soerjomataram, I.; et al. Proportion and Number of Cancer Cases and Deaths Attributable to Potentially Modifiable Risk Factors in the United States. CA Cancer J. Clin. 2018, 68, 31–54. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Luo, G.; Zhang, Y.; Rumgay, H.; Morgan, E.; Langselius, O.; Vignat, J.; Colombet, M.; Bray, F. Estimated Worldwide Variation and Trends in Incidence of Lung Cancer by Histological Subtype in 2022 and over Time: A Population-Based Study. Lancet Respir. Med. 2025, 13, 348–363. [Google Scholar] [CrossRef]
- Zhou, F.; Zhou, C. Lung Cancer in Never Smokers—The East Asian Experience. Transl. Lung Cancer Res. 2018, 7, 450. [Google Scholar] [CrossRef]
- Pandeya, N.; Williams, G.M.; Sadhegi, S.; Green, A.C.; Webb, P.M.; Whiteman, D.C. Associations of Duration, Intensity, and Quantity of Smoking with Adenocarcinoma and Squamous Cell Carcinoma of the Esophagus. Am. J. Epidemiol. 2008, 168, 105–114. [Google Scholar] [CrossRef]
- Wang, J.; Wang, M. OA04.06 The Global Landscape of Lung Squamous Cell Carcinoma, Lung Adenocarcinoma, Small Cell Lung Cancer Incidence in 2020. J. Thorac. Oncol. 2023, 18, S52–S53. [Google Scholar] [CrossRef]
- Zhang, Y.; Vaccarella, S.; Morgan, E.; Li, M.; Etxeberria, J.; Chokunonga, E.; Manraj, S.S.; Kamate, B.; Omonisi, A.; Bray, F. Global Variations in Lung Cancer Incidence by Histological Subtype in 2020: A Population-Based Study. Lancet Oncol. 2023, 24, 1206–1218. [Google Scholar] [CrossRef]
- Mitchell, K.A.; Zingone, A.; Toulabi, L.; Boeckelman, J.; Ryan, B.M. Comparative Transcriptome Profiling Reveals Coding and Noncoding RNA Differences in NSCLC from African Americans and European Americans. Clin. Cancer Res. 2017, 23, 7412–7425. [Google Scholar] [CrossRef]
- Sampson, J.N.; Wheeler, W.A.; Yeager, M.; Panagiotou, O.; Wang, Z.; Berndt, S.I.; Lan, Q.; Abnet, C.C.; Amundadottir, L.T.; Figueroa, J.D.; et al. Analysis of Heritability and Shared Heritability Based on Genome-Wide Association Studies for Thirteen Cancer Types. J. Natl. Cancer Inst. 2015, 107, djv279. [Google Scholar] [CrossRef]
- Zanetti, K.A.; Wang, Z.; Aldrich, M.; Amos, C.I.; Blot, W.J.; Bowman, E.D.; Burdette, L.; Cai, Q.; Caporaso, N.; Chung, C.C.; et al. Genome-Wide Association Study Confirms Lung Cancer Susceptibility Loci on Chromosomes 5p15 and 15q25 in an African-American Population. Lung Cancer 2016, 98, 33–42. [Google Scholar] [CrossRef]
- McKay, J.D.; Hung, R.J.; Han, Y.; Zong, X.; Carreras-Torres, R.; Christiani, D.C.; Caporaso, N.E.; Johansson, M.; Xiao, X.; Li, Y.; et al. Large-Scale Association Analysis Identifies New Lung Cancer Susceptibility Loci and Heterogeneity in Genetic Susceptibility across Histological Subtypes. Nat. Genet. 2017, 49, 1126–1132. [Google Scholar] [CrossRef]
- Adib, E.; Nassar, A.H.; Abou Alaiwi, S.; Groha, S.; Akl, E.W.; Sholl, L.M.; Michael, K.S.; Awad, M.M.; Jänne, P.A.; Gusev, A.; et al. Variation in Targetable Genomic Alterations in Non-Small Cell Lung Cancer by Genetic Ancestry, Sex, Smoking History, and Histology. Genome Med. 2022, 14, 39. [Google Scholar] [CrossRef]
- May, L.; Shows, K.; Nana-Sinkam, P.; Li, H.; Landry, J.W. Sex Differences in Lung Cancer. Cancers 2023, 15, 3111. [Google Scholar] [CrossRef]
- Carrot-Zhang, J.; Soca-Chafre, G.; Patterson, N.; Thorner, A.R.; Nag, A.; Watson, J.; Genovese, G.; Rodriguez, J.; Gelbard, M.K.; Corrales-Rodriguez, L.; et al. Genetic Ancestry Contributes to Somatic Mutations in Lung Cancers from Admixed Latin American Populations. Cancer Discov. 2020, 11, 591. [Google Scholar] [CrossRef]
- Chen, G.; Wei, R.S.; Ma, J.; Li, X.H.; Feng, L.; Yu, J.R. FOXA1 Prolongs S Phase and Promotes Cancer Progression in Non-Small Cell Lung Cancer through Upregulation of CDC5L and Activation of the ERK1/2 and JAK2 Pathways. Kaohsiung J. Med. Sci. 2023, 39, 1077–1086. [Google Scholar] [CrossRef]
- Gu, Y.; Tang, Y.Y.; Wan, J.X.; Zou, J.Y.; Lu, C.G.; Zhu, H.S.; Sheng, S.Y.; Wang, Y.F.; Liu, H.C.; Yang, J.; et al. Sex Difference in the Expression of PD-1 of Non-Small Cell Lung Cancer. Front. Immunol. 2022, 13, 1026214. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, L.; Thaiparambil, J.; Mai, S.; Perera, D.N.; Zhang, J.; Pan, P.Y.; Coarfa, C.; Ramos, K.; Chen, S.H.; et al. Patients with Lung Cancer of Different Racial Backgrounds Harbor Distinct Immune Cell Profiles. Cancer Res. Commun. 2022, 2, 884. [Google Scholar] [CrossRef]
- Ye, Y.; Jing, Y.; Li, L.; Mills, G.B.; Diao, L.; Liu, H.; Han, L. Sex-Associated Molecular Differences for Cancer Immunotherapy. Nat. Commun. 2020, 11, 1779. [Google Scholar] [CrossRef]
- Al Mamun, A.; Sobhan, M.; Tanvir, R.B.; Dimitroff, C.J.; Mondal, A.M. Deep Learning to Discover Cancer Glycome Genes Signifying the Origins of Cancer. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Republic of Korea, 16–19 December 2020; pp. 2425–2431. [Google Scholar] [CrossRef]
- Al Mamun, A.; Tanvir, R.B.; Sobhan, M.; Mathee, K.; Narasimhan, G.; Holt, G.E.; Mondal, A.M. Multi-Run Concrete Autoencoder to Identify Prognostic LncRNAs for 12 Cancers. Int. J. Mol. Sci. 2021, 22, 11919. [Google Scholar] [CrossRef]
- Maharjan, M.; Tanvir, R.B.; Chowdhury, K.; Duan, W.; Mondal, A.M. Computational Identification of Biomarker Genes for Lung Cancer Considering Treatment and Non-Treatment Studies. BMC Bioinform. 2020, 21, 218. [Google Scholar] [CrossRef]
- Sobhan, M.; Mamun, A.A.; Tanvir, R.B.; Alfonso, M.J.; Valle, P.; Mondal, A.M. Deep Learning to Discover Genomic Signatures for Racial Disparity in Lung Cancer. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Republic of Korea, 16–19 December 2020; pp. 2990–2992. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. Adv. Neural Inf. Process. Syst. 2017, 30, 4766–4775. [Google Scholar] [CrossRef]
- Sobhan, M.; Mondal, A.M. Explainable Machine Learning to Identify Patient-Specific Biomarkers for Lung Cancer. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 6–8 December 2022; pp. 3152–3159. [Google Scholar] [CrossRef]
- Captier, N.; Lerousseau, M.; Orlhac, F.; Hovhannisyan-Baghdasarian, N.; Luporsi, M.; Woff, E.; Lagha, S.; Salamoun Feghali, P.; Lonjou, C.; Beaulaton, C.; et al. Integration of Clinical, Pathological, Radiological, and Transcriptomic Data Improves Prediction for First-Line Immunotherapy Outcome in Metastatic Non-Small Cell Lung Cancer. Nat. Commun. 2025, 16, 614. [Google Scholar] [CrossRef]
- Khadirnaikar, S.; Shukla, S.; Prasanna, S.R.M. Machine Learning Based Combination of Multi-Omics Data for Subgroup Identification in Non-Small Cell Lung Cancer. Sci. Rep. 2023, 13, 4636. [Google Scholar] [CrossRef]
- Tanvir, R.B.; Islam, M.M.; Sobhan, M.; Luo, D.; Mondal, A.M. MOGAT: A Multi-Omics Integration Framework Using Graph Attention Networks for Cancer Subtype Prediction. Int. J. Mol. Sci. 2024, 25, 2788. [Google Scholar] [CrossRef]
- Jeong, D.; Koo, B.; Oh, M.; Kim, T.B.; Kim, S. GOAT: Gene-Level Biomarker Discovery from Multi-Omics Data Using Graph ATtention Neural Network for Eosinophilic Asthma Subtype. Bioinformatics 2023, 39, btad582. [Google Scholar] [CrossRef]
- Li, Z.; Katz, S.; Saccenti, E.; Fardo, D.W.; Claes, P.; Martins dos Santos, V.A.P.; Van Steen, K.; Roshchupkin, G.V. Novel Multi-Omics Deconfounding Variational Autoencoders Can Obtain Meaningful Disease Subtyping. Brief. Bioinform. 2024, 25, 512. [Google Scholar] [CrossRef]
- Verma, M. Personalized Medicine and Cancer. J. Pers. Med. 2012, 2, 1–14. [Google Scholar] [CrossRef]
- National Academies of Sciences Engineering and Medicine. Using Population Descriptors in Genetics and Genomics Research: A New Framework for an Evolving Field; National Academies Press: Washington, DC, USA, 2023; ISBN 978-0-309-70062-7. [Google Scholar]
- Grzywa, T.M.; Paskal, W.; Włodarski, P.K. Intratumor and Intertumor Heterogeneity in Melanoma. Transl. Oncol. 2017, 10, 956–975. [Google Scholar] [CrossRef]
- Wang, Q.; Armenia, J.; Zhang, C.; Penson, A.V.; Reznik, E.; Zhang, L.; Minet, T.; Ochoa, A.; Gross, B.E.; Iacobuzio-Donahue, C.A.; et al. Unifying Cancer and Normal RNA Sequencing Data from Different Sources. Sci. Data 2018, 5, 180061. [Google Scholar] [CrossRef]
- Gao, G.F.; Parker, J.S.; Reynolds, S.M.; Silva, T.C.; Wang, L.B.; Zhou, W.; Akbani, R.; Bailey, M.; Balu, S.; Berman, B.P.; et al. Before and After: Comparison of Legacy and Harmonized TCGA Genomic Data Commons’ Data. Cell Syst. 2019, 9, 24–34.e10. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Sobhan, M.; Mondal, A.M. Evaluating SHAP’s Robustness in Precision Medicine: Effect of Filtering and Normalization. In Proceedings of the 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Istanbul, Türkiye, 5–8 December 2023; pp. 3157–3164. [Google Scholar] [CrossRef]
- Soini, Y. Tight Junctions in Lung Cancer and Lung Metastasis: A Review. Int. J. Clin. Exp. Pathol. 2012, 5, 126. [Google Scholar]
- Bayleyegn, B.; Adane, T.; Getawa, S.; Aynalem, M.; Kifle, Z.D. Coagulation Parameters in Lung Cancer Patients: A Systematic Review and Meta-analysis. J. Clin. Lab. Anal. 2022, 36, e24550. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, L.; Ma, Y.; Liu, Y.; Zhu, Q.; Huang, Y.; Feng, W. BACH1 Promotes Lung Adenocarcinoma Cell Metastasis through Transcriptional Activation of ITGA2. Cancer Sci. 2023, 114, 3568–3582. [Google Scholar] [CrossRef]
- Fu, L.; Shi, K.; Wang, J.; Chen, W.; Shi, D.; Tian, Y.; Guo, W.; Yu, W.; Xiao, X.; Kang, T.; et al. TFAP2B Overexpression Contributes to Tumor Growth and a Poor Prognosis of Human Lung Adenocarcinoma through Modulation of ERK and VEGF/PEDF Signaling. Mol. Cancer 2014, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Yano, K. Lipid Metabolic Pathways as Lung Cancer Therapeutic Targets: A Computational Study. Int. J. Mol. Med. 2011, 29, 519. [Google Scholar] [CrossRef]
- Ross, K.E.; Zhang, G.; Akcora, C.; Lin, Y.; Fang, B.; Koomen, J.; Haura, E.B.; Grimes, M. Network Models of Protein Phosphorylation, Acetylation, and Ubiquitination Connect Metabolic and Cell Signaling Pathways in Lung Cancer. PLOS Comput. Biol. 2023, 19, e1010690. [Google Scholar] [CrossRef]
- Vanhove, K.; Graulus, G.J.; Mesotten, L.; Thomeer, M.; Derveaux, E.; Noben, J.P.; Guedens, W.; Adriaensens, P. The Metabolic Landscape of Lung Cancer: New Insights in a Disturbed Glucose Metabolism. Front. Oncol. 2019, 9, 492161. [Google Scholar] [CrossRef]
- Torresano, L.; Santacatterina, F.; Domínguez-Zorita, S.; Nuevo-Tapioles, C.; Núñez-Salgado, A.; Esparza-Moltó, P.B.; González-Llorente, L.; Romero-Carramiñana, I.; Núñez de Arenas, C.; Sánchez-Garrido, B.; et al. Analysis of the Metabolic Proteome of Lung Adenocarcinomas by Reverse-Phase Protein Arrays (RPPA) Emphasizes Mitochondria as Targets for Therapy. Oncogenesis 2022, 11, 24. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Song, C.; Li, Q. Identification of Four Metabolic Subtypes and Key Prognostic Markers in Lung Adenocarcinoma Based on Glycolytic and Glutaminolytic Pathways. BMC Cancer 2023, 23, 152. [Google Scholar] [CrossRef]
- He, L.; Chen, J.; Xu, F.; Li, J. Prognostic Implication of a Metabolism-Associated Gene Signature in Lung Adenocarcinoma. Mol. Ther. Oncolytics 2020, 19, 265. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, S.; Zhang, M. Identification of Urine Biomarkers Associated with Lung Adenocarcinoma. Oncotarget 2017, 8, 38517. [Google Scholar] [CrossRef]
- Anusewicz, D.; Orzechowska, M.; Bednarek, A.K. Lung Squamous Cell Carcinoma and Lung Adenocarcinoma Differential Gene Expression Regulation through Pathways of Notch, Hedgehog, Wnt, and ErbB Signalling. Sci. Rep. 2020, 10, 21128. [Google Scholar] [CrossRef] [PubMed]
- Jyotsana, N.; Ta, K.T.; DelGiorno, K.E. The Role of Cystine/Glutamate Antiporter SLC7A11/XCT in the Pathophysiology of Cancer. Front. Oncol. 2022, 12, 858462. [Google Scholar] [CrossRef]
- Liu, J.; Xia, X.; Huang, P. XCT: A Critical Molecule That Links Cancer Metabolism to Redox Signaling. Mol. Ther. 2020, 28, 2358. [Google Scholar] [CrossRef]
- Carreca, A.P.; Tinnirello, R.; Miceli, V.; Galvano, A.; Gristina, V.; Incorvaia, L.; Pampalone, M.; Taverna, S.; Iannolo, G. Extracellular Vesicles in Lung Cancer: Implementation in Diagnosis and Therapeutic Perspectives. Cancers 2024, 16, 1967. [Google Scholar] [CrossRef]
- Pandya, P.; Al-Qasrawi, D.S.; Klinge, S.; Justilien, V. Extracellular Vesicles in Non-Small Cell Lung Cancer Stemness and Clinical Applications. Front. Immunol. 2024, 15, 1369356. [Google Scholar] [CrossRef]
- Carbone, D.P.; Gandara, D.R.; Antonia, S.J.; Zielinski, C.; Paz-Ares, L. Non-Small-Cell Lung Cancer: Role of the Immune System and Potential for Immunotherapy. J. Thorac. Oncol. 2015, 10, 974–984. [Google Scholar] [CrossRef]
- Yanagi, S.; Kishimoto, H.; Kawahara, K.; Sasaki, T.; Sasaki, M.; Nishio, M.; Yajima, N.; Hamada, K.; Horie, Y.; Kubo, H.; et al. Pten Controls Lung Morphogenesis, Bronchioalveolar Stem Cells, and Onset of Lung Adenocarcinomas in Mice. J. Clin. Investig. 2007, 117, 2929. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Expression of ZNF253 in Cancer—Summary—The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000256771-ZNF253/cancer?utm_source=chatgpt.com (accessed on 16 October 2025).
- Jiang, Y.; Ouyang, W.; Zhang, C.; Yu, Y.; Yao, H. Prognosis and Immunotherapy Response With a Novel Golgi Apparatus Signature-Based Formula in Lung Adenocarcinoma. Front. Cell Dev. Biol. 2022, 9, 817085. [Google Scholar] [CrossRef]
- Sun, T.; Chen, J.; Yang, F.; Zhang, G.; Chen, J.; Wang, X.; Zhang, J. Lipidomics Reveals New Lipid-Based Lung Adenocarcinoma Early Diagnosis Model. EMBO Mol. Med. 2024, 16, 854–869. [Google Scholar] [CrossRef]
- He, J.; Li, W.; Li, Y.; Liu, G. Construction of a Prognostic Model for Lung Adenocarcinoma Based on Bioinformatics Analysis of Metabolic Genes. Transl. Cancer Res. 2020, 9, 3518. [Google Scholar] [CrossRef]
- Chen, M.M.; Guo, W.; Chen, S.M.; Guo, X.Z.; Xu, L.; Ma, X.Y.; Wang, Y.X.; Xie, C.; Meng, L.H. Xanthine Dehydrogenase Rewires Metabolism and the Survival of Nutrient Deprived Lung Adenocarcinoma Cells by Facilitating UPR and Autophagic Degradation. Int. J. Biol. Sci. 2023, 19, 772–788. [Google Scholar] [CrossRef]
- Wang, F.M.; Xu, L.Q.; Zhang, Z.C.; Guo, Q.; Du, Z.P.; Lei, Y.; Han, X.; Wu, C.Y.; Zhao, F.; Chen, J.L. SLC7A8 Overexpression Inhibits the Growth and Metastasis of Lung Adenocarcinoma and Is Correlated with a Dismal Prognosis. Aging 2024, 16, 1605. [Google Scholar] [CrossRef]
- Kuo, T.C.; Huang, K.Y.; Yang, S.C.; Wu, S.; Chung, W.C.; Chang, Y.L.; Hong, T.M.; Wang, S.P.; Chen, H.Y.; Hsiao, T.H.; et al. Monocarboxylate Transporter 4 Is a Therapeutic Target in Non-Small Cell Lung Cancer with Aerobic Glycolysis Preference. Mol. Ther. Oncolytics 2020, 18, 189. [Google Scholar] [CrossRef]
- Eilertsen, M.; Andersen, S.; Al-Saad, S.; Kiselev, Y.; Donnem, T.; Stenvold, H.; Pettersen, I.; Al-Shibli, K.; Richardsen, E.; Busund, L.T.; et al. Monocarboxylate Transporters 1–4 in NSCLC: MCT1 Is an Independent Prognostic Marker for Survival. PLoS ONE 2014, 9, e105038. [Google Scholar] [CrossRef]
- Bharadwaj, R.; Jaiswal, S.; Velarde de la Cruz, E.E.; Thakare, R.P. Targeting Solute Carrier Transporters (SLCs) as a Therapeutic Target in Different Cancers. Diseases 2024, 12, 63. [Google Scholar] [CrossRef]
- Li, C.; Wan, Y.; Deng, W.; Fei, F.; Wang, L.; Qi, F.; Zheng, Z. Promising Novel Biomarkers and Candidate Small-Molecule Drugs for Lung Adenocarcinoma: Evidence from Bioinformatics Analysis of High-Throughput Data. Open Med. 2022, 17, 96–112. [Google Scholar] [CrossRef]
- Summerbell, E.R.; Mouw, J.K.; Bell, J.S.K.; Knippler, C.M.; Pedro, B.; Arnst, J.L.; Khatib, T.O.; Commander, R.; Barwick, B.G.; Konen, J.; et al. Epigenetically Heterogeneous Tumor Cells Direct Collective Invasion through Filopodia-Driven Fibronectin Micropatterning. Sci. Adv. 2020, 6, eaaz6197. [Google Scholar] [CrossRef]
- Snoke, D.B.; Bellefleur, E.; Rehman, H.T.; Carson, J.A.; Poynter, M.E.; Dittus, K.L.; Toth, M.J. Skeletal Muscle Adaptations in Patients with Lung Cancer: Longitudinal Observations from the Whole Body to Cellular Level. J. Cachexia Sarcopenia Muscle 2023, 14, 2579–2590. [Google Scholar] [CrossRef]
- Allegrini, S.; Camici, M.; Garcia-Gil, M.; Pesi, R.; Tozzi, M.G. Interplay between MTOR and Purine Metabolism Enzymes and Its Relevant Role in Cancer. Int. J. Mol. Sci. 2024, 25, 6735. [Google Scholar] [CrossRef]
- Dai, G.; Sun, Y. Knockdown of GNL3 Inhibits LUAD Cell Growth by Regulating Wnt–β-Catenin Pathway. Allergol. Immunopathol. 2024, 52, 46–52. [Google Scholar] [CrossRef]
- Xu, S.; Liu, R.; Da, Y. Comparison of Tumor Related Signaling Pathways with Known Compounds to Determine Potential Agents for Lung Adenocarcinoma. Thorac. Cancer 2018, 9, 974. [Google Scholar] [CrossRef]
- Orstad, G.; Fort, G.; Parnell, T.J.; Jones, A.; Stubben, C.; Lohman, B.; Gillis, K.L.; Orellana, W.; Tariq, R.; Klingbeil, O.; et al. FoxA1 and FoxA2 Control Growth and Cellular Identity in NKX2-1-Positive Lung Adenocarcinoma. Dev. Cell 2022, 57, 1866–1882.e10. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, L.; Ji, L.; Zhang, F.; Chen, D.; Duan, S.; Shen, H.; Liang, Y.; Chen, Y. Potentiated Lung Adenocarcinoma (LUAD) Cell Growth, Migration and Invasion by LncRNA DARS-AS1 via MiR-188-5p/KLF12 Axis. Aging 2021, 13, 23376. [Google Scholar] [CrossRef]
- Song, P.; Rekow, S.S.; Singleton, C.A.; Sekhon, H.S.; Dissen, G.A.; Zhou, M.; Campling, B.; Lindstrom, J.; Spindel, E.R. Choline Transporter-like Protein 4 (CTL4) Links to Non-Neuronal Acetylcholine Synthesis. J. Neurochem. 2013, 126, 451–461. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, R. Cutaneous Metastases from Lung Adenocarcinoma. Case Rep. Dermatol. Med. 2020, 2020, 8880604. [Google Scholar] [CrossRef]
- Hynds, R.E.; Janes, S.M. Airway Basal Cell Heterogeneity and Lung Squamous Cell Carcinoma. Cancer Prev. Res. 2017, 10, 491–493. [Google Scholar] [CrossRef]
- Jiang, S.; Zhu, D.; Wang, Y. Tumor-Infiltrating B Cells in Non-Small Cell Lung Cancer: Current Insights and Future Directions. Cancer Cell Int. 2025, 25, 68. [Google Scholar] [CrossRef]
- Yang, M.; Guo, Y.; Guo, X.; Mao, Y.; Zhu, S.; Wang, N.; Lu, D. Analysis of the Effect of NEKs on the Prognosis of Patients with Non-Small-Cell Lung Carcinoma Based on Bioinformatics. Sci. Rep. 2022, 12, 1705. [Google Scholar] [CrossRef]
- Eldridge, L.; Moldobaeva, A.; Zhong, Q.; Jenkins, J.; Snyder, M.; Brown, R.H.; Mitzner, W.; Wagner, E.M. Bronchial Artery Angiogenesis Drives Lung Tumor Growth. Cancer Res. 2016, 76, 5962. [Google Scholar] [CrossRef]
- Kim, M.S.; Jeong, H.; Choi, B.H.; Park, J.; Shin, G.S.; Jung, J.H.; Shin, H.; Kang, K.W.; Jeon, O.H.; Yu, J.; et al. GCC2 Promotes Non-Small Cell Lung Cancer Progression by Maintaining Golgi Apparatus Integrity and Stimulating EGFR Signaling Pathways. Sci. Rep. 2024, 14, 28926. [Google Scholar] [CrossRef]
- Shi, Y.; Li, Y.; Yan, C.; Su, H.; Ying, K. Identification of Key Genes and Evaluation of Clinical Outcomes in Lung Squamous Cell Carcinoma Using Integrated Bioinformatics Analysis. Oncol. Lett. 2019, 18, 5859–5870. [Google Scholar] [CrossRef]
- Kurmi, K.; Haigis, M.C. Nitrogen Metabolism in Cancer and Immunity. Trends Cell Biol. 2020, 30, 408. [Google Scholar] [CrossRef]
- Niu, Z.; Jin, R.; Zhang, Y.; Li, H. Signaling Pathways and Targeted Therapies in Lung Squamous Cell Carcinoma: Mechanisms and Clinical Trials. Signal Transduct. Target. Ther. 2022, 7, 353. [Google Scholar] [CrossRef]
- Lau, S.C.M.; Pan, Y.; Velcheti, V.; Wong, K.K. Squamous Cell Lung Cancer: Current Landscape and Future Therapeutic Options. Cancer Cell 2022, 40, 1279–1293. [Google Scholar] [CrossRef]

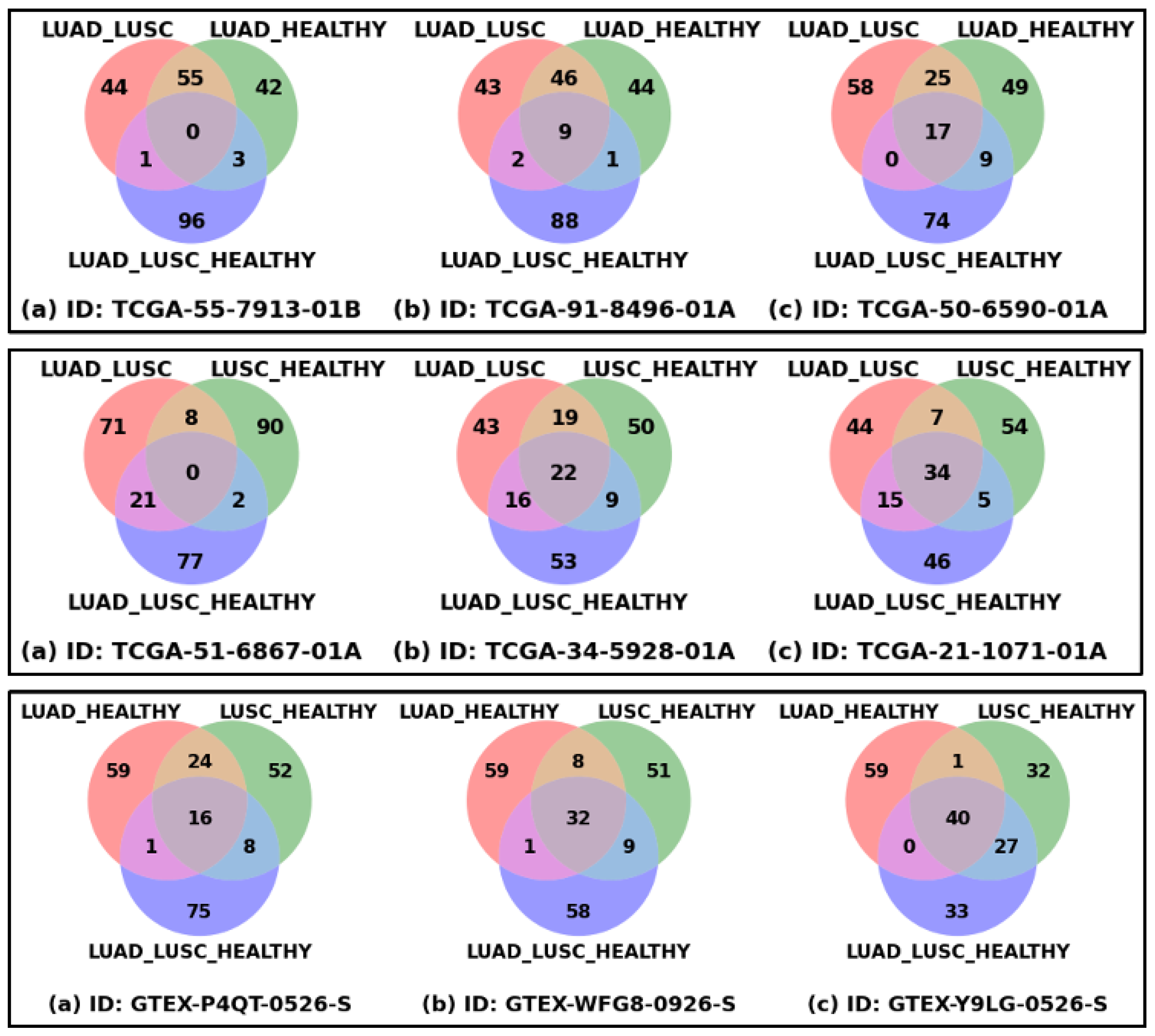
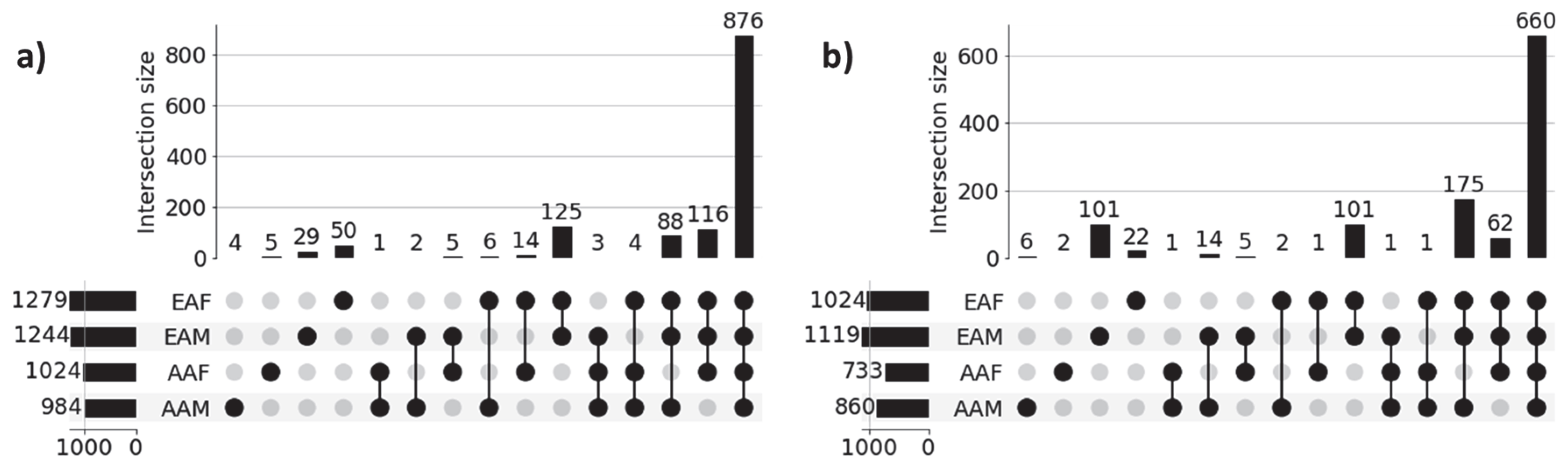
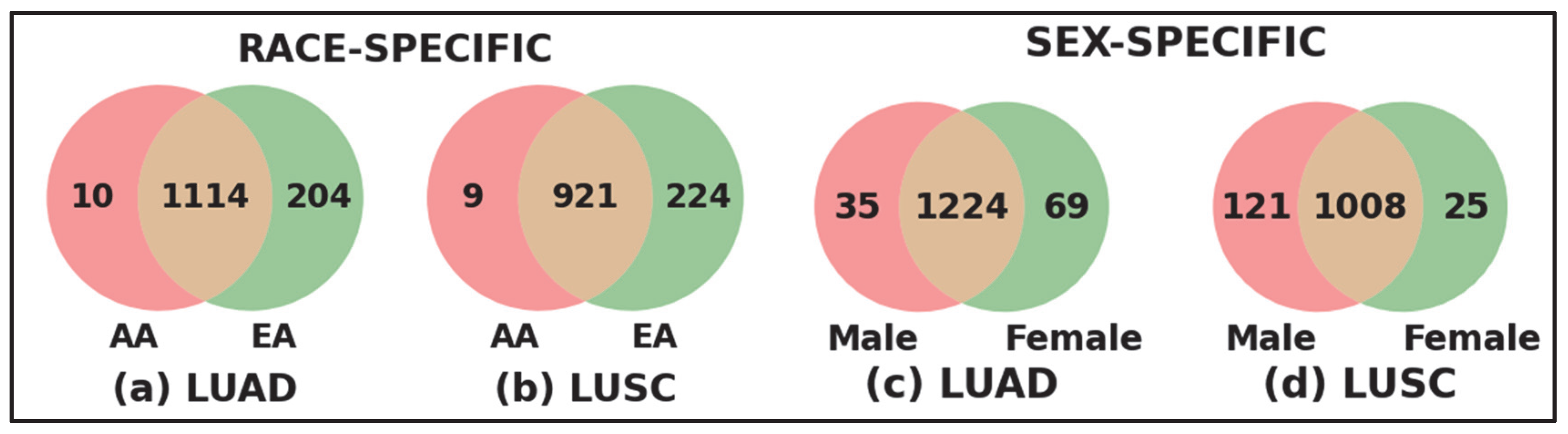
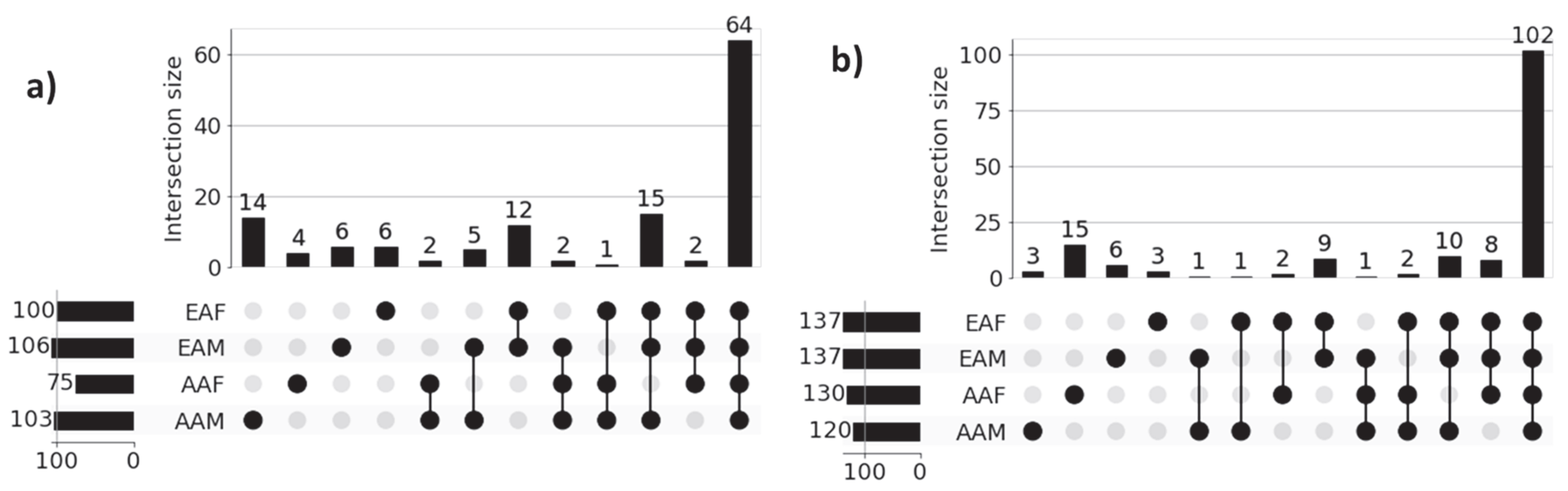
| Disease | Cohort | Sample | |
|---|---|---|---|
| LUAD | AA | EA | |
| Male | 20 | 136 | 156 |
| Female | 27 | 173 | 200 |
| LUSC | |||
| Male | 16 | 195 | 211 |
| Female | 12 | 72 | 84 |
| GTEX Healthy Samples | 313 | ||
| Total Samples | 964 | ||
| Classification Problems | Using Default Values of Hyperparameters | Tuned | |||||
|---|---|---|---|---|---|---|---|
| FCN | LR | NB | SVM | RF | XGBoost | XGBoost | |
| LUAD-LUSC-HEALTHY | 93.36 ± 2.67 | 94.60 ± 0.84 | 65.86 ± 13.29 | 40.77 ± 7.60 | 95.12 ± 1.48 | 96.16 ± 1.29 | 96.37 ± 1.57 |
| LUAD-LUSC | 94.01 ± 1.64 | 93.24 ± 0.57 | 92.33 ± 1.26 | 93.24 ± 1.48 | 94.62 ± 1.29 | 94.77 ± 1.49 | 95.24 ± 0.90 |
| LUAD-HEALTHY | 99.25 ± 0.67 | 99.25 ± 0.82 | 75.78 ± 9.68 | 58.59 ± 10.63 | 98.96 ± 0.60 | 99.10 ± 0.56 | 99.25 ± 0.47 |
| LUSC-HEALTHY | 99.01 ± 0.96 | 99.18 ± 0.52 | 73.88 ± 11.59 | 56.77 ± 10.46 | 99.51 ± 0.66 | 99.51 ± 0.40 | 99.51 ± 0.40 |
| LUAD Cohort | LUSC Cohort | |||||||
|---|---|---|---|---|---|---|---|---|
| Sub-Cohort | Actual | Correct Prediction | Actual | Correct Prediction | ||||
| Proposed | 4-Class: LUAD | 8-Class | Proposed | 4-Class: LUSC | 8-Class | |||
| AAM | 20 | 18 (90%) | 0 (0%) | 0 (0%) | 16 | 14 (88%) | 1 (6%) | 1 (6%) |
| AAF | 27 | 26 (96%) | 2 (7%) | 2 (7%) | 12 | 12 (100%) | 0 (0%) | 2 (16%) |
| EAM | 136 | 129 (95%) | 133 (98%) | 127(93%) | 195 | 182 (93%) | 194 (99%) | 184 (94%) |
| EAF | 173 | 161 (93%) | 172 (99%) | 165 (95%) | 72 | 63 (88%) | 71 (87%) | 63 (88%) |
| Total | 356 | 334 (94%) | 307 (86%) | 294 (83%) | 295 | 271 (92%) | 266 (90%) | 250 (85%) |
| Sub-Cohort | LUAD | LUSC |
|---|---|---|
| AAM | CPT2, ETHE1, TTLL2, XRCC3 | CCL4L2, DNLZ, MAP1LC3A, MLKL, NMRK2, SHKBP1 |
| EAM | ABCB5, CRYBA1, PRKX, RP11-351M8.1, SYCE1L | RAB40A, SUPT5H |
| AAF | AMT, APOBEC3G, ARG2, CIDEC, CMPK2, CYBA, CYP3A5, EIF2AK1, ERVW-1, FUT3, GOLGA8J, MAPK10, MBOAT4, MS4A18, MTHFD1L, NMUR1, OR7A5, PAK2, PEX5, POU2F1, RP11-849F2.7, SLC26A9, SLC32A1, SLC8A2, SMIM7, TNFAIP8L2, TRPS1, ZMYND8, ZNF324B | ADCYAP1, AGPAT4, ANGEL1, APOE, ARG2, ATP2B2, AURKAIP1, BEND5, C10orf10, C10orf90, C12orf40, C16orf74, CAMKK2, CASP1, CASP5, CCAR2, CCDC12, CD276, CD58, CDC42BPA, CELA3A, CEP170B, CPAMD8, CSRP2, CT45A4, CTD-2287O16.3, CXCL3, CYP24A1, DBH, DDX3Y, DMC1, DTYMK, EFR3B, ETHE1, EVC2, F2, FA2H, FAM222B, FER, GPR113, GRIA2, ID1, ITPKB, KRT72, KRTAP13-1, LDB3, LINGO3, LPHN3, MAN1B1, MB21D2, MFAP3, MFAP4, MGRN1, MRPL14, MRPL53, MTFR1, NDST3, NKAPL, NMNAT2, NPIPA5, NR3C1, NYAP1, OR8G5, P2RX5-TAX1BP3, PDE6A, PDIA2, PGLYRP1, PIGW, POU2AF1, PPP1R37, PRAMEF12, PRRG4, PRSS3, PTK2, PXDN, RAB7L1, RAPGEF1, RDH14, RGPD2, RHBDD1, RP11-302B13.5, RP11-794P6.2, SAA4, SCEL, SERTAD1, SFT2D3, SLC26A7, SLC6A7, SMIM7, SNX7, TDRD15, TMEM101, TMEM25, TOR1A, TRIM36, TSHR, UPK1A, YARS, ZNF324B, ZNF514, ZNF766 |
| EAF | AC135983.2, AGPAT4, AL139099.1, ALDOA, BAI1, BRF1, C10orf90, C18orf54, C6orf222, CAMKK2, CCDC168, CEACAM21, CELF5, CNTF, CSRP2, CXCL12, DPRX, DUT, F2, GPR150, GPR84, KCNIP4, KIAA0195, KRTAP13-1, LRRC8E, MAN1C1, MAPK15, MFAP3, MKI67, MYL5, MYO15B, NBPF11, NUTM2B, OR8G5, PRAC1, PSG2, RHBDD1, RP11-77K12.1, SAMD12, SLC1A4, SYN1, TEX101, TLDC1, TMEM130, TMEM17, TMEM25, ZFAND5, ZNF219, ZNF337, ZNF436 | AC026740.1, AHNAK2, C18orf54, C6orf203, CAMSAP2, CD244, DLG5, EDEM1, FBXO11, G6PC2, GATAD1, GFPT2, GPR84, ISM2, KCNK4, LRRC8E, OCSTAMP, PRLH, SBSPON, SKP2, TFDP3, TMEM156 |
| Sub-Cohort | LUAD | LUSC |
|---|---|---|
| AAM | * Anchoring Junction, * Blood Coagulation, * Cytoskeleton Organization, * Factor: AP2BETA; Motif: SCCYCAGGSNN, Formation of The Cornified Envelope, Hindlimb Morphogenesis, Kidney; Distal Tubules [≥low], * Positive Regulation of Cellular Biosynthetic Process, * Positive Regulation of Macromolecule Biosynthetic Process, * Positive Regulation of Macromolecule Metabolic Process, * Protein Metabolic Process, * Regulation of Cellular Component Organization, Tissue Morphogenesis, Tube Morphogenesis | * Choline Transmembrane Transporter Activity, Pancreatic Cancer Subtypes, * Skin Epidermis Development |
| EAM | * Muscle Structure Development, * Purine Nucleoside Monophosphate Catabolic Process, * Purine Ribonucleoside Metabolic Process, * Purine Ribonucleoside Monophosphate Catabolic Process | * Activation of Nima Kinases Nek9, Nek6, Nek7, * Axon, * Circulatory System Process, Developmental Biology, * Golgi Apparatus, Macromolecule Localization, * Mitotic Nuclear Division, Negative Regulation of Chromosome Organization, * Nitrogen Compound Transport, * Positive Regulation of Growth, Positive Regulation of Multicellular Organismal Process, Regulation of Anatomical Structure Size, Regulation of Cellular Component Organization, * Regulation of Growth, Regulation of Organelle Organization |
| AAF | * Antiporter Activity, Defense Response, * Lipid Transport, Regulation of Response to Stimulus, Response to External Stimulus, * Small Molecule Biosynthetic Process | * Basal Part of Cell, * Basal Plasma Membrane, Basolateral Plasma Membrane, Factor: cpbp; Motif: SNCCCNN; Match Class: 1, * Mature B Cell Differentiation Involved in Immune Response, Response to Stress |
| EAF | * Cell Growth, * Cell–cell Signaling, Factor: ZNF253; Motif: SNGNSCGNGGNGCKGNN; Match Class: 1, * Positive Regulation of Growth, Regulation of Anatomical Structure Size, * Regulation of Cell Growth | * Negative Regulation of Cell Cycle Process, RNA Polymerase II-specific DNA Transcription Factor Binding, Skin 2; Cells in Corneal Layer [≥low] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobhan, M.; Islam, M.M.; Trepka, M.J.; Holt, G.E.; Dimitroff, C.J.; Mondal, A.M. TILDA-X: Transcriptome-Informed Lung Cancer Disparities via Explainable AI. Cancers 2025, 17, 3454. https://doi.org/10.3390/cancers17213454
Sobhan M, Islam MM, Trepka MJ, Holt GE, Dimitroff CJ, Mondal AM. TILDA-X: Transcriptome-Informed Lung Cancer Disparities via Explainable AI. Cancers. 2025; 17(21):3454. https://doi.org/10.3390/cancers17213454
Chicago/Turabian StyleSobhan, Masrur, Md Mezbahul Islam, Mary Jo Trepka, Gregory E. Holt, Charles J. Dimitroff, and Ananda M. Mondal. 2025. "TILDA-X: Transcriptome-Informed Lung Cancer Disparities via Explainable AI" Cancers 17, no. 21: 3454. https://doi.org/10.3390/cancers17213454
APA StyleSobhan, M., Islam, M. M., Trepka, M. J., Holt, G. E., Dimitroff, C. J., & Mondal, A. M. (2025). TILDA-X: Transcriptome-Informed Lung Cancer Disparities via Explainable AI. Cancers, 17(21), 3454. https://doi.org/10.3390/cancers17213454







