Predictive Value of the Loss of pRb Expression in the Malignant Transformation Risk of Oral Potentially Malignant Disorders: A Systematic Review and Meta-Analysis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol
2.2. Search Strategy
2.3. Eligibility Criteria
2.4. Study Selection Process
2.5. Data Extraction
2.6. Appraisal of Quality and Risk of Bias
2.7. Statistical Analysis
3. Results
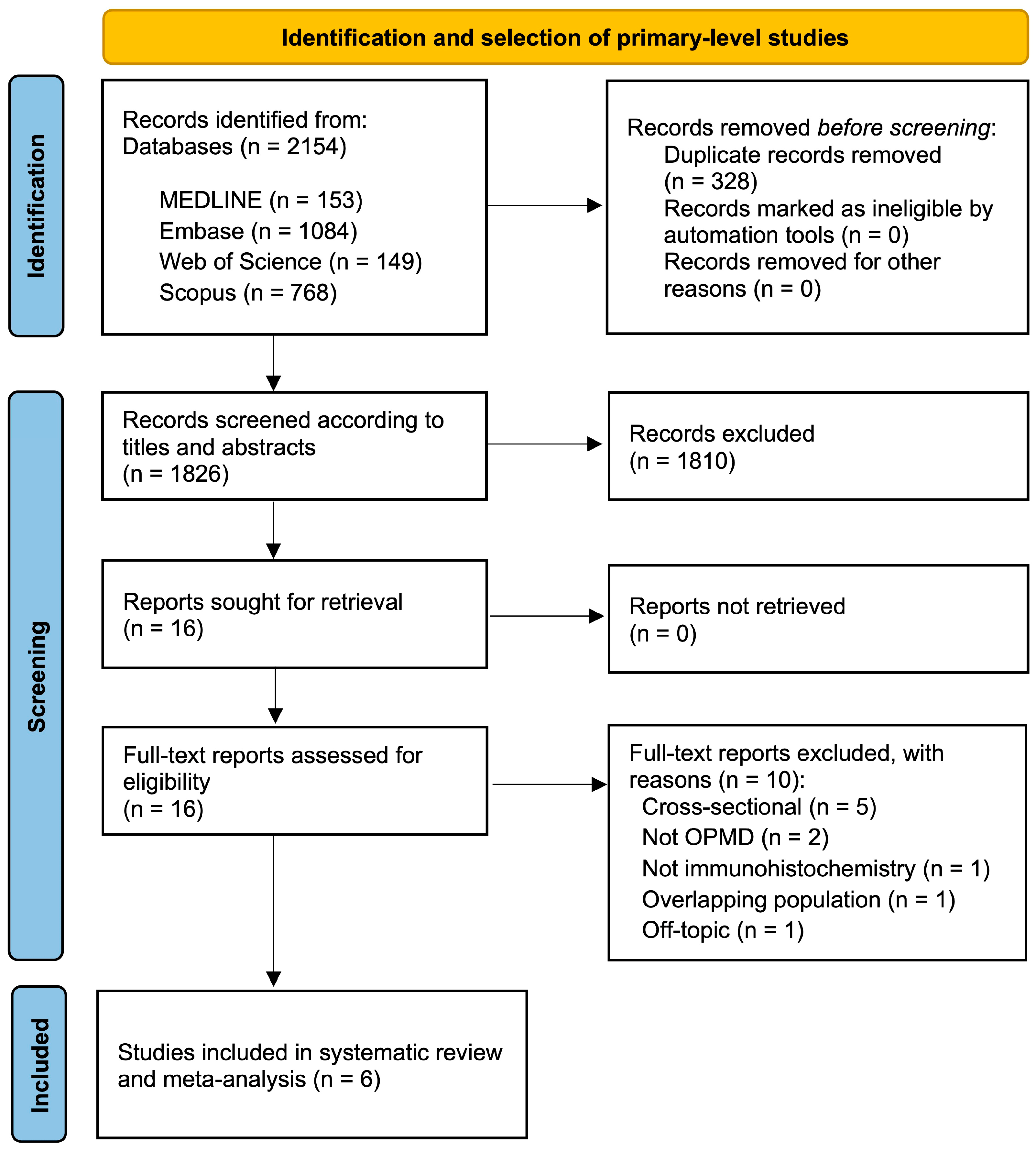
3.1. Results of the Literature Search
3.2. Study Characteristics
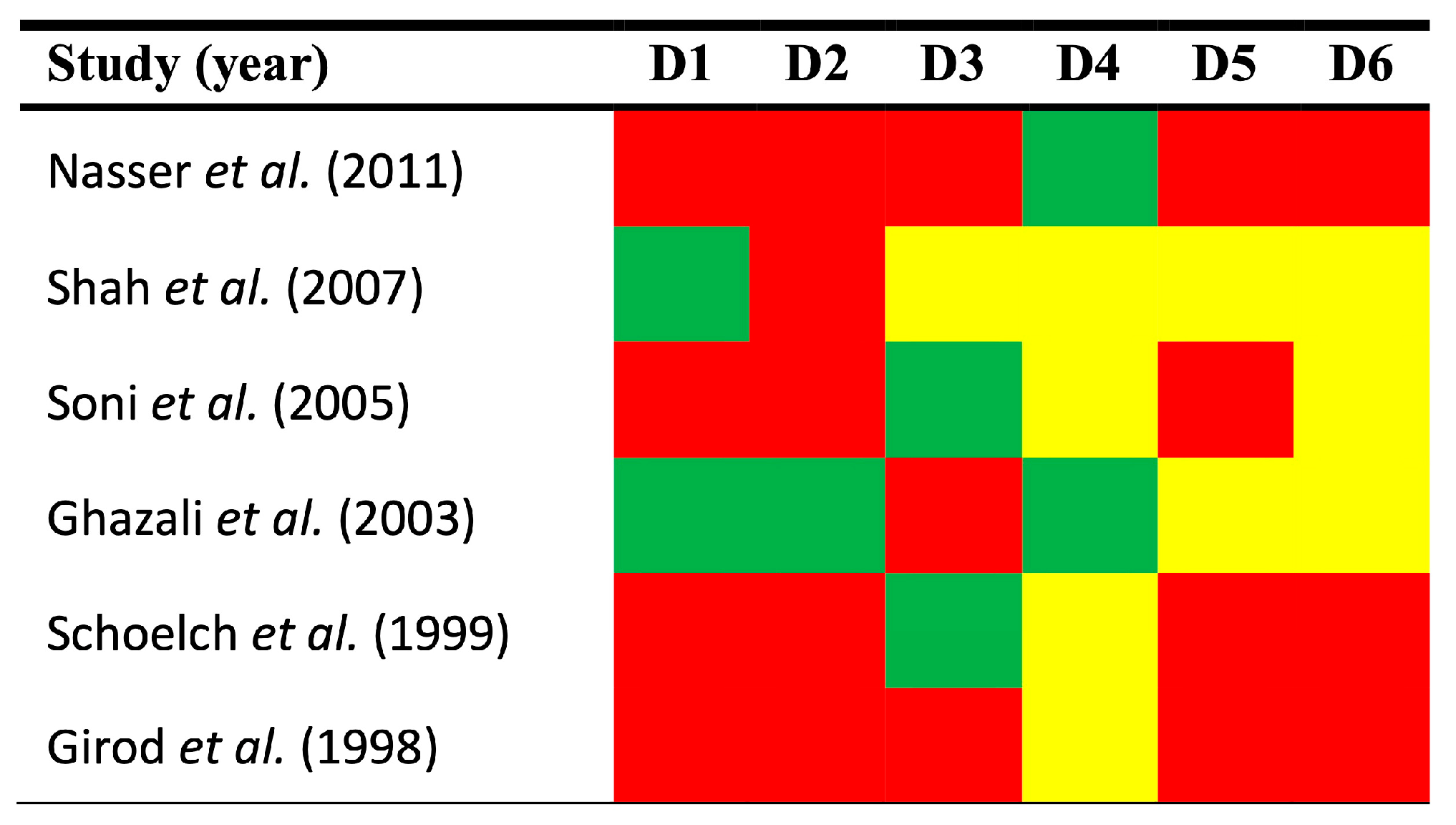
3.3. Qualitative Evaluation
3.4. Quantitative Evaluation (Meta-Analysis)
3.5. Quantitative Secondary Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- González-Moles, M.Á.; Warnakulasuriya, S.; López-Ansio, M.; Ramos-García, P. Hallmarks of Cancer Applied to Oral and Oropharyngeal Carcinogenesis: A Scoping Review of the Evidence Gaps Found in Published Systematic Reviews. Cancers 2022, 14, 3834. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J.; McCormick, F. The RB and p53 pathways in cancer. Cancer Cell 2002, 2, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Dannenberg, J.H.; te Riele, H.P. The retinoblastoma gene family in cell cycle regulation and suppression of tumorigenesis. Results Probl. Cell Differ. 2006, 42, 183–225. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Lee, W.H. The retinoblastoma gene: A prototypic and multifunctional tumor suppressor. Exp. Cell Res. 2001, 264, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, R.; D’Anneo, A.; Tesoriere, G.; Vento, R. RB1 in cancer: Different mechanisms of RB1 inactivation and alterations of pRb pathway in tumorigenesis. J. Cell. Physiol. 2013, 228, 1676–1687. [Google Scholar] [CrossRef] [PubMed]
- Dimaras, H.; Khetan, V.; Halliday, W.; Orlic, M.; Prigoda, N.L.; Piovesan, B.; Marrano, P.; Corson, T.W.; Eagle, R.C.; Squire, J.A.; et al. Loss of RB1 induces non-proliferative retinoma: Increasing genomic instability correlates with progression to retinoblastoma. Hum. Mol. Genet. 2008, 17, 1363–1372. [Google Scholar] [CrossRef]
- Corson, T.W.; Gallie, B.L. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosom. Cancer 2007, 46, 617–634. [Google Scholar] [CrossRef]
- Knudson, A.G. Genetic predisposition to cancer. Cancer Detect. Prev. 1984, 7, 1–8. [Google Scholar]
- Perez-Ordoñez, B.; Beauchemin, M.; Jordan, R.C.K. Molecular biology of squamous cell carcinoma of the head and neck. J. Clin. Pathol. 2006, 59, 445–453. [Google Scholar] [CrossRef]
- Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 2006, 110, 525–541. [Google Scholar] [CrossRef]
- Munakata, T.; Liang, Y.; Kim, S.; McGivern, D.R.; Huibregtse, J.; Nomoto, A.; Lemon, S.M. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007, 3, 1335–1347. [Google Scholar] [CrossRef]
- González-Moles, M.A.; Scully, C.; Ruiz-Ávila, I.; Plaza-Campillo, J.J. The cancer stem cell hypothesis applied to oral carcinoma. Oral Oncol. 2013, 49, 738–746. [Google Scholar] [CrossRef]
- Slack, R.S.; El-Bizri, H.; Wong, J.; Belliveau, D.J.; Miller, F.D. A critical temporal requirement for the retinoblastoma protein family during neuronal determination. J. Cell Biol. 1998, 140, 1497–1509. [Google Scholar] [CrossRef]
- Alaizari, N.A.; Sperandio, M.; Odell, E.W.; Peruzzo, D.; Al-Maweri, S.A. Meta-analysis of the predictive value of DNA aneuploidy in malignant transformation of oral potentially malignant disorders. J. Oral Pathol. Med. 2018, 47, 97–103. [Google Scholar] [CrossRef]
- Odell, E.W. Aneuploidy and loss of heterozygosity as risk markers for malignant transformation in oral mucosa. Oral Dis. 2021, 27, 1993–2007. [Google Scholar] [CrossRef] [PubMed]
- Ramos-García, P.; González-Moles, M.Á.; Ayén, Á.; González-Ruiz, L.; Gil-Montoya, J.A.; Ruiz-Ávila, I. Predictive value of CCND1/cyclin D1 alterations in the malignant transformation of potentially malignant head and neck disorders: Systematic review and meta-analysis. Head Neck 2019, 41, 3395–3407. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Rattay, T.; McConkey, C.; Helliwell, T.; Mehanna, H. Biomarkers in dysplasia of the oral cavity: A systematic review. Oral Oncol. 2009, 45, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Pouso, A.I.; Caponio, V.C.A.; Vieira, E.; Silva, F.F.; Pérez-Jardón, A.; Álvarez-Calderón-Iglesias, Ó.; Gándara-Vila, P.; Pannone, G.; Pérez-Sayáns, M. Predictive value of CDKN2A/p16INK4a expression in the malignant transformation of oral potentially malignant disorders: Systematic review and meta-analysis. Pathol. Res. Pract. 2023, 248, 154656. [Google Scholar] [CrossRef]
- Ramos-García, P.; González-Moles, M.Á.; Warnakulasuriya, S. Significance of p53 overexpression in the prediction of the malignant transformation risk of oral potentially malignant disorders: A systematic review and meta-analysis. Oral Oncol. 2022, 126, 105734. [Google Scholar] [CrossRef]
- Monteiro, L.; Mariano, L.C.; Warnakulasuriya, S. Podoplanin could be a predictive biomarker of the risk of patients with oral leukoplakia to develop oral cancer: A systematic review and meta-analysis. Oral Dis. 2024, 30, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.W.; Kang, J.-H. Antinuclear Positivity and Malignant Transformation Potential of Oral Potentially Malignant Disorder. Oral Dis. 2024. [Google Scholar] [CrossRef] [PubMed]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B. Meta-analysis of observational studies in epidemiology: A proposal for reporting. J. Am. Med. Assoc. 2000, 283, 2008–2012. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Riley, R.D.; Ridley, G.; Williams, K.; Altman, D.G.; Hayden, J.; de Vet, H.C.W. Prognosis research: Toward evidence-based results and a Cochrane methods group. J. Clin. Epidemiol. 2007, 60, 863–865. [Google Scholar] [CrossRef]
- Chandler, J.; Cumpston, M.; Li, T.; Page, M.J.; Welch, V.J.H.W. Cochrane Handbook for Systematic Reviews of Interventions; Higgins, J.P., Green, S., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2008; ISBN 9780470712184. [Google Scholar]
- Shamseer, L.; Moher, D.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A. PRISMA-P Group Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: Elaboration and explanation. BMJ 2015, 350, g7647. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Luo, D.; Wan, X.; Liu, J.; Tong, T. Optimally estimating the sample mean from the sample size, median, mid-range, and/or mid-quartile range. Stat. Methods Med. Res. 2018, 27, 1785–1805. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Wang, W.; Liu, J.; Tong, T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med. Res. Methodol. 2014, 14, 135. [Google Scholar] [CrossRef]
- Hayden, J.A.; Côté, P.; Bombardier, C. Evaluation of the quality of prognosis studies in systematic reviews. Ann. Intern. Med. 2006, 144, 427–437. [Google Scholar] [CrossRef]
- Cívico-Ortega, J.L.; González-Ruiz, I.; Ramos-García, P.; Cruz-Granados, D.; Samayoa-Descamps, V.; González-Moles, M.Á. Prognostic and Clinicopathological Significance of Epidermal Growth Factor Receptor (EGFR) Expression in Oral Squamous Cell Carcinoma: Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2023, 24, 11888. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Viechtbauer, W.; Cheung, M.W.-L. Outlier and influence diagnostics for meta-analysis. Res. Synth. Methods 2010, 1, 112–125. [Google Scholar] [CrossRef]
- Egger, M.; Davey Smith, G.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef]
- Girod, S.C.; Pfeiffer, P.; Ries, J.; Pape, H.D. Proliferative activity and loss of function of tumour suppressor genes as “biomarkers” in diagnosis and prognosis of benign and preneoplastic oral lesions and oral squamous cell carcinoma. Br. J. Oral Maxillofac. Surg. 1998, 36, 252–260. [Google Scholar] [CrossRef]
- Schoelch, M.L.; Regezi, J.A.; Dekker, N.P.; Ng, I.O.L.; McMillan, A.; Ziober, B.L.; Le, Q.T.; Silverman, S.; Fu, K.K. Cell cycle proteins and the development of oral squamous cell carcinoma. Oral Oncol. 1999, 35, 333–342. [Google Scholar] [CrossRef]
- Ghazali, N.; Bakri, M.M.; Zain, R.B. Aggressive, multifocal oral verrucous leukoplakia: Proliferative verrucous leukoplakia or not? J. Oral Pathol. Med. 2003, 32, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Soni, S.; Kaur, J.; Kumar, A.; Chakravarti, N.; Mathur, M.; Bahadur, S.; Shukla, N.K.; Deo, S.V.S.; Ralhan, R. Alterations of rb pathway components are frequent events in patients with oral epithelial dysplasia and predict clinical outcome in patients with squamous cell carcinoma. Oncology 2005, 68, 314–325. [Google Scholar] [CrossRef]
- Shah, N.G.; Trivedi, T.I.; Tankshali, R.A.; Goswami, J.A.; Shah, J.S.; Jetly, D.H.; Kobawala, T.P.; Patel, K.C.; Shukla, S.N.; Shah, P.M.; et al. Molecular alterations in oral carcinogenesis: Significant risk predictors in malignant transformation and tumor progression. Int. J. Biol. Markers 2007, 22, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Nasser, W.; Flechtenmacher, C.; Holzinger, D.; Hofele, C.; Bosch, F.X. Aberrant expression of p53, p16INK4a and Ki-67 as basic biomarker for malignant progression of oral leukoplakias. J. Oral Pathol. Med. 2011, 40, 629–635. [Google Scholar] [CrossRef]
- Altman, D.G.; Lausen, B.; Sauerbrei, W.; Schumacher, M. Dangers of using “optimal” cutpoints in the evaluation of prognostic factors. J. Natl. Cancer Inst. 1994, 86, 829–835. [Google Scholar] [CrossRef]
- Warnakulasuriya, S.; Kujan, O.; Aguirre-Urizar, J.M.; Bagan, J.V.; González-Moles, M.Á.; Kerr, A.R.; Lodi, G.; Mello, F.W.; Monteiro, L.; Ogden, G.R.; et al. Oral potentially malignant disorders: A consensus report from an international seminar on nomenclature and classification, convened by the WHO Collaborating Centre for Oral Cancer. Oral Dis. 2021, 27, 1862–1880. [Google Scholar] [CrossRef] [PubMed]
- Califano, J.; Van Der Riet, P.; Westra, W.; Nawroz, H.; Clayman, G.; Piantadosi, S.; Corio, R.; Lee, D.; Greenberg, B.; Koch, W.; et al. Genetic progression model for head and neck cancer: Implications for field cancerization. Cancer Res. 1996, 56, 2488–2492. [Google Scholar] [CrossRef] [PubMed]
- Tabor, M.P.; Brakenhoff, R.H.; Van Houten, V.M.M.; Kummer, J.A.; Snel, M.H.J.; Snijders, P.J.F.; Snow, G.B.; Leemans, C.R.; Braakhuis, B.J.M. Persistence of genetically altered fields in head and neck cancer patients: Biological and clinical implications. Clin. Cancer Res. 2001, 7, 1523–1532. [Google Scholar] [PubMed]
- Braakhuis, B.J.M.; Tabor, M.P.; Kummer, J.A.; Leemans, C.R.; Brakenhoff, R.H. A genetic explanation of slaughter’s concept of field cancerization: Evidence and clinical implications. Cancer Res. 2003, 63, 1727–1730. [Google Scholar]
- López-Ansio, M.; Ramos-García, P.; González-Moles, M.Á. Prognostic and Clinicopathological Significance of the Loss of Expression of Retinoblastoma Protein (pRb) in Oral Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. Cancers 2023, 15, 3132. [Google Scholar] [CrossRef]

| Total | 6 Studies |
|---|---|
| Year of publication | 1998–2011 |
| Total patients (range) | 330 (9–113) |
| Diagnostic criteria | |
| Clinical lesions | |
| Leukoplakia | 2 studies |
| Proliferative verrucous leukoplakia | 1 study |
| Histopathological lesions | |
| Keratosis, Hyperplasia, Dysplasia | 3 studies |
| pRb immunohistochemical pattern | |
| Nuclear staining | 5 studies |
| Not reported | 1 study |
| Anti-pRb antibody | |
| pRb | 1 study |
| Rb-1 | 2 studies |
| IF-8 | 1 study |
| Ab-1 | 1 study |
| Not reported | 1 study |
| Anti-pRb antibody dilution | |
| ≤1:50 | 4 studies |
| 1:100 | 1 study |
| Not reported | 1 study |
| Anti-pRb antibody incubation time | |
| Overnight | 3 studies |
| 1 h | 1 study |
| Not reported | 2 studies |
| Anti-pRb antibody incubation temperature | |
| 4 °C | 3 studies |
| Not reported | 3 studies |
| Cutoff point for pRb overexpression | |
| 1% | 2 studies |
| 10% | 2 studies |
| Not reported | 2 studies |
| Study design | |
| Retrospective cohorts | 6 studies |
| Geographical region | |
| Asia | 3 studies |
| Europe | 2 studies |
| North America | 1 study |
| Total | 3 continents |
| Table S2 (Supplementary Materials) describes in detail the characteristics of the studies. | |
| Meta-Analyses | No. of Studies | No. of Patients | Stat. Model | Wt | Pooled Data | Heterogeneity | ||
|---|---|---|---|---|---|---|---|---|
| RR (95% CI) | p-Value | Phet | I2 (%) | |||||
| Loss of pRb expression and malignant transformation risk (all) a | 6 | 330 | REM | D-L | 1.92 (1.25–2.94) | 0.003 | 0.58 | 0.0 |
| Subgroup analysis by geographical region b | ||||||||
| Asia | 3 | 159 | REM | D-L | 1.95 (1.23–3.09) | 0.004 | 0.39 | 0.0 |
| Europe | 2 | 153 | REM | D-L | 3.09 (0.72–13.35) | 0.13 | 0.81 | 0.0 |
| North America | 1 | 18 | — | — | 0.58 (0.08–4.16) | 0.59 | — | 0.0 |
| Subgroup analysis by type of diagnostic criteria b | ||||||||
| Oral leukoplakia | 2 | 130 | REM | D-L | 2.00 (1.22–3.29) | 0.006 | 0.55 | 0.0 |
| Proliferative verrucous leukoplakia | 1 | 9 | — | — | 0.86 (0.17–4.36) | 0.86 | — | 0.0 |
| Keratosis, hyperplasia or dysplasia | 3 | 191 | REM | D-L | 2.09 (0.70–6.28) | 0.19 | 0.30 | 16.8 |
| Subgroup analysis by immunohistochemical pattern b | ||||||||
| Nuclear | 5 | 290 | REM | D-L | 1.86 (1.20–2.88) | 0.005 | 0.50 | 0.0 |
| Not reported | 1 | 40 | — | — | 3.73 (0.45–30.81) | 0.22 | — | 0.0 |
| Subgroup analysis by anti-pRb antibody b | ||||||||
| pRb | 1 | 40 | — | — | 3.73 (0.45–30.81) | 0.22 | — | 0.0 |
| Rb-1 | 2 | 78 | REM | D-L | 1.71 (0.27–10.62) | 0.57 | 0.12 | 57.7 |
| IF-8 | 1 | 90 | — | — | 1.93 (1.16–3.22) | 0.01 | — | 0.0 |
| Ab-1 | 1 | 113 | — | — | 2.60 (0.34–19.77) | 0.36 | — | 0.0 |
| Not reported | 1 | 9 | — | — | 0.86 (0.17–4.36) | 0.86 | — | 0.0 |
| Subgroup analysis by anti-pRb antibody dilution b | ||||||||
| ≤1:50 | 4 | 127 | REM | D-L | 1.74 (0.67–4.51) | 0.25 | 0.58 | 0.0 |
| 1:100 | 1 | 90 | — | — | 1.93 (1.16–3.22) | 0.01 | — | 0.0 |
| Not reported | 1 | 113 | — | — | 2.60 (0.34–19.77) | 0.36 | — | 0.0 |
| Subgroup analysis by anti-pRb antibody incubation time b | ||||||||
| Overnight | 3 | 109 | REM | D-L | 2.30 (0.87–6.10) | 0.09 | 0.34 | 6.4 |
| 1 h | 1 | 18 | — | — | 0.58 (0.08–4.16) | 0.59 | — | 0.0 |
| Not reported | 2 | 203 | REM | D-L | 1.96 (1.20–3.22) | 0.008 | 0.78 | 0.0 |
| Subgroup analysis by anti-pRb antibody incubation temperature b | ||||||||
| 4 °C | 3 | 109 | REM | D-L | 2.30 (0.87–6.10) | 0.09 | 0.34 | 6.4 |
| Not reported | 3 | 221 | REM | D-L | 1.83 (1.13–2.95) | 0.01 | 0.48 | 0.0 |
| Subgroup analysis by cutoff point for pRb protein overexpression b | ||||||||
| 1% | 2 | 131 | REM | D-L | 1.20 (0.28–5.23) | 0.80 | 0.30 | 7.5 |
| 10% | 2 | 150 | REM | D-L | 2.10 (1.30–3.38) | 0.002 | 0.36 | 0.0 |
| Not reported | 2 | 49 | REM | D-L | 1.52 (0.37–6.19) | 0.56 | 0.28 | 14.2 |
| Subgroup analysis by overall risk of bias in primary-level studies b | ||||||||
| Low RoB | 3 | 168 | REM | D-L | 1.95 (1.04–3.64) | 0.04 | 0.31 | 15.9 |
| High RoB | 3 | 162 | REM | D-L | 1.74 (0.59–5.17) | 0.32 | 0.50 | 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Ansio, M.; Ramos-García, P.; González-Moles, M.Á. Predictive Value of the Loss of pRb Expression in the Malignant Transformation Risk of Oral Potentially Malignant Disorders: A Systematic Review and Meta-Analysis. Cancers 2025, 17, 329. https://doi.org/10.3390/cancers17020329
López-Ansio M, Ramos-García P, González-Moles MÁ. Predictive Value of the Loss of pRb Expression in the Malignant Transformation Risk of Oral Potentially Malignant Disorders: A Systematic Review and Meta-Analysis. Cancers. 2025; 17(2):329. https://doi.org/10.3390/cancers17020329
Chicago/Turabian StyleLópez-Ansio, María, Pablo Ramos-García, and Miguel Ángel González-Moles. 2025. "Predictive Value of the Loss of pRb Expression in the Malignant Transformation Risk of Oral Potentially Malignant Disorders: A Systematic Review and Meta-Analysis" Cancers 17, no. 2: 329. https://doi.org/10.3390/cancers17020329
APA StyleLópez-Ansio, M., Ramos-García, P., & González-Moles, M. Á. (2025). Predictive Value of the Loss of pRb Expression in the Malignant Transformation Risk of Oral Potentially Malignant Disorders: A Systematic Review and Meta-Analysis. Cancers, 17(2), 329. https://doi.org/10.3390/cancers17020329







