PLASMA: Partial LeAst Squares for Multiomics Analysis
Simple Summary
Abstract
1. Introduction
- Use partial least squares (PLS) with Cox regression analysis [12] to identify factors that can predict time-to-event outcomes in each individual omics data set.
Terminology
- The input data contains a list of omics data sets.
- Each omics data set contains measurements of multiple features.
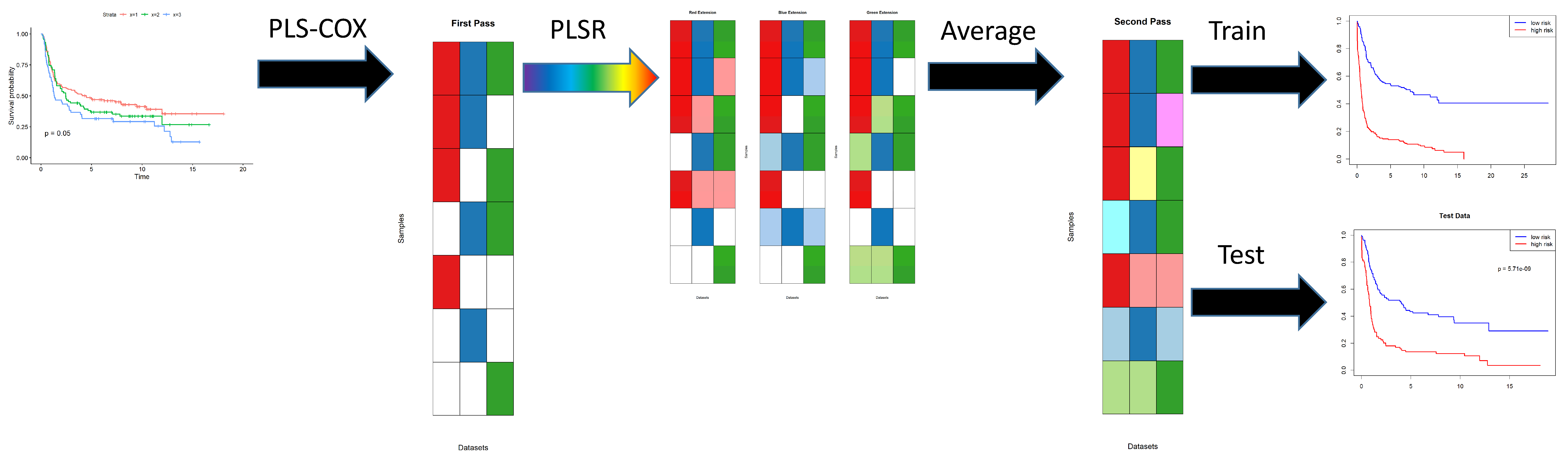
- The first step in the algorithm uses PLS Cox regression to find a set of components. Each component is a linear combination of features. The components are used as predictors in a Cox proportional hazards model, which predicts the log hazard ratio as a linear combination of components.
- The second step in the algorithm creates a secondary layer of components. We do not give these components a separate name. They are not an item of particular focus; we view them as a way to extend the first level components to more samples by “re-interpreting” them in other omics data sets.
2. Methods
2.1. Data
- From TCGA, we obtained 140 columns of clinical, demographic, and laboratory data on 436 patient samples. We removed any columns that always contained the same value, and any columns that were duplicates of other data in the set. We also removed any columns whose values were missing in more than 25% of the patients. We converted categorical variables into sets of binary variables using one-hot-encoding. We then separated the clinical data into three parts:
- Outcome (overall survival)
- Binary covariates (75 columns)
- Continuous covariates (three columns)
- Exome sequencing data for 430 patients with STAD were obtained as mutation allele format (MAF) files. We removed any gene that was mutated in fewer than 4% of the samples. The resulting data set contained 1329 mutated genes.
- Methylation data for 388 STAD patients were obtained as beta values computed by TCGA from Illumina Methylation 450K arrays. We removed any CpG site for which the standard deviation of the beta values was less than 0.25 or for which the mean was within 0.15 of the boundary values of 0 or 1. The resulting data set contained 2291 highly variable CpGs.
- Already normalized sequencing data on 2588 microRNAs (miRs) were obtained for 382 patients. We removed any miR for which the standard deviation of normalized expression was less than 0.10, which left 1064 miRs in the final data set.
- Already normalized sequencing data on 20,531 mRNAs were obtained in 411 patients. We removed any mRNA whose mean normalized expression was less than five or whose standard deviation was less than 1.25. The final data set included 1690 mRNAs.
- Normalized expression data from reverse phase protein arrays (RPPA) were obtained from antibodies targeting 133 proteins in 350 patients. All data were retained for further analysis.
2.2. Imputation
- meanModeImputer will replace any missing data with the mean value of the observed data if there are more than five distinct values; otherwise, it will replace missing data with the mode. This approach works for both continuous data and for binary or small categorical data.
- samplingImputer replaces missing values by sampling randomly from the empirical data distribution.
2.3. Computational Approach
- The plsRcoxmodel contains the coefficients of the components learned by PLS Cox regression. The number of components is determined automatically as a function of the logarithm of the number of features in the omics data set. The output of this model is a continuous prediction of “risk” for the time-to-event outcome of interest.
- Next, two separate models are constructed using the prediction of risk on the training data.
- -
- The riskModel is a coxph model using continuous predicted risk as a single predictor.
- -
- The splitModel is a coxph model using a binary split of the risk (at the median) as the predictor.
2.4. Gene Enrichment
2.5. Preparing the Data
3. Results
3.1. Preprocessing
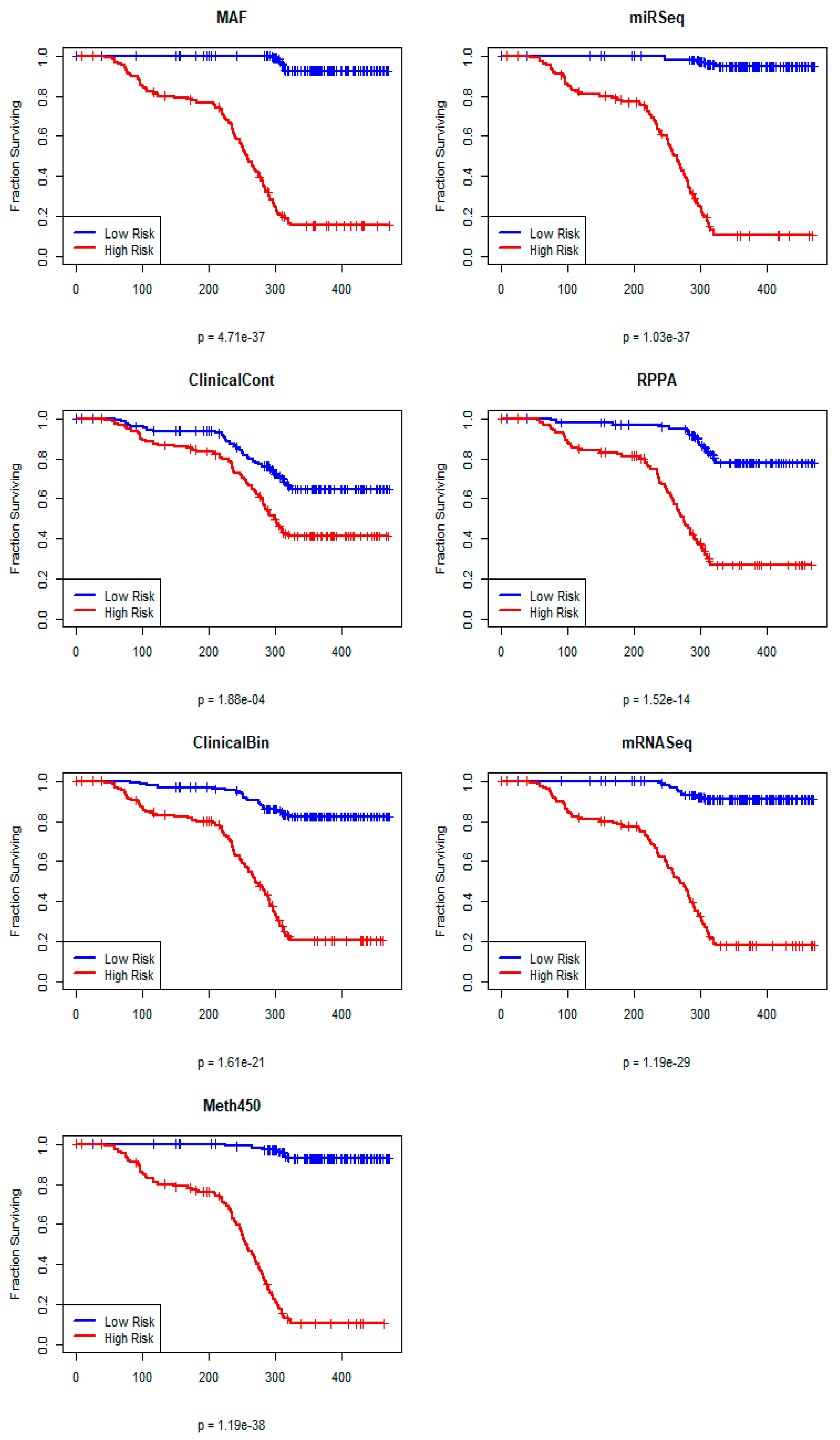
3.2. Individual PLS Cox Regression Models
3.3. Single Omics Predictions on the Test Set
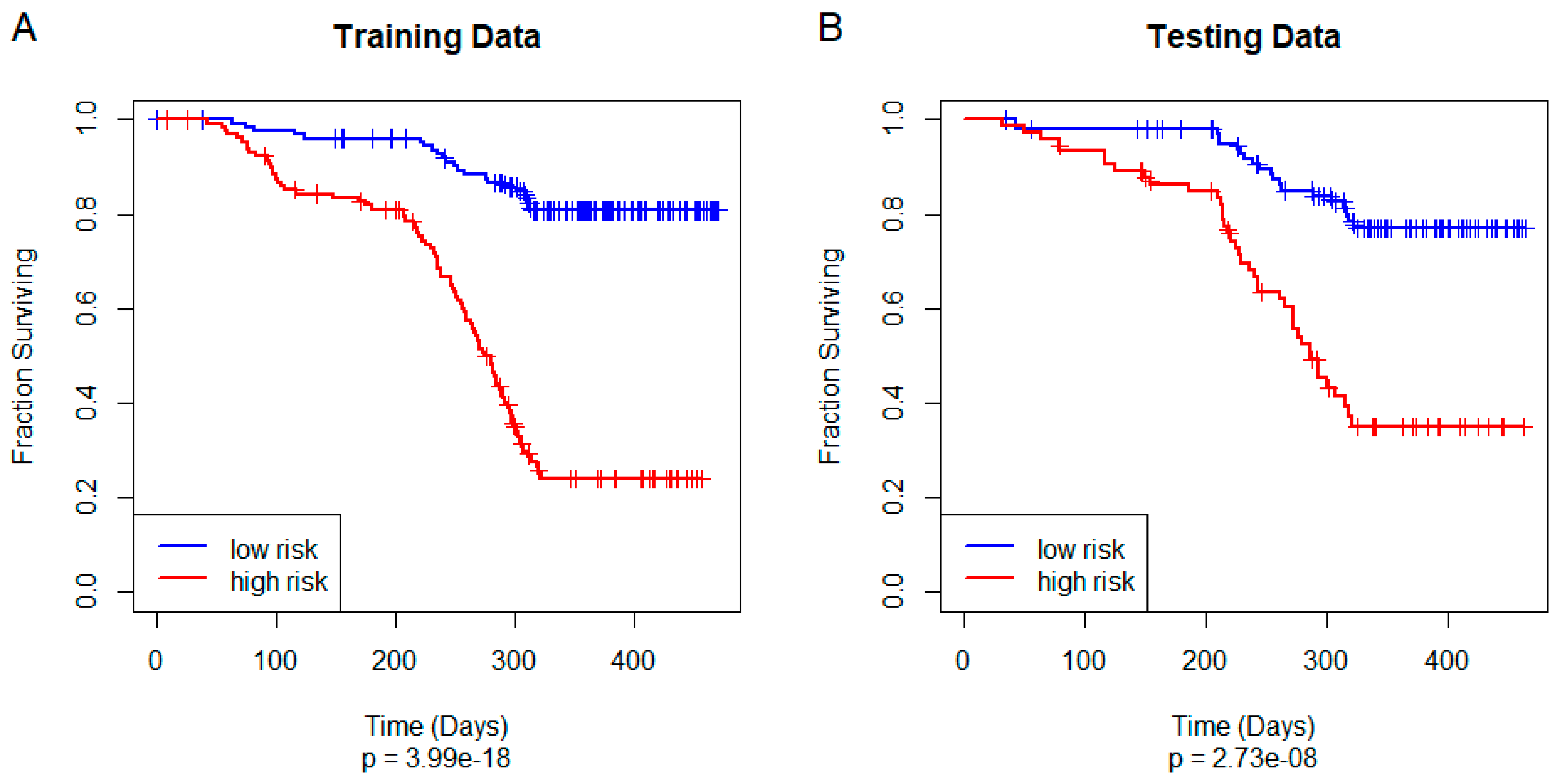
3.4. Training and Testing a Unified Model of Overall Survival
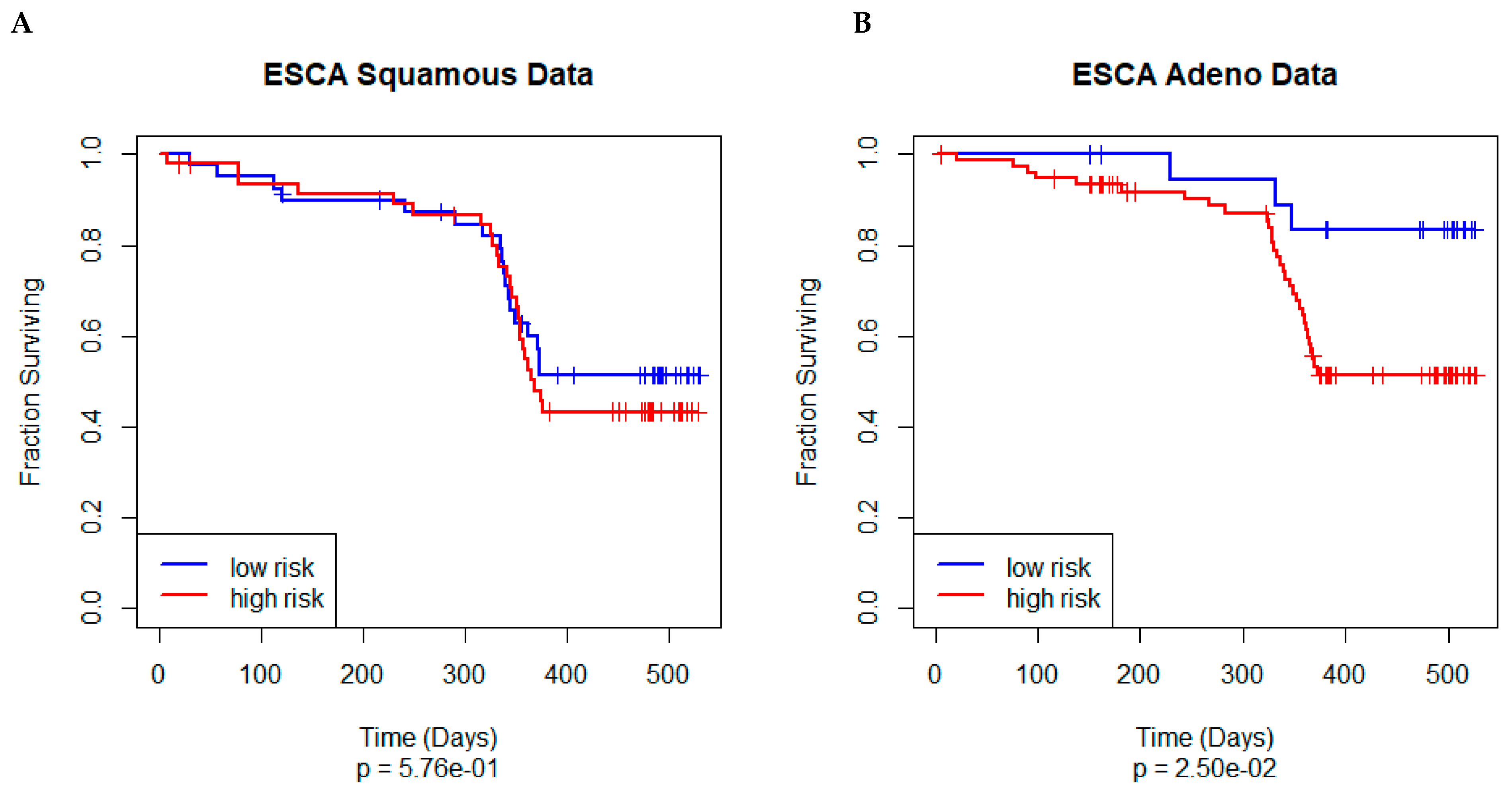
3.5. Independent Validation
3.6. Interpreting the Model
3.7. Final (Composite) Weights
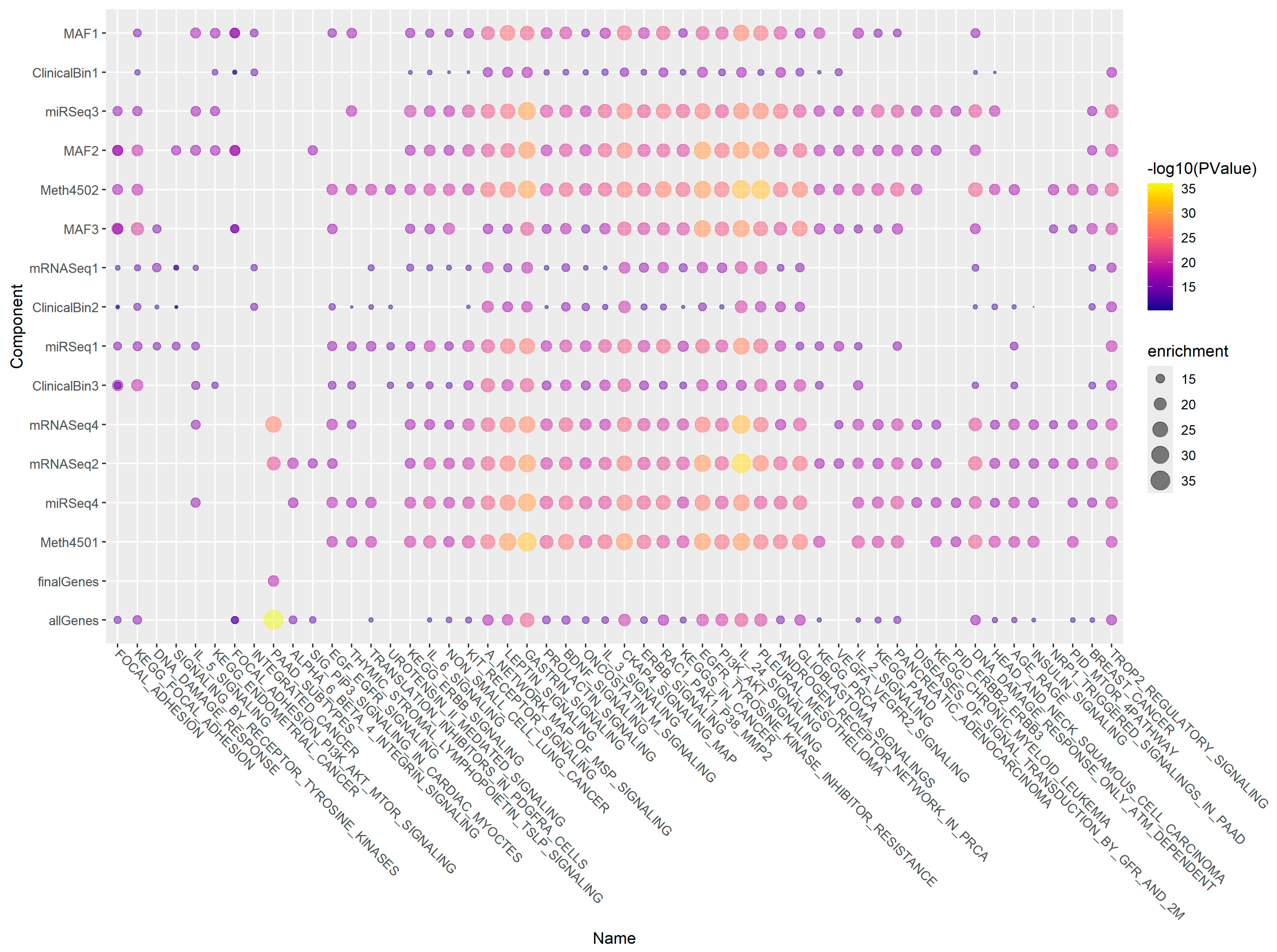
3.8. Gene Enrichment Using ToppGene
4. Discussion
- “Wisdom of the crowd”. There has long been an idea in the machine learning field that combining ensembles of weak models can give rise to a strong model. Well-established examples of this idea are bagging and boosting [24]
- “Out of phase”. Each omics data set may overfit the model in a different way. Instead of reinforcing each other, the extent of overfitting may cancel out.
- “Feature Elimination”. The combined method successfully identifies useful predictive factors. So, we are still able to fit a generalizable Cox model on the final components.
- More research will need to be conducted on a variety of data sets to determine whether this phenomenon is more general.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-Omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef] [PubMed]
- Graw, S.; Chappell, K.; Washam, C.L.; Gies, A.; Bird, J.; Robeson, M.S.; Byrum, S.D. Multi-Omics Data Integration Considerations and Study Design for Biological Systems and Disease. Mol. Omics 2021, 17, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.J.; Hwa, C.; Lee, G.H.; Park, J.M.; An, J.Y. Integrative Multi-Omics Approaches in Cancer Research: From Biological Networks to Clinical Subtypes. Mol. Cells 2021, 44, 433–443. [Google Scholar] [CrossRef]
- Picard, M.; Scott-Boyer, M.P.; Bodein, A.; Perin, O.; Droit, A. Integration Strategies of Multi-Omics Data for Machine Learning Analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3735–3746. [Google Scholar] [CrossRef]
- Reel, P.S.; Reel, S.; Pearson, E.; Trucco, E.; Jefferson, E. Using Machine Learning Approaches for Multi-Omics Data Analysis: A Review. Biotechnol. Adv. 2021, 49, 107739. [Google Scholar] [CrossRef]
- Vlachavas, E.I.; Bohn, J.; Uckert, F.; Nurnberg, S. A Detailed Catalogue of Multi-Omics Methodologies for Identification of Putative Biomarkers and Causal Molecular Networks in Translational Cancer Research. Int. J. Mol. Sci. 2021, 22, 2822. [Google Scholar] [CrossRef] [PubMed]
- Adossa, N.; Khan, S.; Rytkonen, K.T.; Elo, L.L. Computational Strategies for Single-Cell Multi-Omics Integration. Comput. Struct. Biotechnol. J. 2021, 19, 2588–2596. [Google Scholar] [CrossRef]
- Simon, R.M.; Dobbin, K. Experimental Design of DNA Microarray Experiments. Biotechniques 2003, 34 (Suppl. S3), 16–21. [Google Scholar] [CrossRef]
- Halder, R.K.; Uddin, M.N.; Uddin, M.A.; Aryal, S.; Saha, S.; Hossen, R.; Ahmed, S.; Rony, M.A.T.; Akter, M.F. ML-CKDP: Machine Learning-Based Chronic Kidney Disease Prediction with Smart Web Application. J. Pathol. Inform. 2024, 15, 100371. [Google Scholar] [CrossRef]
- Karim, H.T.; Lee, S.; Gerlach, A.; Stinley, M.; Berta, R.; Mahbubani, R.; Tudorascu, D.L.; Butters, M.A.; Gross, J.J.; Andreescu, C. Hippocampal Subfield Volume in Older Adults with and Without Mild Cognitive Impairment: Effects of Worry and Cognitive Reappraisal. Neurobiol. Aging 2024, 141, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Danstrup, C.S.; Andersen, M.; Lundbye-Christensen, S.; Sommer, M.; Lyhne, N.M. Survey of Danish Head and Neck Cancer Patients’ Positions on Personalized Medicine, Gene Tests, and Personalized Follow-Up. J. Pers. Med. 2024, 14, 404. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, F.; Maumy-Bertrand, M. Fitting and Cross-Validating Cox Models to Censored Big Data With Missing Values Using Extensions of Partial Least Squares Regression Models. Front. Big Data 2021, 4, 684794. [Google Scholar] [CrossRef]
- Bastien, P.; Bertrand, F.; Meyer, N.; Maumy-Bertrand, M. Deviance Residuals-Based Sparse PLS and Sparse Kernel PLS Regression for Censored Data. Bioinformatics 2015, 31, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Liland, K.H. Swiss Knife Partial Least Squares (SKPLS): One Tool for Modelling Single Block, Multiblock, Multiway, Multiway Multiblock Including Multi-Responses and Meta Information Under the ROSA Framework. Anal. Chim. Acta 2022, 1206, 339786. [Google Scholar] [CrossRef]
- Argelaguet, R.; Velten, B.; Arnol, D.; Dietrich, S.; Zenz, T.; Marioni, J.C.; Buettner, F.; Huber, W.; Stegle, O. Multi-Omics Factor Analysis-a Framework for Unsupervised Integration of Multi-Omics Data Sets. Mol. Syst. Biol. 2018, 14, e8124. [Google Scholar] [CrossRef] [PubMed]
- Argelaguet, R.; Arnol, D.; Bredikhin, D.; Deloro, Y.; Velten, B.; Marioni, J.C.; Stegle, O. MOFA+: A Statistical Framework for Comprehensive Integration of Multi-Modal Single-Cell Data. Genome Biol. 2020, 21, 111. [Google Scholar] [CrossRef]
- Kim, J.; Bowlby, R.; Mungall, A.J.; Robertson, A.G.; Odze, R.D.; Cherniack, A.D.; Shih, J.; Pedamallu, C.S.; Cibulskis, C.; Dunford, A.; et al. Integrated Genomic Characterization of Oesophageal Carcinoma. Nature 2017, 541, 169–175. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Bragelmann, J.; Kryukov, I.; Saraiva-Agostinho, N.; Perner, S. FirebrowseR: An R Client to the Broad Institute’s Firehose Pipeline. Database 2017, 2017, baw160. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Dietz, K., Gail, M., Krickeberg, K., Samet, J., Tsiatis, A., Eds.; Statistics for Biology and Health; Springer: New York, NY, USA, 2000; ISBN 978-1-4419-3161-0/978-1-4757-3294-8. [Google Scholar]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for Gene List Enrichment Analysis and Candidate Gene Prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- Abrams, Z.B.; Zucker, M.; Wang, M.; Asiaee Taheri, A.; Abruzzo, L.V.; Coombes, K.R. Thirty Biologically Interpretable Clusters of Transcription Factors Distinguish Cancer Type. BMC Genom. 2018, 19, 738. [Google Scholar] [CrossRef]
- Sutton, C.D. Classification and Regression Trees, Bagging, and Boosting. Handb. Stat. 2005, 24, 303–329. [Google Scholar] [CrossRef]
- Arienti, C.; Pignatta, S.; Tesei, A. Epidermal Growth Factor Receptor Family and Its Role in Gastric Cancer. Front. Oncol. 2019, 9, 1308. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Q.; Zeng, Y.; Xie, Y.; Zhou, J. Gastrin/CCK-B Receptor Signaling Promotes Cell Invasion and Metastasis by Upregulating MMP-2 and VEGF Expression in Gastric Cancer. J. Cancer 2022, 13, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-C.; Hsiao, M. Leptin and Cancer: Updated Functional Roles in Carcinogenesis, Therapeutic Niches, and Developments. Int. J. Mol. Sci. 2021, 22, 2870. [Google Scholar] [CrossRef]
- Pan, S.; Tan, J.; Deng, Y.; Wan, B.-H.; Zhang, X.-Y.; Guan, B.-G. KIT Performed as a Driver Gene Candidate Affecting the Survival Status of Patients with Stomach Adenocarcinoma. Oncotarget 2017, 8, 70183–70191. [Google Scholar] [CrossRef]
- Su, L.; Wu, X.; Li, J.; Yu, Z.; Liu, B. Interaction of Oncostatin M and Its Receptor OSMR Promotes Gastric Cancer Progression via STAT3/FAK/Src Signaling. Ann. Oncol. 2018, 29, viii678. [Google Scholar] [CrossRef]
- Luo, M.; Chen, Y.-J.; Xie, Y.; Wang, Q.-R.; Xiang, Y.-N.; Long, N.-Y.; Yang, W.-X.; Zhao, Y.; Zhou, J.-J. Dickkopf-Related Protein 1/Cytoskeleton-Associated Protein 4 Signaling Activation by Helicobacter Pylori-Induced Activator Protein-1 Promotes Gastric Tumorigenesis via the PI3K/AKT/mTOR Pathway. World J. Gastroenterol. 2022, 28, 6769–6787. [Google Scholar] [CrossRef]
- Matsuoka, T.; Yashiro, M. The Role of PI3K/Akt/mTOR Signaling in Gastric Carcinoma. Cancers 2014, 6, 1441–1463. [Google Scholar] [CrossRef] [PubMed]
- Standing, D.; Dandawate, P.; Anant, S. Prolactin Receptor Signaling: A Novel Target for Cancer Treatment—Exploring Anti-PRLR Signaling Strategies. Front. Endocrinol. 2022, 13, 1112987. [Google Scholar] [CrossRef]
- Neradugomma, N.K.; Subramaniam, D.; Tawfik, O.W.; Goffin, V.; Kumar, T.R.; Jensen, R.A.; Anant, S. Prolactin Signaling Enhances Colon Cancer Stemness by Modulating Notch Signaling in a Jak2-STAT3/ERK Manner. Carcinogenesis 2014, 35, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, B.S.; Kljavin, N.M.; Stawiski, E.W.; Chan, E.; Parikh, C.; Durinck, S.; Chaudhuri, S.; Pujara, K.; Guillory, J.; Edgar, K.A.; et al. Oncogenic ERBB3 Mutations in Human Cancers. Cancer Cell 2013, 23, 603–617. [Google Scholar] [CrossRef]
- Moghbeli, M.; Makhdoumi, Y.; Soltani Delgosha, M.; Aarabi, A.; Dadkhah, E.; Memar, B.; Abdollahi, A.; Abbaszadegan, M.R. ErbB1 and ErbB3 Co-over Expression as a Prognostic Factor in Gastric Cancer. Biol. Res. 2019, 52, 2. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, H.; Natsugoe, S.; Ishigami, S.; Okumura, H.; Matsumoto, M.; Hokita, S.; Aikou, T. Clinical Significance of Nuclear Expression of Spleen Tyrosine Kinase (Syk) in Gastric Cancer. Cancer Lett. 2006, 236, 89–94. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sousa, J.F.; Ham, A.-J.L.; Whitwell, C.; Nam, K.T.; Lee, H.-J.; Yang, H.-K.; Kim, W.H.; Zhang, B.; Li, M.; LaFleur, B.; et al. Proteomic Profiling of Paraffin-Embedded Samples Identifies Metaplasia-Specific and Early-Stage Gastric Cancer Biomarkers. Am. J. Pathol. 2012, 181, 1560–1572. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Chen, W.; Chen, X.-Y.; Fan, H.-N.; Zhang, J.; Zhu, J.-S. Dihydroartemisinin Inhibits the Tumorigenesis and Invasion of Gastric Cancer by Regulating STAT1/KDR/MMP9 and P53/BCL2L1/CASP3/7 Pathways. Pathol. Res. Pract. 2021, 218, 153318. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Zhang, Q.; Zhao, Z.; Li, B.; Chen, J.; Wang, Y. OLFM4 Is Associated with Lymph Node Metastasis and Poor Prognosis in Patients with Gastric Cancer. J. Cancer Res. Clin. Oncol. 2011, 137, 1713–1720. [Google Scholar] [CrossRef]
- Fristedt, R.; Gaber, A.; Hedner, C.; Nodin, B.; Uhlén, M.; Eberhard, J.; Jirström, K. Expression and Prognostic Significance of the Polymeric Immunoglobulin Receptor in Esophageal and Gastric Adenocarcinoma. J. Transl. Med. 2014, 12, 83. [Google Scholar] [CrossRef]
- Endo, M.; Kanda, M.; Sawaki, K.; Shimizu, D.; Tanaka, C.; Kobayashi, D.; Hattori, N.; Hayashi, M.; Yamada, S.; Koike, M.; et al. Tissue Expression of Melanoma-associated Antigen A6 and Clinical Characteristics of Gastric Cancer. Anticancer. Res. 2019, 39, 5903–5910. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Xie, R.; Ren, C.; Yang, X. Dickkopf-1 Expression Is a Novel Prognostic Marker for Gastric Cancer. J. Biomed. Biotechnol. 2012, 2012, 804592. [Google Scholar] [CrossRef]
- Zhang, E.-B.; Han, L.; Yin, D.-D.; Kong, R.; De, W.; Chen, J. C-Myc-induced, Long, Noncoding H19 Affects Cell Proliferation and Predicts a Poor Prognosis in Patients with Gastric Cancer. Med. Oncol. 2014, 31, 914. [Google Scholar] [CrossRef] [PubMed]
- Lazăr, D.; Tăban, S.; Sporea, I.; Dema, A.; Cornianu, M.; Lazăr, E.; Goldiş, A.; Raţiu, I.; Vernic, C. The Immunohistochemical Expression of the P53-Protein in Gastric Carcinomas. Correlation with Clinicopathological Factors and Survival of Patients. Rom. J. Morphol. Embryol. Rev. Roum. Morphol. Embryol. 2010, 51, 249–257. [Google Scholar]
- Deng, W.; Hao, Q.; Vadgama, J.; Wu, Y. Wild-Type TP53 Predicts Poor Prognosis in Patients with Gastric Cancer. J. Cancer Sci. Clin. Ther. 2021, 5, 134–153. [Google Scholar] [CrossRef] [PubMed]
- Victorzon, M.; Nordling, S.; Haglund, C.; Lundin, J.; Roberts, P.J. Expression of P53 Protein as a Prognostic Factor in Patients with Gastric Cancer. Eur. J. Cancer 1996, 32A, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Bazas, V.M.; Lukyanova, N.Y.; Lisovenko, G.S.; Rozumiy, D.O.; Potebnya, G.P. Postoperative Autovaccinotherapy for Patients with Gastric Cancer and Expression of Some Proteins in Tumor Tissue. Exp. Oncol. 2009, 31, 182–184. [Google Scholar] [PubMed]
- Gonçalves, A.R.; Carneiro, A.J.V.; Martins, I.; de Faria, P.A.S.; Ferreira, M.A.; de Mello, E.L.R.; Fogaça, H.S.; Elia, C.C.S.; de Souza, H.S.P. Prognostic Significance of P53 Protein Expression in Early Gastric Cancer. Pathol. Oncol. Res. POR 2011, 17, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Ismail, H.M.; Moneer, M.; El-Baradie, M.; Khorshid, O.; Touny, A. Clinicopathologic and Prognostic Significance of Overexpression of Her-2/Neu and P53 Oncoproteins in Gastric Carcinoma Using Tissue Microarray. J. Egypt. Natl. Cancer Inst. 2007, 19, 147–157. [Google Scholar]
- Sezer, C.; Yildirim, M.; Yildiz, M.; Sezgin, A.; Donem Dilli, U.; Goktas, S.; Bulbuller, N. Prognostic Significance of Biological Apoptosis Factors in Gastric Cancer. J BUON 2013, 18, 138–146. [Google Scholar] [PubMed]
- Liu, X.; Yu, H.; Cai, H.; Wang, Y. Expression of CD24, P21, P53, and c-Myc in Alpha-Fetoprotein-Producing Gastric Cancer: Correlation with Clinicopathologic Characteristics and Survival. J. Surg. Oncol. 2014, 109, 859–864. [Google Scholar] [CrossRef]
- Ahadi, M.; Moradi, A.; Musavinejad, L.; Movafagh, A.; Moradi, A. The Expression of P53, CD44, Ki-67, and HER-2/Neu Markers in Gastric Cancer and Its Association with Histopathological Indicators: A Retrospective Study. Asian Pac. J. Cancer Prev. APJCP 2020, 21, 1607–1614. [Google Scholar] [CrossRef] [PubMed]
- Tahara, T.; Shibata, T.; Okamoto, Y.; Yamazaki, J.; Kawamura, T.; Horiguchi, N.; Okubo, M.; Nakano, N.; Ishizuka, T.; Nagasaka, M.; et al. Mutation Spectrum of TP53 Gene Predicts Clinicopathological Features and Survival of Gastric Cancer. Oncotarget 2016, 7, 42252–42260. [Google Scholar] [CrossRef]
- Abdel-Fatah, T.; Arora, A.; Gorguc, I.; Abbotts, R.; Beebeejaun, S.; Storr, S.; Mohan, V.; Hawkes, C.; Soomro, I.; Lobo, D.N.; et al. Are DNA Repair Factors Promising Biomarkers for Personalized Therapy in Gastric Cancer? Antioxid. Redox Signal. 2013, 18, 2392–2398. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Im, S.-A.; Kim, M.A.; Cho, H.J.; Lee, D.W.; Lee, K.-H.; Kim, T.-Y.; Han, S.-W.; Oh, D.-Y.; Lee, H.-J.; et al. Ataxia-Telangiectasia-Mutated Protein Expression with Microsatellite Instability in Gastric Cancer as Prognostic Marker. Int. J. Cancer 2014, 134, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, Z.; Sheng, J.; Yu, Z.; Yao, B.; Huang, K.; Zhou, L.; Qiu, Z.; Huang, C. miR-509-3-5P Inhibits the Invasion and Lymphatic Metastasis by Targeting PODXL and Serves as a Novel Prognostic Indicator for Gastric Cancer. Oncotarget 2017, 8, 34867–34883. [Google Scholar] [CrossRef]
- Sun, J.; Li, J.; Zhang, W.; Zhang, J.; Sun, S.; Li, G.; Song, H.; Wan, D. MicroRNA-509-3p Inhibits Cancer Cell Proliferation and Migration via Upregulation of XIAP in Gastric Cancer Cells. Oncol. Res. 2017, 25, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Yu, M.; Yu, J.; Liu, Z.; Zhou, X.; Liu, Y.; Ge, X.; Gao, H.; Li, M.; Jiang, X.; et al. miR-338-3p Functions as a Tumor Suppressor in Gastric Cancer by Targeting PTP1B. Cell Death Dis. 2018, 9, 522. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Suo, J.; Wang, C.; Sun, X.; Wang, D.; He, L.; Zhang, Y.; Li, W. Downregulation of Tissue miR-338-3p Predicts Unfavorable Prognosis of Gastric Cancer. Cancer Biomark. Sect. A Dis. Markers 2017, 21, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhu, M.; Liao, B.; Tian, T.; Li, M.; Wang, Z.; Chen, G. Upregulation of miR-552 Predicts Unfavorable Prognosis of Gastric Cancer and Promotes the Proliferation, Migration, and Invasion of Gastric Cancer Cells. Oncol. Res. Treat. 2020, 43, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Liu, C.; Shi, E.; Jin, Q.; Zhao, W.; Wang, J.; Ji, R. MiR-105-3p Acts as an Oncogene to Promote the Proliferation and Metastasis of Breast Cancer Cells by Targeting GOLIM4. BMC Cancer 2021, 21, 275. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Xiong, Y.; Xu, W.; Wang, Y.; Chen, F.; Wang, Z.; Yan, Z. A Two-microRNA Signature as a Potential Biomarker for Early Gastric Cancer. Oncol. Lett. 2014, 7, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, X.; Liu, Y.; Ni, Z.; Lin, Y.; Duan, Z.; Shi, Y.; Wang, G.; Li, F. Downregulated miR-31 Level Associates with Poor Prognosis of Gastric Cancer and Its Restoration Suppresses Tumor Cell Malignant Phenotypes by Inhibiting E2F2. Oncotarget 2016, 7, 36577–36589. [Google Scholar] [CrossRef]


| coef | exp(coef) | se(coef) | z | p | |
|---|---|---|---|---|---|
| MAF1 | 1.531 | 4.624 | 0.382 | 4.006 | 0.00006 |
| MAF2 | 1.283 | 3.608 | 0.191 | 6.729 | 0.00000 |
| MAF3 | 6.092 | 442.244 | 1.509 | 4.037 | 0.00005 |
| miRSeq1 | 1.131 | 3.098 | 0.334 | 3.387 | 0.00071 |
| miRSeq3 | 0.263 | 1.301 | 0.104 | 2.532 | 0.01135 |
| miRSeq4 | −0.985 | 0.373 | 0.449 | −2.193 | 0.02833 |
| ClinicalBin1 | −1.700 | 0.183 | 0.955 | −1.780 | 0.07501 |
| ClinicalBin2 | −6.407 | 0.002 | 3.309 | −1.936 | 0.05285 |
| ClinicalBin3 | −4.426 | 0.012 | 1.961 | −2.258 | 0.02397 |
| mRNASeq1 | −0.180 | 0.835 | 0.128 | −1.405 | 0.16005 |
| mRNASeq2 | 0.949 | 2.582 | 0.292 | 3.248 | 0.00116 |
| mRNASeq4 | 0.779 | 2.178 | 0.436 | 1.788 | 0.07385 |
| Meth4501 | −0.095 | 0.909 | 0.041 | −2.301 | 0.02140 |
| Meth4502 | −1.992 | 0.136 | 0.924 | −2.156 | 0.03105 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamaguchi, K.; Abdelbaky, S.; Yu, L.; Oakes, C.C.; Abruzzo, L.V.; Coombes, K.R. PLASMA: Partial LeAst Squares for Multiomics Analysis. Cancers 2025, 17, 287. https://doi.org/10.3390/cancers17020287
Yamaguchi K, Abdelbaky S, Yu L, Oakes CC, Abruzzo LV, Coombes KR. PLASMA: Partial LeAst Squares for Multiomics Analysis. Cancers. 2025; 17(2):287. https://doi.org/10.3390/cancers17020287
Chicago/Turabian StyleYamaguchi, Kyoko, Salma Abdelbaky, Lianbo Yu, Christopher C. Oakes, Lynne V. Abruzzo, and Kevin R. Coombes. 2025. "PLASMA: Partial LeAst Squares for Multiomics Analysis" Cancers 17, no. 2: 287. https://doi.org/10.3390/cancers17020287
APA StyleYamaguchi, K., Abdelbaky, S., Yu, L., Oakes, C. C., Abruzzo, L. V., & Coombes, K. R. (2025). PLASMA: Partial LeAst Squares for Multiomics Analysis. Cancers, 17(2), 287. https://doi.org/10.3390/cancers17020287





