Necroptosis-Related Gene Signature Predicts Prognosis in Patients with Advanced Ovarian Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source and Pre-Processing
2.2. Clustering of Necroptosis-Associated Genes and Construction of a Prognostic Model
2.3. Estimation of Cell Infiltration in the Tumor Immune Microenvironment (TIME)
2.4. Prediction of Immunotherapeutic Efficacy
2.5. Patient Selection and Ethical Approval for Immunohistochemistry
2.6. Tissue Microarray and Immunohistochemistry Analysis
2.7. Statistical Analyses
3. Results
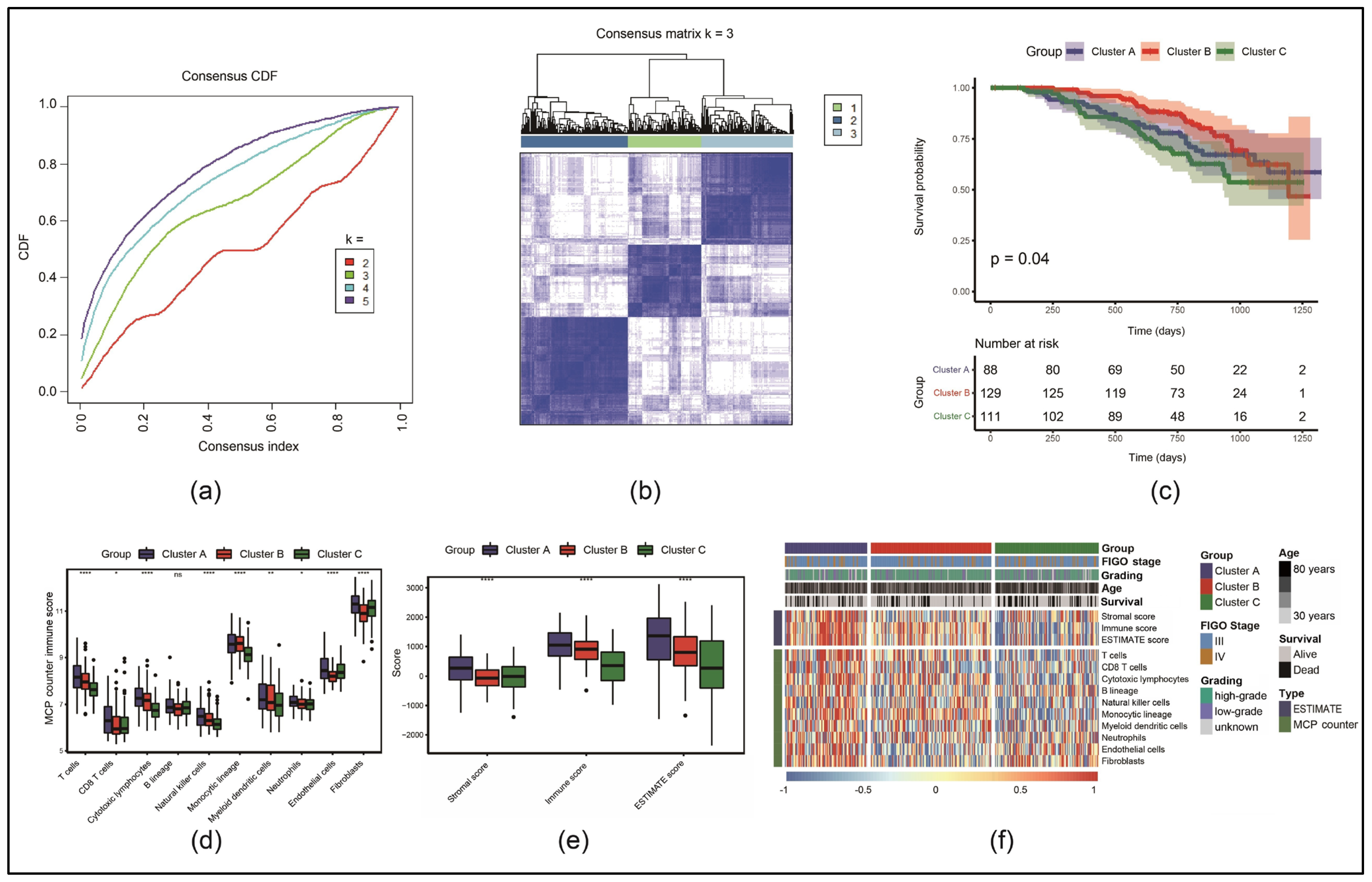
3.1. Identification of Three Molecular Subtypes Based on Necroptosis-Associated Genes
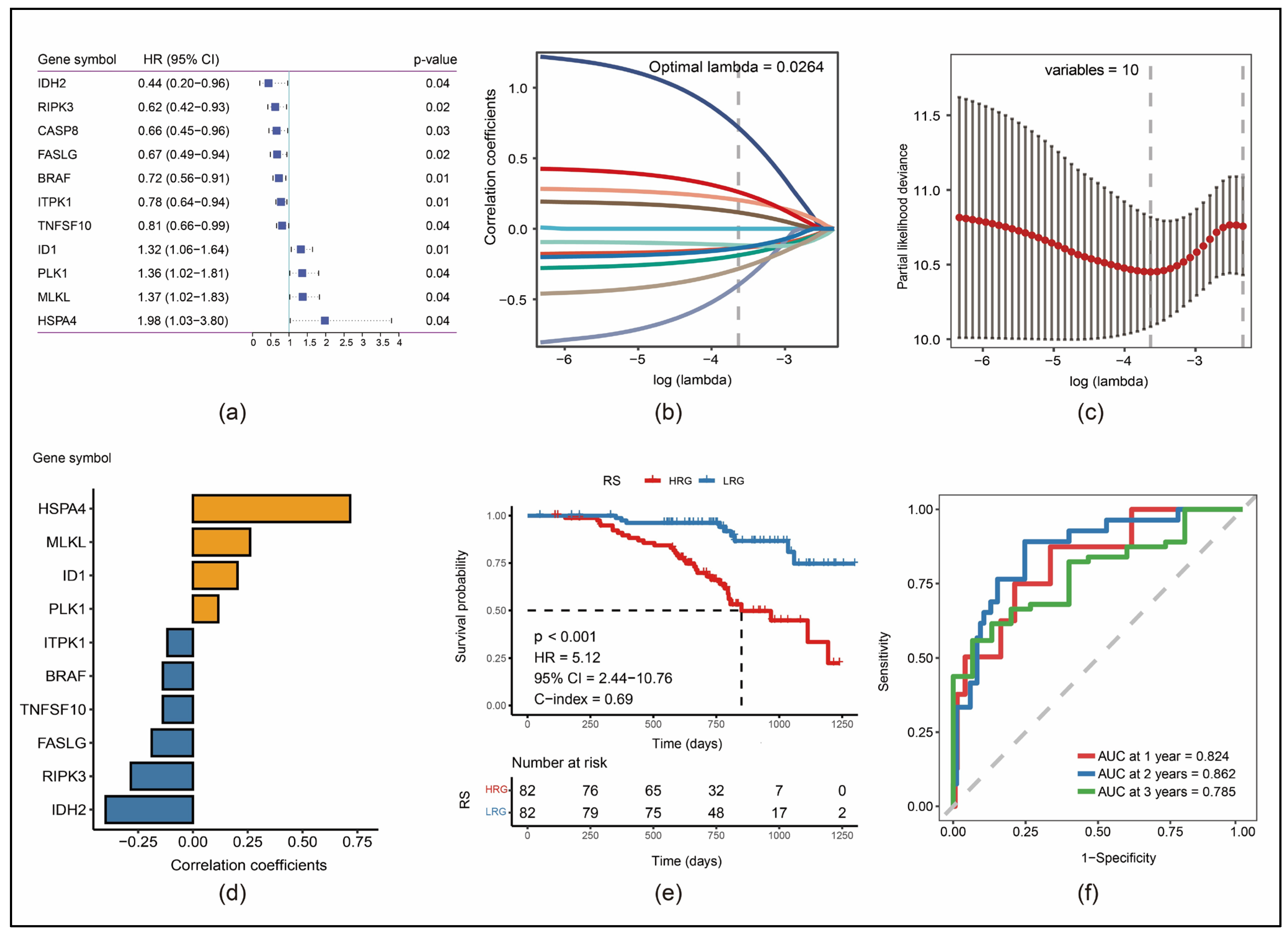
3.2. Construction of a 10-Gene Risk Model
3.3. Robustness of RS in Different Cohorts
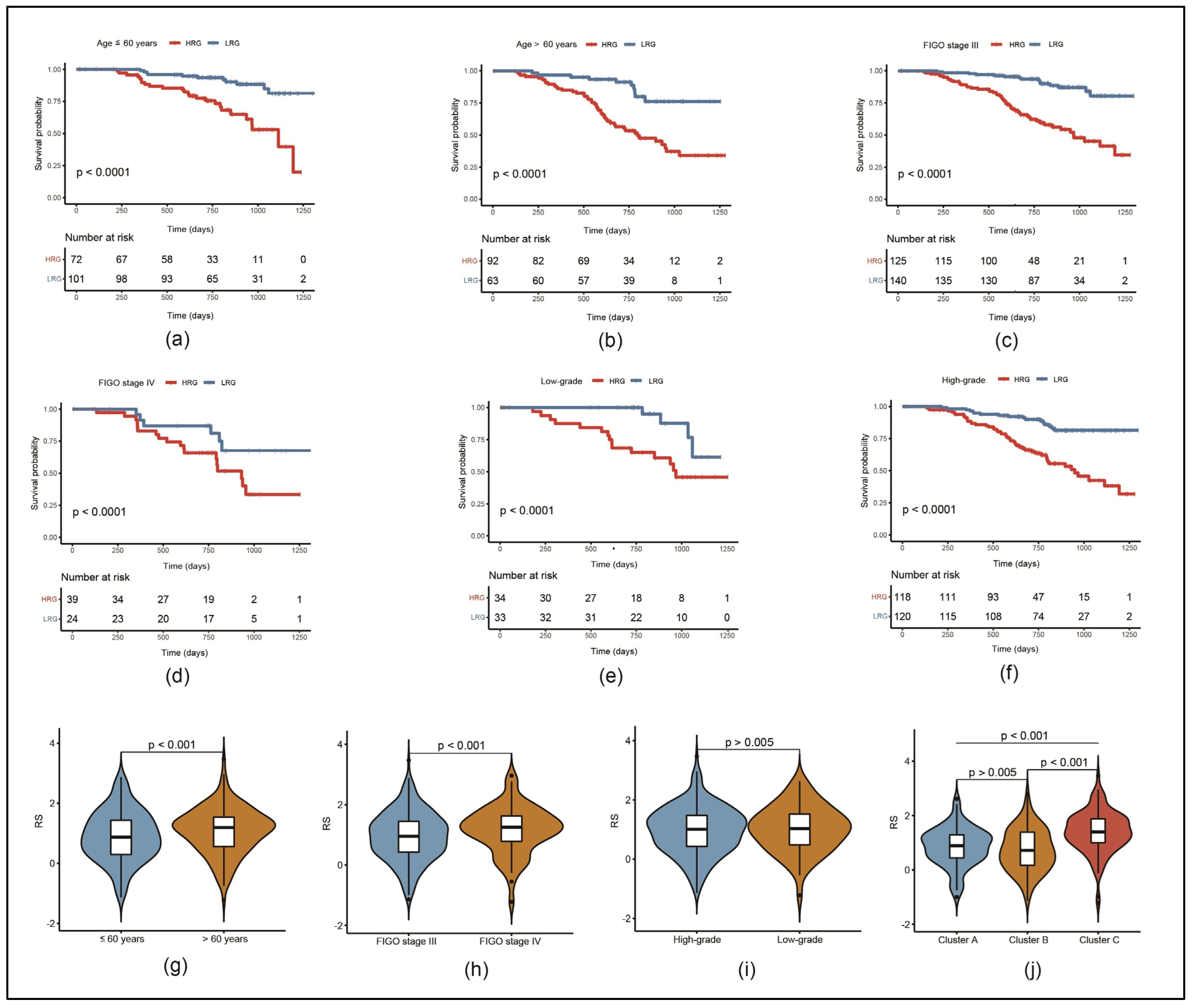
3.4. Risk Model Can Predict the Prognosis of Patients with Different Clinical Characteristics
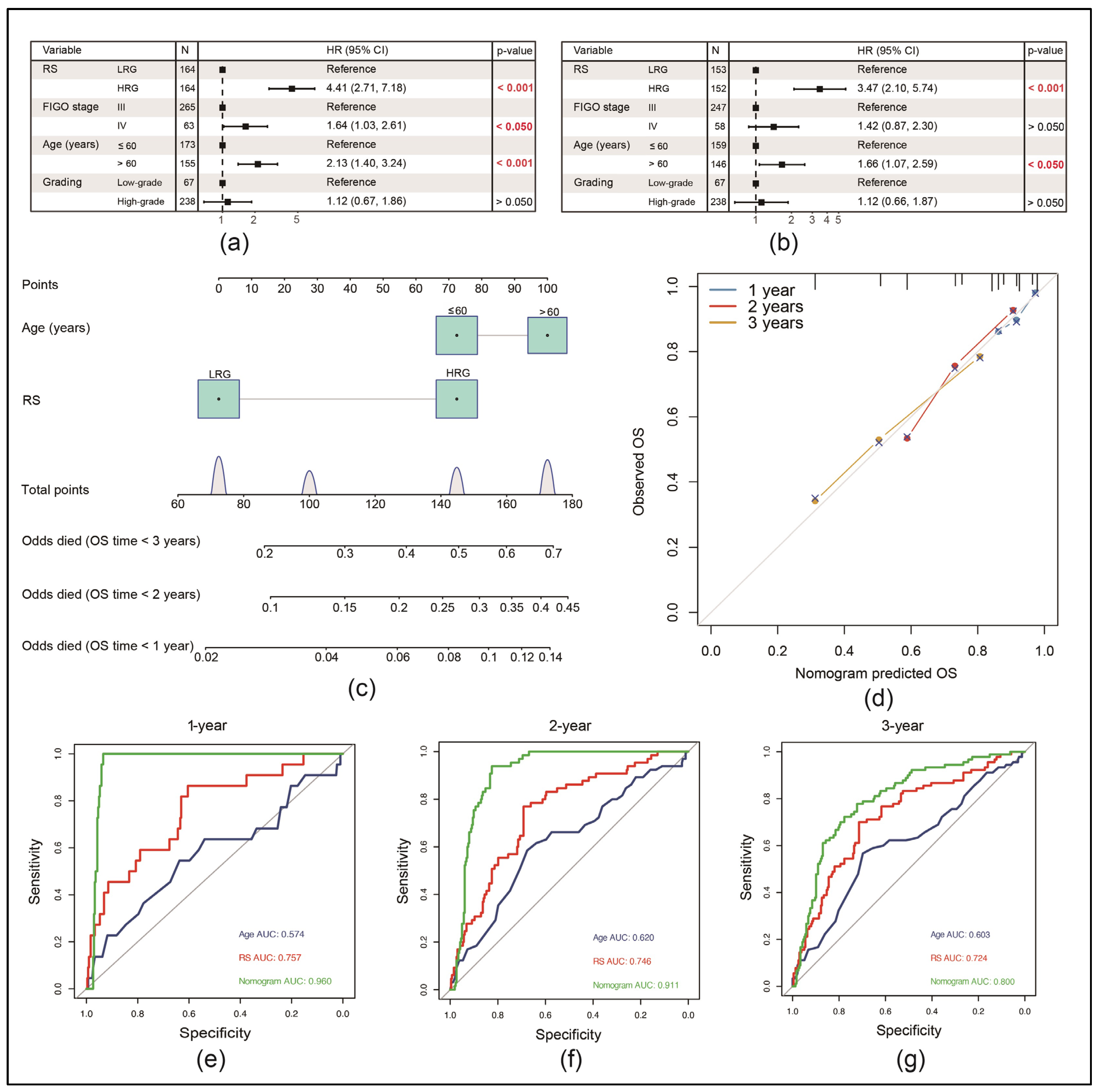
3.5. RS Can Be an Independent Risk Factor for the Prognosis of EOC Patients
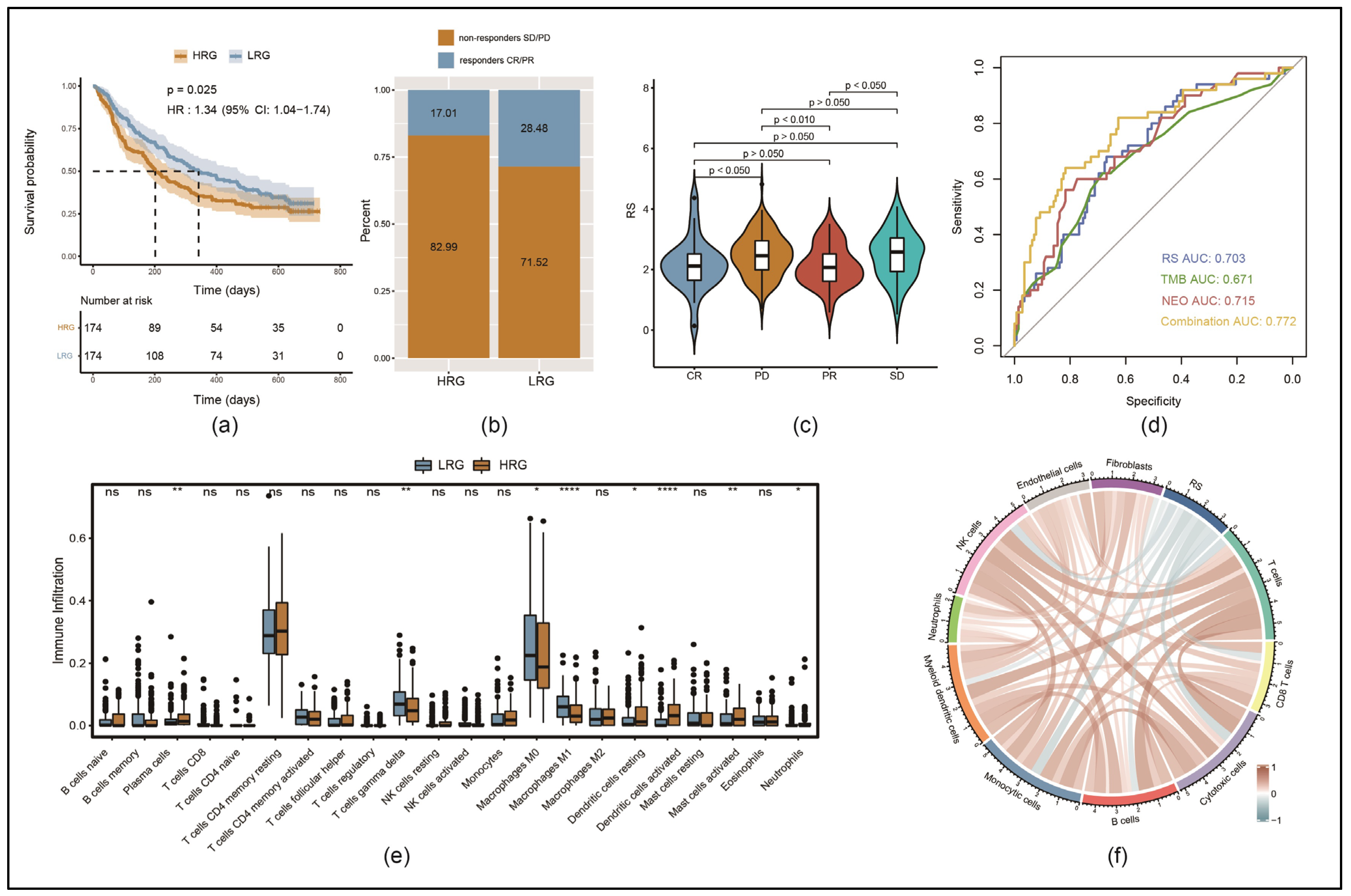
3.6. Immune Cell Infiltration Was Lower in the HRG than in the LRG
3.7. Response to Immunotherapy Was Better in LRG than in HRG
3.8. High Nuclear Expression of RIPK3 Is Associated with Significantly Longer Progression-Free Survival (PFS) and OS in an EOC Patient Cohort
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Tang, R.; Xu, J.; Zhang, B.; Liu, J.; Liang, C.; Hua, J.; Meng, Q.; Yu, X.; Shi, S. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J. Hematol. Oncol. 2020, 13, 110. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhong, C.Q.; Zhang, D.W. Programmed necrosis: Backup to and competitor with apoptosis in the immune system. Nat. Immunol. 2011, 12, 1143–1149. [Google Scholar] [CrossRef]
- Yan, J.; Wan, P.; Choksi, S.; Liu, Z.G. Necroptosis and tumor progression. Trends Cancer 2022, 8, 21–27. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Yatim, N.; Jusforgues-Saklani, H.; Orozco, S.; Schulz, O.; Barreira da Silva, R.; Reis e Sousa, C.; Green, D.R.; Oberst, A.; Albert, M.L. RIPK1 and NF-κB signaling in dying cells determines cross-priming of CD8+ T cells. Science 2015, 350, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Hänggi, K.; Ruffell, B. Cell death, therapeutics, and the immune response in cancer. Trends Cancer 2023, 9, 381–396. [Google Scholar] [CrossRef]
- Liu, S.; Joshi, K.; Denning, M.F.; Zhang, J. RIPK3 signaling and its role in the pathogenesis of cancers. Cell. Mol. Life Sci. 2021, 78, 7199–7217. [Google Scholar] [CrossRef] [PubMed]
- Vergara, G.A.; Eugenio, G.C.; Malheiros, S.M.F.; Victor, E.D.S.; Weinlich, R. RIPK3 is a novel prognostic marker for lower grade glioma and further enriches IDH mutational status subgrouping. J. Neuro-Oncol. 2020, 147, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Wild, C.P.; Weiderpass, E.; Stewart, B.W. World Cancer Report: Cancer Research for Cancer Prevention; International Agency for Research on Cancer: Lyon, France, 2020; pp. 411–420. [Google Scholar]
- Lheureux, S.; Braunstein, M.; Oza, A.M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J. Clin. 2019, 69, 280–304. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Yang, Z.; Xie, L.; DeWitt, J.P.; Chen, Y. Cancer therapy in the necroptosis era. Cell Death Differ. 2016, 23, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, Y.; Inuzuka, H.; Wei, W. Necroptosis pathways in tumorigenesis. Semin. Cancer Biol. 2022, 86, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, K.; Chan, F.K.M. RIP3: A molecular switch for necrosis and inflammation. Genes Dev. 2013, 27, 1640–1649. [Google Scholar] [CrossRef] [PubMed]
- Park, H.H.; Kim, H.R.; Park, S.Y.; Hwang, S.M.; Hong, S.M.; Park, S.; Kang, H.C.; Morgan, M.J.; Cha, J.H.; Lee, D.; et al. RIPK3 activation induces TRIM28 derepression in cancer cells and enhances the anti-tumor microenvironment. Mol. Cancer 2021, 20, 107. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.L.; Yang, J.J.; Wang, Y.Y.; Li, Q.; Song, Y.P.; Su, M.; Li, J.K.; Zhang, L.; Li, Z.P.; Zhou, B.; et al. RIP1 promotes proliferation through G2/M checkpoint progression and mediates cisplatin-induced apoptosis and necroptosis in human ovarian cancer cells. Acta Pharmacol. Sin. 2020, 41, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.Y.; Huang, H.S.; Chao, T.H.; Chou, H.M.; Fang, C.; Qin, C.Z.; Lin, C.Y.; Chu, T.Y.; Zhou, H.H. Progesterone Prevents High-Grade Serous Ovarian Cancer by Inducing Necroptosis of p53-Defective Fallopian Tube Epithelial Cells. Cell Rep. 2017, 18, 2557–2565. [Google Scholar] [CrossRef] [PubMed]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef]
- Kukreja, S.L.; Löfberg, J.; Brenner, M.J. A least absolute shrinkage and selection operator (LASSO) for nonlinear system identification. IFAC Proc. Vol. 2006, 39, 814–819. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; Hoffman-Censits, J.; Powles, T.; van der Heijden, M.S.; Balar, A.V.; Necchi, A.; Dawson, N.; O’Donnell, P.H.; Balmanoukian, A.; Loriot, Y.; et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet 2016, 387, 1909–1920. [Google Scholar] [CrossRef]
- Zhu, J.; Trillsch, F.; Mayr, D.; Kuhn, C.; Rahmeh, M.; Hofmann, S.; Vogel, M.; Mahner, S.; Jeschke, U.; von Schönfeldt, V. EP3 regulates cell proliferation and migration with impact on survival of endometrial cancer patients. Oncotarget 2018, 9, 982–994. [Google Scholar] [CrossRef]
- Heidegger, H.; Dietlmeier, S.; Ye, Y.; Kuhn, C.; Vattai, A.; Aberl, C.; Jeschke, U.; Mahner, S.; Kost, B. The Prostaglandin EP3 Receptor Is an Independent Negative Prognostic Factor for Cervical Cancer Patients. Int. J. Mol. Sci. 2017, 18, 1571. [Google Scholar] [CrossRef] [PubMed]
- Semmlinger, A.; von Schoenfeldt, V.; Wolf, V.; Meuter, A.; Kolben, T.M.; Kolben, T.; Zeder-Goess, C.; Weis, F.; Gallwas, J.; Wuerstlein, R.; et al. EP3 (prostaglandin E2 receptor 3) expression is a prognostic factor for progression-free and overall survival in sporadic breast cancer. BMC Cancer 2018, 18, 431. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, V.P.; Gonen, M.; Smith, J.J.; DeMatteo, R.P. Nomograms in oncology: More than meets the eye. Lancet Oncol. 2015, 16, e173–e180. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Li, L.; Qiu, S.; Lu, Q.; Pan, Q.; Gu, Y.; Luo, J.; Hu, X. Shikonin circumvents cancer drug resistance by induction of a necroptotic death. Mol. Cancer Ther. 2007, 6, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Declercq, W.; Vanden Berghe, T.; Vandenabeele, P. RIP kinases at the crossroads of cell death and survival. Cell 2009, 138, 229–232. [Google Scholar] [CrossRef] [PubMed]
- He, G.W.; Gunther, C.; Thonn, V.; Yu, Y.Q.; Martini, E.; Buchen, B.; Neurath, M.F.; Sturzl, M.; Becker, C. Regression of apoptosis-resistant colorectal tumors by induction of necroptosis in mice. J. Exp. Med. 2017, 214, 1655–1662. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.H.; Son, M.K.; Koh, H.J.; Lee, S.M.; Song, I.H.; Kim, Y.O.; Lee, Y.S.; Jeong, K.S.; Kim, W.B.; Park, J.W.; et al. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 2001, 276, 16168–16176. [Google Scholar] [CrossRef]
- Kil, I.S.; Shin, S.W.; Yeo, H.S.; Lee, Y.S.; Park, J.W. Mitochondrial NADP+-dependent isocitrate dehydrogenase protects cadmium-induced apoptosis. Mol. Pharmacol. 2006, 70, 1053–1061. [Google Scholar] [CrossRef]
- Yang, E.S.; Park, J.W. Regulation of ethanol-induced toxicity by mitochondrial NADP(+)-dependent isocitrate dehydrogenase. Biochimie 2009, 91, 1020–1028. [Google Scholar] [CrossRef]
- Mulay, S.R.; Desai, J.; Kumar, S.V.; Eberhard, J.N.; Thomasova, D.; Romoli, S.; Grigorescu, M.; Kulkarni, O.P.; Popper, B.; Vielhauer, V.; et al. Cytotoxicity of crystals involves RIPK3-MLKL-mediated necroptosis. Nat. Commun. 2016, 7, 10274. [Google Scholar] [CrossRef]
- Rickard, J.A.; O’Donnell, J.A.; Evans, J.M.; Lalaoui, N.; Poh, A.R.; Rogers, T.; Vince, J.E.; Lawlor, K.E.; Ninnis, R.L.; Anderton, H.; et al. RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell 2014, 157, 1175–1188. [Google Scholar] [CrossRef]
- Daniels, B.P.; Kofman, S.B.; Smith, J.R.; Norris, G.T.; Snyder, A.G.; Kolb, J.P.; Gao, X.; Locasale, J.W.; Martinez, J.; Gale, M., Jr.; et al. The Nucleotide Sensor ZBP1 and Kinase RIPK3 Induce the Enzyme IRG1 to Promote an Antiviral Metabolic State in Neurons. Immunity 2019, 50, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Vogelsang, T.L.R.; Kast, V.; Bagnjuk, K.; Eubler, K.; Jeevanandan, S.P.; Schmoeckel, E.; Trebo, A.; Topalov, N.E.; Mahner, S.; Mayr, D.; et al. RIPK1 and RIPK3 are positive prognosticators for cervical cancer patients and C2 ceramide can inhibit tumor cell proliferation in vitro. Front. Oncol 2023, 13, 1110939. [Google Scholar] [CrossRef]
- Kamiya, M.; Mizoguchi, F.; Kawahata, K.; Wang, D.; Nishibori, M.; Day, J.; Louis, C.; Wicks, I.P.; Kohsaka, H.; Yasuda, S. Targeting necroptosis in muscle fibers ameliorates inflammatory myopathies. Nat. Commun. 2022, 13, 166. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Lin, W.; Wu, L.; Xu, A.; Liu, C.; Huang, P. A Novel Prognostic Predictor of Immune Microenvironment and Therapeutic Response in Kidney Renal Clear Cell Carcinoma based on Necroptosis-related Gene Signature. Int. J. Med. Sci. 2022, 19, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.; Wang, X.; Xu, Y.; Li, Y.; Gong, X.; Zeng, Q.; Zhang, B.; Xi, J.; Pei, X.; Yue, W.; et al. Development and Validation of a Novel Survival Model for Cutaneous Melanoma Based on Necroptosis-Related Genes. Front. Oncol. 2022, 12, 852803. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, H.; Therrien, M. Regulation of RAF protein kinases in ERK signalling. Nat. Rev. Mol. Cell. Biol. 2015, 16, 281–298. [Google Scholar] [CrossRef] [PubMed]
- c Xie, T.; Peng, W.; Yan, C.; Wu, J.; Gong, X.; Shi, Y. Structural insights into RIP3-mediated necroptotic signaling. Cell Rep. 2013, 5, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Najafov, A.; Zervantonakis, I.K.; Mookhtiar, A.K.; Greninger, P.; March, R.J.; Egan, R.K.; Luu, H.S.; Stover, D.G.; Matulonis, U.A.; Benes, C.H.; et al. BRAF and AXL oncogenes drive RIPK3 expression loss in cancer. PLoS Biol. 2018, 16, e2005756. [Google Scholar] [CrossRef] [PubMed]
- Shears, S.B. Molecular basis for the integration of inositol phosphate signaling pathways via human ITPK1. Adv. Enzyme Regul. 2009, 49, 87–96. [Google Scholar] [CrossRef]
- Dovey, C.M.; Diep, J.; Clarke, B.P.; Hale, A.T.; McNamara, D.E.; Guo, H.; Brown, N.W., Jr.; Cao, J.Y.; Grace, C.R.; Gough, P.J.; et al. MLKL Requires the Inositol Phosphate Code to Execute Necroptosis. Mol. Cell 2018, 70, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Deeraksa, A.; Pan, J.; Sha, Y.; Liu, X.D.; Eissa, N.T.; Lin, S.H.; Yu-Lee, L.Y. Plk1 is upregulated in androgen-insensitive prostate cancer cells and its inhibition leads to necroptosis. Oncogene 2013, 32, 2973–2983. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Liu, B. PLK1-mediated S369 phosphorylation of RIPK3 during G2 and M phases enables its ripoptosome incorporation and activity. iScience 2021, 24, 102320. [Google Scholar] [CrossRef]
- Liu, Z.G.; Jiao, D. Necroptosis, tumor necrosis and tumorigenesis. Cell Stress 2019, 4, 1–8. [Google Scholar] [CrossRef]
- Hanggi, K.; Vasilikos, L.; Valls, A.F.; Yerbes, R.; Knop, J.; Spilgies, L.M.; Rieck, K.; Misra, T.; Bertin, J.; Gough, P.J.; et al. RIPK1/RIPK3 promotes vascular permeability to allow tumor cell extravasation independent of its necroptotic function. Cell Death Dis. 2017, 8, e2588. [Google Scholar] [CrossRef]
- Van Hoecke, L.; Van Lint, S.; Roose, K.; Van Parys, A.; Vandenabeele, P.; Grooten, J.; Tavernier, J.; De Koker, S.; Saelens, X. Treatment with mRNA coding for the necroptosis mediator MLKL induces antitumor immunity directed against neo-epitopes. Nat. Commun. 2018, 9, 3417. [Google Scholar] [CrossRef]
- Machuca-Aguado, J.; Conde-Martín, A.F.; Alvarez-Muñoz, A.; Rodríguez-Zarco, E.; Polo-Velasco, A.; Rueda-Ramos, A.; Rendón-García, R.; Ríos-Martin, J.J.; Idoate, M.A. Machine Learning Quantification of Intraepithelial Tumor-Infiltrating Lymphocytes as a Significant Prognostic Factor in High-Grade Serous Ovarian Carcinomas. Int. J. Mol. Sci. 2023, 24, 16060. [Google Scholar] [CrossRef] [PubMed]
- Monk, B.J.; Colombo, N.; Oza, A.M.; Fujiwara, K.; Birrer, M.J.; Randall, L.; Poddubskaya, E.V.; Scambia, G.; Shparyk, Y.V.; Lim, M.C.; et al. Chemotherapy with or without avelumab followed by avelumab maintenance versus chemotherapy alone in patients with previously untreated epithelial ovarian cancer (JAVELIN Ovarian 100): An open-label, randomised, phase 3 trial. Lancet Oncol. 2021, 22, 1275–1289. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.N.; Bookman, M.; Sehouli, J.; Miller, A.; Anderson, C.; Scambia, G.; Myers, T.; Taskiran, C.; Robison, K.; Maeenpaeae, J.; et al. Atezolizumab, Bevacizumab, and Chemotherapy for Newly Diagnosed Stage III or IV Ovarian Cancer: Placebo-Controlled Randomized Phase III Trial (IMagyn050/GOG 3015/ENGOT-OV39). J. Clin. Oncol. 2021, 39, 1842–1855. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, J.E.; Pujade-Lauraine, E.; Oaknin, A.; Belin, L.; Leitner, K.; Cibula, D.; Denys, H.; Rosengarten, O.; Rodrigues, M.; de Gregorio, N.; et al. Atezolizumab Combined with Bevacizumab and Platinum-Based Therapy for Platinum-Sensitive Ovarian Cancer: Placebo-Controlled Randomized Phase III ATALANTE/ENGOT-ov29 Trial. J. Clin. Oncol. 2023, 41, 4768–4778. [Google Scholar] [CrossRef]
- Harter, P.; Trillsch, F.; Okamoto, A.; Reuss, A.; Kim, J.W.; Rubio-Pérez, M.J.; Vardar, M.A.; Scambia, G.; Tredan, O.; Nyvang, G.B.; et al. Durvalumab with paclitaxel/carboplatin (PC) and bevacizumab (bev), followed by maintenance durvalumab, bev, and olaparib in patients (pts) with newly diagnosed advanced ovarian cancer (AOC) without a tumor BRCA1/2 mutation (non-tBRCAm): Results from the randomized, placebo (pbo)-controlled phase III DUO-O trial. J. Clin. Oncol. 2023, 41, LBA5506. [Google Scholar] [CrossRef]
- Hardy-Bessard, A.C.; Moore, K.N.; Mirza, M.R.; Asselain, B.; Redondo, A.; Pfisterer, J.; Pignata, S.; Provencher, D.M.; Cibula, D.; Reyners, A.K.L.; et al. ENGOT-OV44/FIRST study: A randomized, double-blind, adaptive, phase III study of platinum-based therapy with dostarlimab (TSR-042) + niraparib versus standard-of-care (SOC) platinum-based therapy as first-line treatment of stage 3/4 non-mucinous epithelial ovarian cancer (OC). J. Clin. Oncol. 2019, 37, TPS5600. [Google Scholar] [CrossRef]
- Monk, B.J.; Coleman, R.L.; Fujiwara, K.; Wilson, M.K.; Oza, A.M.; Oaknin, A.; O’Malley, D.M.; Lorusso, D.; Westin, S.N.; Safra, T.; et al. ATHENA (GOG-3020/ENGOT-ov45): A randomized, phase III trial to evaluate rucaparib as monotherapy (ATHENA-MONO) and rucaparib in combination with nivolumab (ATHENA-COMBO) as maintenance treatment following frontline platinum-based chemotherapy in ovarian cancer. Int. J. Gynecol. Cancer 2021, 31, 1589–1594. [Google Scholar] [CrossRef] [PubMed]
- Vergote, I.; Sehouli, J.; Salutari, V.; Zola, P.; Madry, R.; Wenham, R.M.; Korach, J.; Pautier, P.; Cibula, D.; Lheureux, S.; et al. ENGOT-OV43/KEYLYNK-001: A phase III, randomized, double-blind, active- and placebo-controlled study of pembrolizumab plus chemotherapy with olaparib maintenance for first-line treatment of BRCA-nonmutated advanced epithelial ovarian cancer. J. Clin. Oncol. 2019, 37, TPS5603. [Google Scholar] [CrossRef]
- Qin, Y.; Sheng, Y.; Ren, M.; Hou, Z.; Xiao, L.; Chen, R. Identification of necroptosis-related gene signatures for predicting the prognosis of ovarian cancer. Sci. Rep. 2024, 14, 11133. [Google Scholar] [CrossRef] [PubMed]
- Wieser, V.; Tsibulak, I.; Reimer, D.U.; Zeimet, A.G.; Fiegl, H.; Hackl, H.; Marth, C. An angiogenic tumor phenotype predicts poor prognosis in ovarian cancer. Gynecol. Oncol. 2023, 170, 290–299. [Google Scholar] [CrossRef]
- Zheng, M.; Mullikin, H.; Hester, A.; Czogalla, B.; Heidegger, H.; Vilsmaier, T.; Vattai, A.; Chelariu-Raicu, A.; Jeschke, U.; Trillsch, F.; et al. Development and Validation of a Novel 11-Gene Prognostic Model for Serous Ovarian Carcinomas Based on Lipid Metabolism Expression Profile. Int. J. Mol. Sci. 2020, 21, E9169. [Google Scholar] [CrossRef]
- Zheng, M.; Long, J.; Chelariu-Raicu, A.; Mullikin, H.; Vilsmaier, T.; Vattai, A.; Heidegger, H.H.; Batz, F.; Keckstein, S.; Jeschke, U.; et al. Identification of a Novel Tumor Microenvironment Prognostic Signature for Advanced-Stage Serous Ovarian Cancer. Cancers 2021, 13, 3343. [Google Scholar] [CrossRef] [PubMed]


| Clinical Features | TCGA-OV (n = 347) | GSE140082 (n = 328) |
|---|---|---|
| Age | ||
| ≤60 years | 192 (55.3%) | 173 (52.7%) |
| >60 years | 155 (44.7%) | 155 (47.3%) |
| FIGO stage | ||
| III | 290 (83.6%) | 265 (80.8%) |
| IV | 57 (16.4%) | 63 (19.2%) |
| Grading | ||
| Low | 36 (10.4%) | 67 (20.4%) |
| High | 303 (87.3%) | 238 (72.6%) |
| Unknown | 8 (2.3%) | 23 (7.0%) |
| 3-year survival | ||
| Alive | 125 (36.0%) | 92 (28.0%) |
| Dead | 222 (64.0%) | 236 (72.0%) |
| GSE140082 Training (n = 164) | GSE140082 Testing (n = 164) | p-Value | |
|---|---|---|---|
| Age | |||
| ≤60 years | 92 (56.1%) | 81 (49.4%) | 0.27 |
| >60 years | 72 (43.9%) | 83 (50.6%) | |
| FIGO stage | |||
| III | 128 (78.0%) | 137 (83.5%) | 0.26 |
| IV | 36 (22.0%) | 27 (16.5%) | |
| Grading | |||
| Low | 30 (18.3%) | 37 (22.6%) | 0.33 |
| High | 124 (75.6%) | 114 (69.5%) | |
| Unknown | 10 (6.1%) | 13 (7.9%) | |
| 3-year survival | |||
| Alive | 42 (25.6%) | 50 (30.5%) | 0.39 |
| Dead | 122 (74.4%) | 114 (69.5%) |
| Number of Patients | Percentage | ||
|---|---|---|---|
| Age | |||
| ≤60 years | 84 | 54.2 | |
| >60 years | 71 | 45.8 | |
| FIGO stage | |||
| I | 34 | 21.9 | |
| II | 10 | 6.5 | |
| III | 103 | 66.5 | |
| IV | 3 | 1.9 | |
| Unknown | 5 | 3.2 | |
| Subtype and grading | |||
| Serous | Low grade | 23 | 14.8 |
| High grade | 80 | 51.6 | |
| Unknown | 6 | 3.9 | |
| Total | 109 | 70.3 | |
| Endometrioid | G1 | 6 | 3.9 |
| G2 | 5 | 3.2 | |
| G3 | 8 | 5.2 | |
| Unknown | 2 | 1.3 | |
| Total | 21 | 13.6 | |
| Mucinous | G1 | 6 | 3.9 |
| G2 | 6 | 3.9 | |
| G3 | 0 | 0.0 | |
| Unknown | 1 | 0.6 | |
| Total | 13 | 8.4 | |
| Clear-cell | G1 | 2 | 1.3 |
| G2 | 2 | 1.3 | |
| G3 | 5 | 3.2 | |
| Unknown | 3 | 1.9 | |
| Total | 12 | 7.7 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, M.; Kessler, M.; Jeschke, U.; Reichenbach, J.; Czogalla, B.; Keckstein, S.; Schroeder, L.; Burges, A.; Mahner, S.; Trillsch, F.; et al. Necroptosis-Related Gene Signature Predicts Prognosis in Patients with Advanced Ovarian Cancer. Cancers 2025, 17, 271. https://doi.org/10.3390/cancers17020271
Zheng M, Kessler M, Jeschke U, Reichenbach J, Czogalla B, Keckstein S, Schroeder L, Burges A, Mahner S, Trillsch F, et al. Necroptosis-Related Gene Signature Predicts Prognosis in Patients with Advanced Ovarian Cancer. Cancers. 2025; 17(2):271. https://doi.org/10.3390/cancers17020271
Chicago/Turabian StyleZheng, Mingjun, Mirjana Kessler, Udo Jeschke, Juliane Reichenbach, Bastian Czogalla, Simon Keckstein, Lennard Schroeder, Alexander Burges, Sven Mahner, Fabian Trillsch, and et al. 2025. "Necroptosis-Related Gene Signature Predicts Prognosis in Patients with Advanced Ovarian Cancer" Cancers 17, no. 2: 271. https://doi.org/10.3390/cancers17020271
APA StyleZheng, M., Kessler, M., Jeschke, U., Reichenbach, J., Czogalla, B., Keckstein, S., Schroeder, L., Burges, A., Mahner, S., Trillsch, F., & Kaltofen, T. (2025). Necroptosis-Related Gene Signature Predicts Prognosis in Patients with Advanced Ovarian Cancer. Cancers, 17(2), 271. https://doi.org/10.3390/cancers17020271







