RNF135 Expression Marks Chemokine (C-C Motif) Ligand-Enriched Macrophage–Tumor Interactions in the Glioblastoma Microenvironment
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Data Acquisition and Preprocessing
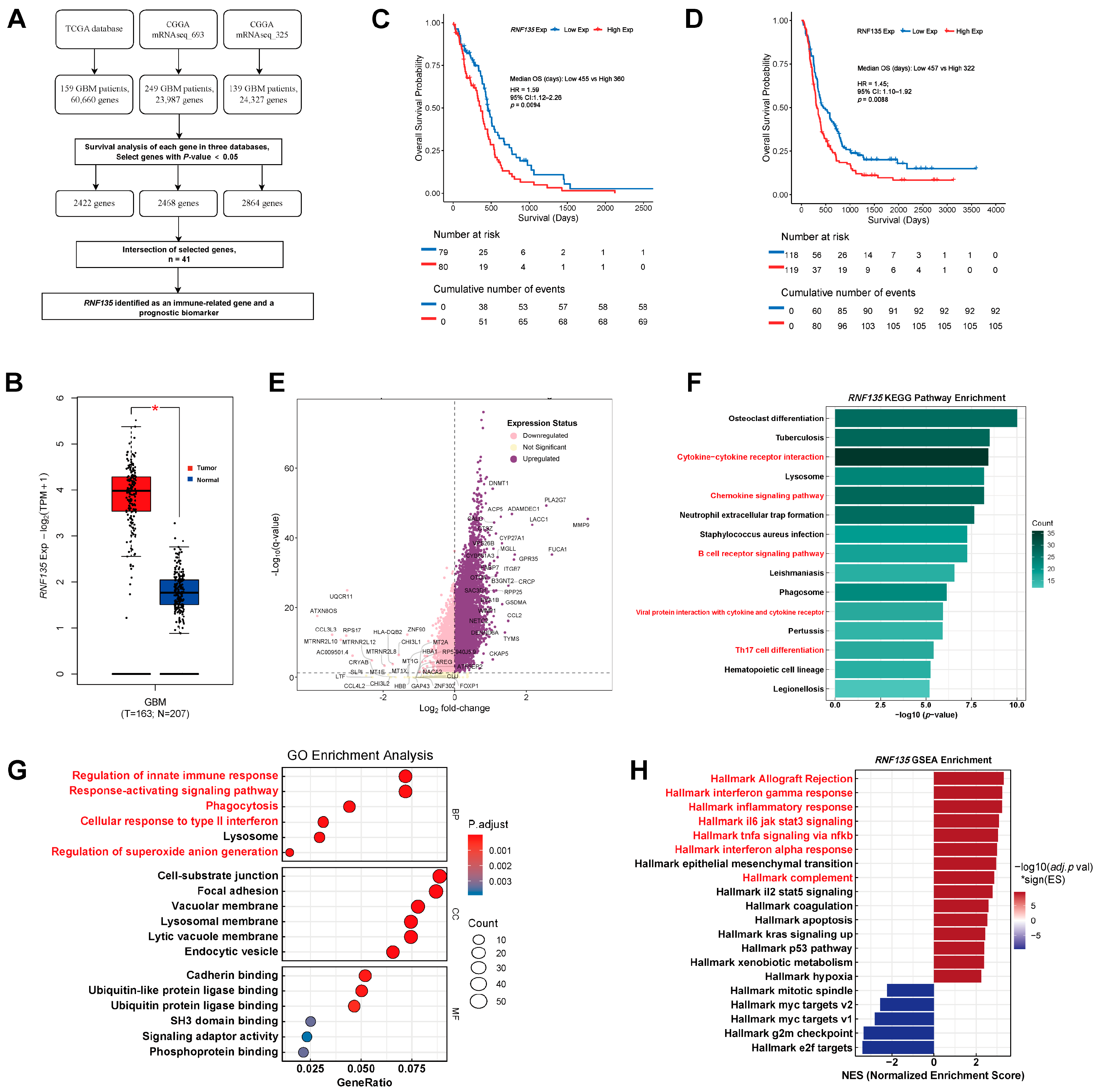
2.2. Differential Expression and Survival Analysis of RNF135
2.3. Functional Enrichment and GSEA
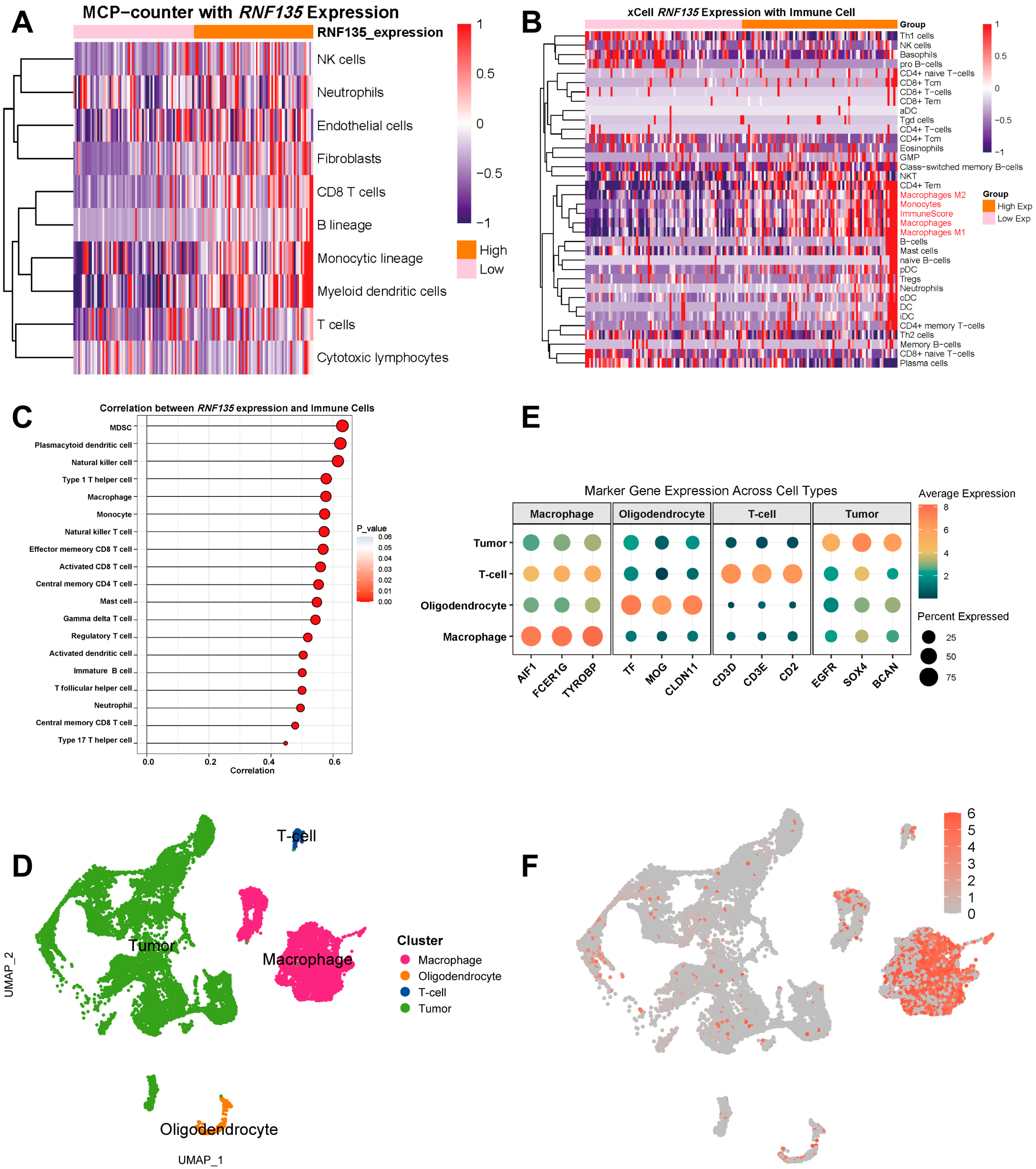
2.4. Immune Infiltration and Correlation Analysis
2.5. Single-Cell RNA-Seq Analysis and Cell-Type Annotation
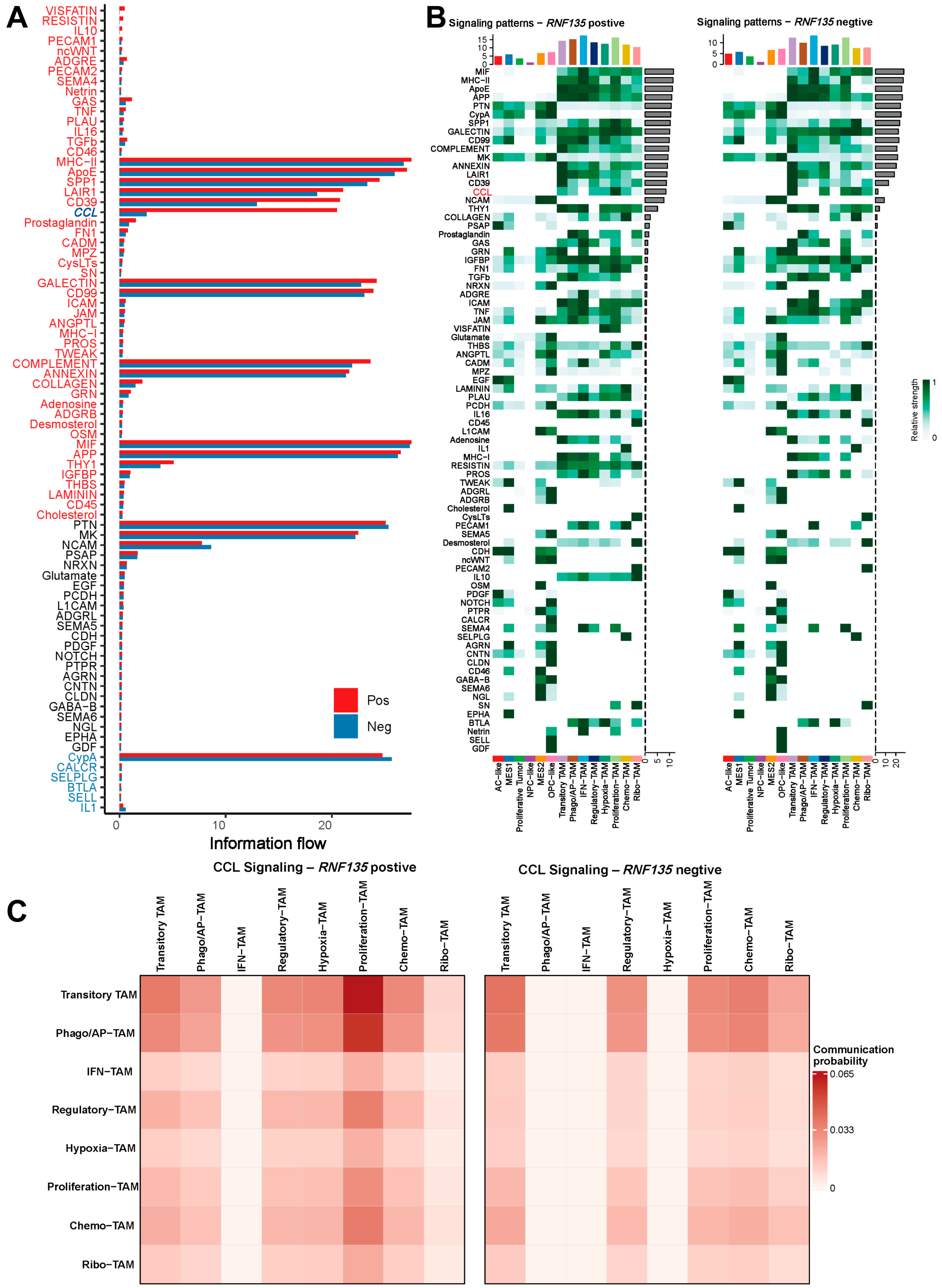
2.6. Cell–Cell Communication Analysis
2.7. Drug Sensitivity Prediction
2.8. Statistical Analysis
3. Results
3.1. RNF135 Is Upregulated in GBM and Predicts Poor Prognosis
3.2. RNF135 Correlates with Immune Cell Infiltration and Is Predominantly Expressed in TAMs
3.3. RNF135 Is Predominantly Expressed in Proliferation and Phagocytic TAM Subsets
3.4. RNF135-Positive TAMs Exhibit Stronger Communication with Aggressive Tumor Cell States
3.5. RNF135-Positive TAMs Exhibit Enhanced CCL Signaling and Broader Communication Profiles
3.6. CCL3/CCL3L3–CCR1 Axis Is a Dominant Contributor to CCL Signaling in RNF135-Positive TAMs
3.7. RNF135 Expression Is Enriched in Microvascular Proliferation Regions and Associated with Differential Drug Sensitivity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef]
- Tan, A.C.; Ashley, D.M.; López, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef]
- Schaff, L.R.; Mellinghoff, I.K. Glioblastoma and other primary brain malignancies in adults: A review. JAMA 2023, 329, 574–587. [Google Scholar] [CrossRef]
- Obrador, E.; Moreno-Murciano, P.; Oriol-Caballo, M.; López-Blanch, R.; Pineda, B.; Gutiérrez-Arroyo, J.L.; Loras, A.; Gonzalez-Bonet, L.G.; Martinez-Cadenas, C.; Estrela, J.M.; et al. Glioblastoma Therapy: Past, Present and Future. Int. J. Mol. Sci. 2024, 25, 2529. [Google Scholar] [CrossRef]
- Losurdo, A.; Di Muzio, A.; Cianciotti, B.C.; Dipasquale, A.; Persico, P.; Barigazzi, C.; Bono, B.; Feno, S.; Pessina, F.; Santoro, A.; et al. T Cell Features in Glioblastoma May Guide Therapeutic Strategies to Overcome Microenvironment Immunosuppression. Cancers 2024, 16, 603. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, J.; Berglund, A.E.; Du, D.; Macaulay, R.J.; Etame, A.B. The reprogramming impact of SMAC-mimetic on glioblastoma stem cells and the immune tumor microenvironment evolution. J. Exp. Clin. Cancer Res. 2025, 44, 191. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, Q.; Berglund, A.E.; Macaulay, R.J.; Mulé, J.J.; Etame, A.B. Tumor-Associated Macrophages in Glioblastoma: Mechanisms of Tumor Progression and Therapeutic Strategies. Cells 2025, 14, 1458. [Google Scholar] [CrossRef] [PubMed]
- Asioli, S.; Gatto, L.; Vardy, U.; Agostinelli, C.; Di Nunno, V.; Righi, S.; Tosoni, A.; Ambrosi, F.; Bartolini, S.; Giannini, C.; et al. Immunophenotypic Profile of Adult Glioblastoma IDH-Wildtype Microenvironment: A Cohort Study. Cancers 2024, 16, 3859. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, S.; To, K.K.; Zhu, S.; Wang, F.; Fu, L. Tumor-associated macrophages remodel the suppressive tumor immune microenvironment and targeted therapy for immunotherapy. J. Exp. Clin. Cancer Res. 2025, 44, 145. [Google Scholar] [CrossRef]
- Pittet, M.J.; Michielin, O.; Migliorini, D. Clinical relevance of tumour-associated macrophages. Nat. Rev. Clin. Oncol. 2022, 19, 402–421. [Google Scholar] [CrossRef]
- Chen, J.; Wu, Q.; Berglund, A.E.; Macaulay, R.J.; Mulé, J.J.; Etame, A.B. Identification of an Immune-Related Gene Signature for Prognostic Prediction in Glioblastoma: Insights from Integrated Bulk and Single-Cell RNA Sequencing. Cancers 2025, 17, 1799. [Google Scholar] [CrossRef]
- Cai, C.; Tang, Y.-D.; Zhai, J.; Zheng, C. The RING finger protein family in health and disease. Signal Transduct. Target. Ther. 2022, 7, 300. [Google Scholar] [CrossRef]
- Wang, M.-X.; Liuyu, T.; Zhang, Z.-D. Multifaceted roles of the E3 ubiquitin ligase RING finger protein 115 in immunity and diseases. Front. Immunol. 2022, 13, 936579. [Google Scholar] [CrossRef]
- Maelfait, J.; Beyaert, R. Emerging role of ubiquitination in antiviral RIG-I signaling. Microbiol. Mol. Biol. Rev. 2012, 76, 33–45. [Google Scholar] [CrossRef]
- Liang, J.; Liao, J.; Chang, R.; Jia, W.; Li, G.; Chen, Z.; Wu, H.; Zhu, C.; Wen, J.; Huang, Q.; et al. Riplet promotes lipid metabolism changes associated with CD8 T cell exhaustion and anti-PD-1 resistance in hepatocellular carcinoma. Sci. Immunol. 2025, 10, eado3485. [Google Scholar] [CrossRef] [PubMed]
- Cadena, C.; Ahmad, S.; Xavier, A.; Willemsen, J.; Park, S.; Park, J.W.; Oh, S.-W.; Fujita, T.; Hou, F.; Binder, M.; et al. Ubiquitin-dependent and -independent roles of E3 ligase RIPLET in innate immunity. Cell 2019, 177, 1187–1200.e1116. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, K.-N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Liu, X.; Li, G.; Chang, X.; Li, S.; Chen, J.; Zhao, Z.; Wang, J.; Jiang, T.; Chai, R. Clinical management and survival outcomes of patients with different molecular subtypes of diffuse gliomas in China (2011–2017): A multicenter retrospective study from CGGA. Cancer Biol. Med. 2022, 19, 1460–1476. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2024, 42, 293–304. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautes-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849.e821. [Google Scholar] [CrossRef]
- Wang, W.; Li, T.; Cheng, Y.; Li, F.; Qi, S.; Mao, M.; Wu, J.; Liu, Q.; Zhang, X.; Li, X.; et al. Identification of hypoxic macrophages in glioblastoma with therapeutic potential for vasculature normalization. Cancer Cell 2024, 42, 815–832.e812. [Google Scholar] [CrossRef]
- Jin, S.; Plikus, M.V.; Nie, Q. CellChat for systematic analysis of cell-cell communication from single-cell transcriptomics. Nat. Protoc. 2025, 20, 180–219. [Google Scholar] [CrossRef] [PubMed]
- Maeser, D.; Gruener, R.F.; Huang, R.S. An R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Briefings Bioinform. 2021, 22, bbab260. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Wang, M.; Wu, Z.; Cai, Y. Ubiquitination of p21 by E3 ligase RNF135 promotes the proliferation of human glioblastoma cells. Oncol. Lett. 2024, 28, 406. [Google Scholar] [CrossRef]
- Zhang, Y.; Sui, R.; Chen, Y.; Liang, H.; Shi, J.; Piao, H. Downregulation of miR-485-3p promotes glioblastoma cell proliferation and migration via targeting RNF135. Exp. Ther. Med. 2019, 18, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Hedayat, M.; Jafari, R.; Majidi Zolbanin, N. Selumetinib: A selective MEK1 inhibitor for solid tumor treatment. Clin. Exp. Med. 2023, 23, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Zlotnik, A.; Yoshie, O. The chemokine superfamily revisited. Immunity 2012, 36, 705–716. [Google Scholar] [CrossRef]
- Trifilo, M.J.; Bergmann, C.C.; Kuziel, W.A.; Lane, T.E. CC Chemokine Ligand 3 (CCL3) Regulates CD8+-T-Cell Effector Function and Migration following Viral Infection. J. Virol. 2003, 77, 4004–4014. [Google Scholar] [CrossRef] [PubMed]
- Zeissig, M.N.; Hewett, D.R.; Panagopoulos, V.; Mrozik, K.M.; To, L.B.; Croucher, P.I.; Zannettino, A.C.W.; Vandyke, K. Expression of the chemokine receptor CCR1 promotes the dissemination of multiple myeloma plasma cells in vivo. Haematologica 2021, 106, 3176–3187. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Wu, Q.; Berglund, A.E.; Macaulay, R.J.; Mulé, J.J.; Etame, A.B. RNF135 Expression Marks Chemokine (C-C Motif) Ligand-Enriched Macrophage–Tumor Interactions in the Glioblastoma Microenvironment. Cancers 2025, 17, 3271. https://doi.org/10.3390/cancers17193271
Chen J, Wu Q, Berglund AE, Macaulay RJ, Mulé JJ, Etame AB. RNF135 Expression Marks Chemokine (C-C Motif) Ligand-Enriched Macrophage–Tumor Interactions in the Glioblastoma Microenvironment. Cancers. 2025; 17(19):3271. https://doi.org/10.3390/cancers17193271
Chicago/Turabian StyleChen, Jianan, Qiong Wu, Anders E. Berglund, Robert J. Macaulay, James J. Mulé, and Arnold B. Etame. 2025. "RNF135 Expression Marks Chemokine (C-C Motif) Ligand-Enriched Macrophage–Tumor Interactions in the Glioblastoma Microenvironment" Cancers 17, no. 19: 3271. https://doi.org/10.3390/cancers17193271
APA StyleChen, J., Wu, Q., Berglund, A. E., Macaulay, R. J., Mulé, J. J., & Etame, A. B. (2025). RNF135 Expression Marks Chemokine (C-C Motif) Ligand-Enriched Macrophage–Tumor Interactions in the Glioblastoma Microenvironment. Cancers, 17(19), 3271. https://doi.org/10.3390/cancers17193271





