Guardians in the Gut: Mechanistic Insights into a Hidden Ally Against Triple-Negative Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
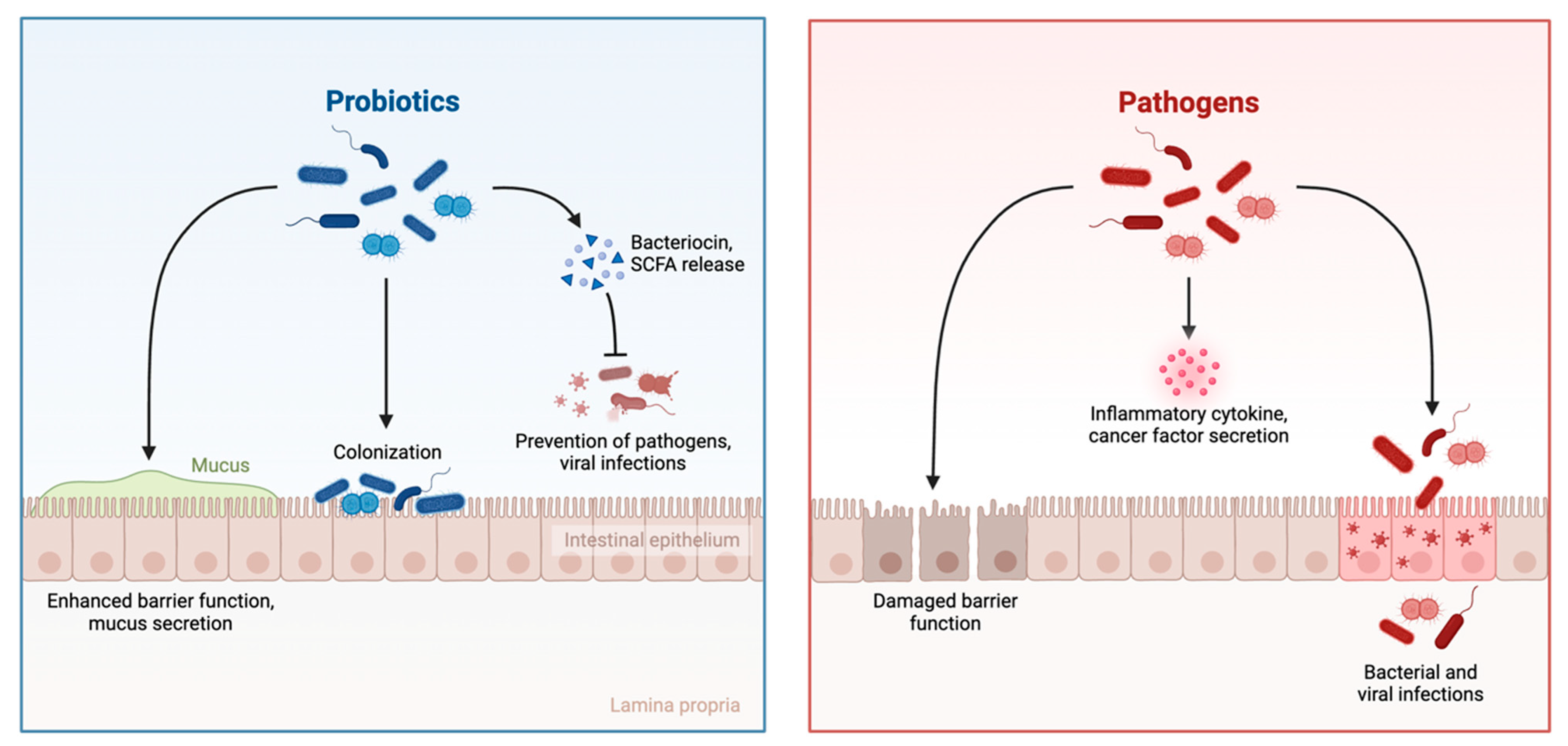
1.1. The Role of the Gut Microbiota in the Maintenance of Host Health
1.2. The Complex Interplay of the Gut–Brain Axis
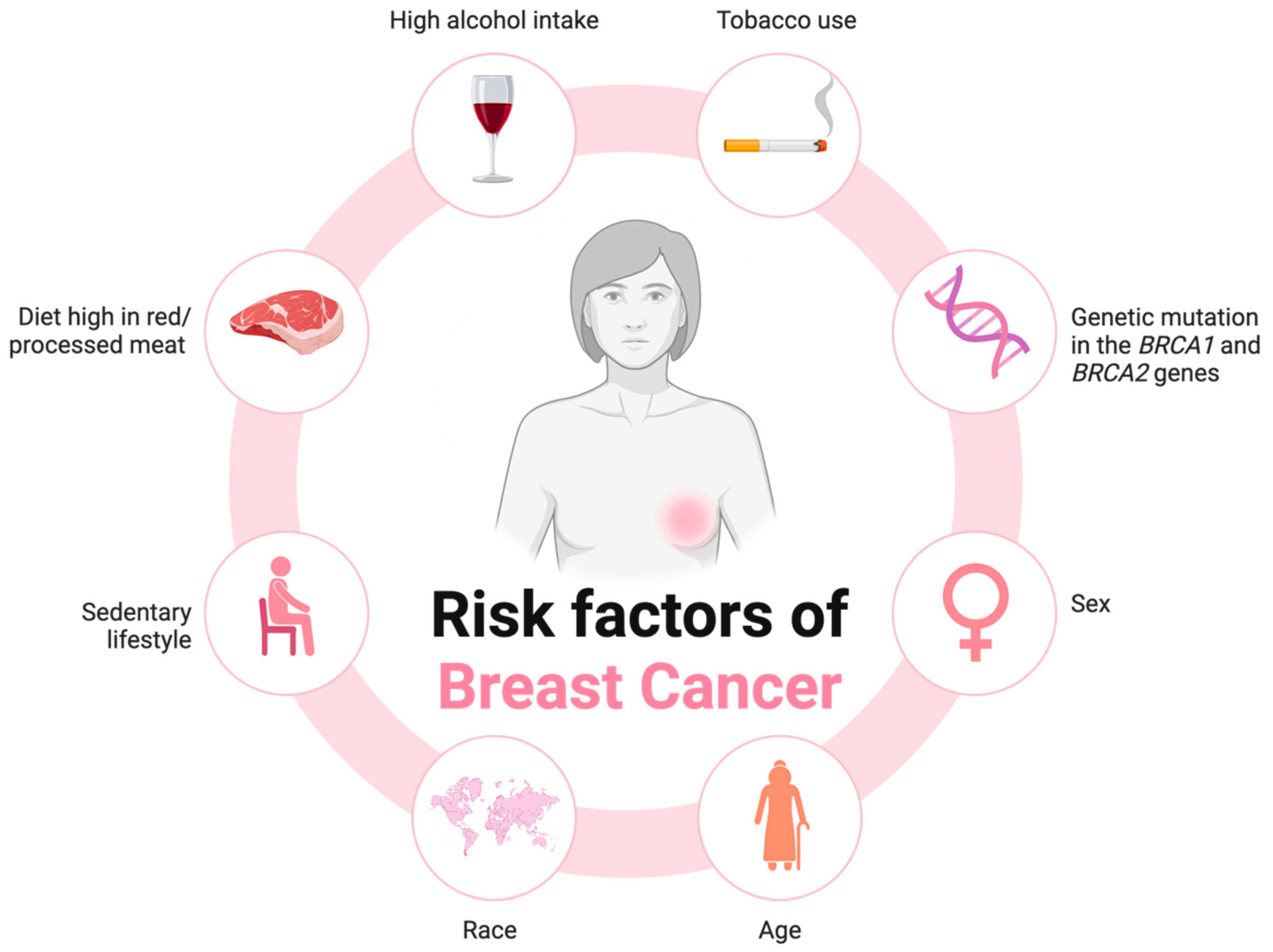
1.3. Gut Dysbiosis and Its Role in Breast Cancer Progression
2. Methods
Search Strategy
3. Results
3.1. Gut Microbial Metabolites and Breast Cancer
3.1.1. Short-Chain Fatty Acids (SCFAs)
3.1.2. Natural Purine Nucleosides
3.1.3. Ellagic Acid Derivatives
3.1.4. Tryptophan Derivatives
3.2. The Association Between the Gut Microbiota and Standard Chemotherapeutic Drugs
3.3. The Gut Microbiota and Epigenetic Modification
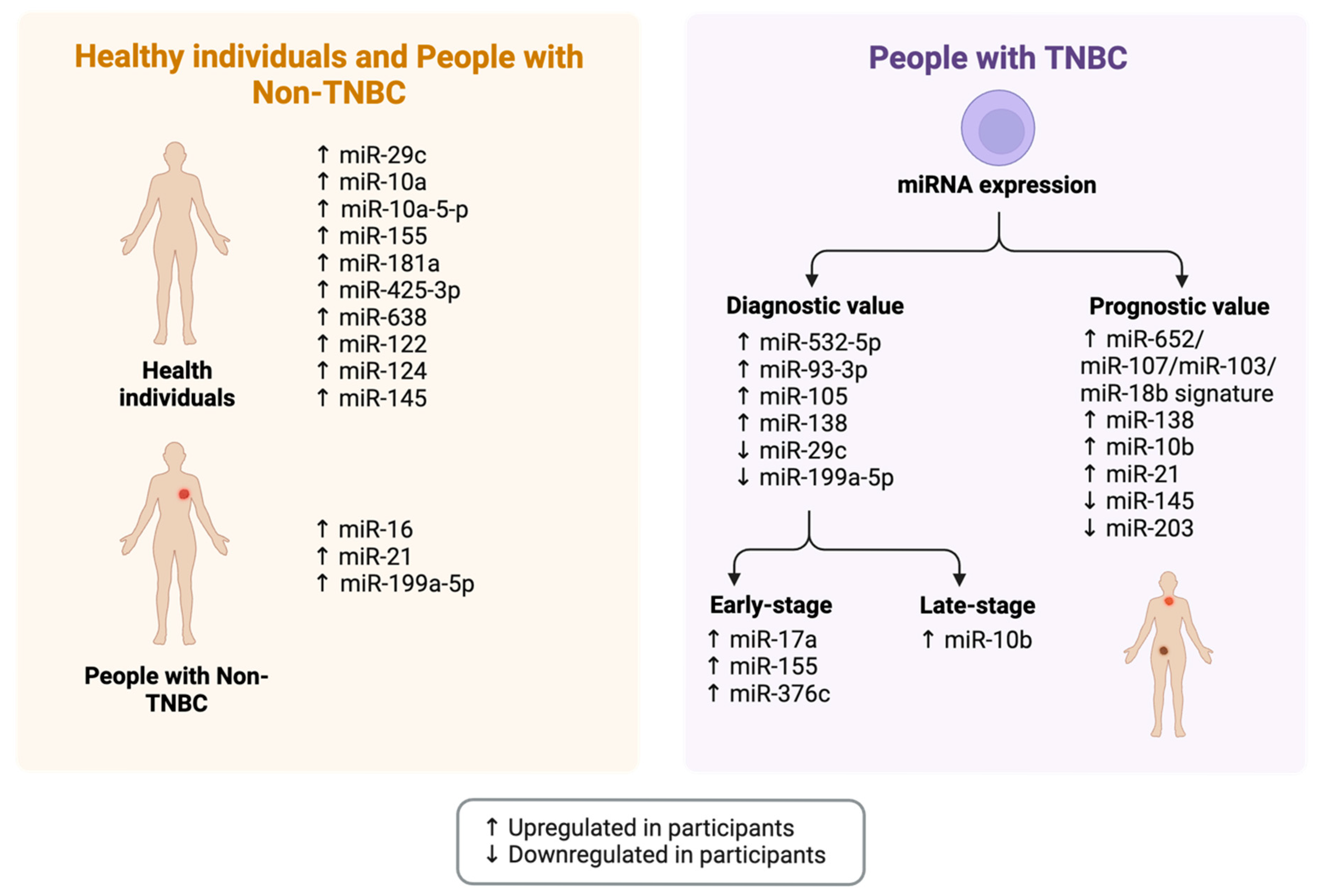
3.3.1. The Gut Microbiota-microRNA Connection
3.3.2. Prevalent miRNAs Associated with the Gut Microbiota as Potential Epigenetic Therapeutic Targets
3.4. The Gut Microbiota-miRNA-Breast Cancer Connection
3.4.1. Prevalent miRNAs Associated with Breast Cancer as Potential Epigenetic Therapeutic Targets
3.4.2. The Association Between the Gut Microbiota, miRNAs, and Breast Cancer
3.5. Additional Diagnostic and Epigenetic Factors in the Management of Disease
Minimal Residual Disease (MRD)
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Cancer Burden Growing, Amidst Mounting Need for Services. Saudi Med. J. 2024, 45, 326–327. [Google Scholar]
- Sedeta, E.T.; Jobre, B.; Avezbakiyev, B. Breast cancer: Global patterns of incidence, mortality, and trends. J. Clin. Oncol. 2023, 41, 10528. [Google Scholar] [CrossRef]
- Altinok Dindar, D.; Chun, B.; Palma, A.; Cheney, J.; Krieger, M.; Kasschau, K.; Stagaman, K.; Mitri, Z.I.; Goodyear, S.M.; Shannon, J. Association between Gut Microbiota Breast Cancer: Diet as a Potential Modulating Factor. Nutrients 2023, 154, 628. [Google Scholar] [CrossRef] [PubMed]
- Garmpis, N.; Damaskos, C.; Garmpi, A.; Kalampokas, E.; Kalampokas, T.; Spartalis, E.; Daskalopoulou, A.; Valsami, S.; Kontos, M.; Nonni, A. Histone deacetylases as new therapeutic targets in triple-negative breast cancer: Progress promises. Cancer Genom. Proteom. 2017, 14, 299–313. [Google Scholar]
- Zhang, J.; Xie, Q.; Huo, X.; Liu, Z.; Da, M.; Yuan, M.; Zhao, Y.; Shen, G. Impact of intestinal dysbiosis on breast cancer metastasis progression. Front. Oncol. 2022, 12, 1037831. [Google Scholar] [CrossRef]
- Cox-York, K.; Stoecker, E.; Hamm, A.K.; Weir, T.L. Microbiome and Cancer. In Microbial Metabolites in Cancer Promotion or Prevention; Current Cancer Research; Springer: Berlin/Heidelberg, Germany, 2019; pp. 317–346. [Google Scholar]
- Jaye, K.; Li, C.G.; Bhuyan, D.J. The complex interplay of gut microbiota with the five most common cancer types: From carcinogenesis to therapeutics to prognoses. Crit. Rev. Oncol. 2021, 165, 103429. [Google Scholar] [CrossRef] [PubMed]
- Jaye, K.; Li, C.G.; Chang, D.; Bhuyan, D.J. The role of key gut microbial metabolites in the development treatment of cancer. Gut Microbes 2022, 14, 2038865. [Google Scholar] [CrossRef]
- Jeong, S. Factors influencing development of the infant microbiota: From prenatal period to early infancy. Clin. Exp. Pediatr. 2022, 65, 438–447. [Google Scholar] [CrossRef]
- Thu, M.S.; Chotirosniramit, K.; Nopsopon, T.; Hirankarn, N.; Pongpirul, K. Human gut breast oral microbiome in breast cancer: Asystematic review meta-analysis. Front. Oncol. 2023, 13, 1144021. [Google Scholar] [CrossRef]
- Ruo, S.W.; Alkayyali, T.; Win, M.; Tara, A.; Joseph, C.; Kannan, A.; Srivastava, K.; Ochuba, O.; Sandhu, J.K.; Went, T.R.; et al. Role of Gut Microbiota Dysbiosis in Breast Cancer Novel Approaches in Prevention Diagnosis Treatment. Cureus 2021, 13, e17472. [Google Scholar] [CrossRef]
- Dukowicz, A.C.; E Lacy, B.; Levine, G.M. Small intestinal bacterial overgrowth: A comprehensive review. Gastroenterol. Hepatol. 2007, 3, 112. [Google Scholar]
- Song, P.; Peng, Z.; Guo, X. Gut microbial metabolites in cancer therapy. Trends Endocrinol. Metab. 2024, 36, 55–69. [Google Scholar] [CrossRef]
- Jaye, K.; Chang, D.; Li, C.G.; Bhuyan, D.J. Gut Metabolites Breast Cancer: The Continuum of Dysbiosis Breast Cancer Risk Potential Breast Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 9490. [Google Scholar] [CrossRef]
- Asadi, A.; Shadab Mehr, N.; Mohamadi, M.H.; Shokri, F.; Heidary, M.; Sadeghifard, N.; Khoshnood, S. Obesity gut-microbiota-brain axis: Anarrative review. J. Clin. Lab. Anal. 2022, 36, e24420. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Wang, X.; Guo, Y.; Yan, J.; Abuduwaili, A.; Aximujiang, K.; Yan, J.; Wu, M. Gut microbiota influence tumor development and Alter interactions with the human immune system. J. Exp. Clin. Cancer Res. 2021, 40, 42. [Google Scholar] [CrossRef]
- Sheflin, A.M.; Whitney, A.K.; Weir, T.L. Cancer-Promoting Effects of Microbial Dysbiosis. Curr. Oncol. Rep. 2014, 16, 406. [Google Scholar] [CrossRef] [PubMed]
- Miko, E.; Vida, A.; Kovacs, T.; Ujlaki, G.; Trencsenyi, G.; Marton, J.; Sari, Z.; Kovacs, P.; Boratko, A.; Hujber, Z.; et al. Lithocholic acid a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim. Biophys. Acta-Bioenerg. 2018, 1859, 958–974. [Google Scholar] [CrossRef]
- Bobin-Dubigeon, C.; Luu, H.T.; Leuillet, S.; Lavergne, S.N.; Carton, T.; Le Vacon, F.; Michel, C.; Nazih, H.; Bard, J.M. Faecal Microbiota Composition Varies between Patients with Breast Cancer and Healthy Women: A Comparative Case-Control Study. Nutrients 2021, 13, 2705. [Google Scholar] [CrossRef] [PubMed]
- Bard, J.M.; Luu, H.T.; Dravet, F.; Michel, C.; Moyon, T.; Pagniez, A.; Nazih, H.; Bobin-Dubigeon, C. Relationship between intestinal microbiota and clinical characteristics of patients with early stage breast cancer. FASEB J. 2015, 29, 914.2. [Google Scholar] [CrossRef]
- Yang, P.; Wang, Z.; Peng, Q.; Lian, W.; Chen, D. Comparison of the Gut Microbiota in Patients with Benign and Malignant Breast Tumors: A Pilot Study. Evol. Bioinform. 2021, 17, 11769343211057573. [Google Scholar] [CrossRef]
- Plaza, D.J.; Alvarez-Mercado, A.I.; Ruiz-Marin, C.M.; Reina-Perez, I.; Perez-Alonso, A.J.; Sanchez-Andujar, M.B.; Torne, P.; Gallart-Aragon, T.; Sanchez-Barron, M.T.; Reyes Lartategui, S.; et al. Association of breast gut microbiota dysbiosis the risk of breast cancer: Acase-control clinical study. BMC Cancer 2019, 19, 495. [Google Scholar]
- Goedert, J.J.; Jones, G.; Hua, X.; Xu, X.; Yu, G.; Flores, R.; Falk, R.T.; Gail, M.H.; Shi, J.; Ravel, J.; et al. Investigation of the Association Between the Fecal Microbiota and Breast Cancer in Postmenopausal Women: A Population-Based Case-Control Pilot Study. J. Natl. Cancer Inst. 2015, 107, jv147. [Google Scholar] [CrossRef] [PubMed]
- Zidi, O.; Souai, N.; Raies, H.; Ben Ayed, F.; Mezlini, A.; Mezrioui, S.; Tranchida, F.; Sabatier, J.M.; Mosbah, A.; Cherif, A.; et al. Fecal Metabolic Profiling of Breast Cancer Patients during Neoadjuvant Chemotherapy Reveals Potential Biomarkers. Molecules 2021, 26, 2266. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H.; Vigen, C.; Tseng, C.; Garcia, A.A.; Spicer, D. Effect of Chemotherapy on the Gut Microbiome of Breast Cancer Patients During the First Year of Treatment. Breast Cancer Targets Ther. 2022, 14, 433–451. [Google Scholar] [CrossRef] [PubMed]
- Nakkarach, A.; Foo, H.L.; Song, A.A.L.; Mutalib, N.E.A.; Nitisinprasert, S.; Withayagiat, U. Anti-cancer anti-inflammatory effects elicited by short chain fatty acids produced by Escherichia coli isolated from healthy human gut microbiota. Microb. Cell Factories 2021, 20, 36. [Google Scholar] [CrossRef]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; De Los Reyes-gavilán, C.G.; Salazar, N. Intestinal short chain fatty acids their link with diet human health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef]
- Lallemand, F.; Courilleau, D.; Buquet-Fagot, C.; Atfi, A.; Montagne, M.N.; Mester, J. Sodium butyrate induces G2 arrest in the human breast cancer cells MDA- MB-231, renders them competent for DNArereplication Experimental. Cell Res. 1999, 247, 432–440. [Google Scholar] [CrossRef]
- Jaye, K.; Alsherbiny, M.A.; Chang, D.; Li, C.G.; Bhuyan, D.J. Mechanistic Insights into the Anti-Proliferative Action of Gut Microbial Metabolites against Breast Adenocarcinoma Cells. Int. J. Mol. Sci. 2023, 24, 15053. [Google Scholar] [CrossRef]
- Wang, Z.T.; Chen, Z.J.; Jiang, G.M.; Wu, Y.M.; Liu, T.; Yi, Y.M.; Zeng, J.; Du, J.; Wang, H.S. Histone deacetylase inhibitors suppress mutant p53 transcription via HDAC8/YY1 signals in triple negative breast cancer cells. Cell. Signal. 2016, 28, 506–515. [Google Scholar] [CrossRef]
- Park, H.S.; Han, J.H.; Park, J.W.; Lee, D.H.; Jang, K.W.; Lee, M.; Heo, K.S.; Myung, C.S. Sodium propionate exerts anticancer effect in mice bearing breast cancer cell xenograft by regulating JAK2/STAT3/ROS/p38 MAPKsignaling. Acta Pharmacol. Sin. 2021, 42, 1311–1323. [Google Scholar] [CrossRef]
- Thirunavukkarasan, M.; Wang, C.; Rao, A.; Hind, T.; Teo, Y.R.; Siddiquee, A.A.-M.; Goghari, M.A.I.; Kumar, A.P.; Herr, D.R. Short-chain fatty acid receptors inhibit invasive phenotypes in breast cancer cells. PLoS ONE 2017, 12, e0186334. [Google Scholar] [CrossRef] [PubMed]
- Semaan, J.; El-Hakim, S.; Ibrahim, J.N.; Safi, R.; Elnar, A.A.; El Boustany, C. Comparative effect of sodium butyrate sodium propionate on proliferation cell cycle apoptosis in human breast cancer cells MCF-7. Breast Cancer 2020, 27, 696–705. [Google Scholar] [CrossRef]
- Soares, A.S.; Costa, V.M.; Diniz, C.; Fresco, P. Inosine Strongly Enhances Proliferation of Human C32 Melanoma Cells through PLC-PKC-MEK1/2-ERK1/2, PI3KPathways. Basic Clin. Pharmacol. Toxicol. 2015, 116, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef]
- Smith, D.M. Inosine Production and Cytoprotective Activity in a Gradient Model of Breast Cancer Hypoxia Nutrient Exclusion (CHyNE). Ph.D. Thesis, Texas Tech University, Lubbock, TX, USA, 2018. [Google Scholar]
- Panth, N.; Manandhar, B.; Paudel, K.R. Anticancer Activity of Punica granatum (Pomegranate): A Review. Phytother. Res. 2017, 31, 568–578. [Google Scholar] [CrossRef]
- Chen, P.; Guo, Z.; Chen, F.; Wu, Y.; Zhou, B. Recent Advances Perspectives on the Health Benefits of Urolithin B, ABioactive Natural Product Derived From Ellagitannins. Front. Pharmacol. 2022, 13, 917266. [Google Scholar]
- N Syed, D.; Chamcheu, J.-C.; M Adhami, V.; Mukhtar, H. Pomegranate extracts cancer prevention: Molecular cellular activities. Anti-Cancer Agents Med. Chem. 2013, 13, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Al-Maghout, T.; Cao, H.; Pelzl, L.; Salker, M.S.; Veldhoen, M.; Cheng, A.; Lang, F.; Singh, Y. Gut Bacterial Metabolite Urolithin A(UA) Mitigates Ca2+ Entry in TCells by Regulating miR-10a-5p. Front. Immunol. 2019, 10, 1737. [Google Scholar] [CrossRef]
- Gonzalez-Sarrias, A.; Miguel, V.; Merino, G.; Lucas, R.; Morales, J.C.; Tomas-Barberan, F.; Alvarez, A.I.; Espin, J.C. The gut microbiota ellagic acid-derived metabolite urolithin a its sulfate conjugate are substrates for the drug efflux transporter breast cancer resistance protein (ABCG2/BCRP). J. Agric. Food Chem. 2013, 61, 4352–4359. [Google Scholar] [CrossRef]
- Han, L.; Zhao, D.; Li, Y.; Jin, J.; El-kott, A.F.; Al-Saeed, F.A.; Eldib, A.M. Assessment of the Anti-Breast Cancer Effects of Urolithin with Molecular Docking Studies in the In Vitro Condition: Introducing a Novel Chemotherapeutic Drug. Mol. Biotechnol. 2024, 66, 554–566. [Google Scholar] [CrossRef]
- Vini, R.; Jaikumar, V.S.; Remadevi, V.; Ravindran, S.; Azeez, J.M.; Sasikumar, A.; Sundaram, S.; Sreeja, S. Urolithin A: Apromising selective estrogen receptor modulator 27-hydroxycholesterol attenuator in breast cancer. Phytother. Res. 2023, 37, 4504–4521. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Wang, Y.; Zhou, B.; Qian, F.; Liu, D.; Ye, D.; Zhou, X.; Fang, L. Urolithin A inhibits breast cancer progression via activating TFEB-mediated mitophagy in tumor macrophages. J. Adv. Res. 2024, 69, 125–138. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Jiang, H.; Chen, C.; Gao, J. Extensive Summary of the Important Roles of Indole Propionic Acid, a Gut Microbial Metabolite in Host Health and Disease. Nutrients 2023, 15, 151. [Google Scholar] [CrossRef]
- Sari, Z.; Miko, E.; Kovacs, T.; Janko, L.; Csonka, T.; Lente, G.; Sebo, E.; Toth, J.; Toth, D.; Arkosy, P.; et al. Indolepropionic acid a metabolite of the microbiome has cytostatic properties in breast cancer by activating ahr pxr receptors inducing oxidative stress. Cancers 2020, 12, 2411. [Google Scholar] [CrossRef]
- Hussain, A.; Xie, L.; Deng, G.; Kang, X. Common alterations in plasma free amino acid profiles gut microbiota-derived tryptophan metabolites of five types of cancer patients. Amino Acids 2023, 55, 1189–1200. [Google Scholar] [CrossRef]
- Auslander, N.; Yizhak, K.; Weinstock, A.; Budhu, A.; Tang, W.; Wang, X.W.; Ambs, S.; Ruppin, E. A joint analysis of transcriptomic metabolomic data uncovers enhanced enzyme-metabolite coupling in breast cancer. Sci. Rep. 2016, 6, 29662. [Google Scholar] [CrossRef]
- Tang, X.; Lin, C.-C.; Spasojevic, I.; Iversen, E.S.; Chi, J.-T.; Marks, J.R. Ajoint analysis of metabolomics genetics of breast cancer. Breast Cancer Res. 2014, 16, 415. [Google Scholar] [CrossRef] [PubMed]
- Devoy, C.; Bueso, Y.F.; Tangney, M. Tangney, Understanding and harnessing triple-negative breast cancer-related microbiota in oncology. Front. Oncol. 2022, 12, 1020121. [Google Scholar] [CrossRef]
- Kovács, T.; Mikó, E.; Ujlaki, G.; Yousef, H.; Csontos, V.; Uray, K.; Bai, P. The involvement of oncobiosis bacterial metabolite signaling in metastasis formation in breast cancer Cancer. Metastasis Rev. 2021, 40, 1223–1249. [Google Scholar] [CrossRef] [PubMed]
- Kovács, T.; Mikó, E.; Vida, A.; Sebő, É.; Toth, J.; Csonka, T.; Boratkó, A.; Ujlaki, G.; Lente, G.; Kovács, P. Cadaverine a metabolite of the microbiome reduces breast cancer aggressiveness through trace amino acid receptors. Sci. Rep. 2019, 9, 1300. [Google Scholar] [CrossRef]
- Li, Y.; Dong, B.; Wu, W.; Wang, J.; Jin, H.; Chen, K.; Huang, K.; Huang, S.; Yao, Y. Metagenomic Analyses Reveal Distinct Gut Microbiota Signature for Predicting the Neoadjuvant Chemotherapy Responsiveness in Breast Cancer Patients. Front. Oncol. 2022, 12, 865121. [Google Scholar] [CrossRef]
- Deng, Y.; Hou, X.; Wang, H.; Du, H.; Liu, Y. Influence of Gut Microbiota-Mediated Immune Regulation on Response to Chemotherapy. Pharmaceuticals 2024, 17, 604. [Google Scholar] [CrossRef]
- Gori, S.; Inno, A.; Belluomini, L.; Bocus, P.; Bisoffi, Z.; Russo, A.; Arcaro, G. Gut microbiota cancer: How gut microbiota modulates activity efficacy toxicity of antitumoral therapy. Crit. Rev. Oncol. Hematol. 2019, 143, 139–147. [Google Scholar] [CrossRef]
- Terrisse, S.; Derosa, L.; Iebba, V.; Ghiringhelli, F.; Vaz-Luis, I.; Kroemer, G.; Fidelle, M.; Christodoulidis, S.; Segata, N.; Thomas, A.M.; et al. Intestinal microbiota influences clinical outcome side effects of early breast cancer treatment. Cell Death Differ. 2021, 28, 2778–2796. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, B.; Zheng, Q.; Li, H.; Meng, X.; Zhou, F.; Zhang, L. A review of gut microbiota-derived metabolites in tumor progression cancer therapy. Adv. Sci. 2023, 10, 2207366. [Google Scholar] [CrossRef]
- Bilenduke, E.; Sterrett, J.D.; Ranby, K.W.; Borges, V.F.; Grigsby, J.; Carr, A.L.; Kilbourn, K.; Lowry, C.A. Impacts of breast cancer chemotherapy on gut microbiome cognitive functioning mood relative to healthy controls. Sci. Rep. 2022, 12, 19547. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, N.; Nakajo, T.; Katayoshi, T.; Tsuji-Naito, K. Nucleotide-binding oligomerization domain protein-1 is expressed involved in the inflammatory response in human sebocytes. Biochem. Biophys. Rep. 2023, 36, 101561. [Google Scholar] [CrossRef]
- Cui, B.; Luo, H.; He, B.; Liu, X.; Lv, D.; Zhang, X.; Su, K.; Zheng, S.; Lu, J.; Wang, C. Gut dysbiosis conveys psychological stress to activate LRP5/β-catenin pathway promoting cancer stemness Signal Transduction. Target. Ther. 2025, 10, 79. [Google Scholar]
- Sevcikova, A.; Izoldova, N.; Stevurkova, V.; Kasperova, B.; Chovanec, M.; Ciernikova, S.; Mego, M. The impact of the microbiome on resistance to cancer treatment with chemotherapeutic agents immunotherapy. Int. J. Mol. Sci. 2022, 23, 488. [Google Scholar] [CrossRef] [PubMed]
- Velloso, F.J.; Sogayar, M.C.; Correa, R.G. Expression and in vitro assessment of tumorigenicity for NOD1 and NOD2 receptors in breast cancer cell lines. BMC Res. Notes 2018, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Sonoyama, K.; Ohsaka, F. Role of microRNAs in the crosstalk between the gut microbiota and intestinal immune system. Biosci. Microbiota 2023, 42, 222–228. [Google Scholar]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Xue, X.; Feng, T.; Yao, S.; Wolf, K.J.; Liu, C.-G.; Liu, X.; Elson, C.O.; Cong, Y. Microbiota downregulates dendritic cell expression of miR-10a which targets IL-12/IL-23p40. J. Immunol. 2011, 187, 5879–5886. [Google Scholar] [CrossRef]
- Runtsch, M.C.; Round, J.L.; O’Connell, R.M. MicroRNAs and the regulation of intestinal homeostasis. Front. Genet. 2014, 5, 347. [Google Scholar] [CrossRef]
- Prukpitikul, P.; Sirivarasai, J.; Sutjarit, N. The molecular mechanisms underlying gut microbiota-miRNA interaction in metabolic disorders. Benef. Microbes 2024, 15, 83–96. [Google Scholar] [CrossRef]
- Zhao, L.; Ye, Y.; Gu, L.; Jian, Z.; Stary, C.M.; Xiong, X. Extracellular vesicle-derived miRNAas a novel regulatory system for bi-directional communication in gut-brain-microbiota axis. J. Transl. Med. 2021, 19, 202. [Google Scholar] [CrossRef]
- Das, K.; Rao, L.V.M. The role of microRNAs in inflammation. Int. J. Mol. Sci. 2022, 23, 15479. [Google Scholar] [CrossRef] [PubMed]
- Behrouzi, A.; Ashrafian, F.; Mazaheri, H.; Lari, A.; Nouri, M.; Riazi Rad, F.; Hoseini Tavassol, Z.; Siadat, S.D. The importance of interaction between MicroRNAs gut microbiota in several pathways. Microb. Pathog. 2020, 144, 104200. [Google Scholar] [CrossRef]
- Guo, Y.; Zhu, X.; Zeng, S.; He, M.; Xing, X.; Wang, C. MiRNA-10a-5p Alleviates Insulin Resistance Maintains Diurnal Patterns of Triglycerides Gut Microbiota in High-Fat Diet-Fed Mice. Mediat. Inflamm. 2020, 2020, 8192187. [Google Scholar]
- Bhardwaj, A.; Tachibana, K.; Ganesan, N.; Pan, Y.; Rajapakshi, K.; Coarfa, C.; Gunaratne, P.H.; Bedrosian, I. Targeting of miRNAnetworks for prevention of basal-like breast cancers. Cancer Res. 2015, 75, 3982. [Google Scholar] [CrossRef]
- Kleivi Sahlberg, K.; Bottai, G.; Naume, B.; Burwinkel, B.; Calin, G.A.; Børresen-Dale, A.-L.; Santarpia, L. Aserum microRNAsignature predicts tumor relapse survival in triple-negative breast cancer patients. Clin. Cancer Res. 2015, 21, 1207–1214. [Google Scholar] [CrossRef]
- Piasecka, D.; Braun, M.; Kordek, R.; Sadej, R.; Romanska, H. MicroRNAs in regulation of triple-negative breast cancer progression. J. Cancer Res. Clin. Oncol. 2018, 144, 1401–1411. [Google Scholar] [CrossRef]
- Tsai, H.-P.; Huang, S.-F.; Li, C.-F.; Chien, H.-T.; Chen, S.-C. Differential microRNAexpression in breast cancer with different onset age. PLoS ONE 2018, 13, e0191195. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, J.; Shahid, S.; Shahzadi, S.; Akhtar, M.W.; Sadaf, S. Identification of circulating miRNAs as non-invasive biomarkers of triple negative breast cancer in the population of Pakistan Pakistan. J. Zool. 2019, 51, 1113. [Google Scholar] [CrossRef]
- Shin, V.; Siu, J.; Cheuk, I.; Ng, E.; Kwong, A. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer British. J. Cancer 2015, 112, 1751–1759. [Google Scholar] [CrossRef]
- Nama, S.; Muhuri, M.; Di Pascale, F.; Quah, S.; Aswad, L.; Fullwood, M.; Sampath, P. MicroRNA-138 is a prognostic biomarker for triple-negative breast cancer promotes tumorigenesis via TUSC2 repression. Sci. Rep. 2019, 9, 12718. [Google Scholar] [CrossRef]
- Li, H.-Y.; Liang, J.-L.; Kuo, Y.-L.; Lee, H.-H.; Calkins, M.J.; Chang, H.-T.; Lin, F.-C.; Chen, Y.-C.; Hsu, T.-I.; Hsiao, M. miR-105/93–3p promotes chemoresistance circulating miR-105/93–3p acts as a diagnostic biomarker for triple negative breast cancer Breast. Cancer Res. 2017, 19, 133. [Google Scholar] [CrossRef]
- De Silva, S.; Tennekoon, K.H.; Karunanayake, E.H. Interaction of gut microbiome and host micrornas with the occurrence of colorectal and breast cancer and their impact on patient immunity. Onco Targets Ther. 2021, 14, 5115–5129. [Google Scholar] [CrossRef] [PubMed]
- Ahani, S.L.; Niknejad, A.; Amini, E. Sodium Butyrate as Histone Deacetylase Inhibitor Can Alter miR-101, ZEB1, ZEB2, and E-cadherin Expression in MDA-MB-468 Cells as Triple Negative Breast Cancer Cells. Int. J. Cancer Manag. 2023, 16, e139329. [Google Scholar]
- Liu, X.; Tang, H.; Chen, J.; Song, C.; Yang, L.; Liu, P.; Wang, N.; Xie, X.; Lin, X.; Xie, X. MicroRNA-101 inhibits cell progression increases paclitaxel sensitivity by suppressing MCL-1 expression in human triple-negative breast cancer. Oncotarget 2015, 6, 20070. [Google Scholar] [CrossRef]
- dos Santos Ferreira, A.C.; Robaina, M.C.; de Rezende, L.M.M.; Severino, P.; Klumb, C.E. Histone deacetylase inhibitor prevents cell growth in Burkitt’s lymphoma by regulating PI3K/Akt pathways leads to upregulation of miR-143, miR-145, miR-101. Ann. Hematol. 2014, 93, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.-H.; Dimri, M.; Dimri, G.P. MicroRNA-31 is a transcriptional target of histone deacetylase inhibitors and a regulator of cellular senescence. J. Biol. Chem. 2015, 290, 10555–10567. [Google Scholar]
- Gasparini, P.; Lovat, F.; Fassan, M.; Casadei, L.; Cascione, L.; Jacob, N.K.; Carasi, S.; Palmieri, D.; Costinean, S.; Shapiro, C.L. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc. Natl. Acad. Sci. USA 2014, 111, 4536–4541. [Google Scholar] [CrossRef]
- Laborda-Illanes, A.; Aranega-Martín, L.; Sánchez-Alcoholado, L.; Boutriq, S.; Plaza-Andrades, I.; Peralta-Linero, J.; Garrido Ruiz, G.; Pajares-Hachero, B.; Álvarez, M.; Alba, E. Exploring the Relationship between MicroRNAs Intratumoral Microbiota Breast Cancer Progression in Patients with without Metastasis. Int. J. Mol. Sci. 2024, 25, 7091. [Google Scholar] [CrossRef]
- German, R.; Marino, N.; Hemmerich, C.; Podicheti, R.; Rusch, D.B.; Stiemsma, L.T.; Gao, H.; Xuei, X.; Rockey, P.; Storniolo, A.M. Exploring breast tissue microbial composition the association with breast cancer risk factors Breast. Cancer Res. 2023, 25, 82. [Google Scholar] [CrossRef]
- Tachtsidis, A.; McInnes, L.M.; Jacobsen, N.; Thompson, E.; Saunders, C.M. Minimal residual disease in breast cancer: An overview of circulating disseminated tumour cells Clinical. Clin. Exp. Metastasis 2016, 33, 521–550. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, S.; Cristofanilli, M.; Singh, B.; Reuben, J.; Gao, H.; Cohen, E.N.; Andreopoulou, E.; Hall, C.S.; Lodhi, A.; Jackson, S. Detection of minimal residual disease in blood bone marrow in early stage breast cancer. Cancer 2010, 116, 3330–3337. [Google Scholar] [CrossRef]
- Parsons, H.A.; Rhoades, J.; Reed, S.C.; Gydush, G.; Ram, P.; Exman, P.; Xiong, K.; Lo, C.C.; Li, T.; Fleharty, M. Sensitive detection of minimal residual disease in patients treated for early-stage breast cancer. Clin. Cancer Res. 2020, 26, 2556–2564. [Google Scholar] [CrossRef]
- Tarallo, S.; Ferrero, G.; De Filippis, F.; Francavilla, A.; Pasolli, E.; Panero, V.; Cordero, F.; Segata, N.; Grioni, S.; Pensa, R.G.; et al. Stool microRNAprofiles reflect different dietary gut microbiome patterns in healthy individuals. Gut 2022, 71, 302–1314. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaye, K.; Alsherbiny, M.A.; Chang, D.; Li, C.-G.; Bhuyan, D.J. Guardians in the Gut: Mechanistic Insights into a Hidden Ally Against Triple-Negative Breast Cancer. Cancers 2025, 17, 3248. https://doi.org/10.3390/cancers17193248
Jaye K, Alsherbiny MA, Chang D, Li C-G, Bhuyan DJ. Guardians in the Gut: Mechanistic Insights into a Hidden Ally Against Triple-Negative Breast Cancer. Cancers. 2025; 17(19):3248. https://doi.org/10.3390/cancers17193248
Chicago/Turabian StyleJaye, Kayla, Muhammad A. Alsherbiny, Dennis Chang, Chun-Guang Li, and Deep Jyoti Bhuyan. 2025. "Guardians in the Gut: Mechanistic Insights into a Hidden Ally Against Triple-Negative Breast Cancer" Cancers 17, no. 19: 3248. https://doi.org/10.3390/cancers17193248
APA StyleJaye, K., Alsherbiny, M. A., Chang, D., Li, C.-G., & Bhuyan, D. J. (2025). Guardians in the Gut: Mechanistic Insights into a Hidden Ally Against Triple-Negative Breast Cancer. Cancers, 17(19), 3248. https://doi.org/10.3390/cancers17193248








