1. Introduction
The existing evidence confirms that long non-coding RNAs (lncRNAs) can function as competitive endogenous RNAs (ceRNAs) by modulating microRNAs (miRNAs), thereby influencing tumor cell migration and proliferation [
1]. Long intergenic non-protein coding
RNA 02381 (
LINC02381), a member of the lincRNA family, has emerged a crucial regulator in oncogenesis. Recent studies have indicated that lincRNAs are actively involved in transcriptional reprogramming and play essential roles in malignant progression [
1]. Aberrant
LINC02381 expression has been documented in various malignancies. For instance, it is significantly overexpressed in esophageal squamous cell carcinoma tissues compared to normal esophageal mucosa [
2], and elevated levels have also been observed in glioma [
3] and cervical carcinoma cells, where it exerts oncogenic functions contributing to tumor progression [
1]. Notably, Xia Bian et al. reported that
LINC02381 knockdown suppresses osteosarcoma (OS) cell invasion, proliferation, and migration [
4]. However, the precise molecular mechanisms through which
LINC02381 functions in OS remain largely undefined.
To explore its downstream regulatory network, we focused on miRNAs potentially interacting with
LINC02381. Among them,
microRNA-let-7g-5p (
let-7g-5p) attracted our attention due to its established tumor-suppressive role. As a member of the let-7 family,
let-7g-5p is known for its low expression in cancers such as nasopharyngeal carcinoma and cholangiocarcinoma [
5,
6]. Meanwhile, reduced
let-7g-5p levels have been detected in osteoporotic fractures [
7], although its role in OS remains poorly understood. Emerging evidence suggests that
let-7g-5p may target oncogenes like
HMGA2 and
MAP3K1 [
8,
9], indicating its potential involvement in OS progression [
10]. Bioinformatic predictions reveal potential binding sites between
let-7g-5p and Thrombospondin-1 (
THBS1), a stromal glycoprotein involved in tumor-associated processes such as cell–matrix interactions, extracellular matrix remodeling, and angiogenesis [
11,
12].
THBS1 is frequently overexpressed in gastric and colorectal cancers and is related to poor prognosis [
13,
14]. Furthermore, elevated
THBS1 levels have been linked to osteoporosis [
15], and its upregulation in OS has been correlated with aggressive tumor behavior and poor clinical outcomes [
16,
17,
18]. Mechanistically,
THBS1 is believed to promote tumor progression by modulating the extracellular matrix, stimulating angiogenesis, and activating the
transforming growth factor beta (
TGF-β) signaling pathway [
19]. However, the direct regulatory relationship between
THBS1 and
let-7g-5p has not yet been established.
Based on these molecular insights, we propose that LINC02381, let-7g-5p, and THBS1 may constitute a cascade regulatory network in OS. Supported by bioinformatic predictions and preliminary experimental data, we hypothesize that LINC02381 modulates let-7g-5p activity via a ceRNA mechanism, thereby regulating THBS1 expression and contributing to OS pathogenesis. Accordingly, this study aims to investigate the role of the LINC02381/let-7g-5p/THBS1 axis in OS cell viability and tumor progression.
2. Materials and Methods
2.1. Ethics Statement
All experimental procedures were authorized by the Research Ethics Committee of Shanghai Changzheng Hospital, Navy Medical University in compliance with international ethical guidelines (approval number: 2017052, ethics approval data: 20170523). Written informed consent was obtained from all patients. All animal procedures were authorized by the Animal Ethics Review Board of the same institution (approval number: 2017122, ethics approval data: 20171225) and conducted following the NIH Guide for the Care and Use of Laboratory Animals as well as relevant local animal welfare regulations.
2.2. Experimental Subjects
A total of 71 patients with OS who underwent surgical resection at Shanghai Changzheng Hospital, Navy Medical University from July 2017 to July 2022 were included. Paired OS tumor tissues and paracancerous tissues were collected. Clinicopathological variables such as gender (male/female), age (≥20 vs. <20 years), tumor size (≥3 cm vs. <3 cm), TNM stage (I–II vs. III–IV), and lymph node metastasis (yes/no) were systematically recorded. Detailed information is presented in
Table 1. All clinical definitions adhered to the
AJCC Cancer Staging Manual, 8th Edition, and institutional diagnostic criteria established by the pathology and radiology departments. Data were independently reviewed and validated by two researchers. None of the enrolled subjects had received prior radiotherapy or chemotherapy. Collected tissues were labeled and cryopreserved at −70 °C for subsequent experiments.
To further investigate the correlation between molecular expression and treatment response, follow-up was conducted for all 71 patients who received postoperative adjuvant chemotherapy. According to RECIST criteria, patients were categorized into chemotherapy-sensitive (CR/PR) and chemotherapy-insensitive (SD/PD) groups based on radiological evaluations.
2.3. Cell Culture
Human OS cell lines (143B, U-2OS, Saos-2, MNNG-HOS, MG-63) and the human normal osteoblast cell line hFOB1.19 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). 143B and MNNG-HOS cells were cultured in RPMI-1640; Saos-2 in α-minimum Eagle’s medium (α-MEM); MG-63 in MEM; and U-2OS and hFOB1.19 in Dulbecco’s modified Eagle medium (DMEM), all supplied by Gibco (Grand Island, NY, USA). Media were supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic–antimycotic solution (100 U/mL penicillin and 100 U/mL streptomycin). Cells were maintained in a 37 °C incubator with 5% CO2, with medium changes every two days. Subculturing was performed when cell confluence reached 80–90%.
Cells in the logarithmic growth phase after 2–3 stable passages were used for experiments. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA concentration and purity were assessed using a NanoDrop 2000 spectrophotometer (acceptable A260/A280 ratio: 1.8–2.1). The integrity of 28S/18S ribosomal RNA was confirmed by agarose gel electrophoresis to ensure RNA quality for downstream reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis. The mRNA expression levels of LINC02381, let-7g-5p, and THBS1 were detected by RT-qPCR, and THBS1 protein levels were assessed by Western blotting. Based on relative expression differences, U-2OS and Saos-2 cells—both exhibiting the greatest divergence from the normal osteoblast hFOB1.19—were selected for subsequent functional experiments.
2.4. Cell Grouping and Transfection
U-2OS and SaOS-2 cells in the logarithmic growth phase were seeded into 6-well plates at a density of 2 × 105 cells per well. Transfection was initiated when cell confluence reached approximately 80%. Lipofectamine™ 2000 (Catalog No.: 11668-027, Invitrogen, Carlsbad, CA, USA) was used for transfection, strictly following the manufacturer’s protocol. Briefly, each oligonucleotide (final concentration: 50 nM; all synthesized by Shanghai GenePharma Co., Ltd., Shanghai, China) was diluted in 250 μL of serum-free DMEM and incubated at room temperature for 5 min. Separately, 5 μL of Lipofectamine™ 2000 was diluted in 250 μL of serum-free DMEM and incubated for another 5 min. The two solutions were then combined and allowed to form complexes at room temperature for 20 min before being added to the wells. Cells were incubated at 37 °C in a humidified incubator with 5% CO2 for 6 h. Afterward, the transfection medium was replaced with DMEM containing 10% FBS, and cells were cultured further for downstream applications.
Cells were divided into the following eight groups, with three replicates per group. Transfection efficiency was verified by RT-qPCR and/or Western blot prior to functional assays: short hairpin RNA (sh)-negative control (NC) group (transfected with the negative control of LINC02381 knockdown vector), sh-LINC02381 group (transfected with LINC02381 knockdown vector), mimic-NC group (transfected with the negative control of let-7g-5p mimic), let-7g-5p mimic group (transfected with let-7g-5p mimic), sh-LINC02381 + inhibitor-NC group (transfected with sh-LINC02381 background with negative control miRNA inhibitor), sh-LINC02381 + let-7g-5p inhibitor group (transfected with sh-LINC02381 background with let-7g-5p inhibitor), sh-LINC02381 + overexpression (oe)-NC group (co-transfected with the negative control of THBS1 overexpression vector and LINC02381 knockdown vector), and sh-LINC02381 + oe-THBS1 group (co-transfected with the THBS1 overexpression plasmid and LINC02381 knockdown vector).
2.5. Cell Counting Kit-8 (CCK-8) Assay
The proliferative capacity of U-2OS and SaOS-2 cells was assessed using a CCK-8 assay kit (Dojindo Molecular Technologies, Kumamoto, Japan). Cells from each group were seeded into 96-well plates at a density of 1 × 104 cells per well, with three replicates per group and one blank well as an NC. Cells were incubated at 37 °C in a 5% CO2 atmosphere for 24, 48, and 72 h. At each time point, 10 μL of CCK-8 working solution was added directly to each well (without medium replacement), followed by a 4 h incubation under the same conditions. The optical density (OD) at 450 nm was measured using a microplate reader (Bio-Rad, Hercules, CA, USA). Cell proliferation curves were plotted accordingly. All procedures strictly adhered to the manufacturer’s instructions, and all experiments were independently repeated at least three times to ensure data reliability and reproducibility.
2.6. Colony Formation Assay
A plate colony formation assay was performed to evaluate the clonogenic potential of U-2OS and SaOS-2 cells in each experimental group. Cells in the logarithmic growth phase were seeded into 6 cm culture dishes at a density of 200 cells per dish (1 × 103/mL, in 200 μL of cell suspension). The dishes were gently swirled to evenly distribute the cells and then incubated at 37 °C in a humidified incubator with 5% CO2 for 10 days.
Once visible colonies (each > 50 μm in diameter) were observed by the naked eye, the culture was terminated. The supernatant was carefully aspirated, and cells were washed twice with phosphate-buffered saline (PBS). Colonies were fixed with 96% ethanol for 10 min, stained with 1% crystal violet solution for 5 min, rinsed gently with tap water, and air-dried for approximately 1 h. The dishes were then inverted, and colonies were counted using a transparent grid film and an inverted microscope (Olympus Corporation, Tokyo, Japan). Each group included three replicates, and all experiments were repeated at least three times. The average number of colonies and colony formation rate (number of colonies/number of seeded cells × 100%) were recorded.
2.7. Transwell Assay
Transwell chambers with an 8.0 μm pore size (Corning, Corning, USA) were used to assess the migratory ability of U-2OS and SaOS-2 cells. At 24 h post-transfection, cells were digested, resuspended, and adjusted to a concentration of 1 × 105/mL. A 200 μL serum-free cell suspension was added to the upper chamber, while 600 μL of complete medium containing 10% FBS was added to the lower chamber as a chemoattractant. The cells were incubated at 37 °C with 5% CO2 for 24 h.
After incubation, non-migrated cells on the upper surface of the membrane were carefully removed with a cotton swab. The inserts were then fixed with 4% paraformaldehyde for 30 min, washed twice with PBS, and stained with 0.5% crystal violet for 20 min. Following staining, the membranes were rinsed thoroughly and air-dried. Five randomly selected non-overlapping fields per chamber were imaged and counted under an inverted microscope (×200 magnification, Olympus, Tokyo, Japan). Each group included three parallel replicates, and the experiments were independently repeated at least three times. The mean number of migrated cells was used for statistical analysis.
2.8. RT-qPCR
Total RNA was extracted from tissues and cells using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. RNA concentration and purity were assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), with an A260/A280 ratio between 1.8 and 2.1 considered acceptable. RNA integrity was confirmed by 1% agarose gel electrophoresis through visualization of intact 28S and 18S rRNA bands and evaluation of the 28S:18S ratio.
Complementary DNA (cDNA) was synthesized using the PrimeScript RT Master Mix Perfect Real Time kit (Takara, Kyoto, Japan). Each 10 μL reaction contained 500 ng of total RNA. The reaction conditions included denaturation at 70 °C for 5 min to eliminate secondary structures, cooling on ice for 3 min, reverse transcription at 37 °C for 60 min, and enzyme inactivation at 95 °C for 10 min. The resulting cDNA was stored at −20 °C for further analysis.
Quantitative real-time PCR was performed using SYBR Green chemistry. Each 20 μL reaction mixture consisted of 10 μL of 2× Taq MasterMix, 0.4 μL each of forward and reverse primers (5 μM), 1 μL of cDNA template, 0.4 μL of ROX Reference Dye, and nuclease-free water. Primer sequences were synthesized by Shanghai GenePharma Co., Ltd., Shanghai, China (
Table 2).
The qPCR protocol was as follows: initial denaturation at 95 °C for 5 min, followed by 35 cycles of 94 °C for 45 s, 56 °C for 45 s, and 72 °C for 45 s, with a final extension at 72 °C for 5 min. Amplification specificity was verified using melting curve analysis. GAPDH was used as the internal control for LINC02381 and THBS1, and U6 for let-7g-5p. Each sample was analyzed in triplicate. Relative gene expression was calculated using the 2^ΔΔCt method. A no-template control was included in each run to monitor for contamination.
2.9. Western Blot Assay
Tissues and cells were lysed using RIPA lysis buffer (Cat# P0013C, Beyotime Biotechnology, Jiangsu, China), and total protein was collected by centrifugation at 12,000× g for 15 min at 4 °C. Protein concentrations were determined with a BCA protein assay kit (Beyotime, Jiangsu, China). For each sample, 30 μg of protein in 20 μL loading buffer was separated on 15% SDS-PAGE gels under the following conditions: pre-electrophoresis at 80 V for 40 min, followed by separation at 120 V for 70 min.
Proteins were transferred onto PVDF membranes (Millipore, Burlington, MA, USA) using a Bio-Rad (Hercules, CA, USA) electrotransfer system at 100 V for 90 min. The membranes were blocked in 5% non-fat milk (prepared in TBST buffer) at room temperature for 1 h, then incubated overnight at 4 °C with the following primary antibodies: anti-THBS1 (ab267388, Abcam, Cambridge, UK; 1:1000) and anti-GAPDH (sc-32233, Santa Cruz Biotechnology, Dallas, TX, USA; 1:1000). After three 10 min washes with TBST, membranes were incubated for 1 h at room temperature with HRP-conjugated secondary antibodies (Goat anti-Rabbit IgG-HRP, Bio-Rad, Hercules, CA, USA; 1:5000).
Protein bands were visualized using an enhanced chemiluminescence (ECL) kit (Millipore, Burlington, MA, USA) and exposed to X-ray film (Kodak, Rochester, NY, USA). Band intensities were analyzed using ImageJ software (version 1.53k, NIH, Bethesda, MD, USA), and target protein levels were normalized to GAPDH.
2.10. Dual Luciferase Reporter Assay
The binding sites between
LINC02381 and
let-7g-5p were predicted using the Starbase database (
https://starbase.sysu.edu.cn/agoClipRNA.php). The wild-type (Wt) sequence of
LINC02381 containing the predicted binding site for
let-7g-5p was synthesized and cloned into the psiCHECK2 luciferase reporter vector (Promega, WI, USA), generating the LINC02381-Wt construct. A mutant version (LINC02381-Mut) was created using the QuikChange XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). U-2OS and SaOS-2 cells at 70–80% confluence were co-transfected with either
let-7g-5p mimic or mimic negative control (mimic-NC) along with LINC02381-Wt or LINC02381-Mut using Lipofectamine
TM 2000 (Invitrogen, Carlsbad, CA, USA). After 48 h, luciferase activity was measured using a Dual-Luciferase
® Reporter Assay System (Promega, Madison, WI, USA).
The same method was employed to verify the targeting relationship between let-7g-5p and the 3′UTR of THBS1.
2.11. RNA-Pull Down Assay
To validate the direct interaction between let-7g-5p and LINC02381, biotin-labeled wild-type (Wt) and mutant (Mut) probes of let-7g-5p were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China) U-2OS and SaOS-2 cells (2 × 105 cells/well in 6-well plates) were transfected with these probes using Lipofectamine™ 2000. After 48 h of incubation, cells were lysed with RNA pull-down lysis buffer (Ambion, Austin, TX, USA) on ice for 10 min. Lysates were centrifuged, and supernatants were incubated with streptavidin-coated magnetic beads (M-280 Dynabeads, Sigma-Aldrich, Saint Louis, MO, USA) pre-blocked with 0.1 mg/mL yeast tRNA and 0.1 mg/mL RNase-free BSA for 30 min. Bead-lysate mixtures were rotated at 4 °C for 3 h.
The beads were then washed sequentially: twice with cold lysis buffer, three times with low-salt buffer (50 mM NaCl), and once with high-salt buffer (500 mM NaCl) to eliminate nonspecific binding. The unlabeled Mut probe served as the negative control. RNA bound to the beads was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA), reverse transcribed (PrimeScript RT Master Mix, Takara, Kyoto, Japan), and LINC02381 enrichment was quantified via qPCR. Each group was tested in triplicate, and experiments were independently repeated three times.
2.12. Tumor Xenografts in Nude Mice
Male BALB/c nude mice (18–20 g and 4–5 weeks old) were maintained under specific pathogen-free conditions. U-2OS and SaOS-2 cells stably transfected with sh-NC, sh-LINC02381, mimic-NC, or let-7g-5p mimic were subcutaneously injected into the flanks (2 × 106 cells per mouse, with 7 nude mice in each group). Tumor volumes were measured every 7 days and calculated using the formula: (length × width2)/2. On day 28, mice were euthanized, tumors were harvested, excised, and weighed. Power analysis using G*Power v3.1 software (α = 0.05, effect size f = 0.8) confirmed that 7 mice per group would achieve >85% power for detecting differences in tumor volume and weight via one-way analysis of variance (ANOVA).
2.13. Statistical Analysis
Data analysis was carried out using SPSS v21.0 (IBM, Armonk, NY, USA). Measurement data following a normal distribution were presented as mean ± standard deviation. Differences between two groups were analyzed using the independent samples t-test, while multiple-group comparisons were performed via one-way ANOVA with Tukey’s post-hoc test. Fisher’s exact test was used to assess the relationships between LINC02381/let-7g-5p expression and clinicopathological characteristics. Quality control measures included assessment of data normality using the Shapiro–Wilk test, homogeneity of variance via the Levene test, and outlier detection using the Grubbs test (α = 0.05). Abnormal results due to technical error were excluded and experiments repeated. Animal grouping was randomized, and both tumor measurements and endpoint evaluations were conducted in a blinded manner. Meanwhile, in vitro experiments such as cell proliferation, migration, and dual-luciferase assays were also implemented and interpreted independently to minimize bias. A p-value below 0.05 was considered statistically significant.
3. Results
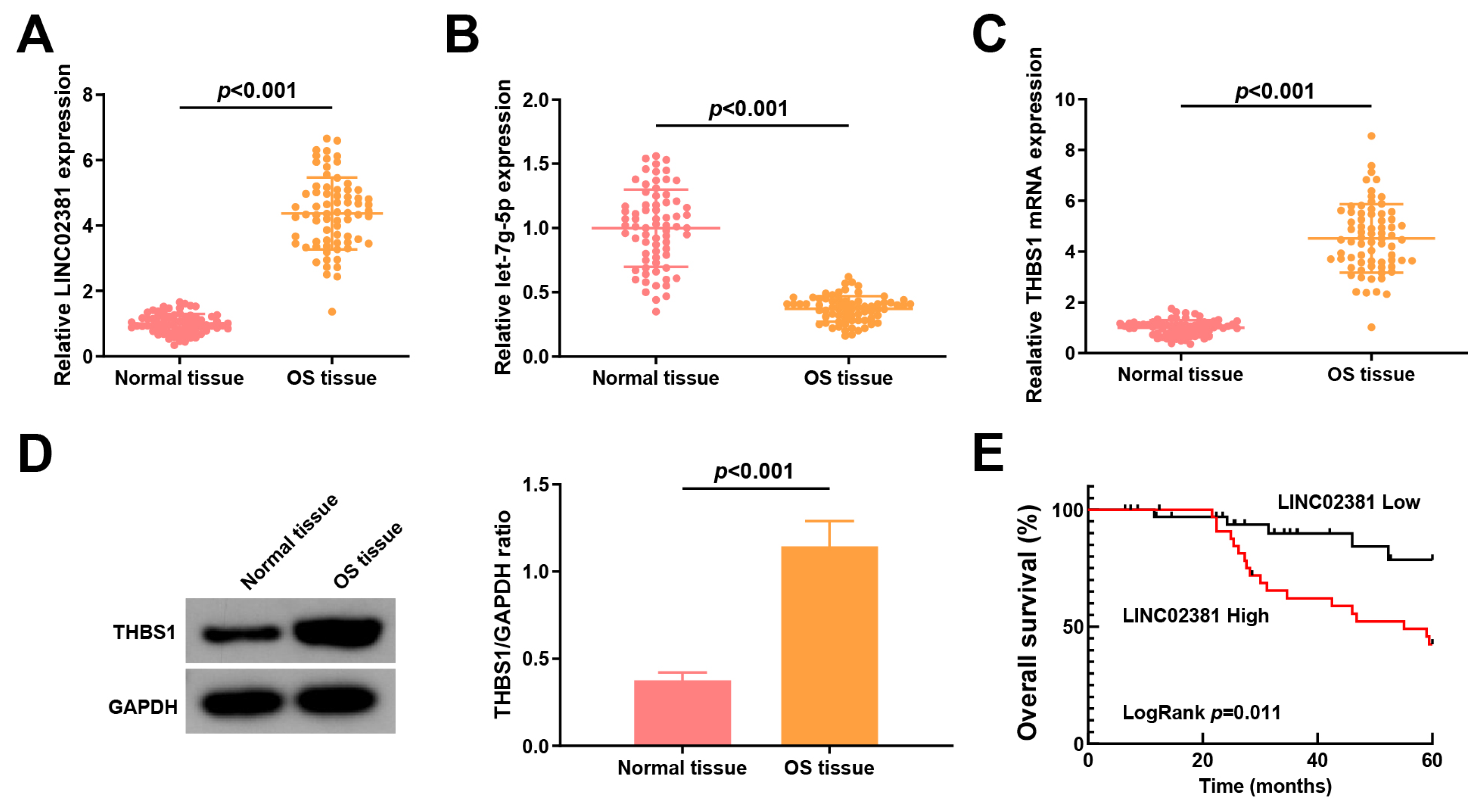
LINC02381 and let-7g-5p expression are linked to lymph node metastasis (LNM) and tumor node metastasis (TNM) staging of patients with OS
Initially,
LINC02381,
let-7g-5p, and
THBS1 expression levels in adjacent normal tissues and OS tissues were measured.
LINC02381 was upregulated in OS tissues (fold change ≈ 3.77,
p < 0.001,
Figure 1A), while
let-7g-5p was downregulated (fold change ≈ 0.38,
p ≤ 0.001,
Figure 1B), suggesting a potential regulatory interaction.
Furthermore,
THBS1, a predicted target of
let-7g-5p, was significantly upregulated at both the mRNA (fold change ≈ 2.77,
p < 0.001,
Figure 1C) and protein level (fold change ≈ 3.03,
p < 0.001,
Figure 1D) in OS tissues. Kaplan–Meier survival analysis revealed that high
LINC02381 expression was associated with shorter overall survival (Log-rank
p = 0.0448,
Figure 1E).
Patients with OS were stratified into low- and high-expression groups based on the mean levels of
LINC02381 or
let-7g-5p. High
LINC02381 or low
let-7g-5p expression was significantly associated with advanced TNM stage (III–IV) and the presence of LNM (all
p < 0.05), whereas no significant correlation was observed with gender, tumor size, or age (all
p > 0.05) (
Table 1).
To assess the relationship between molecular expression and therapeutic response, follow-up analysis was performed in 71 OS patients who underwent postoperative adjuvant chemotherapy. Based on RECIST criteria, patients were divided into chemotherapy-sensitive (CR/PR) and chemotherapy-insensitive (SD/PD) groups. Notably, the proportion of chemotherapy-insensitive patients was higher in the high
LINC02381 expression group (
p = 0.008), while elevated
let-7g-5p expression was associated with improved chemotherapy response (
Table 3).
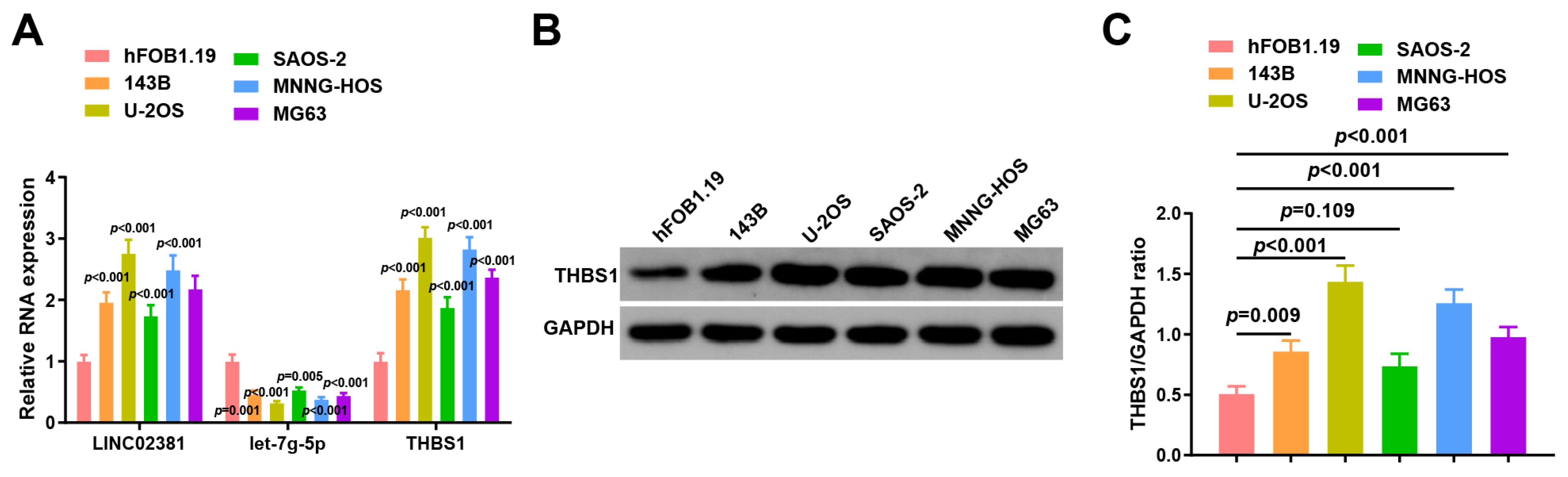
3.1. LINC02381 and THBS1 Are Upregulated and let-7g-5p Is Downregulated in OS Cell Lines
To further validate these findings, we assessed the expression of
LINC02381,
let-7g-5p, and
THBS1 in human normal osteoblasts (hFOB1.19) and five OS cell lines (143B, U-2OS, SaOS-2, MNNG-HOS, and MG63) using RT-qPCR and Western blot. Compared to hFOB1.19,
LINC02381 was significantly upregulated in all OS cell lines (
p < 0.001,
p < 0.001,
p < 0.001,
p = 0.001,
p < 0.001), while
let-7g-5p was consistently downregulated (
p < 0.001,
p < 0.001,
p = 0.005,
p < 0.001,
p < 0.001). Correspondingly,
THBS1 expression was increased across all OS cell lines (
p < 0.001,
p < 0.001,
p < 0.001,
p < 0.001,
p < 0.001) (
Figure 2A).
At the protein level, the expression of
THBS1 was markedly higher in U-2OS cells (
p = 0.009), 143B, SAOS-2, and MNNG-HOS cells (
p < 0.001) (
Figure 2B,C).
Based on these results, U-2OS and SAOS-2 cells with a high expression of LINC02381 and THBS1 and a low expression of let-7g-5p were finally selected for subsequent functional experiments.
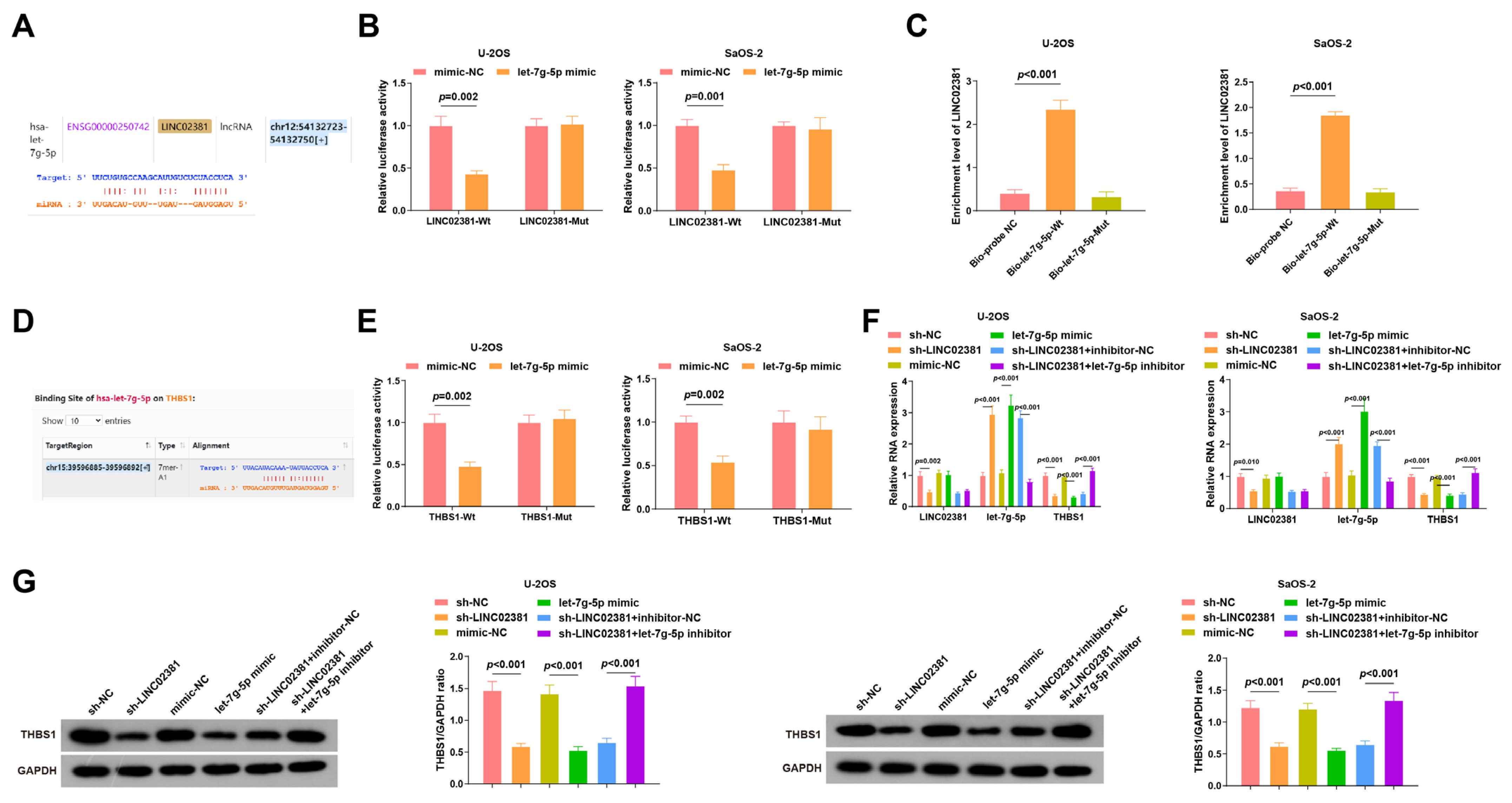
3.2. LINC02381 Regulates THBS1 Expression by Targeting let-7g-5p
Previous studies have demonstrated that
LINC02381 regulates the process of cancer cells by interacting with miRNAs. For example, it promotes the viability and migration of cervical cancer cells by targeting
miR-133b [
1]. Based on this evidence, we speculated that
LINC02381 may exert regulatory effects in OS by regulating
let-7g-5p.
Bioinformatic analysis predicted potential binding sites between
LINC02381 and
let-7g-5p (
Figure 3A). Dual-luciferase reporter assays in U-2OS and SaOS-2 cells confirmed this interaction: co-transfection with LINC02381-Wt and let-7g-5p mimic significantly reduced luciferase activity compared to the mimic-NC group (U-2OS:
p = 0.002; SaOS-2:
p < 0.001), whereas no significant change was observed in the LINC02381-Mut group (
p > 0.05) (
Figure 3B). These findings indicate specific binding between
let-7g-5p and
LINC02381. The interaction was further validated via RNA pull-down assays, which showed significant enrichment of
LINC02381 by the biotin-labeled
let-7g-5p probe (U-2OS:
p < 0.001; SaOS-2:
p < 0.001) (
Figure 3C).
Target prediction tools identified
THBS1 as a putative downstream target of
let-7g-5p (
Figure 3D). This relationship was confirmed by dual-luciferase assays: co-transfection of THBS1-Wt with the let-7g-5p mimic led to reduced luciferase activity (U-2OS:
p = 0.002; SaOS-2:
p = 0.002), whereas THBS1-Mut showed no significant change (
p > 0.05), indicating that
THBS1 is a direct target of
let-7g-5p (
Figure 3E).
Subsequent RT-qPCR and Western blot analyses demonstrated that either knockdown of
LINC02381 or overexpression of
let-7g-5p significantly downregulated THBS1 expression (all
p < 0.01,
Figure 3F,G). In addition, sh-
LINC02381 elevated
let-7g-5p levels, an effect that was reversed by co-transfection with the
let-7g-5p inhibitor (
p < 0.01), further proving that
LINC02381 can promote the expression of
THBS1 by acting as a molecular sponge for
let-7g-5p.
3.3. LINC02381 Modulates Cell Proliferation and Migration via let-7g-5p
To investigate the functional consequences of the
LINC02381/let-7g-5p axis in OS, we performed CCK-8 and colony formation assays. In both U-2OS and SaOS-2 cells, knockdown of
LINC02381 or overexpression of
let-7g-5p significantly inhibited cell proliferation (U-2OS:
p < 0.001; SaOS-2:
p < 0.001) and reduced colony formation (U-2OS:
p < 0.001; SaOS-2:
p = 0.001). Notably, co-treatment with the
let-7g-5p inhibitor reversed the suppressive effects of sh-LINC02381 on proliferation and colony formation (U-2OS:
p < 0.001; SaOS-2:
p = 0.001) (
Figure 4A–C).
Cell migration capacity, assessed using Transwell migration assays, revealed similar trends. Silencing
LINC02381 or overexpressing
let-7g-5p markedly inhibited migration in both cell lines (U-2OS:
p = 0.002 and
p < 0.001; SaOS-2:
p < 0.001 and
p = 0.001), while co-transfection with the
let-7g-5p inhibitor rescued this migratory defect (U-2OS:
p < 0.001; SaOS-2:
p = 0.005) (
Figure 4D).
Collectively, these findings suggest that LINC02381 regulates THBS1 by targeting let-7g-5p, thereby affecting the proliferation and migration ability of OS cells.
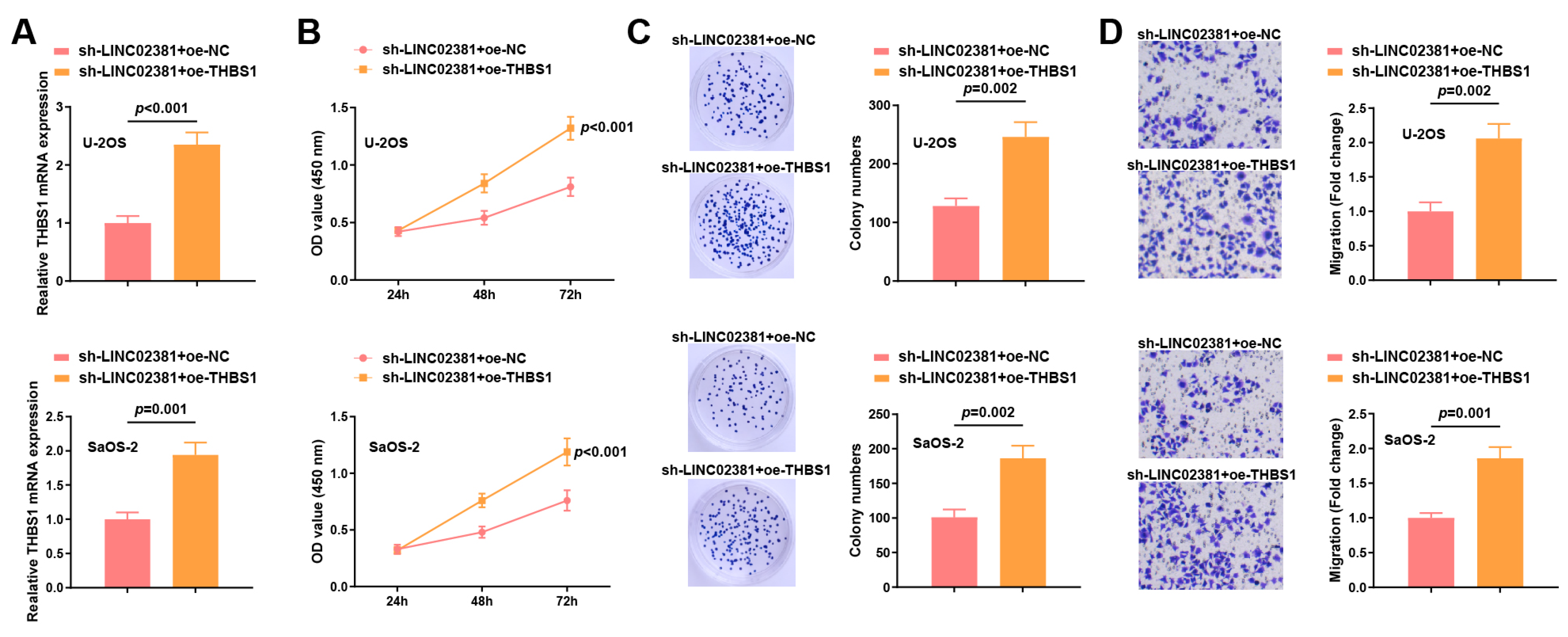
3.4. THBS1 Reverses the Effects of Both LINC02381 and let-7g-5p in OS Cells
To clarify whether THBS1 can mediate the regulatory effects of the LINC02381/let-7g-5p axis on the biological functions of OS cells, rescue experiments were conducted in U-2OS and SaOS-2 cells by co-transfecting sh-LINC02381 with either oe-NC or oe-THBS1.
qPCR results showed that compared with the sh-
LINC02381 + oe-NC group, the expression of
THBS1 in the sh-
LINC02381 + oe-
THBS1 group was increased (U-2OS:
p < 0.001; SaOS-2:
p = 0.001) (
Figure 5A).
Functional assays showed that
THBS1 overexpression significantly reversed the inhibitory effects of
LINC02381 silencing. CCK-8 and colony formation assays revealed restored proliferative capacity (U-2OS:
p < 0.001; SaOS-2:
p < 0.001) (U-2OS:
p = 0.002; SaOS-2:
p = 0.002) (
Figure 5B,C). Transwell migration assays further demonstrated that
THBS1 overexpression significantly rescued the migration ability suppressed by
LINC02381 knockdown (U-2OS:
p = 0.002; SaOS-2:
p = 0.001) (
Figure 5D).
These findings indicate that THBS1, as a downstream effector, plays a crucial role in mediating the pro-tumorigenic functions of the LINC02381/let-7g-5p axis in OS cells.
3.5. Xenograft Tumor Experiment in Nude Mice
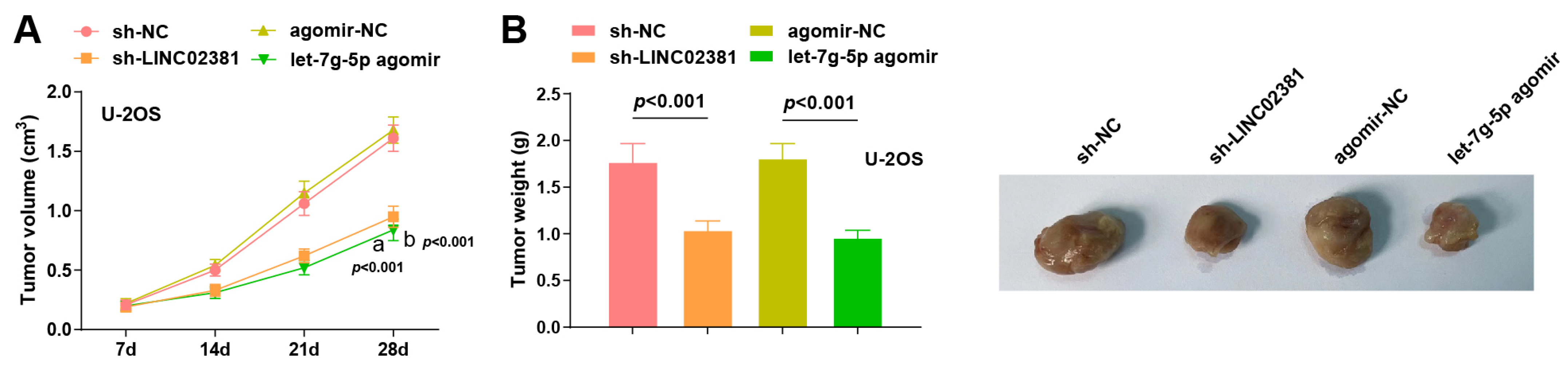
To further validate the role of LINC02381 and let-7g-5p in vivo, U-2OS cells from the sh-NC, sh-LINC02381, agomir-NC, and let-7g-5p agomir groups (2 × 106 cells/mouse) were subcutaneously injected into the flanks of 4–5-week-old male nude mice. Tumor dimensions were measured every 7 days to calculate volume and plot growth curves.
Tumors began to visibly grow by day 7 post-inoculation in all groups. From day 14 onward, tumor volumes in both the sh-
LINC02381 group and the
let-7g-5p agomir group were notably smaller than their respective controls. On day 28, tumor volume was significantly reduced in the sh-
LINC02381 group compared to the sh-NC group (
p < 0.001), and similarly in the
let-7g-5p agomir group versus the agomir-NC group (
p < 0.001) (
Figure 6A).
Following euthanasia on day 28, tumor weights were recorded. Both
LINC02381 knockdown and
let-7g-5p overexpression significantly reduced tumor weight compared to their controls (
p < 0.001) (
Figure 6B).
Collectively, these in vivo findings confirm that both LINC02381 silencing and let-7g-5p upregulation effectively inhibit OS tumor growth, reinforcing their critical roles in OS progression and potential as therapeutic target.
4. Discussion
OS is a prevalent primary malignant bone tumor, predominantly affecting adolescents [
20]. In this study, we demonstrated that
LINC02381 is significantly involved in the pathogenesis of OS through modulation of the
let-7g-5p/
THBS1 axis. Mechanistically,
LINC02381 acts as a molecular sponge for
let-7g-5p that sequesters
THBS1 thereby relieving its suppressive effect on
THBS1, and consequently enhancing OS cell proliferation, migration, and invasion in vitro and in vivo.
Our study revealed upregulated
LINC02381 and
THBS1 expression, alongside downregulated
let-7g-5p levels in OS tissues compared to normal controls. Furthermore,
LINC02381 and
let-7g-5p expression levels correlated with TNM staging and LNM in OS patients, indicating their clinical relevance in OS progression. These findings are consistent with prior reports showing that
LINC02381 is aberrantly expressed in various malignancies [
2,
3], with its overexpression linked to advanced disease stages and poor outcomes [
4]. Additionally,
let-7g-5p downregulation has been observed in multiple cancers, including cholangiocarcinoma and glioblastoma [
6,
9], and its re-expression can inhibit cancer cell proliferation, migration, and invasion [
10]. The overexpression of
THBS1 in OS tissues, as previously reported [
17], further reinforces its oncogenic role.
Through dual-luciferase reporter and RNA pull-down assays, we validated the direct interaction between
let-7g-5p and the 3′UTRs of both
LINC02381 and
THBS1, confirming a ceRNA regulatory mechanism. Functional assays demonstrated that
INC02381 could regulate
THBS1 expression by targeting
let-7g-5p, thereby influencing OS cell proliferation and migration. Importantly, overexpression of
THBS1 mitigated the inhibitory effects of
LINC02381 knockdown, highlighting
THBS1 as a critical downstream effector of this axis. These findings align with existing evidence that lncRNAs can function as ceRNAs in tumorigenesis. For instance,
LINC02381 has been shown to promote cervical cancer progression through sponging
miR-133b [
1], and to upregulate
CTNNB1 in endometriosis via
miR-27b-3p [
21]. In OS specifically,
LINC02381 was reported to promote malignancy by downregulating
CDCA4 via sponging
miR-503-5p [
4]. Together, these data suggest that
LINC02381 may operate through multiple ceRNA pathways, possibly exhibiting functional redundancy across different oncogenic contexts.
Notably,
THBS1 is a multifunctional matricellular protein that relies on downstream signaling pathways for its tumor-promoting activities. The literature indicates that
THBS1 regulates cell migration by binding to integrin receptors, as well as interacting with
CD47 and
CD36, which modulate integrin-mediated signaling [
22].
THBS1 is also known to activate latent TGF-β1, contributing to bone metastasis in prostate cancer by facilitating TGF-β signaling [
23]. These mechanisms are highly relevant to OS, particularly in the context of lung metastasis, as integrin β3 has been implicated in metastatic OS cell lines, and its silencing significantly impairs invasion and metastasis [
24]. Moreover,
TGF-β pathway overactivation has been associated with chemoresistance in OS [
25]. Based on these insights, future research should incorporate phosphoproteomic profiling to assess downstream signaling alterations following
THBS1 overexpression and apply pharmacological inhibitors of integrin or
TGF-β receptors to further dissect
THBS1-mediated pathways.