An Update on Single-Cell RNA Sequencing in Illuminating Disease Mechanisms of Cutaneous T-Cell Lymphoma
Simple Summary
Abstract
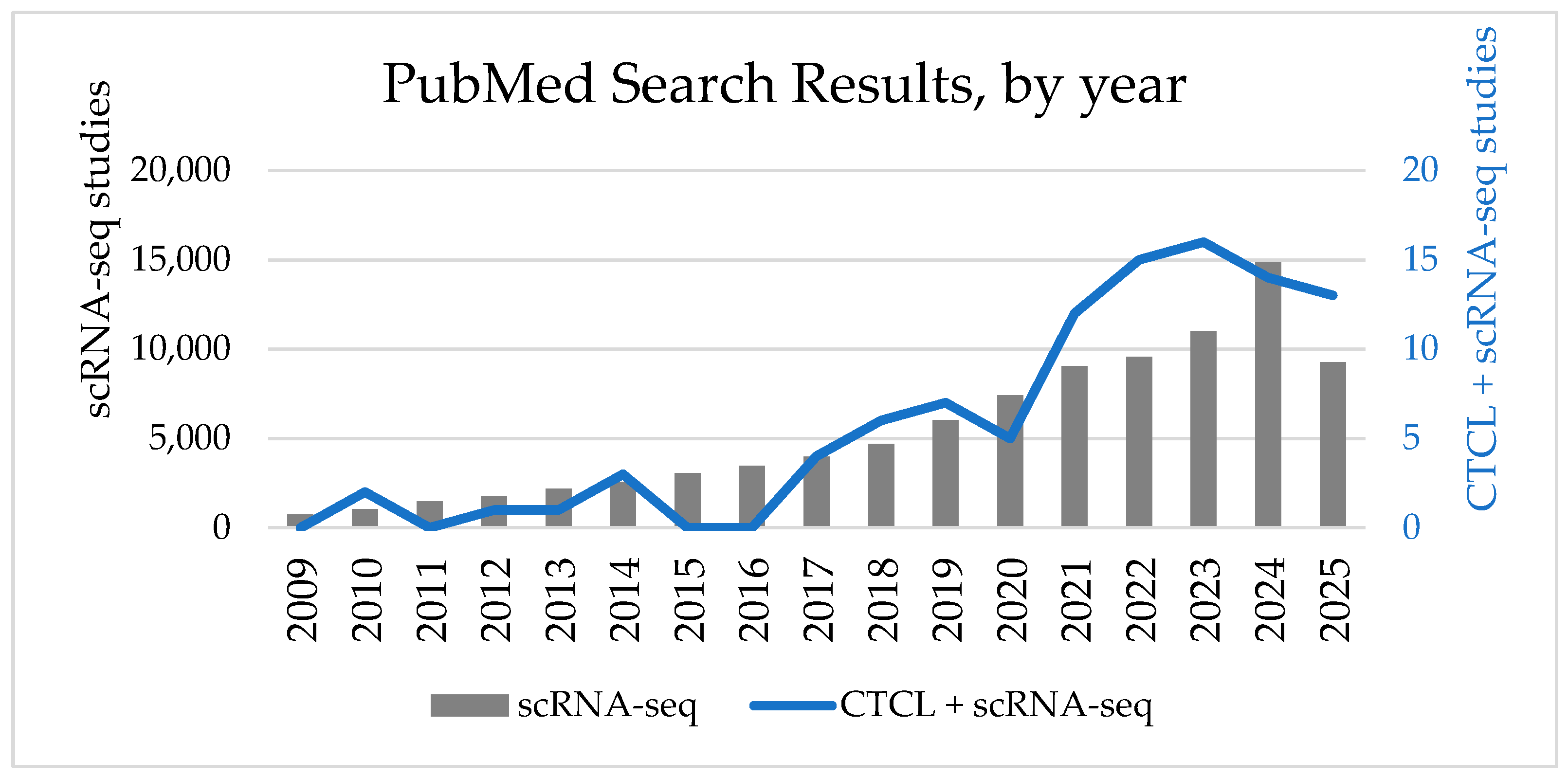
1. Introduction
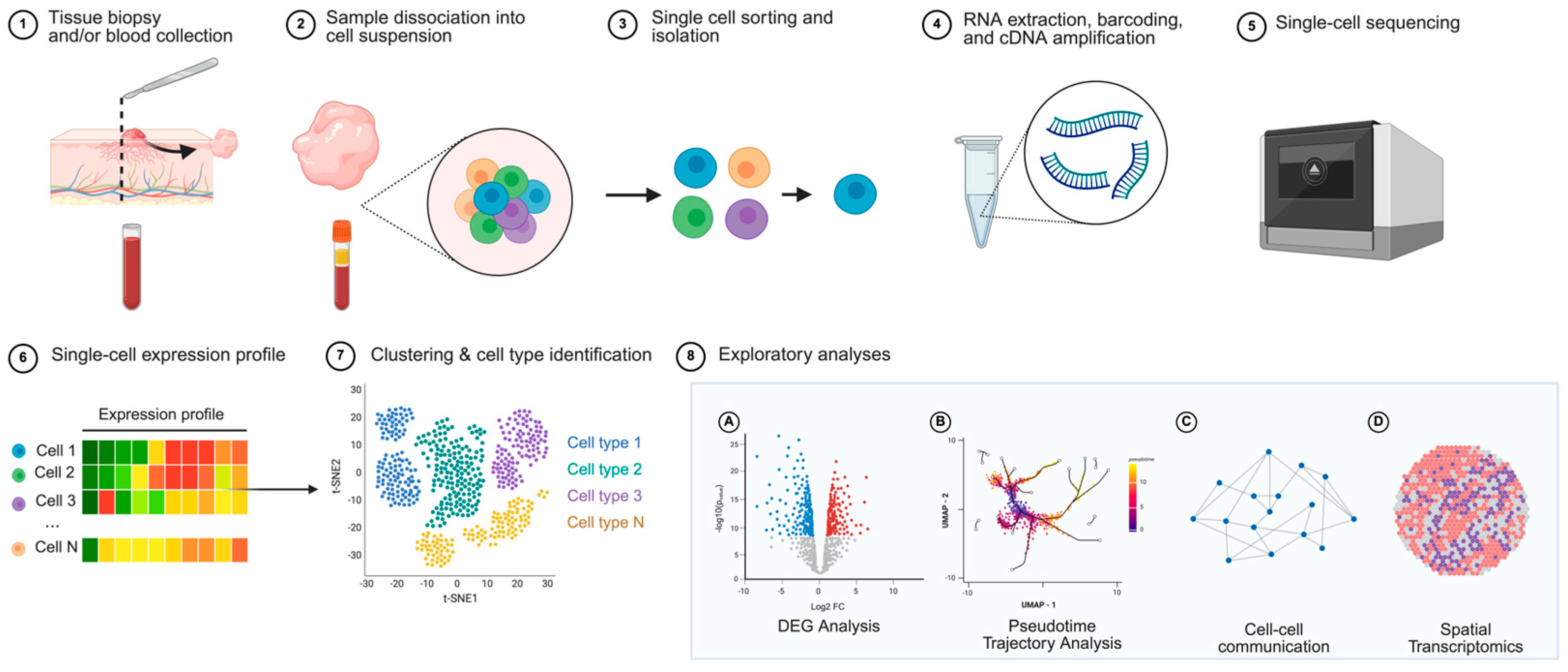
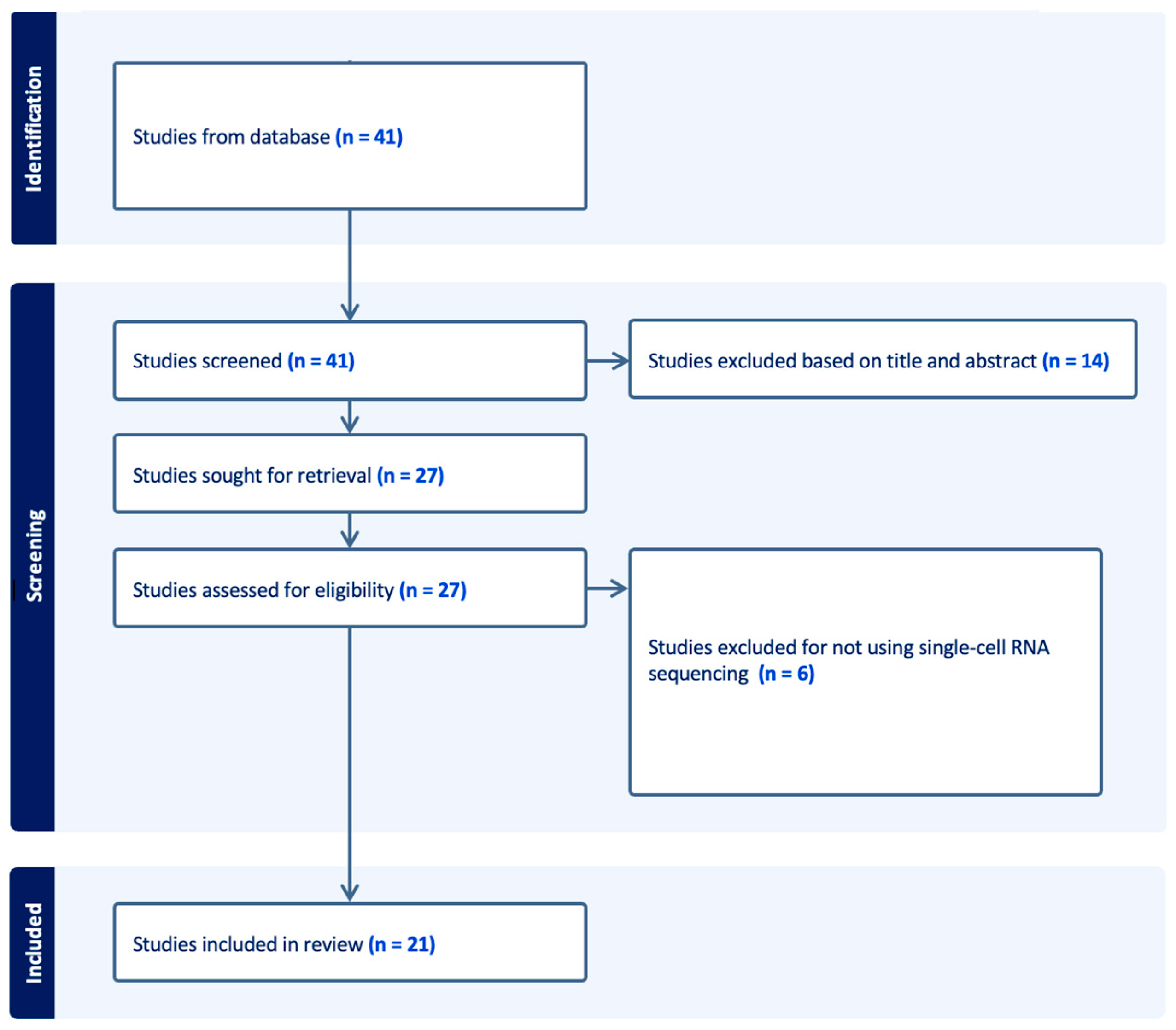
2. Methods
3. Findings from Single-Cell RNA Sequencing Studies in CTCL
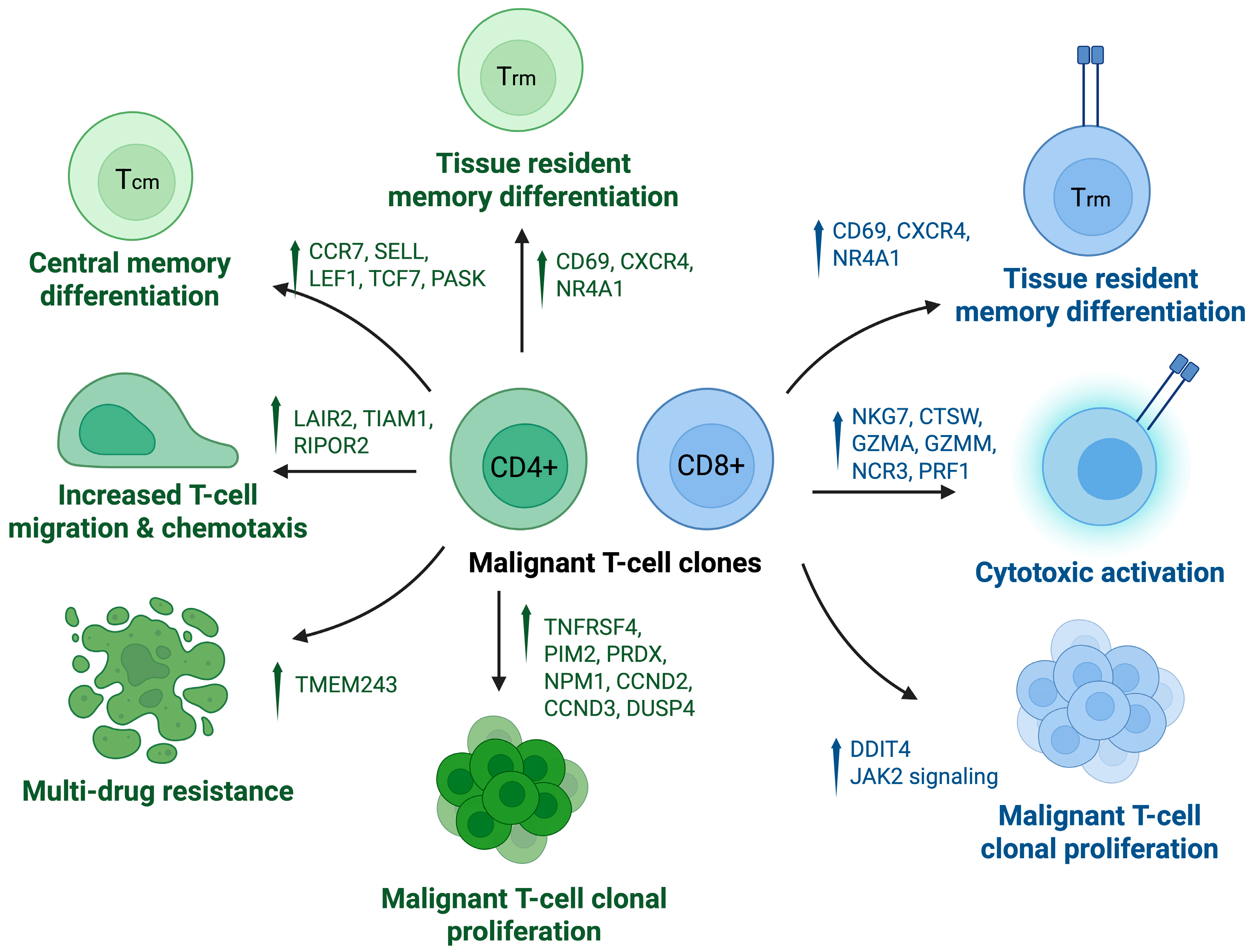
3.1. CTCL Pathogenesis
3.2. Characterizing the Tumor Microenvironment
3.2.1. T Lymphocytes
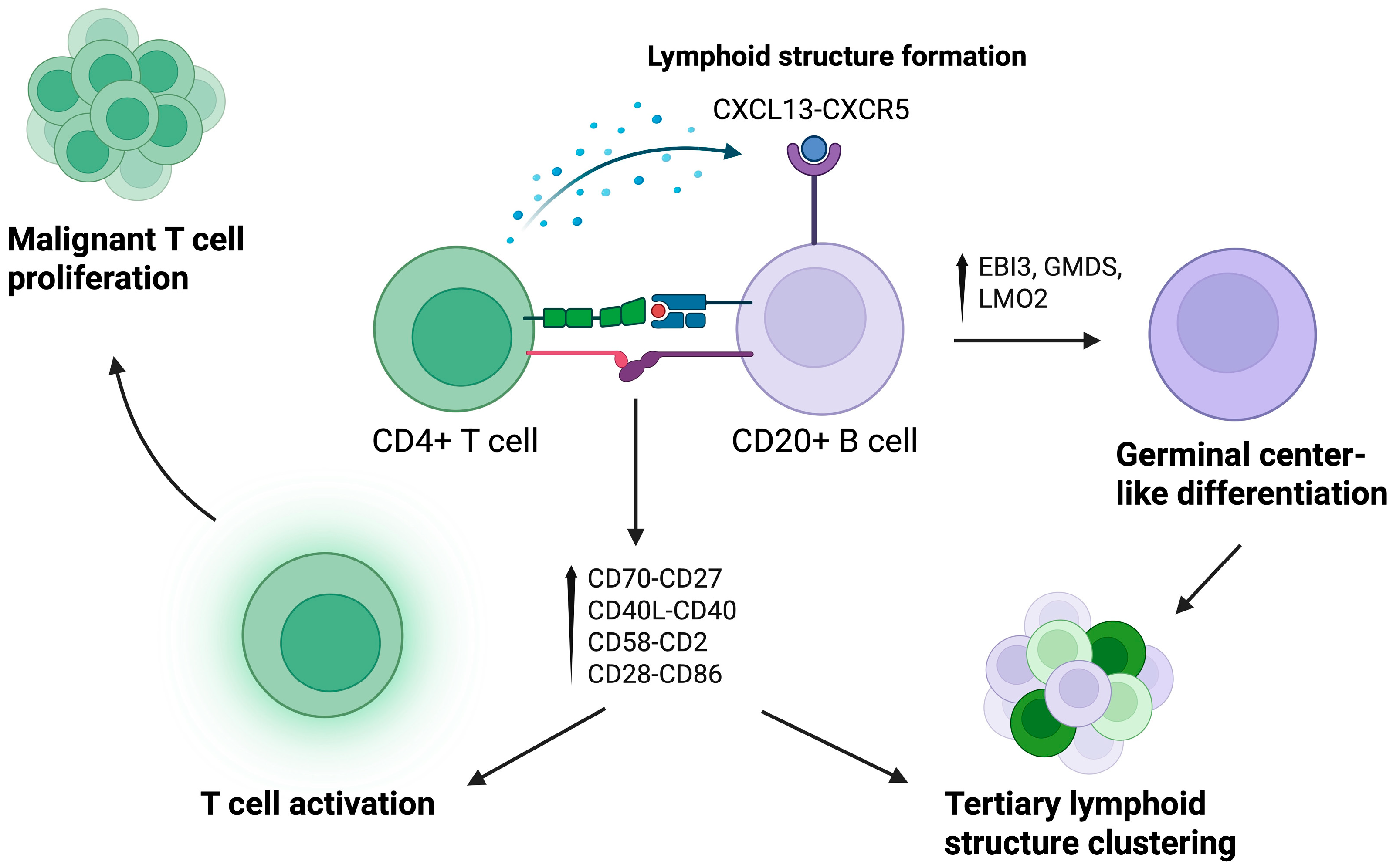
3.2.2. B Lymphocytes
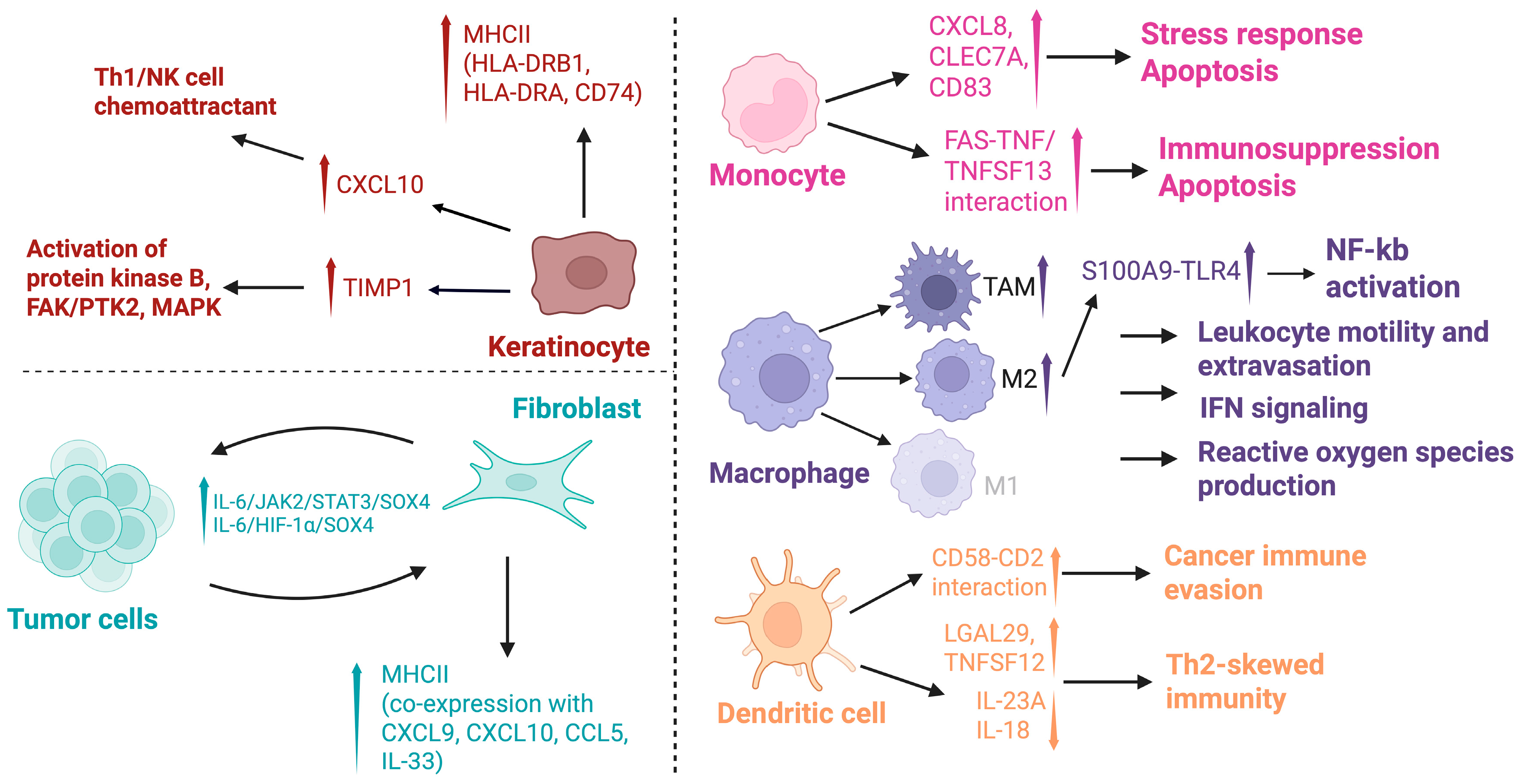
3.2.3. Keratinocytes
3.2.4. Fibroblasts
3.2.5. Myeloid Cells
3.3. Drug Interactions
3.4. Additional Works
3.5. Challenges and Future Directions
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUC | Area Under the Curve |
| AI | Artificial Intelligence |
| AD | Atopic Dermatitis |
| CD | Cluster of Differentiation (e.g., CD4, CD8, CD20) |
| cDC | Conventional Dendritic Cell |
| pDC | Plasmacytoid Dendritic Cell |
| CTCL | Cutaneous T-cell Lymphoma |
| ECCITE-seq | Expanded CRISPR-compatible Cellular Indexing of Transcriptomes and Epitopes by sequencing |
| FAK/PTK2 | Focal Adhesion Kinase/Protein Tyrosine Kinase 2 |
| HUWE1 | HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1 |
| HIF-1α | Hypoxia-inducible factor 1-alpha |
| IFN-γ | Interferon gamma |
| JAK2 | Janus Kinase 2 |
| KLHL42 | Kelch-like family member 42 |
| KCs | Keratinocytes |
| MHC II | Major histocompatibility complex class II |
| MAPKs | Mitogen-Activated Protein Kinases |
| MF | Mycosis Fungoides |
| NGS | Next-generation sequencing |
| NF-κB | Nuclear Factor kappa-light-chain-enhancer of activated B cells |
| (pcAECyTCL) | Primary Cutaneous CD8+ Aggressive Epidermotropic Cytotoxic T-cell Lymphoma |
| PBMCs | Peripheral blood mononuclear cells |
| PRKCQ | Protein Kinase C theta (gene) |
| PKCθ | Protein Kinase C theta (protein) |
| RARα | Retinoic Acid Receptor alpha |
| SS | Sézary Syndrome |
| scRNA-seq | Single-cell RNA sequencing |
| SPTCL | Subcutaneous Panniculitis-like T-cell Lymphoma |
| Th1/Th2/Th17 | T helper cell types 1, 2, and 17 |
| TCR | T-cell receptor |
| TME | Tumor microenvironment |
| TLS | Tertiary Lymphoid Structures |
| TNF-α | Tumor Necrosis Factor alpha |
Appendix A. Search Strategy
| Study | CTCL Types | # CTCL Patients in Single-Cell Analysis | Key Analysis Types | Key Findings |
|---|---|---|---|---|
| Cabrera-Perez et al., 2025 [40] | Advanced CTCL | 5 | scRNA-seq Bulk RNA-seq |
|
| Costanza et al., 2025 [52] | SS, MF, LyP, pcALCL | 10 | scRNA-seq TCR-seq, PCR, IHC, IF, FC, CODEX, DNA methylation profiling |
|
| Chennareddy et al., 2025 [35] | Erythrodermic MF, SS | 8 | scRNA-seq, TCR-seq, IF |
|
| Jung et al., 2025 [36] | MF (CD4+, patch stage) | 8 | scRNA-Seq, CosMx spatial molecular imager, IHC, protein–protein interaction, GSEA |
|
| Alkon et al., 2025 [24] | MF (Advanced & early stage) | 7 | scRNA-seq, TCR-seq |
|
| Chennareddy et al., 2025 [34] | CD4+ MF, TCR-γδ+ MF, pcAECyTCL | 14 | scRNAseq, TCR-seq, trajectory analysis |
|
| Zhao et al., 2024 [41] | Advanced MF | 2 | scRNA-seq, CNV, trajectory analysis, SCENIC analysis, cell–cell communication, IHC, IF |
|
| Luo et al., 2024 [37] | MF, SS | 20 | scRNA-seq, Bulk RNA-seq, proteomic screening |
|
| Li et al., 2024 [39] | MF | 45 | scRNA-seq, TCR-seq, Bulk RNA-seq, WGS, Spatial transcriptomics, IF, IHC, CNV, cell–cell interactions, druggable target prediction |
|
| Jiang et al., 2024 [27] | MF, SS | 25 | scRNA-seq, TCR-seq, custom gene panel, trajectory analysis, SMART-seq, Cell proliferation |
|
| Jiang et al., 2024 [42] | MF, SS (Relapsed/ Refractory) | 9 | scRNA-seq, TCR-seq, custom gene panel, Monocyte phagocytosis assay, FC |
|
| Shi et al., 2024 [28] | MF, FMF | 2 | scRNA-seq |
|
| Calugareanu et al., 2023 [30] | Advanced MF, eMF, SS, pcAECyTCL | 5 | scRNA-seq, cell–cell communication |
|
| Gaydosik et al., 2023 [31] | Advanced MF | 10 | scRNA-seq, pathway analysis, cell–cell communication, IHC |
|
| Harro et al., 2023 [23] | MF, SS | 7 | scRNA-seq, TCR-seq, scATAC-seq, qPCR, WES |
|
| Gao et al., 2023 [44] | Advanced MF | 1 | scRNA-seq, TCR-seq, WGS, WES, FC, ex vivo cell assay |
|
| Ren et al., 2023 [26] | MF, SS | 11 | scRNA-seq, TCR-seq, PHATE, CNV, SNV, WES, WGS |
|
| Borcherding et al., 2023 [25] | SS | 6 | scRNA-seq, TCR-seq, Bulk RNA-seq, IHC, FC |
|
| Du et al., 2022 [43] | SS, LCT | 5 | scRNA-seq, trajectory analysis, CNV, cell–cell communication |
|
| Xue et al., 2022 [32] | SS | 1 | scRNA-seq, scTCR-seq, scATAC-seq, validation with qRT-PCR, FC, IHC, in vitro cell culture |
|
| Su et al., 2022 [53] | SS | 6 | scRNA-seq, Bulk RNA-seq, TCR-seq, trajectory analysis, SNV, CNV, Flow cytometry |
|
References
- Willemze, R.; Cerroni, L.; Kempf, W.; Berti, E.; Facchetti, F.; Swerdlow, S.H.; Jaffe, E.S. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood 2019, 133, 1703–1714. [Google Scholar] [CrossRef]
- Campo, E.; Jaffe, E.S.; Cook, J.R.; Quintanilla-Martinez, L.; Swerdlow, S.H.; Anderson, K.C.; Brousset, P.; Cerroni, L.; de Leval, L.; Dirnhofer, S.; et al. The International Consensus Classification of Mature Lymphoid Neoplasms: A report from the Clinical Advisory Committee. Blood 2022, 140, 1229–1253. [Google Scholar] [CrossRef] [PubMed]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Hristov, A.C.; Tejasvi, T.; Wilcox, R.A. Mycosis Fungoides, Sézary Syndrome, and Cutaneous B-Cell Lymphomas: 2025 Update on Diagnosis, Risk-Stratification, and Management. Am. J. Hematol. 2025, 100, 1603–1628. [Google Scholar] [CrossRef]
- Cai, Z.R.; Chen, M.L.; Weinstock, M.A.; Kim, Y.H.; Novoa, R.A.; Linos, E. Incidence Trends of Primary Cutaneous T-Cell Lymphoma in the US From 2000 to 2018: A SEER Population Data Analysis. JAMA Oncol. 2022, 8, 1690–1692. [Google Scholar] [CrossRef]
- Tensen, C.P.; Quint, K.D.; Vermeer, M.H. Genetic and epigenetic insights into cutaneous T-cell lymphoma. Blood 2022, 139, 15–33. [Google Scholar] [CrossRef]
- Park, J.; Daniels, J.; Wartewig, T.; Ringbloom, K.G.; Martinez-Escala, M.E.; Choi, S.; Thomas, J.J.; Doukas, P.G.; Yang, J.; Snowden, C.; et al. Integrated genomic analyses of cutaneous T-cell lymphomas reveal the molecular bases for disease heterogeneity. Blood 2021, 138, 1225–1236. [Google Scholar] [CrossRef]
- Bakr, F.S.; Whittaker, S.J. Advances in the understanding and treatment of Cutaneous T-cell Lymphoma. Front. Oncol. 2022, 12, 1043254. [Google Scholar] [CrossRef]
- Hristov, A.C.; Tejasvi, T.; Wilcox, R.A. Cutaneous T-cell lymphomas: 2021 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2021, 96, 1313–1328. [Google Scholar] [CrossRef]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., II; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Steen, C.B.; Luca, B.A.; Esfahani, M.S.; Azizi, A.; Sworder, B.J.; Nabet, B.Y.; Kurtz, D.M.; Liu, C.L.; Khameneh, F.; Advani, R.H.; et al. The landscape of tumor cell states and ecosystems in diffuse large B cell lymphoma. Cancer Cell 2021, 39, 1422–1437.e10. [Google Scholar] [CrossRef]
- Andor, N.; Simonds, E.F.; Czerwinski, D.K.; Chen, J.; Grimes, S.M.; Wood-Bouwens, C.; Zheng, G.X.Y.; Kubit, M.A.; Greer, S.; Weiss, W.A.; et al. Single-cell RNA-Seq of follicular lymphoma reveals malignant B-cell types and coexpression of T-cell immune checkpoints. Blood 2019, 133, 1119–1129. [Google Scholar] [CrossRef]
- Aoki, T.; Chong, L.C.; Takata, K.; Milne, K.; Hav, M.; Colombo, A.; Chavez, E.A.; Nissen, M.; Wang, X.; Miyata-Takata, T.; et al. Single-Cell Transcriptome Analysis Reveals Disease-Defining T-cell Subsets in the Tumor Microenvironment of Classic Hodgkin Lymphoma. Cancer Discov. 2020, 10, 406–421. [Google Scholar] [CrossRef]
- Gaydosik, A.M.; Tabib, T.; Geskin, L.J.; Bayan, C.A.; Conway, J.F.; Lafyatis, R.; Fuschiotti, P. Single-Cell Lymphocyte Heterogeneity in Advanced Cutaneous T-cell Lymphoma Skin Tumors. Clin. Cancer Res. 2019, 25, 4443–4454. [Google Scholar] [CrossRef]
- Mimitou, E.P.; Cheng, A.; Montalbano, A.; Hao, S.; Stoeckius, M.; Legut, M.; Roush, T.; Herrera, A.; Papalexi, E.; Ouyang, Z.; et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 2019, 16, 409–412. [Google Scholar] [CrossRef]
- Rindler, K.; Jonak, C.; Alkon, N.; Thaler, F.M.; Kurz, H.; Shaw, L.E.; Stingl, G.; Weninger, W.; Halbritter, F.; Bauer, W.M.; et al. Single-cell RNA sequencing reveals markers of disease progression in primary cutaneous T-cell lymphoma. Mol. Cancer 2021, 20, 124. [Google Scholar] [CrossRef]
- Herrera, A.; Cheng, A.; Mimitou, E.P.; Seffens, A.; George, D.; Bar-Natan, M.; Heguy, A.; Ruggles, K.V.; Scher, J.U.; Hymes, K.; et al. Multimodal single-cell analysis of cutaneous T-cell lymphoma reveals distinct subclonal tissue-dependent signatures. Blood 2021, 138, 1456–1464. [Google Scholar] [CrossRef]
- Jonak, C.; Alkon, N.; Rindler, K.; Rojahn, T.B.; Shaw, L.E.; Porkert, S.; Weninger, W.; Trautinger, F.; Stingl, G.; Tschandl, P.; et al. Single-cell RNA sequencing profiling in a patient with discordant primary cutaneous B-cell and T-cell lymphoma reveals micromilieu-driven immune skewing. Br. J. Dermatol. 2021, 185, 1013–1025. [Google Scholar] [CrossRef]
- Li, Z.; Wang, H.; Dong, R.; Man, J.; Sun, L.; Qian, X.; Zhu, X.; Cao, P.; Yu, Y.; Le, J.; et al. Single-Cell RNA-seq Reveals Characteristics of Malignant Cells and Immune Microenvironment in Subcutaneous Panniculitis-Like T-Cell Lymphoma. Front. Oncol. 2021, 11, 611580. [Google Scholar] [CrossRef]
- Borcherding, N.; Voigt, A.P.; Liu, V.; Link, B.K.; Zhang, W.; Jabbari, A. Single-Cell Profiling of Cutaneous T-Cell Lymphoma Reveals Underlying Heterogeneity Associated with Disease Progression. Clin. Cancer Res. 2019, 25, 2996–3005. [Google Scholar] [CrossRef]
- Buus, T.B.; Willerslev-Olsen, A.; Fredholm, S.; Blümel, E.; Nastasi, C.; Gluud, M.; Hu, T.; Lindahl, L.M.; Iversen, L.; Fogh, H.; et al. Single-cell heterogeneity in Sézary syndrome. Blood Adv. 2018, 2, 2115–2126. [Google Scholar] [CrossRef]
- Harro, C.M.; Sprenger, K.B.; Chaurio, R.A.; Powers, J.J.; Innamarato, P.; Anadon, C.M.; Zhang, Y.; Biswas, S.; Mandal, G.; Mine, J.A.; et al. Sézary syndrome originates from heavily mutated hematopoietic progenitors. Blood Adv. 2023, 7, 5586–5602. [Google Scholar] [CrossRef]
- Alkon, N.; Chennareddy, S.; Cohenour, E.R.; Ruggiero, J.R.; Stingl, G.; Bangert, C.; Rindler, K.; Bauer, W.M.; Weninger, W.; Griss, J.; et al. Single-cell sequencing delineates T-cell clonality and pathogenesis of the parapsoriasis disease group. J. Allergy Clin. Immunol. 2025, 155, 461–478. [Google Scholar] [CrossRef]
- Borcherding, N.; Severson, K.J.; Henderson, N.; Ortolan, L.S.; Rosenthal, A.C.; Bellizzi, A.M.; Liu, V.; Link, B.K.; Mangold, A.R.; Jabbari, A. Single-cell analysis of Sézary syndrome reveals novel markers and shifting gene profiles associated with treatment. Blood Adv. 2023, 7, 321–335. [Google Scholar] [CrossRef]
- Ren, J.; Qu, R.; Rahman, N.T.; Lewis, J.M.; King, A.L.O.; Liao, X.; Mirza, F.N.; Carlson, K.R.; Huang, Y.; Gigante, S.; et al. Integrated transcriptome and trajectory analysis of cutaneous T-cell lymphoma identifies putative precancer populations. Blood Adv. 2023, 7, 445–457. [Google Scholar] [CrossRef]
- Jiang, T.T.; Cao, S.; Kruglov, O.; Virmani, A.; Geskin, L.J.; Falo, L.D.; Akilov, O.E. Deciphering Tumor Cell Evolution in Cutaneous T-Cell Lymphomas: Distinct Differentiation Trajectories in Mycosis Fungoides and Sézary Syndrome. J. Investig. Dermatol. 2024, 144, 1088–1098. [Google Scholar] [CrossRef]
- Shi, H.Z.; Tian, C.C.; Kong, Y.Q.; Sun, J.F.; Chen, H. Single-cell RNA sequencing reveals the underlying mechanism of folliculotropism in folliculotropic mycosis fungoides. Exp. Dermatol. 2024, 33, e14992. [Google Scholar] [CrossRef] [PubMed]
- Kwantwi, L.B.; Rosen, S.T.; Querfeld, C. The Tumor Microenvironment as a Therapeutic Target in Cutaneous T Cell Lymphoma. Cancers 2024, 16, 3368. [Google Scholar] [CrossRef] [PubMed]
- Calugareanu, A.; de Masson, A.; Battistella, M.; Michel, L.; Ram-Wolff, C.; Bouaziz, J.D.; Peltier, S.; Bensussan, A.; Bagot, M.; Dobos, G. Exploring the Nonlymphocytic Cutaneous Microenvironment in Advanced Cutaneous T-Cell Lymphomas using Single-Cell RNA Sequencing. J. Investig. Dermatol. 2023, 143, 2078–2082.e4. [Google Scholar] [CrossRef] [PubMed]
- Gaydosik, A.M.; Stonesifer, C.J.; Tabib, T.; Lafyatis, R.; Geskin, L.J.; Fuschiotti, P. The mycosis fungoides cutaneous microenvironment shapes dysfunctional cell trafficking, antitumor immunity, matrix interactions, and angiogenesis. JCI Insight 2023, 8, e170015. [Google Scholar] [CrossRef]
- Xue, X.; Wang, Z.; Mi, Z.; Liu, T.; Wang, C.; Shi, P.; Sun, L.; Yang, Y.; Li, W.; Wang, Z.; et al. Single-cell analyses reveal novel molecular signatures and pathogenesis in cutaneous T cell lymphoma. Cell Death Dis. 2022, 13, 970. [Google Scholar] [CrossRef]
- Hirahara, K.; Poholek, A.; Vahedi, G.; Laurence, A.; Kanno, Y.; Milner, J.D.; O’Shea, J.J. Mechanisms underlying helper T-cell plasticity: Implications for immune-mediated disease. J. Allergy Clin. Immunol. 2013, 131, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Chennareddy, S.; Rindler, K.; Ruggiero, J.R.; Alkon, N.; Cohenour, E.R.; Tran, S.; Weninger, W.; Griss, J.; Jonak, C.; Brunner, P.M. Single-cell RNA sequencing comparison of CD4+, CD8+ and T-cell receptor γδ+ cutaneous T-cell lymphomas reveals subset-specific molecular phenotypes. Br. J. Dermatol. 2025, 192, 269–282. [Google Scholar] [CrossRef]
- Chennareddy, S.; Rindler, K.; Meledathu, S.; Naidu, M.P.; Alkon, N.; Ruggiero, J.R.; Szmolyan, L.; Weninger, W.; Bauer, W.M.; Griss, J.; et al. Single-cell RNA sequencing of chronic idiopathic erythroderma defines disease-specific markers. J. Allergy Clin. Immunol. 2025, 155, 892–908. [Google Scholar] [CrossRef]
- Jung, J.M.; Won, C.H.; Chang, S.E.; Lee, M.W.; Lee, W.J. Spatially Resolved Single-Cell Transcriptome Analysis of Mycosis Fungoides Reveals Distinct Biomarkers GNLY and FYB1 Compared With Psoriasis and Chronic Spongiotic Dermatitis. Mod. Pathol. 2025, 38, 100681. [Google Scholar] [CrossRef]
- Luo, C.H.; Hu, L.H.; Liu, J.Y.; Xia, L.; Zhou, L.; Sun, R.H.; Lin, C.C.; Qiu, X.; Jiang, B.; Yang, M.Y.; et al. CDK9 recruits HUWE1 to degrade RARα and offers therapeutic opportunities for cutaneous T-cell lymphoma. Nat. Commun. 2024, 15, 10594. [Google Scholar] [CrossRef]
- Srinivas, N.; Peiffer, L.; Horny, K.; Lei, K.C.; Buus, T.B.; Kubat, L.; Luo, M.; Yin, M.; Spassova, I.; Sucker, A.; et al. Single-cell RNA and T-cell receptor sequencing unveil mycosis fungoides heterogeneity and a possible gene signature. Front. Oncol. 2024, 14, 1408614. [Google Scholar] [CrossRef]
- Li, R.; Strobl, J.; Poyner, E.F.M.; Balbaa, A.; Torabi, F.; Mazin, P.V.; Chipampe, N.J.; Stephenson, E.; Ramírez-Suástegi, C.; Shanmugiah, V.B.M.; et al. Cutaneous T cell lymphoma atlas reveals malignant T(H)2 cells supported by a B cell-rich tumor microenvironment. Nat. Immunol. 2024, 25, 2320–2330. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Perez, J.S.; Carey, V.J.; Odejide, O.O.; Singh, S.; Kupper, T.S.; Pillai, S.S.; Weiss, S.T.; Akenroye, A. Integrative epidemiology and immunotranscriptomics uncover a risk and potential mechanism for cutaneous lymphoma unmasking or progression with dupilumab therapy. J. Allergy Clin. Immunol. 2025, 155, 1584–1594. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, Y.; Wang, P.; Zhu, M.; Wang, J.; Xie, B.; Tang, C.; Ma, Y.; Wang, S.; Jin, S.; et al. The cancer-associated fibroblasts interact with malignant T cells in mycosis fungoides and promote the disease progression. Front. Immunol. 2024, 15, 1474564. [Google Scholar] [CrossRef]
- Jiang, T.T.; Kruglov, O.; Akilov, O.E. Unleashed monocytic engagement in Sézary syndrome during the combination of anti-CCR4 antibody with type I interferon. Blood Adv. 2024, 8, 2384–2397. [Google Scholar] [CrossRef]
- Du, Y.; Cai, Y.; Lv, Y.; Zhang, L.; Yang, H.; Liu, Q.; Hong, M.; Teng, Y.; Tang, W.; Ma, R.; et al. Single-cell RNA sequencing unveils the communications between malignant T and myeloid cells contributing to tumor growth and immunosuppression in cutaneous T-cell lymphoma. Cancer Lett. 2022, 551, 215972. [Google Scholar] [CrossRef]
- Gao, Y.; Hu, S.; Li, R.; Jin, S.; Liu, F.; Liu, X.; Li, Y.; Yan, Y.; Liu, W.; Gong, J.; et al. Hyperprogression of cutaneous T cell lymphoma after anti-PD-1 treatment. JCI Insight 2023, 8, e164793. [Google Scholar] [CrossRef]
- Roccuzzo, G.; Giordano, S.; Fava, P.; Pileri, A.; Guglielmo, A.; Tonella, L.; Sanlorenzo, M.; Ribero, S.; Fierro, M.T.; Quaglino, P. Immune Check Point Inhibitors in Primary Cutaneous T-Cell Lymphomas: Biologic Rationale, Clinical Results and Future Perspectives. Front. Oncol. 2021, 11, 733770. [Google Scholar] [CrossRef]
- Lesokhin, A.M.; Ansell, S.M.; Armand, P.; Scott, E.C.; Halwani, A.; Gutierrez, M.; Millenson, M.M.; Cohen, A.D.; Schuster, S.J.; Lebovic, D.; et al. Nivolumab in Patients With Relapsed or Refractory Hematologic Malignancy: Preliminary Results of a Phase Ib Study. J. Clin. Oncol. 2016, 34, 2698–2704. [Google Scholar] [CrossRef] [PubMed]
- Narducci, M.G.; Tosi, A.; Frezzolini, A.; Scala, E.; Passarelli, F.; Bonmassar, L.; Monopoli, A.; Accetturi, M.P.; Cantonetti, M.; Antonini Cappellini, G.C.; et al. Reduction of T Lymphoma Cells and Immunological Invigoration in a Patient Concurrently Affected by Melanoma and Sezary Syndrome Treated With Nivolumab. Front. Immunol. 2020, 11, 579894. [Google Scholar] [CrossRef]
- Khodadoust, M.S.; Rook, A.H.; Porcu, P.; Foss, F.; Moskowitz, A.J.; Shustov, A.; Shanbhag, S.; Sokol, L.; Fling, S.P.; Ramchurren, N.; et al. Pembrolizumab in Relapsed and Refractory Mycosis Fungoides and Sézary Syndrome: A Multicenter Phase II Study. J. Clin. Oncol. 2020, 38, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Marchi, E.; Ma, H.; Montanari, F.; Sawas, A.; Lue, J.K.; Deng, C.; Whitfield, K.T.; Klein, S.; Scotto, L.; Jain, S.S.; et al. The Integration of PD1 blockade with epigenetic therapy is highly active and safe in heavily treated patients with T-cell lymphoma (PTCL) and cutaneous T-cell lymphoma (CTCL). J. Clin. Oncol. 2020, 38, 8049. [Google Scholar] [CrossRef]
- Querfeld, C.; Palmer, J.; Han, Z.; Wu, X.; Yuan, Y.C.; Chen, M.H.; Su, C.; Tsai, N.C.; Smith, D.L.; Hammond, S.N.; et al. Phase 1 trial of durvalumab (anti-PD-L1) combined with lenalidomide in relapsed/refractory cutaneous T-cell lymphoma. Blood Adv. 2025, 9, 2247–2260. [Google Scholar] [CrossRef] [PubMed]
- Querfeld, C.; Thompson, J.A.; Taylor, M.H.; DeSimone, J.A.; Zain, J.M.; Shustov, A.R.; Johns, C.; McCann, S.; Lin, G.H.Y.; Petrova, P.S.; et al. Intralesional TTI-621, a novel biologic targeting the innate immune checkpoint CD47, in patients with relapsed or refractory mycosis fungoides or Sézary syndrome; syndrome: A multicentre, phase 1 study. Lancet Haematol. 2021, 8, e808–e817. [Google Scholar] [CrossRef]
- Costanza, M.; Giordano, C.; von Brünneck, A.-C.; Zhao, J.; Makky, A.; Vinh, K.; Montes-Mojarro, I.A.; Reisinger, F.; Forchhammer, S.; Witalisz-Siepracka, A.; et al. Preclinical in vitro and in vivo evidence for targeting CD74 as an effective treatment strategy for cutaneous T-cell lymphomas. Br. J. Dermatol. 2025, 192, 883–895. [Google Scholar] [CrossRef]
- Su, T.; Duran, G.E.; Kwang, A.C.; Ramchurren, N.; Fling, S.P.; Kim, Y.H.; Khodadoust, M.S. Single-cell RNA-sequencing reveals predictive features of response to pembrolizumab in Sézary syndrome. OncoImmunology 2022, 11, 2115197. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Billi, A.; Runge, J.; Wasikowski, R.; Li, Q.; Wang, Y.; Sartor, M.; Harms, P.W.; Gudjonsson, J.; Hristov, A.; et al. 311 Single-cell transcriptomics reveals distinct molecular programs in folliculotropic mycosis fungoides. J. Investig. Dermatol. 2023, 143, S53. [Google Scholar] [CrossRef]
- Wang, C.; Wolfe, A.; Geng, X.; Wilcox, R.A. Single Cell Atlas of Cutaneous T Cell Lymphomas Reveals XPO1 Dependency. Blood 2024, 144, 858. [Google Scholar] [CrossRef]
- Childs, B.A.; Elghonaimy, E.; Aguilera, T.; Goff, H.W. 0767 Characterizing malignant T cells in CTCL to inform personalized therapy: A scRNAseq study. J. Investig. Dermatol. 2025, 145, S133. [Google Scholar] [CrossRef]
- Childs, B.; Elghonaimy, E.; Aguilera, T.; Goff, H. 64086 Dissecting Malignant T-Cell Behavior in CTCL: A Single-Cell RNA Sequencing Comparison with Benign T Cells in the Immune Microenvironment. J. Am. Acad. Dermatol. 2025, 93, AB37. [Google Scholar] [CrossRef]
- Childs, B.; Elghonaimy, E.; Ramakrishnan Geethakumari, P.; Aguilera, T.; Goff, H.W. Single-Cell Insights into Cellular Heterogeneity and Immune Dynamics in Cutaneous T-Cell Lymphoma. Blood 2024, 144, 6268. [Google Scholar] [CrossRef]
- Johnson, C.; Solhjoo, S.; Li, W.; Ali, I.; Nash, K.; Hicks, S.; Timp, W. 099 Single-cell RNA sequencing reveals differential gene expression of cancer-associated fibroblast markers in mycosis fungoides by stage and race. J. Investig. Dermatol. 2024, 144, S18. [Google Scholar] [CrossRef]
- Rassek, K.; Iżykowska, K. Single-Cell Heterogeneity of Cutaneous T-Cell Lymphomas Revealed Using RNA-Seq Technologies. Cancers 2020, 12, 2129. [Google Scholar] [CrossRef]
- Haque, A.; Engel, J.; Teichmann, S.A.; Lönnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017, 9, 75. [Google Scholar] [CrossRef]
- Luo, Y.; De Gruijl, F.R.; Vermeer, M.H.; Tensen, C.P. “Next top” mouse models advancing CTCL research. Front. Cell Dev. Biol. 2024, 12, 1372881. [Google Scholar] [CrossRef] [PubMed]
- Marx, V. Method of the Year: Spatially resolved transcriptomics. Nat. Methods 2021, 18, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Deconinck, L.; Cannoodt, R.; Saelens, W.; Deplancke, B.; Saeys, Y. Recent advances in trajectory inference from single-cell omics data. Curr. Opin. Syst. Biol. 2021, 27, 100344. [Google Scholar] [CrossRef]
- Lähnemann, D.; Köster, J.; Szczurek, E.; McCarthy, D.J.; Hicks, S.C.; Robinson, M.D.; Vallejos, C.A.; Campbell, K.R.; Beerenwinkel, N.; Mahfouz, A.; et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020, 21, 31. [Google Scholar] [CrossRef]
- Liu, X.; Jin, S.; Hu, S.; Li, R.; Pan, H.; Liu, Y.; Lai, P.; Xu, D.; Sun, J.; Liu, Z.; et al. Single-cell transcriptomics links malignant T cells to the tumor immune landscape in cutaneous T cell lymphoma. Nat. Commun. 2022, 13, 1158. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suhl, S.; Kaminsky, A.; Chen, C.; Lapolla, B.A.; Zhou, M.H.; Kent, J.; Marx, A.; Nebo, I.D.; Ramush, G.; Luyten, S.; et al. An Update on Single-Cell RNA Sequencing in Illuminating Disease Mechanisms of Cutaneous T-Cell Lymphoma. Cancers 2025, 17, 2921. https://doi.org/10.3390/cancers17172921
Suhl S, Kaminsky A, Chen C, Lapolla BA, Zhou MH, Kent J, Marx A, Nebo ID, Ramush G, Luyten S, et al. An Update on Single-Cell RNA Sequencing in Illuminating Disease Mechanisms of Cutaneous T-Cell Lymphoma. Cancers. 2025; 17(17):2921. https://doi.org/10.3390/cancers17172921
Chicago/Turabian StyleSuhl, Sara, Alexander Kaminsky, Caroline Chen, Brigit A. Lapolla, Maggie H. Zhou, Joshua Kent, Abigail Marx, Ikenna David Nebo, Geat Ramush, Sophia Luyten, and et al. 2025. "An Update on Single-Cell RNA Sequencing in Illuminating Disease Mechanisms of Cutaneous T-Cell Lymphoma" Cancers 17, no. 17: 2921. https://doi.org/10.3390/cancers17172921
APA StyleSuhl, S., Kaminsky, A., Chen, C., Lapolla, B. A., Zhou, M. H., Kent, J., Marx, A., Nebo, I. D., Ramush, G., Luyten, S., Sacknovitz, Y., Sung, J., Bear, C. M., Schreidah, C. M., Gru, A., & Geskin, L. J. (2025). An Update on Single-Cell RNA Sequencing in Illuminating Disease Mechanisms of Cutaneous T-Cell Lymphoma. Cancers, 17(17), 2921. https://doi.org/10.3390/cancers17172921






