Melanoma Skin Cancer: A Comprehensive Review of Current Knowledge
Simple Summary
Abstract
1. Introduction
2. From Melanocytes to Melanoma: Molecular Biology and Genetics
3. Pathophysiology
4. Subtypes: Morphology and Histopathology
4.1. Superficial Spreading Melanoma
4.2. Lentigo Maligna Melanoma
4.3. Nodular Melanoma
4.4. Acral Lentiginous Melanoma
4.5. The 2022 World Health Organization Classification—Fifth Ed
5. Epidemiology
6. Diagnosis
6.1. Clinical Diagnosis
6.2. Diagnosis Confirmation
7. Staging
7.1. T Category: Primary Tumor
7.2. N Category: Nodal Involvement
7.3. M Category: Distant Metastasis
8. Prognosis
Prognostic Factors
9. Management
9.1. Imaging and Laboratory Studies
9.2. Surgery
9.3. Non-Surgical Approaches
9.4. Lymphatic Mapping and Sentinel Lymph Node Biopsy
9.5. Additional Treatments
9.5.1. Negative Sentinel Lymph Node Biopsy
9.5.2. Positive Sentinel Lymph Node Biopsy
9.5.3. Clinically Detected Regional Lymph Nodes
9.5.4. Distant Metastatic Disease
9.6. Surveillance
10. Genetic Counseling
11. Future Directions
11.1. Risk Prediction Models
11.2. Artificial Intelligence-Based Screening Algorithms
11.3. Molecular Tests with Prognostic Value
11.3.1. Gene Expression Profile
11.3.2. microRNA Expression Profile
11.3.3. Circulating Tumor DNA
11.4. Additional Biomarkers with Prognostic Value
11.5. Vaccine as Adjuvant Therapy
12. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AJCC | American Joint Committee on Cancer |
| AI | Artificial intelligence |
| AMS | Atypical mole syndrome |
| BIN | BAP1-inactivated nevus |
| CDS | Cumulative solar damage |
| CLND | Complete lymph node resection |
| CNS | Central nervous system |
| ctDNA | Circulating tumor DNA |
| cTNM | Clinical stage group |
| CT Scan | Computed tomography |
| DPN | Deep penetrating nevus |
| FAMMM | Familial atypical multiple mole and melanoma |
| FISH | Fluorescence in situ hybridization |
| GEP | Gene expression profiling |
| HAV | Herpes simplex virus |
| IAMP | Intraepidermal atypical melanocytic proliferation |
| IAMPUS | Intraepidermal atypical melanocytic proliferation of uncertain significance |
| IMP | Intraepidermal melanocytic proliferation without atypia |
| LDH | Lactate dehydrogenase |
| LM | Lentigo maligna |
| LMM | Lentigo maligna melanoma |
| MAPK | Mitogen-activated protein kinase |
| MELTUMP | Melanocytic tumor of uncertain malignant potential |
| miRNAs | MicroRNAs |
| MIS | Melanoma in situ |
| MMS | Mohs micrographic surgery |
| MRI | Resonance imaging |
| NNMT | N-methyltransferase |
| PD-1 | Programmed cell death protein 1 |
| PD-L1 | Programmed cell death-ligand 1 |
| PET-CT | Positron emission tomography |
| PEM | Pigmented epithelioid melanocytoma |
| PI3K | Phosphatidyl inositol 3-kinase |
| PON2 | Paraoxonase-2 |
| pTNM | Pathological stage group |
| RGP | Radial growth phase |
| SLN | Sentinel lymph node |
| SLNB | Sentinel lymph node biopsy |
| SSM | Superficial spreading melanoma |
| STUMP | Spitzoid tumor of uncertain malignant potential |
| T-VEC | Talimogene laherparepvec |
| UV | Ultraviolet |
| VGP | Vertical growth phase |
| WHO | World Health Organization |
References
- National Cancer Institute Surveillance, Epidemiology, and End Results Program. Cancer Stat Facts: Melanoma of the Skin. 2025. Available online: https://seer.cancer.gov/statfacts/html/melan.html (accessed on 28 May 2025).
- Guy, G.P.; Thomas, C.C.; Thompson, T.; Watson, M.; Massetti, G.M.; Richardson, L.C. Centers for Disease Control and Prevention (CDC). Vital signs: Melanoma incidence and mortality trends and projections-United States, 1982–2030. MMWR Morb. Mortal. Wkly. Rep. 2015, 64, 591–596. [Google Scholar]
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040. JAMA Dermatol. 2022, 158, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef]
- Sommer, L. Generation of melanocytes from neural crest cells. Pigment Cell Melanoma Res. 2011, 24, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Snyman, M.; Walsdorf, R.E.; Wix, S.N.; Gill, J.G. The metabolism of melanin synthesis—From melanocytes to melanoma. Pigment Cell Melanoma Res. 2024, 37, 438–452. [Google Scholar] [CrossRef]
- Bastian, B.C. The molecular pathology of melanoma: An integrated taxonomy of melanocytic neoplasia. Annu. Rev. Pathol. 2014, 9, 239–271. [Google Scholar] [CrossRef]
- Carr, J.; Mackie, R.M. Point mutations in the N-ras oncogene in malignant melanoma and congenital naevi. Br. J. Dermatol. 1994, 131, 72–77. [Google Scholar] [CrossRef]
- Pollock, P.M.; Harper, U.L.; Hansen, K.S.; Yudt, L.M.; Stark, M.; Robbins, C.M.; Moses, T.Y.; Hostetter, G.; Wagner, U.; Kakareka, J.; et al. High frequency of BRAF mutations in nevi. Nat. Genet. 2003, 33, 19–20. [Google Scholar] [CrossRef]
- Van Raamsdonk, C.D.; Bezrookove, V.; Green, G.; Bauer, J.; Gaugler, L.; O’Brien, J.M.; Simpson, E.M.; Barsh, G.S.; Bastian, B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009, 457, 599–602. [Google Scholar] [CrossRef]
- Curtin, J.A.; Busam, K.; Pinkel, D.; Bastian, B.C. Somatic activation of KIT in distinct subtypes of melanoma. J. Clin. Oncol. 2006, 24, 4340–4346. [Google Scholar] [CrossRef] [PubMed]
- Curtin, J.A.; Fridlyand, J.; Kageshita, T.; Patel, H.N.; Busam, K.J.; Kutzner, H.; Cho, K.H.; Aiba, S.; Bröcker, E.B.; LeBoit, P.E.; et al. Distinct Sets of Genetic Alterations in Melanoma. N. Engl. J. Med. 2005, 353, 2135–2147. [Google Scholar] [CrossRef]
- Long, G.V.; Swetter, S.M.; Menzies, A.M.; Gershenwald, J.E.; Scolyer, R.A. Cutaneous melanoma. Lancet 2023, 402, 485–502. [Google Scholar] [CrossRef]
- Wiesner, T.; Obenauf, A.C.; Murali, R.; Fried, I.; Griewank, K.G.; Ulz, P.; Windpassinger, C.; Wackernagel, W.; Loy, S.; Wolf, I.; et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat. Genet. 2011, 43, 1018–1021. [Google Scholar] [CrossRef]
- Wiesner, T.; Murali, R.; Fried, I.; Cerroni, L.; Busam, K.; Kutzner, H.; Bastian, B.C. A distinct subset of atypical Spitz tumors is characterized by BRAF mutation and loss of BAP1 expression. Am. J. Surg. Pathol. 2012, 36, 818–830. [Google Scholar] [CrossRef] [PubMed]
- Krauthammer, M.; Kong, Y.; Ha, B.H.; Evans, P.; Bacchiocchi, A.; McCusker, J.P.; Cheng, E.; Davis, M.J.; Goh, G.; Choi, M.; et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 2012, 44, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Soura, E.; Eliades, P.J.; Shannon, K.; Stratigos, A.J.; Tsao, H. Hereditary melanoma: Update on syndromes and management. J. Am. Acad. Dermatol. 2016, 74, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Elder, D.E.; Barnhill, R.L.; Bastian, B.C.; Cook, M.G.; de la Fouchardiere, A.; Gerami, P.; Lazar, A.J.; Massi, D.; Mihm, M.C.J.; Nagore, E. Melanocytic Tumour Classification and the Pathway Concept of Melanoma Pathogenesis. In WHO Classification of Skin Tumours; World Health Organization: Geneva, Switzerland, 2018; Volume 11, pp. 66–71. [Google Scholar]
- Motwani, J.; Eccles, M.R. Genetic and Genomic Pathways of Melanoma Development, Invasion and Metastasis. Genes 2021, 12, 1543. [Google Scholar] [CrossRef]
- Clark, W.H.; Elder, D.E.; Guerry, D.; Braitman, L.E.; Trock, B.J.; Schultz, D.; Synnestvedt, M.; Halpern, A.C. Model predicting survival in stage I melanoma based on tumor progression. J. Natl. Cancer Inst. 1989, 81, 1893–1904. [Google Scholar] [CrossRef]
- Roncati, L.; Piscioli, F.; Pusiol, T.; Maiorana, A. Microinvasive Radial Growth Phase of Cutaneous Melanoma: A Histopathological and Immunohistochemical Study with Diagnostic Implications. Acta Dermatovenerol. Croat. 2017, 25, 39–45. [Google Scholar]
- Bandarchi, B.; Ma, L.; Navab, R.; Seth, A.; Rasty, G. From melanocyte to metastatic malignant melanoma. Dermatol. Res. Pract. 2010, 2010, 583748. [Google Scholar] [CrossRef]
- Clark, W.H.; Elder, D.E.; Guerry, D.; Epstein, M.N.; Greene, M.H.; Van Horn, M. A study of tumor progression: The precursor lesions of superficial spreading and nodular melanoma. Hum. Pathol. 1984, 15, 1147–1165. [Google Scholar] [CrossRef]
- Clark, W.H.; Elder, D.E.; Van Horn, M. The biologic forms of malignant melanoma. Hum. Pathol. 1986, 17, 443–450. [Google Scholar] [CrossRef]
- Slominski, A.; Wortsman, J.; Carlson, A.J.; Matsuoka, L.Y.; Balch, C.M.; Mihm, M.C. Malignant Melanoma. Arch. Pathol. Lab. Med. 2001, 125, 1295–1306. [Google Scholar] [CrossRef]
- Clark, W.H.; From, L.; Bernardino, E.A.; Mihm, M.C. The histogenesis and biologic behavior of primary human malignant melanomas of the skin. Cancer Res. 1969, 29, 705–727. [Google Scholar]
- Breslow, A. Thickness, cross-sectional areas and depth of invasion in the prognosis of cutaneous melanoma. Ann. Surg. 1970, 172, 902–908. [Google Scholar] [CrossRef] [PubMed]
- Morton, D.L.; Davtyan, D.G.; Wanek, L.A.; Foshag, L.J.; Cochran, A.J. Multivariate analysis of the relationship between survival and the microstage of primary melanoma by clark level and breslow thickness. Cancer 1993, 71, 3737–3743. [Google Scholar] [CrossRef] [PubMed]
- Balch, C.M.; Murad, T.M.; Soong, S.J.; Ingalls, A.L.; Halpern, N.B.; Maddox, W.A. A Multifactorial Analysis of Melanoma. Ann. Surg. 1978, 188, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Edge, S.B.; Greene, F.L.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. AJCC Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2017. [Google Scholar]
- Massone, C.; Hofman-Wellenhof, R.; Chiodi, S.; Sola, S. Dermoscopic Criteria, Histopathological Correlates and Genetic Findings of Thin Melanoma on Non-Volar Skin. Genes 2021, 12, 1288. [Google Scholar] [CrossRef]
- Cohen, L.M. Lentigo maligna and lentigo maligna melanoma. J. Am. Acad. Dermatol. 1995, 33, 923–936. [Google Scholar] [CrossRef]
- Huang, K.; Fan, J.; Misra, S. Acral Lentiginous Melanoma: Incidence and Survival in the United States, 2006–2015, an Analysis of the SEER Registry. J. Surg Res. 2020, 251, 329–339. [Google Scholar] [CrossRef]
- Shaikh, W.R. The Contribution of Nodular Subtype to Melanoma Mortality in the United States, 1978 to 2007. Arch Dermatol. 2012, 148, 30. [Google Scholar] [CrossRef]
- Saida, T. Histogenesis of cutaneous malignant melanoma: The vast majority do not develop from melanocytic nevus but arise de novo as melanoma in situ. J. Dermatol. 2019, 46, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Whiteman, D.C.; Pavan, W.J.; Bastian, B.C. The melanomas: A synthesis of epidemiological, clinical, histopathological, genetic, and biological aspects, supporting distinct subtypes, causal pathways, and cells of origin. Pigment Cell Melanoma Res. 2011, 24, 879–897. [Google Scholar] [CrossRef]
- Smoller, B.R. Histologic criteria for diagnosing primary cutaneous malignant melanoma. Mod. Pathol. 2006, 19, S34–S40. [Google Scholar] [CrossRef]
- Pampena, R.; Kyrgidis, A.; Lallas, A.; Moscarella, E.; Argenziano, G.; Longo, C. A meta-analysis of nevus-associated melanoma: Prevalence and practical implications. J. Am. Acad. Dermatol. 2017, 77, 938–945.e4. [Google Scholar] [CrossRef] [PubMed]
- Elwood, J.M.; Gallagher, R.P.; Worth, A.J.; Wood, W.S.; Pearson, J.C. Etiological differences between subtypes of cutaneous malignant melanoma: Western Canada Melanoma Study. J. Natl. Cancer Inst. 1987, 78, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Clark, W.H.; Mihm, M.C. Lentigo maligna and lentigo-maligna melanoma. Am. J. Pathol. 1969, 55, 39–67. [Google Scholar]
- Prieto-Granada, C.; Howe, N.; McCardle, T. Melanoma Pathology. In Melanoma; Oxford University Press: New York, NY, USA, 2015; pp. 10–30. [Google Scholar]
- Weinstock, M.A.; Sober, A.J. The risk of progression of lentigo maligna to lentigo maligna melanoma. Br. J. Dermatol. 1987, 116, 303–310. [Google Scholar] [CrossRef]
- Durbec, F.; Martin, L.; Derancourt, C.; Grange, F. Melanoma of the hand and foot: Epidemiological, prognostic and genetic features. A systematic review. Br. J. Dermatol. 2012, 166, 727–739. [Google Scholar] [CrossRef]
- Basurto-Lozada, P.; Molina-Aguilar, C.; Castaneda-Garcia, C.; Vázquez-Cruz, M.E.; Garcia-Salinas, O.I.; Álvarez-Cano, A.; Martínez-Said, H.; Roldán-Marín, R.; Adams, D.J.; Possik, P.A.; et al. Acral lentiginous melanoma: Basic facts, biological characteristics and research perspectives of an understudied disease. Pigment Cell Melanoma Res. 2021, 34, 59–71. [Google Scholar] [CrossRef]
- Conway, J.; Bellet, J.S.; Rubin, A.I.; Lipner, S.R. Adult and Pediatric Nail Unit Melanoma: Epidemiology, Diagnosis, and Treatment. Cells 2023, 12, 964. [Google Scholar] [CrossRef]
- Magro, C.M.; Crowson, A.N.; Mihm, M.C. Unusual variants of malignant melanoma. Mod. Pathol. 2006, 19, S41–S70. [Google Scholar] [CrossRef]
- Druskovich, C.; Kelley, J.; Aubrey, J.; Palladino, L.; Wright, G.P. A Review of Melanoma Subtypes: Genetic and Treatment Considerations. J. Surg. Oncol. 2025, 131, 356–364. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. WHO Classification of Tumours Series, 5th ed.; Skin Tumours [Internet; Beta Version Ahead of Print]; WHO Classification of Tumours Editorial Board: Lyon, France, 2023; Volume 12, Available online: https://tumourclassification.iarc.who.int/chapters/64. (accessed on 20 July 2025).
- Elder, D.E.; Bastian, B.C.; Cree, I.A.; Massi, D.; Scolyer, R.A. The 2018 World Health Organization Classification of Cutaneous, Mucosal, and Uveal Melanoma: Detailed Analysis of 9 Distinct Subtypes Defined by Their Evolutionary Pathway. Arch. Pathol. Lab. Med. 2020, 144, 500–522. [Google Scholar] [CrossRef] [PubMed]
- Saginala, K.; Barsouk, A.; Aluru, J.S.; Rawla, P.; Barsouk, A. Epidemiology of Melanoma. Med. Sci. 2021, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Aitken, J.F.; Youlden, D.R.; Baade, P.D.; Soyer, H.P.; Green, A.C.; Smithers, B.M. Generational shift in melanoma incidence and mortality in Queensland, Australia, 1995–2014. Int. J. Cancer 2018, 142, 1528–1535. [Google Scholar] [CrossRef] [PubMed]
- Paulson, K.G.; Gupta, D.; Kim, T.S.; Veatch, J.R.; Byrd, D.R.; Bhatia, S.; Wojcik, K.; Chapuis, A.G.; Thompson, J.A.; Madeleine, M.M.; et al. Age-Specific Incidence of Melanoma in the United States. JAMA Dermatol. 2020, 156, 57–64. [Google Scholar] [CrossRef]
- Arnold, M.; de Vries, E.; Whiteman, D.C.; Jemal, A.; Bray, F.; Parkin, D.M.; Soerjomataram, I. Global burden of cutaneous melanoma attributable to ultraviolet radiation in 2012. Int. J. Cancer 2018, 143, 1305–1314. [Google Scholar] [CrossRef]
- Gandini, S.; Sera, F.; Cattaruzza, M.S.; Pasquini, P.; Picconi, O.; Boyle, P.; Melchi, C.F. Meta-analysis of risk factors for cutaneous melanoma: II. Sun exposure. Eur. J. Cancer 2005, 41, 45–60. [Google Scholar] [CrossRef]
- Elwood, J.M.; Jopson, J. Melanoma and sun exposure: An overview of published studies. Int. J. Cancer 1997, 73, 198–203. [Google Scholar] [CrossRef]
- Ghiasvand, R.; Robsahm, T.E.; Green, A.C.; Rueegg, C.S.; Weiderpass, E.; Lund, E.; Veierød, M.B. Association of Phenotypic Characteristics and UV Radiation Exposure With Risk of Melanoma on Different Body Sites. JAMA Dermatol. 2019, 155, 39–49. [Google Scholar] [CrossRef]
- An, S.; Kim, K.; Moon, S.; Ko, K.P.; Kim, I.; Lee, J.E.; Park, S.K. Indoor Tanning and the Risk of Overall and Early-Onset Melanoma and Non-Melanoma Skin Cancer: Systematic Review and Meta-Analysis. Cancers 2021, 13, 5940. [Google Scholar] [CrossRef]
- Dennis, L.K.; Vanbeek, M.J.; Beane Freeman, L.E.; Smith, B.J.; Dawson, D.V.; Coughlin, J.A. Sunburns and Risk of Cutaneous Melanoma: Does Age Matter? A Comprehensive Meta-Analysis. Ann. Epidemiol. 2008, 18, 614–627. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer Working Group on artificial ultraviolet (UV) light and skin cancer. The association of use of sunbeds with cutaneous malignant melanoma and other skin cancers: A systematic review. Int. J. Cancer 2007, 120, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Gandini, S.; Sera, F.; Cattaruzza, M.S.; Pasquini, P.; Zanetti, R.; Masini, C.; Boyle, P.; Melchi, C.F. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur. J. Cancer 2005, 41, 2040–2059. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, T.B. The validity and practicality of sun-reactive skin types I through VI. Arch. Dermatol. 1988, 124, 869–871. [Google Scholar] [CrossRef] [PubMed]
- Mangione, C.M.; Barry, M.J.; Nicholson, W.K.; Chelmow, D.; Coker, T.R.; Davis, E.M.; Donahue, K.E.; Jaén, C.R.; Kubik, M.; Li, L.; et al. Screening for Skin Cancer. JAMA 2023, 329, 1290. [Google Scholar] [CrossRef]
- Hollenbeak, C.S.; Todd, M.M.; Billingsley, E.M.; Harper, G.; Dyer, A.M.; Lengerich, E.J. Increased incidence of melanoma in renal transplantation recipients. Cancer 2005, 104, 1962–1967. [Google Scholar] [CrossRef]
- Kubica, A.W.; Brewer, J.D. Melanoma in immunosuppressed patients. Mayo Clin. Proc. 2012, 87, 991–1003. [Google Scholar] [CrossRef]
- Wheless, L.; Black, J.; Alberg, A.J. Nonmelanoma skin cancer and the risk of second primary cancers: A systematic review. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1686–1695. [Google Scholar] [CrossRef]
- Van Der Leest, R.J.T.; Flohil, S.C.; Arends, L.R.; De Vries, E.; Nijsten, T. Risk of subsequent cutaneous malignancy in patients with prior melanoma: A systematic review and meta-analysis. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 1053–1062. [Google Scholar] [CrossRef]
- Ni, Y.; Watts, C.G.; Scolyer, R.A.; Madronio, C.; Armstrong, B.K.; Morton, R.L.; Menzies, S.W.; Mann, G.J.; Thompson, J.F.; Lo, S.N.; et al. Risk of developing a second primary melanoma after a first primary melanoma in a population-based Australian cohort. Br. J. Dermatol. 2023, 188, 814–816. [Google Scholar] [CrossRef]
- Beroukhim, K.; Pourang, A.; Eisen, D.B. Risk of second primary cutaneous and noncutaneous melanoma after cutaneous melanoma diagnosis: A population-based study. J. Am. Acad. Dermatol. 2020, 82, 683–689. [Google Scholar] [CrossRef]
- Lynch, H.T.; Frichot, B.C.; Lynch, J.F. Familial atypical multiple mole-melanoma syndrome. J. Med. Genet. 1978, 15, 352–356. [Google Scholar] [CrossRef]
- Gandini, S.; Sera, F.; Cattaruzza, M.S.; Pasquini, P.; Abeni, D.; Boyle, P.; Melchi, C.F. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur. J. Cancer 2005, 41, 28–44. [Google Scholar] [CrossRef]
- Bataille, V.; Bishop, J.A.; Sasieni, P.; Swerdlow, A.J.; Pinney, E.; Griffiths, K.; Cuzick, J. Risk of cutaneous melanoma in relation to the numbers, types and sites of naevi: A case-control study. Br. J. Cancer 1996, 73, 1605–1611. [Google Scholar] [CrossRef]
- Silva, J.H.; de Sá, B.C.S.; de Ávila, A.L.R.; Landman, G.; Neto, J.P.D. Atypical mole syndrome and dysplastic nevi: Identification of populations at risk for developing melanoma-review article. Clinics 2011, 66, 493–499. [Google Scholar] [CrossRef]
- Goldsmith, L.A. Diagnosis and Treatment of Early Melanoma. JAMA 1992, 268, 1314. [Google Scholar] [CrossRef]
- Melanoma Skin Cancer Statistics. Cancer Research, U.K. 2023. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/melanoma-skin-cancer (accessed on 24 June 2024).
- The Australasian College of Dermatologists Position Statement: Population-Based Screening for Melanoma. 2024. Available online: https://www.dermcoll.edu.au/about/position-statements/ (accessed on 28 August 2024).
- Ward, E.M.; Burnett, C.A.; Ruder, A.; Davis-King, K. Industries and cancer. Cancer Causes Control 1997, 8, 356–370. [Google Scholar] [CrossRef]
- Inzelberg, R.; Israeli-Korn, S.D. The particular relationship between Parkinson’s disease and malignancy: A focus on skin cancers. J. Neural Transm. 2009, 116, 1503–1507. [Google Scholar] [CrossRef]
- Bertoni, J.M.; Arlette, J.P.; Fernandez, H.H.; Fitzer-Attas, C.; Frei, K.; Hassan, M.N.; Isaacson, S.H.; Lew, M.F.; Molho, E.; Ondo, W.G.; et al. Increased melanoma risk in Parkinson disease: A prospective clinicopathological study. Arch. Neurol. 2010, 67, 347–352. [Google Scholar] [CrossRef]
- Olsen, J.H.; Friis, S.; Frederiksen, K. Malignant melanoma and other types of cancer preceding Parkinson disease. Epidemiology 2006, 17, 582–587. [Google Scholar] [CrossRef]
- Kvaskoff, M.; Mesrine, S.; Fournier, A.; Boutron-Ruault, M.C.; Clavel-Chapelon, F. Personal history of endometriosis and risk of cutaneous melanoma in a large prospective cohort of French women. Arch. Intern. Med. 2007, 167, 2061–2065. [Google Scholar] [CrossRef]
- Breitbart, E.W.; Waldmann, A.; Nolte, S.; Capellaro, M.; Greinert, R.; Volkmer, B.; Katalinic, A. Systematic skin cancer screening in Northern Germany. J. Am. Acad. Dermatol. 2012, 66, 201–211. [Google Scholar] [CrossRef]
- Görig, T.; Schneider, S.; Breitbart, E.W.; Diehl, K. Is the quality of skin cancer screening in Germany related to the specialization of the physician who performs it?: Results of a nationwide survey among participants of skin cancer screening. Photodermatol. Photoimmunol. Photomed. 2021, 37, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Datzmann, T.; Schoffer, O.; Meier, F.; Seidler, A.; Schmitt, J. Are patients benefiting from participation in the German skin cancer screening programme? A large cohort study based on administrative data. Br. J. Dermatol. 2022, 186, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Katalinic, A.; Eisemann, N.; Waldmann, A. Skin Cancer Screening in Germany. Documenting Melanoma Incidence and Mortality From 2008 to 2013. Dtsch. Ärzteblatt Int. 2015, 112, 629–634. [Google Scholar]
- Rigel, D.S.; Friedman, R.J.; Kopf, A.W.; Polsky, D. ABCDE--an evolving concept in the early detection of melanoma. Arch. Dermatol. 2005, 141, 1032–1034. [Google Scholar] [CrossRef]
- Thomas, L.; Tranchand, P.; Berard, F.; Secchi, T.; Colin, C.; Moulin, G. Semiological Value of ABCDE Criteria in the Diagnosis of Cutaneous Pigmented Tumors. Dermatology 1998, 197, 11–17. [Google Scholar] [CrossRef]
- Abbasi, N.R.; Yancovitz, M.; Gutkowicz-Krusin, D.; Panageas, K.S.; Mihm, M.C.; Googe, P.; King, R.; Prieto, V.; Osman, I.; Friedman, R.J.; et al. Utility of Lesion Diameter in the Clinical Diagnosis of Cutaneous Melanoma. Arch Dermatol. 2008, 144, 469–474. [Google Scholar] [CrossRef][Green Version]
- Chamberlain, A.J.; Fritschi, L.; Kelly, J.W. Nodular melanoma: Patients’ perceptions of presenting features and implications for earlier detection. J. Am. Acad. Dermatol. 2003, 48, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Jaimes, N.; Chen, L.; Dusza, S.W.; Carrera, C.; Puig, S.; Thomas, L.; Kelly, J.W.; Dang, L.; Zalaudek, I.; Braun, R.P.; et al. Clinical and Dermoscopic Characteristics of Desmoplastic Melanomas. JAMA Dermatol. 2013, 149, 413. [Google Scholar] [CrossRef]
- Jaimes, N.; Braun, R.P.; Thomas, L.; Marghoob, A.A. Clinical and dermoscopic characteristics of amelanotic melanomas that are not of the nodular subtype. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Grob, J.J.; Bonerandi, J.J. The “ugly duckling” sign: Identification of the common characteristics of nevi in an individual as a basis for melanoma screening. Arch. Dermatol. 1998, 134, 103–104. [Google Scholar] [CrossRef]
- MacKie, R.M. Malignant Melanoma. A Guide to Early Diagnosis; Duphar Laboratories: London, UK, 1989. [Google Scholar]
- MacKie, R.M. Clinical recognition of early invasive malignant melanoma. Br. Med. J. 1990, 301, 1005–1006. [Google Scholar] [CrossRef]
- Gachon, J.; Beaulieu, P.; Sei, J.F.; Gouvernet, J.; Claudel, J.P.; Lemaitre, M.; Richard, M.A.; Grob, J.J. First Prospective Study of the Recognition Process of Melanoma in Dermatological Practice. Arch Dermatol. 2005, 141, 434–438. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Grainge, M.J.; Chuchu, N.; Ferrante di Ruffano, L.; Matin, R.N.; Thomson, D.R.; Wong, K.Y.; Aldridge, R.B.; Abbott, R.; et al. Visual inspection for diagnosing cutaneous melanoma in adults. Cochrane Database Syst. Rev. 2018, 12, CD013194. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Chuchu, N.; Ferrante di Ruffano, L.; Matin, R.N.; Thomson, D.R.; Wong, K.Y.; Aldridge, R.B.; Abbott, R.; Fawzy, M.; et al. Dermoscopy, with and without visual inspection, for diagnosing melanoma in adults. Cochrane Database Syst. Rev. 2018, 12, CD011902. [Google Scholar] [CrossRef]
- Williams, N.M.; Rojas, K.D.; Reynolds, J.M.; Kwon, D.; Shum-Tien, J.; Jaimes, N. Assessment of Diagnostic Accuracy of Dermoscopic Structures and Patterns Used in Melanoma Detection. JAMA Dermatol. 2021, 157, 1078. [Google Scholar] [CrossRef] [PubMed]
- Nazzaro, G.; Maronese, C.A.; Casazza, G.; Giacalone, S.; Spigariolo, C.B.; Roccuzzo, G.; Avallone, G.; Guida, S.; Brancaccio, G.; Broganelli, P.; et al. Dermoscopic predictors of melanoma in small diameter melanocytic lesions (mini-melanoma): A retrospective multicentric study of 269 cases. Int. J. Dermatol. 2023, 62, 1040–1049. [Google Scholar] [CrossRef]
- Nazzaro, G.; Passoni, E.; Pozzessere, F.; Maronese, C.A.; Marzano, A.V. Dermoscopy Use Leads to Earlier Cutaneous Melanoma Diagnosis in Terms of Invasiveness and Size? A Single-Center, Retrospective Experience. J. Clin. Med. 2022, 11, 4912. [Google Scholar] [CrossRef]
- Meng, X.; Chen, J.; Zhang, Z.; Li, K.; Li, J.; Yu, Z.; Zhang, Y. Non-invasive optical methods for melanoma diagnosis. Photodiagnosis Photodyn. Ther. 2021, 34, 102266. [Google Scholar] [CrossRef]
- Pezzini, C.; Kaleci, S.; Chester, J.; Farnetani, F.; Longo, C.; Pellacani, G. Reflectance confocal microscopy diagnostic accuracy for malignant melanoma in different clinical settings: Systematic review and meta-analysis. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 2268–2279. [Google Scholar] [CrossRef]
- Ferrante di Ruffano, L.; Dinnes, J.; Deeks, J.J.; Chuchu, N.; Bayliss, S.E.; Davenport, C.; Takwoingi, Y.; Godfrey, K.; O’Sullivan, C.; Matin, R.N.; et al. Optical coherence tomography for diagnosing skin cancer in adults. Cochrane Database Syst. Rev. 2018, 12, CD013189. [Google Scholar] [CrossRef]
- Razi, S.; Kuo, Y.H.; Pathak, G.; Agarwal, P.; Horgan, A.; Parikh, P.; Deshmukh, F.; Rao, B.K. Line-Field Confocal Optical Coherence Tomography for the Diagnosis of Skin Tumors: A Systematic Review and Meta-Analysis. Diagnostics 2024, 14, 1522. [Google Scholar] [CrossRef]
- Balu, M.; Kelly, K.M.; Zachary, C.B.; Harris, R.M.; Krasieva, T.B.; König, K.; Durkin, A.J.; Tromberg, B.J. Distinguishing between Benign and Malignant Melanocytic Nevi by In Vivo Multiphoton Microscopy. Cancer Res. 2014, 74, 2688–2697. [Google Scholar] [CrossRef]
- Forschner, A.; Keim, U.; Hofmann, M.; Spänkuch, I.; Lomberg, D.; Weide, B.; Tampouri, I.; Eigentler, T.; Fink, C.; Garbe, C.; et al. Diagnostic accuracy of dermatofluoroscopy in cutaneous melanoma detection: Results of a prospective multicentre clinical study in 476 pigmented lesions. Br. J. Dermatol. 2018, 179, 478–485. [Google Scholar] [CrossRef]
- Swetter, S.M.; Tsao, H.; Bichakjian, C.K.; Curiel-Lewandrowski, C.; Elder, D.E.; Gershenwald, J.E.; Guild, V.; Grant-Kels, J.M.; Halpern, A.C.; Johnson, T.M.; et al. Guidelines of care for the management of primary cutaneous melanoma. J. Am. Acad. Dermatol. 2019, 80, 208–250. [Google Scholar] [CrossRef]
- NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Melanoma: Cutaneous Version 2.2025 [Internet]. 2025. Available online: https://www.nccn.org/professionals/physician_gls/pdf/cutaneous_melanoma.pdf (accessed on 27 May 2025).
- Scolyer, R.A.; Rawson, R.V.; Gershenwald, J.E.; Ferguson, P.M.; Prieto, V.G. Melanoma pathology reporting and staging. Mod. Pathol. 2020, 33, 15–24. [Google Scholar] [CrossRef]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef]
- Reinhart, J.P.; Campbell, E.H.; Proffer, S.L.; Crum, O.M.; Todd, A.; Gibson, L.E.; Brewer, J.D.; Demer, A.M. Incidence and mortality trends of primary cutaneous melanoma: A 50-year Rochester Epidemiologic Project study. JAAD Int. 2024, 16, 144–154. [Google Scholar] [CrossRef]
- Berk-Krauss, J.; Stein, J.A.; Weber, J.; Polsky, D.; Geller, A.C. New Systematic Therapies and Trends in Cutaneous Melanoma Deaths Among US Whites, 1986–2016. Am. J. Public Health 2020, 110, 731–733. [Google Scholar] [CrossRef]
- Balch, C.M.; Buzaid, A.C.; Soong, S.J.; Atkins, M.B.; Cascinelli, N.; Coit, D.G.; Fleming, I.D.; Gershenwald, J.E.; Houghton, A.; Kirkwood, J.M.; et al. Final Version of the American Joint Committee on Cancer Staging System for Cutaneous Melanoma. J. Clin. Oncol. 2001, 19, 3635–3648. [Google Scholar] [CrossRef]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015, 372, 30–39. [Google Scholar] [CrossRef]
- Balch, C.M.; Soong, S.; Gershenwald, J.E.; Thompson, J.F.; Coit, D.G.; Atkins, M.B.; Ding, S.; Cochran, A.J.; Eggermont, A.M.M.; Flaherty, K.T.; et al. Age as a prognostic factor in patients with localized melanoma and regional metastases. Ann. Surg. Oncol. 2013, 20, 3961–3968. [Google Scholar] [CrossRef]
- Joosse, A.; Collette, S.; Suciu, S.; Nijsten, T.; Patel, P.M.; Keilholz, U.; Eggermont, A.M.M.; Coebergh, J.W.W.; de Vries, E. Sex is an independent prognostic indicator for survival and relapse/progression-free survival in metastasized stage III to IV melanoma: A pooled analysis of five European organisation for research and treatment of cancer randomized controlled trials. J. Clin. Oncol. 2013, 31, 2337–2346. [Google Scholar] [CrossRef]
- Balch, C.M.; Gershenwald, J.E.; Soong, S.J.; Thompson, J.F.; Atkins, M.B.; Byrd, D.R.; Buzaid, A.C.; Cochran, A.J.; Coit, D.G.; Ding, S.; et al. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009, 27, 6199–6206. [Google Scholar] [CrossRef]
- Callender, G.G.; Egger, M.E.; Burton, A.L.; Scoggins, C.R.; Ross, M.I.; Stromberg, A.J.; Hagendoorn, L.; Martin, R.C.G.; McMasters, K.M. Prognostic implications of anatomic location of primary cutaneous melanoma of 1 mm or thicker. Am. J. Surg. 2011, 202, 659–665. [Google Scholar] [CrossRef]
- Lee, J.H.; Saw, R.P.; Thompson, J.F.; Lo, S.; Spillane, A.J.; Shannon, K.F.; Stretch, J.R.; Howle, J.; Menzies, A.M.; Carlino, M.S.; et al. Pre-operative ctDNA predicts survival in high-risk stage III cutaneous melanoma patients. Ann. Oncol. 2019, 30, 815–822. [Google Scholar] [CrossRef]
- Lucci, A.; Hall, C.S.; Patel, S.P.; Narendran, B.; Bauldry, J.B.; Royal, R.E.; Karhade, M.; Upshaw, J.R.; Wargo, J.A.; Glitza, I.C.; et al. Circulating Tumor Cells and Early Relapse in Node-positive Melanoma. Clin. Cancer Res. 2020, 26, 1886–1895. [Google Scholar] [CrossRef]
- Long, G.V.; Desai, K.; Tang, T.; Weber, J.S.; Dolfi, S.; Ritchings, C.; Huang, S.P.; Bolisetty, M.; Sausen, M.; Del Vecchio, M.; et al. 788O Association of pre-treatment ctDNA with disease recurrence and clinical and translational factors in patients with stage IIIB-D/IV melanoma treated with adjuvant immunotherapy (CheckMate 915). Ann. Oncol. 2022, 33, S904. [Google Scholar] [CrossRef]
- Greenhaw, B.N.; Covington, K.R.; Kurley, S.J.; Yeniay, Y.; Cao, N.A.; Plasseraud, K.M.; Cook, R.W.; Hsueh, E.C.; Gastman, B.R.; Wei, M.L. Molecular risk prediction in cutaneous melanoma: A meta-analysis of the 31-gene expression profile prognostic test in 1,479 patients. J. Am. Acad. Dermatol. 2020, 83, 745–753. [Google Scholar] [CrossRef]
- Podlipnik, S.; Carrera, C.; Boada, A.; Richarz, N.A.; López-Estebaranz, J.L.; Pinedo-Moraleda, F.; Elosua-González, M.; Martín-González, M.M.; Carrillo-Gijón, R.; Redondo, P.; et al. Early outcome of a 31-gene expression profile test in 86 AJCC stage IB-II melanoma patients. A prospective multicentre cohort study. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 857–862. [Google Scholar] [CrossRef]
- Eggermont, A.M.M.; Bellomo, D.; Arias-Mejias, S.M.; Quattrocchi, E.; Sominidi-Damodaran, S.; Bridges, A.G.; Lehman, J.S.; Hieken, T.J.; Jakub, J.W.; Murphree, D.H.; et al. Identification of stage I/IIA melanoma patients at high risk for disease relapse using a clinicopathologic and gene expression model. Eur. J. Cancer 2020, 140, 11–18. [Google Scholar] [CrossRef]
- Grossman, D.; Okwundu, N.; Bartlett, E.K.; Marchetti, M.A.; Othus, M.; Coit, D.G.; Hartman, R.I.; Leachman, S.A.; Berry, E.G.; Korde, L.; et al. Prognostic Gene Expression Profiling in Cutaneous Melanoma. JAMA Dermatol. 2020, 156, 1004. [Google Scholar] [CrossRef]
- Marchetti, M.A.; Coit, D.G.; Dusza, S.W.; Yu, A.; McLean, L.; Hu, Y.; Nanda, J.K.; Matsoukas, K.; Mancebo, S.E.; Bartlett, E.K. Performance of Gene Expression Profile Tests for Prognosis in Patients with Localized Cutaneous Melanoma. JAMA Dermatol. 2020, 156, 953. [Google Scholar] [CrossRef]
- Haydu, L.E.; Scolyer, R.A.; Lo, S.; Quinn, M.J.; Saw, R.P.M.; Shannon, K.F.; Spillane, A.J.; Stretch, J.R.; McCarthy, W.H.; Thompson, J.F. Conditional Survival: An Assessment of the Prognosis of Patients at Time Points After Initial Diagnosis and Treatment of Locoregional Melanoma Metastasis. J. Clin. Oncol. 2017, 35, 1721–1729. [Google Scholar] [CrossRef]
- Garbe, C.; Amaral, T.; Peris, K.; Hauschild, A.; Arenberger, P.; Basset-Seguin, N.; Bastholt, L.; Bataille, V.; Brochez, L.; del Marmol, V.; et al. European consensus-based interdisciplinary guideline for melanoma. Part 1: Diagnostics—Update 2024. Eur. J. Cancer 2025, 215, 115152. [Google Scholar] [CrossRef]
- Garbe, C.; Amaral, T.; Peris, K.; Hauschild, A.; Arenberger, P.; Basset-Seguin, N.; Bastholt, L.; Bataille, V.; Brochez, L.; del Marmol, V.; et al. European consensus-based interdisciplinary guideline for melanoma. Part 2: Treatment—Update 2024. Eur. J. Cancer 2025, 215, 115153. [Google Scholar]
- Grotz, T.E.; Markovic, S.N.; Erickson, L.A.; Harmsen, W.S.; Huebner, M.; Farley, D.R.; Pockaj, B.A.; Donohue, J.H.; Sim, F.H.; Grant, C.S.; et al. Mayo Clinic Consensus Recommendations for the Depth of Excision in Primary Cutaneous Melanoma. Mayo Clin. Proc. 2011, 86, 522–528. [Google Scholar] [CrossRef]
- Swetter, S.M.; Thompson, J.A.; Albertini, M.R.; Barker, C.A.; Baumgartner, J.; Boland, G.; Chmielowski, B.; DiMaio, D.; Durham, A.; Fields, R.C.; et al. NCCN Guidelines® Insights: Melanoma: Cutaneous, Version 2.2021. J. Natl. Compr. Cancer Netw. 2021, 19, 364–376. [Google Scholar] [CrossRef]
- Bricca, G.M.; Brodland, D.G.; Ren, D.; Zitelli, J.A. Cutaneous head and neck melanoma treated with Mohs micrographic surgery. J. Am. Acad. Dermatol. 2005, 52, 92–100. [Google Scholar] [CrossRef]
- Mohs, F.E. Chemosurgery. Clin. Plast. Surg. 1980, 7, 349–360. [Google Scholar] [CrossRef]
- Mohs, F.E. Chemosurgery for the microscopically controlled excision of cutaneous cancer. Head Neck Surg. 1978, 1, 150–166. [Google Scholar] [CrossRef]
- Wong, E.; Axibal, E.; Brown, M. Mohs Micrographic Surgery. Facial Plast. Surg. Clin. N. Am. 2019, 27, 15–34. [Google Scholar] [CrossRef]
- Hong, A.M.; Lo, S.N.; Fogarty, G.B.; Stretch, J.; Wang, W.; Penas, P.F.; Martin, R.C.; Foote, M.C.; Soyer, H.P.; Ruben, J.; et al. A randomised, controlled, multicentre trial of imiquimod versus radiotherapy for lentigo maligna. J. Clin. Oncol. 2024, 42 (Suppl. 16), 9502. [Google Scholar] [CrossRef]
- Vaienti, S.; Calzari, P.; Nazzaro, G. Topical Treatment of Melanoma In Situ, Lentigo Maligna, and Lentigo Maligna Melanoma with Imiquimod Cream: A Systematic Review of the Literature. Dermatol. Ther. 2023, 13, 2187–2215. [Google Scholar] [CrossRef]
- Lallas, A.; Moscarella, E.; Kittler, H.; Longo, C.; Thomas, L.; Zalaudek, I.; Kyrgidis, A.; Manoli, S.M.; Meo, N.; Papageorgiou, C.; et al. Real-world experience of off-label use of imiquimod 5% as an adjuvant therapy after surgery or as a monotherapy for lentigo maligna. Br. J. Dermatol. 2021, 185, 675–677. [Google Scholar] [CrossRef]
- Luke, J.J.; Rutkowski, P.; Queirolo, P.; Del Vecchio, M.; Mackiewicz, J.; Chiarion-Sileni, V.; de la Cruz Merino, L.; Khattak, M.A.; Schadendorf, D.; Long, G.V.; et al. Pembrolizumab versus placebo as adjuvant therapy in completely resected stage IIB or IIC melanoma (KEYNOTE-716): A randomised, double-blind, phase 3 trial. Lancet 2022, 399, 1718–1729. [Google Scholar] [CrossRef]
- Kirkwood, J.M.; Del Vecchio, M.; Weber, J.; Hoeller, C.; Grob, J.J.; Mohr, P.; Loquai, C.; Dutriaux, C.; Chiarion-Sileni, V.; Mackiewicz, J.; et al. Adjuvant nivolumab in resected stage IIB/C melanoma: Primary results from the randomized, phase 3 CheckMate 76K trial. Nat. Med. 2023, 29, 2835–2843. [Google Scholar] [CrossRef]
- Schadendorf, D.; Luke, J.J.; Ascierto, P.A.; Long, G.V.; Rutkowski, P.; Khattak, A.; Del Vecchio, M.; de la Cruz-Merino, L.; Mackiewicz, J.; Sileni, V.C.; et al. Pembrolizumab versus placebo as adjuvant therapy in resected stage IIB or IIC melanoma: Outcomes in histopathologic subgroups from the randomized, double-blind, phase 3 KEYNOTE-716 trial. J. Immunother. Cancer 2024, 12, e007501. [Google Scholar] [CrossRef]
- Leiter, U.; Stadler, R.; Mauch, C.; Hohenberger, W.; Brockmeyer, N.H.; Berking, C.; Sunderkötter, C.; Kaatz, M.; Schatton, K.; Lehmann, P.; et al. Final Analysis of DeCOG-SLT Trial: No Survival Benefit for Complete Lymph Node Dissection in Patients with Melanoma with Positive Sentinel Node. J. Clin. Oncol. 2019, 37, 3000–3008. [Google Scholar] [CrossRef]
- Faries, M.B.; Thompson, J.F.; Cochran, A.J.; Andtbacka, R.H.; Mozzillo, N.; Zager, J.S.; Jahkola, T.; Bowles, T.L.; Testori, A.; Beitsch, P.D.; et al. Completion Dissection or Observation for Sentinel-Node Metastasis in Melanoma. N. Engl. J. Med. 2017, 376, 2211–2222. [Google Scholar] [CrossRef]
- Moncrieff, M.D.; Lo, S.N.; Scolyer, R.A.; Heaton, M.J.; Nobes, J.P.; Snelling, A.P.; Carr, M.J.; Nessim, C.; Wade, R.; Peach, A.H.; et al. Clinical Outcomes and Risk Stratification of Early-Stage Melanoma Micrometastases from an International Multicenter Study: Implications for the Management of American Joint Committee on Cancer IIIA Disease. J. Clin. Oncol. 2022, 40, 3940–3951. [Google Scholar] [CrossRef]
- Amaral, T.; Nanz, L.; Stadler, R.; Berking, C.; Ulmer, A.; Forschner, A.; Meiwes, A.; Wolfsperger, F.; Meraz-Torres, F.; Chatziioannou, E.; et al. Isolated melanoma cells in sentinel lymph node in stage IIIA melanoma correlate with a favorable prognosis similar to stage IB. Eur. J. Cancer 2024, 201, 113912. [Google Scholar] [CrossRef]
- Larkin, J.; Del Vecchio, M.; Mandalá, M.; Gogas, H.; Arance Fernandez, A.M.; Dalle, S.; Cowey, C.L.; Schenker, M.; Grob, J.J.; Chiarion-Sileni, V.; et al. Adjuvant Nivolumab versus Ipilimumab in Resected Stage III/IV Melanoma: 5-Year Efficacy and Biomarker Results from CheckMate 238. Clin. Cancer Res. 2023, 29, 3352–3361. [Google Scholar] [CrossRef]
- Eggermont, A.M.M.; Blank, C.U.; Mandala, M.; Long, G.V.; Atkinson, V.G.; Dalle, S.; Haydon, A.M.; Meshcheryakov, A.; Khattak, A.; Carlino, M.S.; et al. Longer Follow-Up Confirms Recurrence-Free Survival Benefit of Adjuvant Pembrolizumab in High-Risk Stage III Melanoma: Updated Results From the EORTC 1325-MG/KEYNOTE-054 Trial. J. Clin. Oncol. 2020, 38, 3925–3936. [Google Scholar] [CrossRef]
- Long, G.V.; Hauschild, A.; Santinami, M.; Atkinson, V.; Mandalà, M.; Chiarion-Sileni, V.; Larkin, J.; Nyakas, M.; Dutriaux, C.; Haydon, A.; et al. Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N. Engl. J. Med. 2017, 377, 1813–1823. [Google Scholar] [CrossRef]
- Long, G.V.; Hauschild, A.; Santinami, M.; Kirkwood, J.M.; Atkinson, V.; Mandala, M.; Merelli, B.; Sileni, V.C.; Nyakas, M.; Haydon, A.; et al. Final Results for Adjuvant Dabrafenib plus Trametinib in Stage III Melanoma. N. Engl. J. Med. 2024, 391, 1709–1720. [Google Scholar] [CrossRef]
- Reijers, I.L.M.; Menzies, A.M.; van Akkooi, A.C.J.; Versluis, J.M.; van den Heuvel, N.M.J.; Saw, R.P.M.; Pennington, T.E.; Kapiteijn, E.; van der Veldt, A.A.M.; Suijkerbuijk, K.P.M.; et al. Personalized response-directed surgery and adjuvant therapy after neoadjuvant ipilimumab and nivolumab in high-risk stage III melanoma: The PRADO trial. Nat. Med. 2022, 28, 1178–1188. [Google Scholar] [CrossRef]
- Versluis, J.M.; Reijers, I.L.M.; Rozeman, E.A.; Menzies, A.M.; van Akkooi, A.C.J.; Wouters, M.W.; Ch’ng, S.; Saw, R.P.M.; Scolyer, R.A.; van de Wiel, B.A.; et al. Neoadjuvant ipilimumab plus nivolumab in synchronous clinical stage III melanoma. Eur. J. Cancer 2021, 148, 51–57. [Google Scholar] [CrossRef]
- Rozeman, E.A.; Menzies, A.M.; van Akkooi, A.C.J.; Adhikari, C.; Bierman, C.; van de Wiel, B.A.; Scolyer, R.A.; Krijgsman, O.; Sikorska, K.; Eriksson, H.; et al. Identification of the optimal combination dosing schedule of neoadjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma (OpACIN-neo): A multicentre, phase 2, randomised, controlled trial. Lancet Oncol. 2019, 20, 948–960. [Google Scholar] [CrossRef] [PubMed]
- Blank, C.U.; Lucas, M.W.; Scolyer, R.A.; van de Wiel, B.A.; Menzies, A.M.; Lopez-Yurda, M.; Hoeijmakers, L.L.; Saw, R.P.M.; Lijnsvelt, J.M.; Maher, N.G.; et al. Neoadjuvant Nivolumab and Ipilimumab in Resectable Stage III Melanoma. N. Engl. J. Med. 2024, 391, 1696–1708. [Google Scholar] [CrossRef]
- Patel, S.P.; Othus, M.; Chen, Y.; Wright, G.P.; Yost, K.J.; Hyngstrom, J.R.; Hu-Lieskovan, S.; Lao, C.D.; Fecher, L.A.; Truong, T.G.; et al. Neoadjuvant-Adjuvant or Adjuvant-Only Pembrolizumab in Advanced Melanoma. N. Engl. J. Med. 2023, 388, 813–823. [Google Scholar] [CrossRef]
- Amaria, R.N.; Postow, M.; Burton, E.M.; Tetzlaff, M.T.; Ross, M.I.; Torres-Cabala, C.; Glitza, I.C.; Duan, F.; Milton, D.R.; Busam, K.; et al. Neoadjuvant relatlimab and nivolumab in resectable melanoma. Nature 2022, 611, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Menzies, A.M.; Amaria, R.N.; Rozeman, E.A.; Huang, A.C.; Tetzlaff, M.T.; van de Wiel, B.A.; Lo, S.; Tarhini, A.A.; Burton, E.M.; Pennington, T.E.; et al. Pathological response and survival with neoadjuvant therapy in melanoma: A pooled analysis from the International Neoadjuvant Melanoma Consortium (INMC). Nat. Med. 2021, 27, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Lebbé, C.; Meyer, N.; Mortier, L.; Marquez-Rodas, I.; Robert, C.; Rutkowski, P.; Menzies, A.M.; Eigentler, T.; Ascierto, P.A.; Smylie, M.; et al. Evaluation of Two Dosing Regimens for Nivolumab in Combination with Ipilimumab in Patients with Advanced Melanoma: Results from the Phase IIIb/IV CheckMate 511 Trial. J. Clin. Oncol. 2019, 37, 867–875. [Google Scholar] [CrossRef]
- Livingstone, E.; Zimmer, L.; Hassel, J.C.; Fluck, M.; Eigentler, T.K.; Loquai, C.; Haferkamp, S.; Gutzmer, R.; Meier, F.; Mohr, P.; et al. Adjuvant nivolumab plus ipilimumab or nivolumab alone versus placebo in patients with resected stage IV melanoma with no evidence of disease (IMMUNED): Final results of a randomised, double-blind, phase 2 trial. Lancet 2022, 400, 1117–1129. [Google Scholar] [CrossRef]
- Robert, C.; Carlino, M.S.; McNeil, C.; Ribas, A.; Grob, J.J.; Schachter, J.; Nyakas, M.; Kee, D.; Petrella, T.M.; Blaustein, A.; et al. Seven-Year Follow-Up of the Phase III KEYNOTE-006 Study: Pembrolizumab Versus Ipilimumab in Advanced Melanoma. J. Clin. Oncol. 2023, 41, 3998–4003. [Google Scholar] [CrossRef]
- Ascierto, P.A.; Long, G.V.; Robert, C.; Brady, B.; Dutriaux, C.; Di Giacomo, A.M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; et al. Survival Outcomes in Patients with Previously Untreated BRAF Wild-Type Advanced Melanoma Treated With Nivolumab Therapy: Three-Year Follow-up of a Randomized Phase 3 Trial. JAMA Oncol. 2019, 5, 187–194. [Google Scholar] [CrossRef]
- Wolchok, J.D.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Long-Term Outcomes with Nivolumab Plus Ipilimumab or Nivolumab Alone Versus Ipilimumab in Patients With Advanced Melanoma. J. Clin. Oncol. 2022, 40, 127–137. [Google Scholar] [CrossRef]
- Mantia, C.M.; Werner, L.; Stwalley, B.; Ritchings, C.; Tarhini, A.A.; Atkins, M.B.; McDermott, D.F.; Regan, M.M. Sensitivity of treatment-free survival to subgroup analyses in patients with advanced melanoma treated with immune checkpoint inhibitors. Melanoma Res. 2022, 32, 35–44. [Google Scholar] [CrossRef]
- Tawbi, H.A.; Schadendorf, D.; Lipson, E.J.; Ascierto, P.A.; Matamala, L.; Castillo Gutiérrez, E.; Rutkowski, P.; Gogas, H.J.; Lao, C.D.; De Menezes, J.J.; et al. Relatlimab and Nivolumab versus Nivolumab in Untreated Advanced Melanoma. N. Engl. J. Med. 2022, 386, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Stephen Hodi, F.; Lipson, E.J.; Schadendorf, D.; Ascierto, P.A.; Matamala, L.; Salman, P.; Castillo Gutiérrez, E.; Rutkowski, P.; Gogas, H.J.; et al. Overall Survival and Response with Nivolumab and Relatlimab in Advanced Melanoma. NEJM Evid. 2023, 2, EVIDoa2200239. [Google Scholar] [CrossRef]
- Ascierto, P.A.; Dummer, R.; Gaudy-Marqueste, C.; Bowyer, S.; Lipson, E.J.; Ghisoni, E.; Middleton, M.R.; Ratto, B.; Jackson, W.J.; Cheong, A.; et al. Efficacy and safety of triplet nivolumab, relatlimab, and ipilimumab (NIVO + RELA + IPI) in advanced melanoma: Results from RELATIVITY-048. J. Clin. Oncol. 2024, 42 (Suppl. 16), 9504. [Google Scholar] [CrossRef]
- Dugan, M.M.; Shannon, A.B.; DePalo, D.K.; Perez, M.C.; Zager, J.S. Intralesional and Infusional Updates for Metastatic Melanoma. Cancers 2024, 16, 1957. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, S.; Galenkamp, A.L.; Singh, R.; Khosla, A.A.; McGranaghan, P.; Cintolo-Gonzalez, J. Talimogene laherparepvec (T-VEC) and Emerging Intralesional Immunotherapies for Metastatic Melanoma: A Review. Curr. Oncol. Rep. 2024, 26, 1651–1663. [Google Scholar] [CrossRef] [PubMed]
- Andtbacka, R.H.I.; Collichio, F.; Harrington, K.J.; Middleton, M.R.; Downey, G.; Öhrling, K.; Kaufman, H.L. Final analyses of OPTiM: A randomized phase III trial of talimogene laherparepvec versus granulocyte-macrophage colony-stimulating factor in unresectable stage III-IV melanoma. J. Immunother. Cancer 2019, 7, 145. [Google Scholar] [CrossRef]
- Stahlie, E.H.A.; Mulder, E.E.A.P.; Reijers, S.; Balduzzi, S.; Zuur, C.L.; Klop, W.M.C.; van der Hiel, B.; Van de Wiel, B.A.; Wouters, M.W.J.M.; Schrage, Y.M.; et al. Single agent Talimogene Laherparepvec for stage IIIB-IVM1c melanoma patients: A systematic review and meta-analysis. Crit. Rev. Oncol. Hematol. 2022, 175, 103705. [Google Scholar] [CrossRef]
- Chesney, J.; Puzanov, I.; Collichio, F.; Singh, P.; Milhem, M.M.; Glaspy, J.; Hamid, O.; Ross, M.; Friedlander, P.; Garbe, C.; et al. Randomized, Open-Label Phase II Study Evaluating the Efficacy and Safety of Talimogene Laherparepvec in Combination with Ipilimumab Versus Ipilimumab Alone in Patients With Advanced, Unresectable Melanoma. J. Clin. Oncol. 2018, 36, 1658–1667. [Google Scholar] [CrossRef]
- Chesney, J.A.; Puzanov, I.; Collichio, F.A.; Singh, P.; Milhem, M.M.; Glaspy, J.; Hamid, O.; Ross, M.; Friedlander, P.; Garbe, C.; et al. Talimogene laherparepvec in combination with ipilimumab versus ipilimumab alone for advanced melanoma: 5-year final analysis of a multicenter, randomized, open-label, phase II trial. J. Immunother. Cancer 2023, 11, e006270. [Google Scholar] [CrossRef]
- Chesney, J.A.; Ribas, A.; Long, G.V.; Kirkwood, J.M.; Dummer, R.; Puzanov, I.; Hoeller, C.; Gajewski, T.F.; Gutzmer, R.; Rutkowski, P.; et al. Randomized, Double-Blind, Placebo-Controlled, Global Phase III Trial of Talimogene Laherparepvec Combined With Pembrolizumab for Advanced Melanoma. J. Clin. Oncol. 2023, 41, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Gastman, B.; Gogas, H.; Rutkowski, P.; Long, G.V.; Chaney, M.F.; Joshi, H.; Lin, Y.L.; Snyder, W.; Chesney, J.A. Open-label, phase II study of talimogene laherparepvec plus pembrolizumab for the treatment of advanced melanoma that progressed on prior anti-PD-1 therapy: MASTERKEY-115. Eur. J. Cancer 2024, 207, 114120. [Google Scholar] [CrossRef] [PubMed]
- Rohaan, M.W.; Stahlie, E.H.A.; Franke, V.; Zijlker, L.P.; Wilgenhof, S.; van der Noort, V.; van Akkooi, A.C.J.; Haanen, J.B.A.G. Neoadjuvant nivolumab + T-VEC combination therapy for resectable early stage or metastatic (IIIB-IVM1a) melanoma with injectable disease: Study protocol of the NIVEC trial. BMC Cancer 2022, 22, 851. [Google Scholar] [CrossRef]
- Franke, V.; Stahlie, E.H.A.; van der Hiel, B.; van de Wiel, B.A.; Wouters, M.W.J.M.; van Houdt, W.J.; van Akkooi, A.C.J. Re-introduction of T-VEC Monotherapy in Recurrent Melanoma is Effective. J. Immunother. 2022, 45, 263–266. [Google Scholar] [CrossRef]
- Dummer, R.; Gyorki, D.E.; Hyngstrom, J.; Berger, A.C.; Conry, R.; Demidov, L.; Sharma, A.; Treichel, S.A.; Radcliffe, H.; Gorski, K.S.; et al. Neoadjuvant talimogene laherparepvec plus surgery versus surgery alone for resectable stage IIIB-IVM1a melanoma: A randomized, open-label, phase 2 trial. Nat. Med. 2021, 27, 1789–1796. [Google Scholar] [CrossRef] [PubMed]
- Dummer, R.; Gyorki, D.E.; Hyngstrom, J.R.; Ning, M.; Lawrence, T.; Ross, M.I. Final 5-Year Follow-Up Results Evaluating Neoadjuvant Talimogene Laherparepvec Plus Surgery in Advanced Melanoma: A Randomized Clinical Trial. JAMA Oncol. 2023, 9, 1457–1459. [Google Scholar] [CrossRef]
- Neoadjuvant Combination Immunotherapy for Stage III Melanoma. 2024. Available online: https://clinicaltrials.gov/study/NCT03842943?term=T-VEC%20neoadjuvant%20melanoma&rank=1 (accessed on 16 July 2025).
- Wong, M.K.; Milhem, M.M.; Sacco, J.J.; Michels, J.; In, G.K.; Couselo, E.M.; Schadendorf, D.; Beasley, G.M.; Niu, J.; Chmielowski, B.; et al. RP1 Combined with Nivolumab in Advanced Anti–PD-1–Failed Melanoma (IGNYTE). J. Clin. Oncol. 2025. [Google Scholar] [CrossRef]
- Andtbacka, R.H.I.; Ross, M.I.; Agarwala, S.S.; Taylor, M.H.; Vetto, J.T.; Neves, R.I.; Daud, A.; Khong, H.T.; Ungerleider, R.S.; Tanaka, M.; et al. Final results of a phase II multicenter trial of HF10, a replication-competent HSV-1 oncolytic virus, and ipilimumab combination treatment in patients with stage IIIB-IV unresectable or metastatic melanoma. J. Clin. Oncol. 2017, 35 (Suppl. 15), 9510. [Google Scholar] [CrossRef]
- Curti, B.D.; Richards, J.; Hyngstrom, J.R.; Daniels, G.A.; Faries, M.; Feun, L.; Margolin, K.A.; Hallmeyer, S.; Grose, M.; Zhang, Y.; et al. Intratumoral oncolytic virus V937 plus ipilimumab in patients with advanced melanoma: The phase 1b MITCI study. J. Immunother. Cancer 2022, 10, e005224. [Google Scholar] [CrossRef]
- Beasley, G.M.; Nair, S.K.; Farrow, N.E.; Landa, K.; Selim, M.A.; Wiggs, C.A.; Jung, S.H.; Bigner, D.D.; True Kelly, A.; Gromeier, M.; et al. Phase I trial of intratumoral PVSRIPO in patients with unresectable, treatment-refractory melanoma. J. Immunother Cancer 2021, 9, e002203. [Google Scholar] [CrossRef] [PubMed]
- Kähler, K.C.; Hassel, J.C.; Ziemer, M.; Rutkowski, P.; Meier, F.; Flatz, L.; Gaudy-Marqueste, C.; Zimmer, L.; Santinami, M.; Russano, F.; et al. Neoadjuvant Intralesional Targeted Immunocytokines (Daromun) in Stage III Melanoma. Ann. Oncol. 2025. [Google Scholar] [CrossRef] [PubMed]
- Study of the Efficacy of Intratumoral L19IL2 or L19TNF or L19IL2/L19TNF, in Combination with Pembrolizumab, in Unresectable Melanoma Patients (INTACT/MeRCI). 2024. Available online: https://clinicaltrials.gov/study/NCT06284590?term=Daromun%20and%20melanoma&rank=4 (accessed on 16 July 2025).
- Helvind, N.M.; Brinch-Møller Weitemeyer, M.; Chakera, A.H.; Hendel, H.W.; Ellebæk, E.; Svane, I.M.; Kjærskov, M.W.; Bojesen, S.; Skyum, H.; Petersen, S.K.; et al. Stage-Specific Risk of Recurrence and Death from Melanoma in Denmark, 2008–2021. JAMA Dermatol. 2023, 159, 1213. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.M.; Geller, A.C.; Warton, E.M.; Schwalbe, J.; Asgari, M.M. Multiple primary melanomas among 16,570 patients with melanoma diagnosed at Kaiser Permanente Northern California, 1996 to 2011. J. Am. Acad. Dermatol. 2015, 73, 630–636. [Google Scholar] [CrossRef]
- Podlipnik, S.; Moreno-Ramírez, D.; Carrera, C.; Barreiro, A.; Manubens, E.; Ferrandiz-Pulido, L.; Sánchez, M.; Vidal-Sicart, S.; Malvehy, J.; Puig, S. Cost-effectiveness analysis of imaging strategy for an intensive follow-up of patients with American Joint Committee on Cancer stage IIB, IIC and III malignant melanoma. Br. J. Dermatol. 2019, 180, 1190–1197. [Google Scholar] [CrossRef]
- Podlipnik, S.; Carrera, C.; Sánchez, M.; Arguis, P.; Olondo, M.L.; Vilana, R.; Rull, R.; Vidal-Sicart, S.; Vilalta, A.; Conill, C.; et al. Performance of diagnostic tests in an intensive follow-up protocol for patients with American Joint Committee on Cancer (AJCC) stage IIB, IIC, and III localized primary melanoma: A prospective cohort study. J. Am. Acad. Dermatol. 2016, 75, 516–524. [Google Scholar] [CrossRef]
- Trotter, S.C.; Sroa, N.; Winkelmann, R.R.; Olencki, T.; Bechtel, M. A Global Review of Melanoma Follow-up Guidelines. J. Clin. Aesthet. Dermatol. 2013, 6, 18–26. [Google Scholar]
- Weide, B.; Elsässer, M.; Büttner, P.; Pflugfelder, A.; Leiter, U.; Eigentler, T.K.; Bauer, J.; Witte, M.; Meier, F.; Garbe, C. Serum markers lactate dehydrogenase and S100B predict independently disease outcome in melanoma patients with distant metastasis. Br. J. Cancer 2012, 107, 422–428. [Google Scholar] [CrossRef]
- Gebhardt, C.; Lichtenberger, R.; Utikal, J. Biomarker value and pitfalls of serum S100B in the follow-up of high-risk melanoma patients. J. Dtsch. Dermatol. Ges. 2016, 14, 158–164. [Google Scholar] [CrossRef]
- Leachman, S.A.; Carucci, J.; Kohlmann, W.; Banks, K.C.; Asgari, M.M.; Bergman, W.; Bianchi-Scarrà, G.; Brentnall, T.; Bressac-de Paillerets, B.; Bruno, W.; et al. Selection criteria for genetic assessment of patients with familial melanoma. J. Am. Acad. Dermatol. 2009, 61, 677.e1–677.e14. [Google Scholar] [CrossRef]
- Kraemer, K.H.; Lee, M.-M.; Andrews, A.D.; Clark Lambert, W. The Role of Sunlight and DNA Repair in Melanoma and Nonmelanoma Skin Cancer: The Xeroderma Pigmentosum Paradigm. Arch. Dermatol. 1994, 130, 1018–1021. [Google Scholar] [CrossRef] [PubMed]
- Curiel-Lewandrowski, C.; Speetzen, L.S.; Cranmer, L.; Warneke, J.A.; Loescher, L.J. Multiple primary cutaneous melanomas in Li-Fraumeni syndrome. Arch. Dermatol. 2011, 147, 248–250. [Google Scholar] [CrossRef] [PubMed]
- Robles-Espinoza, C.D.; Harland, M.; Ramsay, A.J.; Aoude, L.G.; Quesada, V.; Ding, Z.; Pooley, K.A.; Pritchard, A.L.; Tiffen, J.C.; Petljak, M.; et al. POT1 loss-of-function variants predispose to familial melanoma. Nat. Genet. 2014, 46, 478–481. [Google Scholar] [CrossRef]
- Bertolotto, C.; Lesueur, F.; Giuliano, S.; Strub, T.; de Lichy, M.; Bille, K.; Dessen, P.; d’Hayer, B.; Mohamdi, H.; Remenieras, A.; et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011, 480, 94–98. [Google Scholar] [CrossRef]
- Puntervoll, H.E.; Yang, X.R.; Vetti, H.H.; Bachmann, I.M.; Avril, M.F.; Benfodda, M.; Catricalà, C.; Dalle, S.; Duval-Modeste, A.B.; Ghiorzo, P.; et al. Melanoma prone families with CDK4 germline mutation: Phenotypic profile and associations with MC1R variants. J. Med. Genet. 2013, 50, 264–270. [Google Scholar] [CrossRef]
- Leachman, S.A.; Lucero, O.M.; Sampson, J.E.; Cassidy, P.; Bruno, W.; Queirolo, P.; Ghiorzo, P. Identification, genetic testing, and management of hereditary melanoma. Cancer Metastasis Rev. 2017, 36, 77–90. [Google Scholar] [CrossRef]
- Ransohoff, K.J.; Jaju, P.D.; Tang, J.Y.; Carbone, M.; Leachman, S.; Sarin, K.Y. Familial skin cancer syndromes: Increased melanoma risk. J. Am. Acad. Dermatol. 2016, 74, 423–434, quiz 435–436. [Google Scholar] [CrossRef]
- Funchain, P.; Ni, Y.; Heald, B.; Bungo, B.; Arbesman, M.; Behera, T.R.; McCormick, S.; Song, J.M.; Kennedy, L.B.; Nielsen, S.M.; et al. Germline cancer susceptibility in individuals with melanoma. J. Am. Acad. Dermatol. 2024, 91, 265–272. [Google Scholar] [CrossRef]
- Lochrin, S.E.; Kemel, Y.; Stuart, J.; Smithy, J.W.; Mandelker, D.; Momtaz, P.; Chatila, W.K.; Schultz, N.; Postow, M.A.; Stadler, Z.K.; et al. Germline pathogenic variants in a large convenience cohort of multiple melanoma subtypes. J. Clin. Oncol. 2024, 42 (Suppl. 16), 9595. [Google Scholar] [CrossRef]
- Olsen, C.M.; Pandeya, N.; Thompson, B.S.; Dusingize, J.C.; Webb, P.M.; Green, A.C.; Neale, R.E.; Whiteman, D.C.; Study, Q. Risk Stratification for Melanoma: Models Derived and Validated in a Purpose-Designed Prospective Cohort. J. Natl. Cancer Inst. 2018, 110, 1075–1083. [Google Scholar] [CrossRef]
- Vuong, K.; Armstrong, B.K.; Weiderpass, E.; Lund, E.; Adami, H.O.; Veierod, M.B.; Barrett, J.H.; Davies, J.R.; Bishop, D.T.; Whiteman, D.C.; et al. Development and External Validation of a Melanoma Risk Prediction Model Based on Self-assessed Risk Factors. JAMA Dermatol. 2016, 152, 889–896. [Google Scholar] [CrossRef]
- Kaiser, I.; Pfahlberg, A.B.; Uter, W.; Heppt, M.V.; Veierød, M.B.; Gefeller, O. Risk Prediction Models for Melanoma: A Systematic Review on the Heterogeneity in Model Development and Validation. Int. J. Environ. Res. Public Health 2020, 17, 7919. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.N.; Ma, J.; Scolyer, R.A.; Haydu, L.E.; Stretch, J.R.; Saw, R.P.M.; Nieweg, O.E.; Shannon, K.F.; Spillane, A.J.; Ch’ng, S.; et al. Improved Risk Prediction Calculator for Sentinel Node Positivity in Patients with Melanoma: The Melanoma Institute Australia Nomogram. J. Clin. Oncol. 2020, 38, 2719–2727. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.L.; Kattan, M.W.; McMasters, K.M.; Coit, D.G. A nomogram that predicts the presence of sentinel node metastasis in melanoma with better discrimination than the American Joint Committee on Cancer staging system. Ann. Surg. Oncol. 2005, 12, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Friedman, C.; Lyon, M.; Torphy, R.J.; Thieu, D.; Hosokawa, P.; Gonzalez, R.; Lewis, K.D.; Medina, T.M.; Rioth, M.J.; Robinson, W.A.; et al. A nomogram to predict node positivity in patients with thin melanomas helps inform shared patient decision making. J. Surg. Oncol. 2019, 120, 1276–1283. [Google Scholar] [CrossRef]
- Penn, L.A.; Qian, M.; Zhang, E.; Ng, E.; Shao, Y.; Berwick, M.; Lazovich, D.; Polsky, D. Development of a melanoma risk prediction model incorporating MC1R genotype and indoor tanning exposure: Impact of mole phenotype on model performance. PLoS ONE 2014, 9, e101507. [Google Scholar] [CrossRef]
- Quéreux, G.; Moyse, D.; Lequeux, Y.; Jumbou, O.; Brocard, A.; Antonioli, D.; Dréno, B.; Nguyen, J.M. Development of an individual score for melanoma risk. Eur. J. Cancer Prev. 2011, 20, 217–224. [Google Scholar] [CrossRef]
- Guther, S.; Ramrath, K.; Dyall-Smith, D.; Landthaler, M.; Stolz, W. Development of a targeted risk-group model for skin cancer screening based on more than 100,000 total skin examinations. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 86–94. [Google Scholar] [CrossRef]
- Williams, L.H.; Shors, A.R.; Barlow, W.E.; Solomon, C.; White, E. Identifying Persons at Highest Risk of Melanoma Using Self-Assessed Risk Factors. J. Clin. Exp. Dermatol. Res. 2011, 2, 1000129. [Google Scholar]
- Goldberg, M.S.; Doucette, J.T.; Lim, H.W.; Spencer, J.; Carucci, J.A.; Rigel, D.S. Risk factors for presumptive melanoma in skin cancer screening: American Academy of Dermatology National Melanoma/Skin Cancer Screening Program experience 2001–2005. J. Am. Acad. Dermatol. 2007, 57, 60–66. [Google Scholar] [CrossRef]
- Mar, V.; Wolfe, R.; Kelly, J.W. Predicting melanoma risk for the Australian population. Australas. J. Dermatol. 2011, 52, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Fortes, C.; Mastroeni, S.; Bakos, L.; Antonelli, G.; Alessandroni, L.; Pilla, M.A.; Alotto, M.; Zappalà, A.; Manoorannparampill, T.; Bonamigo, R.; et al. Identifying individuals at high risk of melanoma: A simple tool. Eur. J. Cancer Prev. 2010, 19, 393–400. [Google Scholar] [CrossRef]
- Cho, E.; Rosner, B.A.; Feskanich, D.; Colditz, G.A. Risk factors and individual probabilities of melanoma for whites. J. Clin. Oncol. 2005, 23, 2669–2675. [Google Scholar] [CrossRef]
- Usher-Smith, J.A.; Emery, J.; Kassianos, A.P.; Walter, F.M. Risk Prediction Models for Melanoma: A Systematic Review. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1450–1463. [Google Scholar] [CrossRef]
- Maddineni, S.; Dizon, M.P.; Muralidharan, V.; Young, L.A.; Sunwoo, J.B.; Baik, F.M.; Swetter, S.M. Validation of the Melanoma Institute of Australia’s Sentinel Lymph Node Biopsy Risk Prediction Tool for Cutaneous Melanoma. Ann. Surg. Oncol. 2024, 31, 2737–2746. [Google Scholar] [CrossRef]
- Drebin, H.M.; Hosein, S.; Kurtansky, N.R.; Nadelmann, E.; Moy, A.P.; Ariyan, C.E.; Bello, D.M.; Brady, M.S.; Coit, D.G.; Marchetti, M.A.; et al. Clinical Utility of Melanoma Sentinel Lymph Node Biopsy Nomograms. J. Am. Coll. Surg. 2024, 238, 23–31. [Google Scholar] [CrossRef]
- Olofsson Bagge, R.; Mikiver, R.; Marchetti, M.A.; Lo, S.N.; van Akkooi, A.C.J.; Coit, D.G.; Ingvar, C.; Isaksson, K.; Scolyer, R.A.; Thompson, J.F.; et al. Population-Based Validation of the MIA and MSKCC Tools for Predicting Sentinel Lymph Node Status. JAMA Surg. 2024, 159, 260–268. [Google Scholar] [CrossRef]
- Miller, I.; Rosic, N.; Stapelberg, M.; Hudson, J.; Coxon, P.; Furness, J.; Walsh, J.; Climstein, M. Performance of Commercial Dermatoscopic Systems That Incorporate Artificial Intelligence for the Identification of Melanoma in General Practice: A Systematic Review. Cancers 2024, 16, 1443. [Google Scholar] [CrossRef] [PubMed]
- Haggenmüller, S.; Maron, R.C.; Hekler, A.; Utikal, J.S.; Barata, C.; Barnhill, R.L.; Beltraminelli, H.; Berking, C.; Betz-Stablein, B.; Blum, A.; et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts. Eur. J. Cancer 2021, 156, 202–216. [Google Scholar] [CrossRef]
- Ferrante di Ruffano, L.; Takwoingi, Y.; Dinnes, J.; Chuchu, N.; Bayliss, S.E.; Davenport, C.; Matin, R.N.; Godfrey, K.; O’Sullivan, C.; Gulati, A.; et al. Computer-assisted diagnosis techniques (dermoscopy and spectroscopy-based) for diagnosing skin cancer in adults. Cochrane Database Syst. Rev. 2018, 12, CD013186. [Google Scholar] [CrossRef] [PubMed]
- Ertürk Zararsız, G.; Yerlitaş Taştan, S.I.; Çelik Gürbulak, E.; Erakcaoğlu, A.; Yılmaz Işıkhan, S.; Demirbaş, A.; Ertaş, R.; Eroğlu, İ.; Korkmaz, S.; Elmas, Ö.F.; et al. Diagnosis melanoma with artificial intelligence systems: A meta-analysis study and systematic review. J. Eur. Acad. Dermatol. Venereol. 2025, 00, 1–11. [Google Scholar] [CrossRef]
- Soyer, H.P.; Shumack, S. Position Statement: Population-Based Screening for Melanoma. 2024. Available online: https://www.dermcoll.edu.au/wp-content/uploads/2024/01/ACD-Position-Statement-Population-screening-for-melanoma-January-2024.pdf (accessed on 28 May 2025).
- Sun, J.; Karasaki, K.M.; Farma, J.M. The Use of Gene Expression Profiling and Biomarkers in Melanoma Diagnosis and Predicting Recurrence: Implications for Surveillance and Treatment. Cancers 2024, 16, 583. [Google Scholar] [CrossRef]
- Amaral, T.; Sinnberg, T.; Chatziioannou, E.; Niessner, H.; Leiter, U.; Keim, U.; Forschner, A.; Dwarkasing, J.; Tjien-Fooh, F.; Wever, R.; et al. Identification of stage I/II melanoma patients at high risk for recurrence using a model combining clinicopathologic factors with gene expression profiling (CP-GEP). Eur. J. Cancer 2023, 182, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Yousaf, A.; Tjien-Fooh, F.J.; Rentroia-Pacheco, B.; Quattrocchi, E.; Kobic, A.; Tempel, D.; Kolodney, M.; Meves, A. Validation of CP-GEP (Merlin Assay) for predicting sentinel lymph node metastasis in primary cutaneous melanoma patients: A US cohort study. Int. J. Dermatol. 2021, 60, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Mulder, E.E.A.P.; Dwarkasing, J.T.; Tempel, D.; van der Spek, A.; Bosman, L.; Verver, D.; Mooyaart, A.L.; van der Veldt, A.A.M.; Verhoef, C.; Nijsten, T.E.C.; et al. Validation of a clinicopathological and gene expression profile model for sentinel lymph node metastasis in primary cutaneous melanoma. Br. J. Dermatol. 2021, 184, 944–951. [Google Scholar] [CrossRef] [PubMed]
- Zager, J.S.; Gastman, B.R.; Leachman, S.; Gonzalez, R.C.; Fleming, M.D.; Ferris, L.K.; Ho, J.; Miller, A.R.; Cook, R.W.; Covington, K.R.; et al. Performance of a prognostic 31-gene expression profile in an independent cohort of 523 cutaneous melanoma patients. BMC Cancer 2018, 18, 130. [Google Scholar] [CrossRef]
- Friedman, E.B.; Shang, S.; de Miera, E.V.S.; Fog, J.U.; Teilum, M.W.; Ma, M.W.; Berman, R.S.; Shapiro, R.L.; Pavlick, A.C.; Hernando, E.; et al. Serum microRNAs as biomarkers for recurrence in melanoma. J. Transl. Med. 2012, 10, 155. [Google Scholar] [CrossRef]
- Gracie, L.; Pan, Y.; Atenafu, E.G.; Ward, D.G.; Teng, M.; Pallan, L.; Stevens, N.M.; Khoja, L. Circulating tumour DNA (ctDNA) in metastatic melanoma, a systematic review and meta-analysis. Eur. J. Cancer 2021, 158, 191–207. [Google Scholar] [CrossRef]
- Aoude, L.G.; Brosda, S.; Ng, J.; Lonie, J.M.; Belle, C.J.; Patel, K.; Koufariotis, L.T.; Wood, S.; Atkinson, V.; Smithers, B.M.; et al. Circulating Tumor DNA: A Promising Biomarker for Predicting Recurrence in Patients with BRAF-Negative Melanoma. J. Mol. Diagn. 2023, 25, 771–781. [Google Scholar] [CrossRef]
- Gerami, P.; Cook, R.W.; Wilkinson, J.; Russell, M.C.; Dhillon, N.; Amaria, R.N.; Gonzalez, R.; Lyle, S.; Johnson, C.E.; Oelschlager, K.M.; et al. Development of a prognostic genetic signature to predict the metastatic risk associated with cutaneous melanoma. Clin. Cancer Res. 2015, 21, 175–183. [Google Scholar] [CrossRef]
- Durgham, R.A.; Nassar, S.I.; Gun, R.; Nguyen, S.A.; Asarkar, A.A.; Nathan, C.A.O. The Prognostic Value of the 31-Gene Expression Profile Test in Cutaneous Melanoma: A Systematic Review and Meta-Analysis. Cancers 2024, 16, 3714. [Google Scholar] [CrossRef]
- Whitman, E.D.; Koshenkov, V.P.; Gastman, B.R.; Lewis, D.; Hsueh, E.C.; Pak, H.; Trezona, T.P.; Davidson, R.S.; McPhee, M.; Guenther, J.M.; et al. Integrating 31-Gene Expression Profiling with Clinicopathologic Features to Optimize Cutaneous Melanoma Sentinel Lymph Node Metastasis Prediction. JCO Precis. Oncol. 2021, 5, 1466–1479. [Google Scholar] [CrossRef]
- Jarell, A.; Gastman, B.R.; Dillon, L.D.; Hsueh, E.C.; Podlipnik, S.; Covington, K.R.; Cook, R.W.; Bailey, C.N.; Quick, A.P.; Martin, B.J.; et al. Optimizing treatment approaches for patients with cutaneous melanoma by integrating clinical and pathologic features with the 31-gene expression profile test. J. Am. Acad. Dermatol. 2022, 87, 1312–1320. [Google Scholar] [CrossRef]
- Gerami, P.; Yao, Z.; Polsky, D.; Jansen, B.; Busam, K.; Ho, J.; Martini, M.; Ferris, L.K. Development and validation of a noninvasive 2-gene molecular assay for cutaneous melanoma. J. Am. Acad. Dermatol. 2017, 76, 114–120.e2. [Google Scholar] [CrossRef]
- Estrada, S.; Shackelton, J.; Cleaver, N.; Depcik-Smith, N.; Cockerell, C.; Lencioni, S.; Martin, H.; Wilkinson, J.; Meldi Sholl, L.; Berg, M.; et al. Development and Validation of a Diagnostic 35-Gene Expression Profile Test for Ambiguous or Difficult-to-Diagnose Suspicious Pigmented Skin Lesions. Ski. J. Cutan. Med. 2020, 4, 506–522. [Google Scholar] [CrossRef]
- Clarke, L.E.; Warf, M.B.; Flake, D.D.; Hartman, A.R.; Tahan, S.; Shea, C.R.; Gerami, P.; Messina, J.; Florell, S.R.; Wenstrup, R.J.; et al. Clinical validation of a gene expression signature that differentiates benign nevi from malignant melanoma. J. Cutan. Pathol. 2015, 42, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, E.K.; O’Donoghue, C.; Boland, G.; Bowles, T.; Delman, K.A.; Hieken, T.J.; Moncrieff, M.; Wong, S.; White, R.L.; Karakousis, G. Society of Surgical Oncology Consensus Statement: Assessing the Evidence for and Utility of Gene Expression Profiling of Primary Cutaneous Melanoma. Ann. Surg. Oncol. 2025, 32, 1429–1442. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, M.S.; Polivka, J.; Treskova, I.; Houfkova, K.; Knizkova, T.; Woznica, V.; Fikrle, T.; Pivovarcikova, K.; Svaton, M.; Shetti, D.; et al. Preoperative Plasma miRNA Levels Predict Prognosis in Early-stage Malignant Melanoma. Anticancer Res. 2023, 43, 695–706. [Google Scholar] [CrossRef]
- Tian, R.; Liu, T.; Qiao, L.; Gao, M.; Li, J. Decreased serum microRNA-206 level predicts unfavorable prognosis in patients with melanoma. Int. J. Clin. Exp. Pathol. 2015, 8, 3097–3103. [Google Scholar]
- Love, C.G.; Coombs, L.; Van Laar, R. RNA-seq validation of microRNA expression signatures for precision melanoma diagnosis and prognostic stratification. BMC Med. Genom. 2024, 17, 256. [Google Scholar] [CrossRef]
- Li, P.; He, Q.Y.; Luo, C.Q.; Qian, L.Y. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med. Sci. Monit. 2014, 20, 2472–2477. [Google Scholar]
- Bustos, M.A.; Tran, K.D.; Rahimzadeh, N.; Gross, R.; Lin, S.Y.; Shoji, Y.; Murakami, T.; Boley, C.L.; Tran, L.T.; Cole, H.; et al. Integrated Assessment of Circulating Cell-Free MicroRNA Signatures in Plasma of Patients with Melanoma Brain Metastasis. Cancers 2020, 12, 1692. [Google Scholar] [CrossRef]
- Guo, W.; Wang, H.; Yang, Y.; Guo, S.; Zhang, W.; Liu, Y.; Yi, X.; Ma, J.; Zhao, T.; Liu, L.; et al. Down-regulated miR-23a Contributes to the Metastasis of Cutaneous Melanoma by Promoting Autophagy. Theranostics 2017, 7, 2231–2249. [Google Scholar] [CrossRef]
- Jones, N.; Nonaka, T. Circulating miRNAs as biomarkers for the diagnosis in patients with melanoma: Systematic review and meta-analysis. Front. Genet. 2024, 15, 1339357. [Google Scholar] [CrossRef] [PubMed]
- Van Laar, R.; Lincoln, M.; Van Laar, B. Development and validation of a plasma-based melanoma biomarker suitable for clinical use. Br. J. Cancer 2018, 118, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, S.; Fukushima, S.; Okada, E.; Morinaga, J.; Kubo, Y.; Tokuzumi, A.; Matsumoto, S.; Tsuruta-Kadohisa, M.; Kimura, T.; Kuriyama, H.; et al. MicroRNAs that predict the effectiveness of anti-PD-1 therapies in patients with advanced melanoma. J. Dermatol. Sci. 2020, 97, 77–79. [Google Scholar] [CrossRef]
- Huber, V.; Vallacchi, V.; Fleming, V.; Hu, X.; Cova, A.; Dugo, M.; Shahaj, E.; Sulsenti, R.; Vergani, E.; Filipazzi, P.; et al. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J. Clin. Investig. 2018, 128, 5505–5516. [Google Scholar] [CrossRef] [PubMed]
- Fattore, L.; Ruggiero, C.F.; Pisanu, M.E.; Liguoro, D.; Cerri, A.; Costantini, S.; Capone, F.; Acunzo, M.; Romano, G.; Nigita, G.; et al. Reprogramming miRNAs global expression orchestrates development of drug resistance in BRAF mutated melanoma. Cell Death Differ. 2019, 26, 1267–1282. [Google Scholar] [CrossRef]
- Gaponova, S.; Patutina, O.; Sen’kova, A.; Burakova, E.; Savin, I.; Markov, A.; Shmendel, E.; Maslov, M.; Stetsenko, D.; Vlassov, V.; et al. Single Shot vs. Cocktail: A Comparison of Mono- and Combinative Application of miRNA-Targeted Mesyl Oligonucleotides for Efficient Antitumor Therapy. Cancers 2022, 14, 4396. [Google Scholar] [CrossRef]
- Huynh, C.; Segura, M.F.; Gaziel-Sovran, A.; Menendez, S.; Darvishian, F.; Chiriboga, L.; Levin, B.; Meruelo, D.; Osman, I.; Zavadil, J.; et al. Efficient in vivo microRNA targeting of liver metastasis. Oncogene 2011, 30, 1481–1488. [Google Scholar] [CrossRef][Green Version]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wu, Z.; Chen, J.; Guo, S.; You, W.; Wang, S.; Ma, J.; Wang, H.; Wang, X.; Wang, H.; et al. Nanoparticle delivery of miR-21-3p sensitizes melanoma to anti-PD-1 immunotherapy by promoting ferroptosis. J. Immunother. Cancer 2022, 10, e004381. [Google Scholar] [CrossRef]
- Lim, S.Y.; Boyd, S.C.; Diefenbach, R.J.; Rizos, H. Circulating MicroRNAs: Functional biomarkers for melanoma prognosis and treatment. Mol. Cancer 2025, 24, 99. [Google Scholar] [CrossRef] [PubMed]
- Segal, M.; Slack, F.J. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin. Drug Discov. 2020, 15, 987–992. [Google Scholar] [CrossRef]
- Eroglu, Z.; Krinshpun, S.; Kalashnikova, E.; Sudhaman, S.; Ozturk Topcu, T.; Nichols, M.; Martin, J.; Bui, K.M.; Palsuledesai, C.C.; Malhotra, M.; et al. Circulating tumor DNA-based molecular residual disease detection for treatment monitoring in advanced melanoma patients. Cancer 2023, 129, 1723–1734. [Google Scholar] [CrossRef]
- Syeda, M.M.; Wiggins, J.M.; Corless, B.C.; Long, G.V.; Flaherty, K.T.; Schadendorf, D.; Nathan, P.D.; Robert, C.; Ribas, A.; Davies, M.A.; et al. Circulating tumour DNA in patients with advanced melanoma treated with dabrafenib or dabrafenib plus trametinib: A clinical validation study. Lancet Oncol. 2021, 22, 370–380. [Google Scholar] [CrossRef]
- Syeda, M.M.; Long, G.V.; Garrett, J.; Atkinson, V.; Santinami, M.; Schadendorf, D.; Hauschild, A.; Millward, M.; Mandala, M.; Chiarion-Sileni, V.; et al. Clinical validation of droplet digital PCR assays in detecting BRAFV600-mutant circulating tumour DNA as a prognostic biomarker in patients with resected stage III melanoma receiving adjuvant therapy (COMBI-AD): A biomarker analysis from a double-blind, randomised phase 3 trial. Lancet Oncol. 2025, 26, 641–653. [Google Scholar]
- Tan, L.; Sandhu, S.; Lee, R.J.; Li, J.; Callahan, J.; Ftouni, S.; Dhomen, N.; Middlehurst, P.; Wallace, A.; Raleigh, J.; et al. Prediction and monitoring of relapse in stage III melanoma using circulating tumor DNA. Ann. Oncol. 2019, 30, 804–814. [Google Scholar] [CrossRef]
- Gandini, S.; Zanna, I.; Angelis, S.P.D.; Cocorocchio, E.; Queirolo, P.; Lee, J.H.; Carlino, M.S.; Mazzarella, L.; Achutti Duso, B.; Palli, D.; et al. Circulating tumour DNA and melanoma survival: A systematic literature review and meta-analysis. Crit. Rev. Oncol. Hematol. 2021, 157, 103187. [Google Scholar] [CrossRef]
- Marsavela, G.; Lee, J.; Calapre, L.; Wong, S.Q.; Pereira, M.R.; McEvoy, A.C.; Reid, A.L.; Robinson, C.; Warburton, L.; Abed, A.; et al. Circulating Tumor DNA Predicts Outcome from First-, but not Second-line Treatment and Identifies Melanoma Patients Who May Benefit from Combination Immunotherapy. Clin. Cancer Res. 2020, 26, 5926–5933. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Menzies, A.M.; Carlino, M.S.; McEvoy, A.C.; Sandhu, S.; Weppler, A.M.; Diefenbach, R.J.; Dawson, S.J.; Kefford, R.F.; Millward, M.J.; et al. Longitudinal Monitoring of ctDNA in Patients with Melanoma and Brain Metastases Treated with Immune Checkpoint Inhibitors. Clin. Cancer Res. 2020, 26, 4064–4071. [Google Scholar] [CrossRef] [PubMed]
- Ganzetti, G.; Sartini, D.; Campanati, A.; Rubini, C.; Molinelli, E.; Brisigotti, V.; Cecati, M.; Pozzi, V.; Campagna, R.; Offidani, A.; et al. Nicotinamide N-methyltransferase: Potential involvement in cutaneous malignant melanoma. Melanoma Res. 2018, 28, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Sartini, D.; Molinelli, E.; Pozzi, V.; Campagna, R.; Salvolini, E.; Rubini, C.; Goteri, G.; Simonetti, O.; Campanati, A.; Offidani, A.; et al. Immunohistochemical expression of nicotinamide N-methyltransferase in lymph node metastases from cutaneous malignant melanoma. Hum. Cell 2023, 36, 480–482. [Google Scholar] [CrossRef]
- Campagna, R.; Salvolini, E.; Pompei, V.; Pozzi, V.; Salvucci, A.; Molinelli, E.; Brisigotti, V.; Sartini, D.; Campanati, A.; Offidani, A.; et al. Nicotinamide N-methyltransferase gene silencing enhances chemosensitivity of melanoma cell lines. Pigment Cell Melanoma Res. 2021, 34, 1039–1048. [Google Scholar] [CrossRef]
- Bacchetti, T.; Salvolini, E.; Pompei, V.; Campagna, R.; Molinelli, E.; Brisigotti, V.; Togni, L.; Lucarini, G.; Sartini, D.; Campanati, A.; et al. Paraoxonase-2: A potential biomarker for skin cancer aggressiveness. Eur. J. Clin. Investig. 2021, 51, e13452. [Google Scholar] [CrossRef] [PubMed]
- Campagna, R.; Bacchetti, T.; Salvolini, E.; Pozzi, V.; Molinelli, E.; Brisigotti, V.; Sartini, D.; Campanati, A.; Ferretti, G.; Offidani, A.; et al. Paraoxonase-2 Silencing Enhances Sensitivity of A375 Melanoma Cells to Treatment with Cisplatin. Antioxidants 2020, 9, 1238. [Google Scholar] [CrossRef]
- Hu, Z.; Leet, D.E.; Allesøe, R.L.; Oliveira, G.; Li, S.; Luoma, A.M.; Liu, J.; Forman, J.; Huang, T.; Iorgulescu, J.B.; et al. Personal neoantigen vaccines induce persistent memory T cell responses and epitope spreading in patients with melanoma. Nat. Med. 2021, 27, 515–525. [Google Scholar] [CrossRef]
- Mehta, A.; Motavaf, M.; Nebo, I.; Luyten, S.; Osei-Opare, K.D.; Gru, A.A. Advancements in Melanoma Treatment: A Review of PD-1 Inhibitors, T-VEC, mRNA Vaccines, and Tumor-Infiltrating Lymphocyte Therapy in an Evolving Landscape of Immunotherapy. J. Clin. Med. 2025, 14, 1200. [Google Scholar] [CrossRef]
- Weber, J.S.; Carlino, M.S.; Khattak, A.; Meniawy, T.; Ansstas, G.; Taylor, M.H.; Kim, K.B.; McKean, M.; Long, G.V.; Sullivan, R.J.; et al. Individualised neoantigen therapy mRNA-4157 (V940) plus pembrolizumab versus pembrolizumab monotherapy in resected melanoma (KEYNOTE-942): A randomised, phase 2b study. Lancet 2024, 403, 632–644. [Google Scholar] [CrossRef]
- Ninmer, E.K.; Zhu, H.; Chianese-Bullock, K.A.; von Mehren, M.; Haas, N.B.; Ross, M.I.; Dengel, L.T.; Slingluff, C.L. Multipeptide vaccines for melanoma in the adjuvant setting: Long-term survival outcomes and post-hoc analysis of a randomized phase II trial. Nat. Commun. 2024, 15, 2570. [Google Scholar] [CrossRef]
- A Clinical Study of V940 Plus Pembrolizumab in People with High-Risk Melanoma (V940-001). 2024. Available online: https://clinicaltrials.gov/study/NCT05933577 (accessed on 28 May 2025).
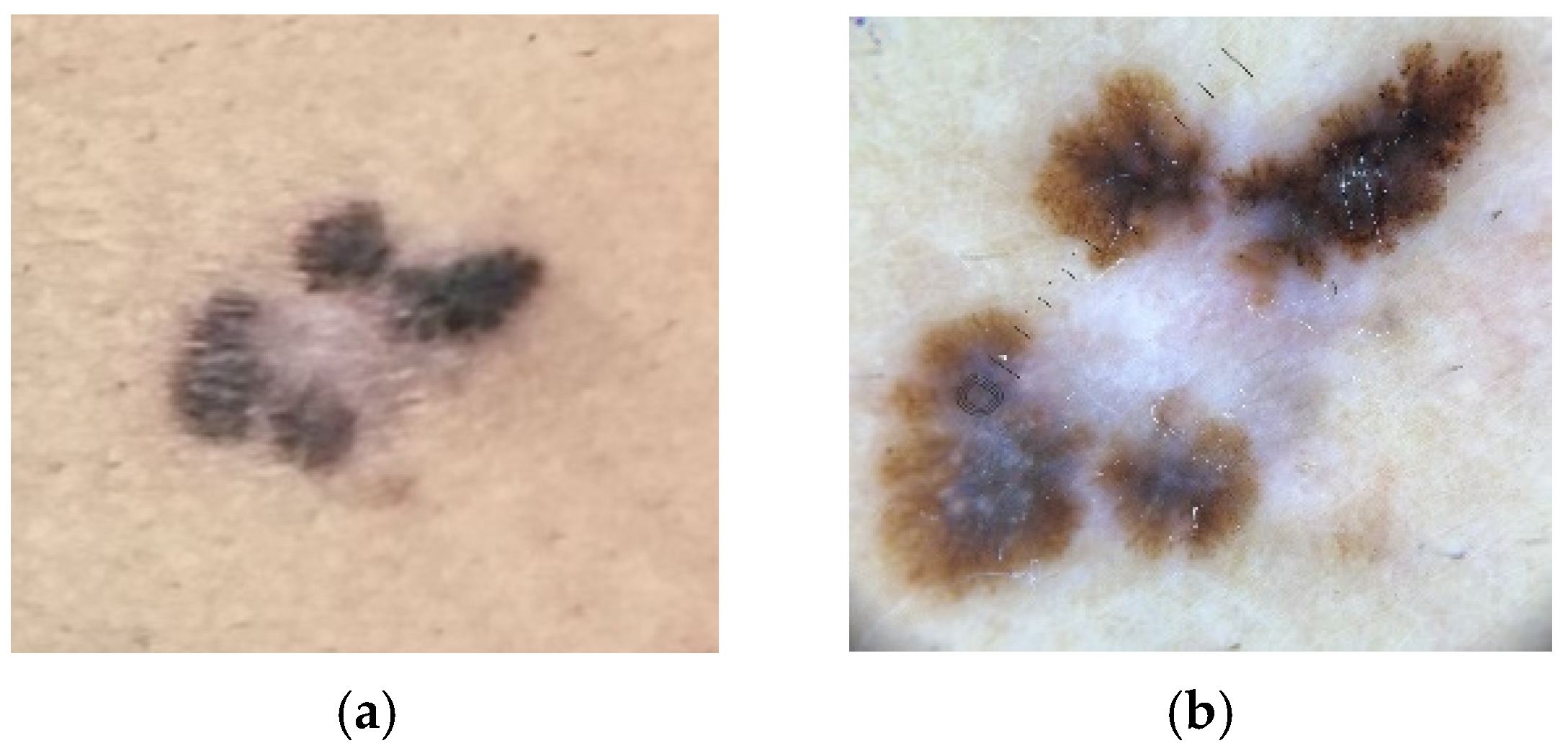
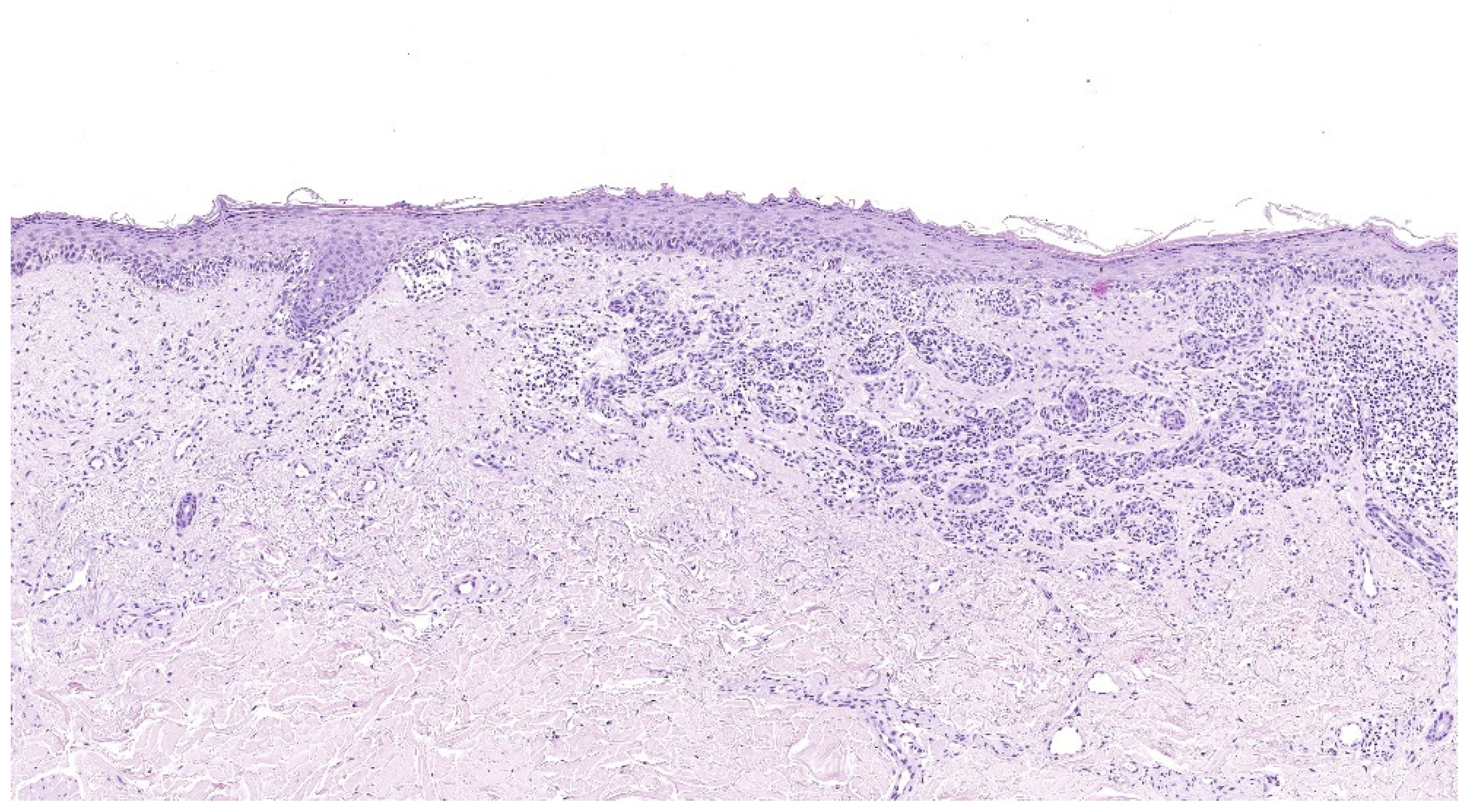
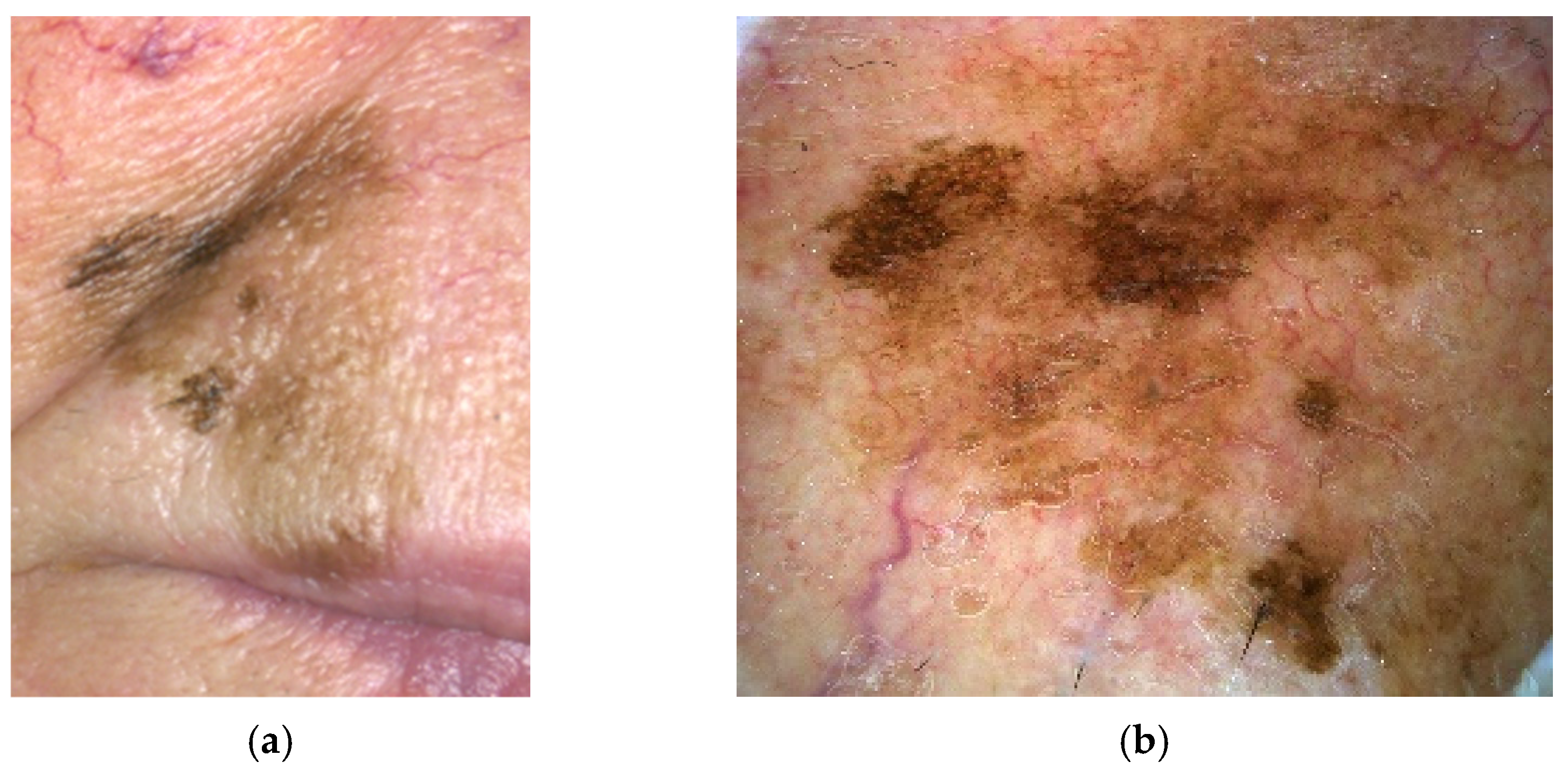
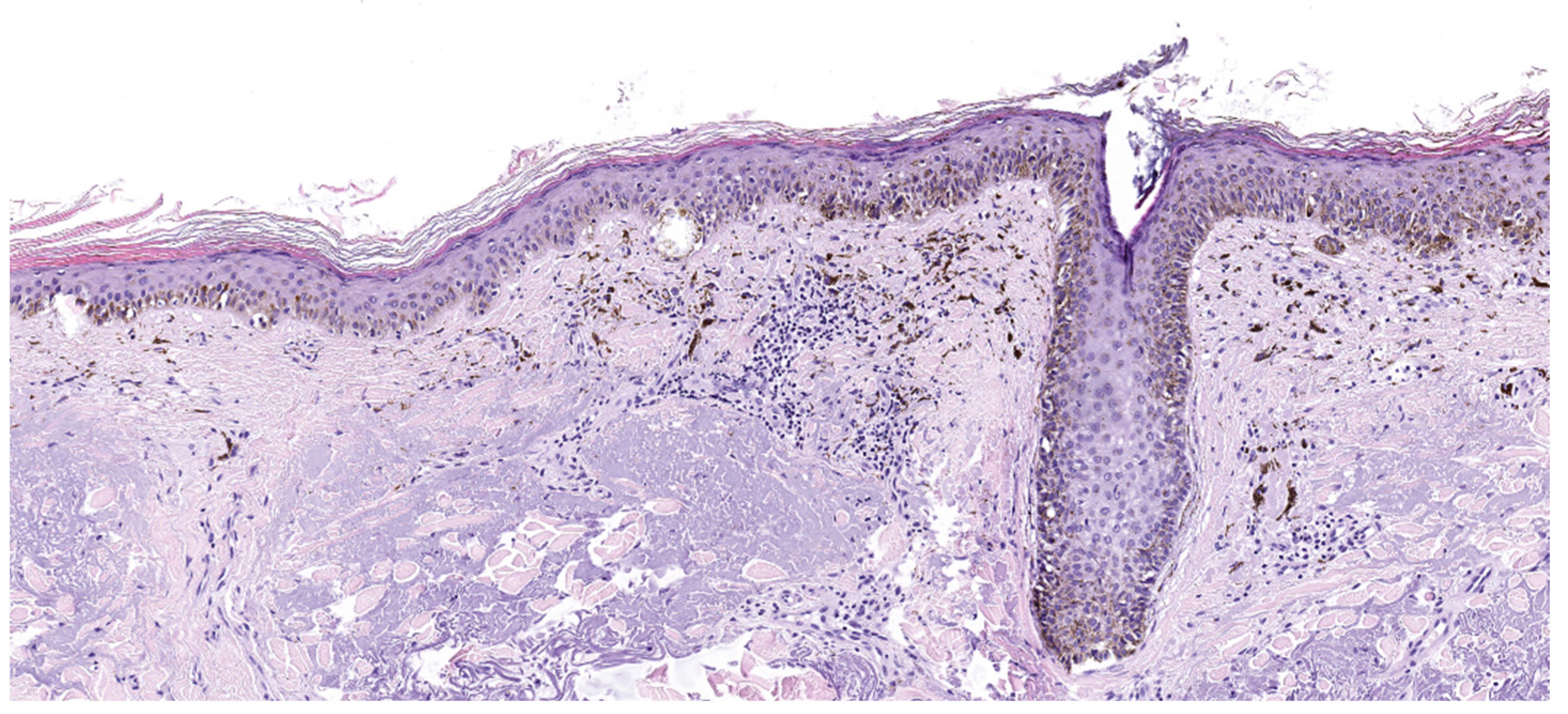
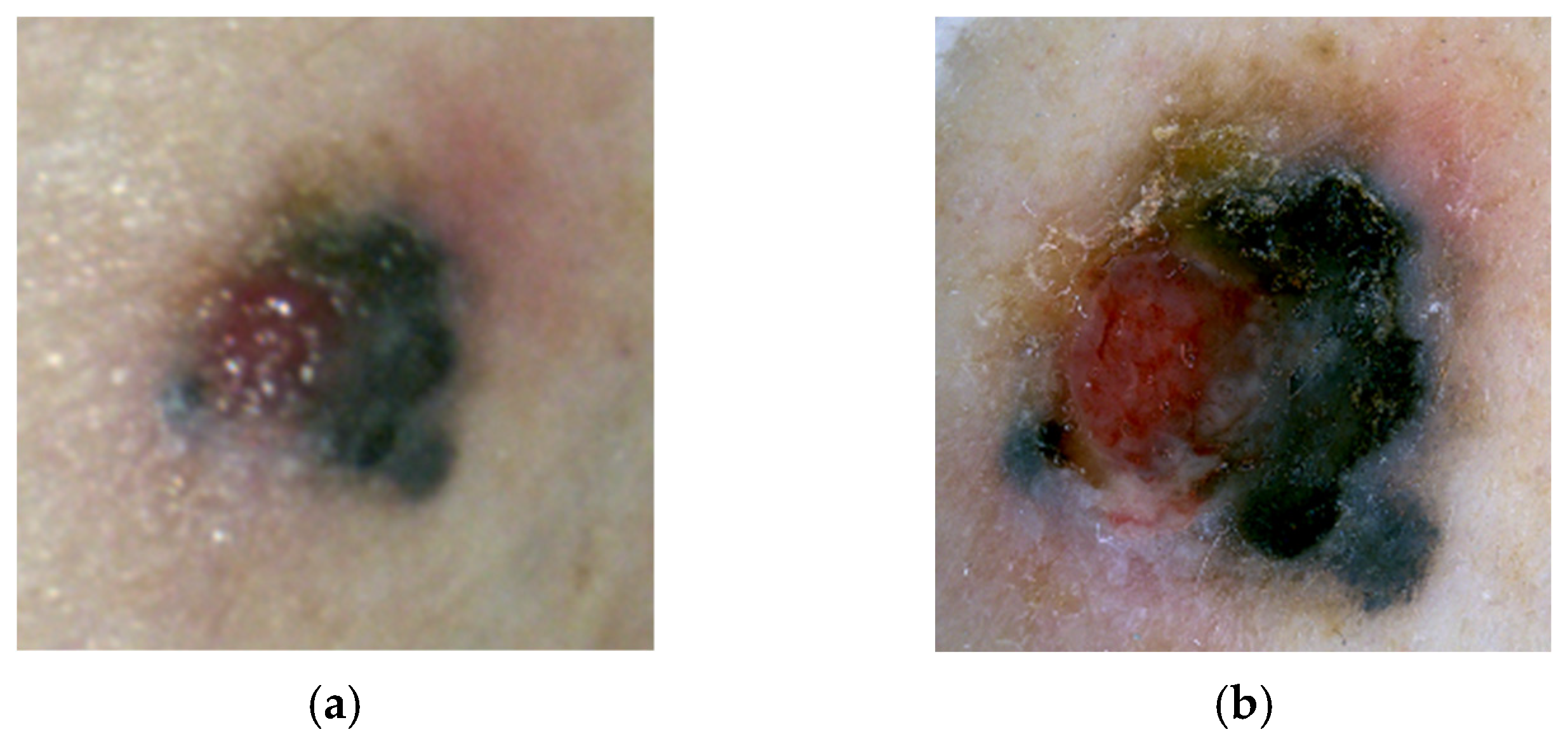
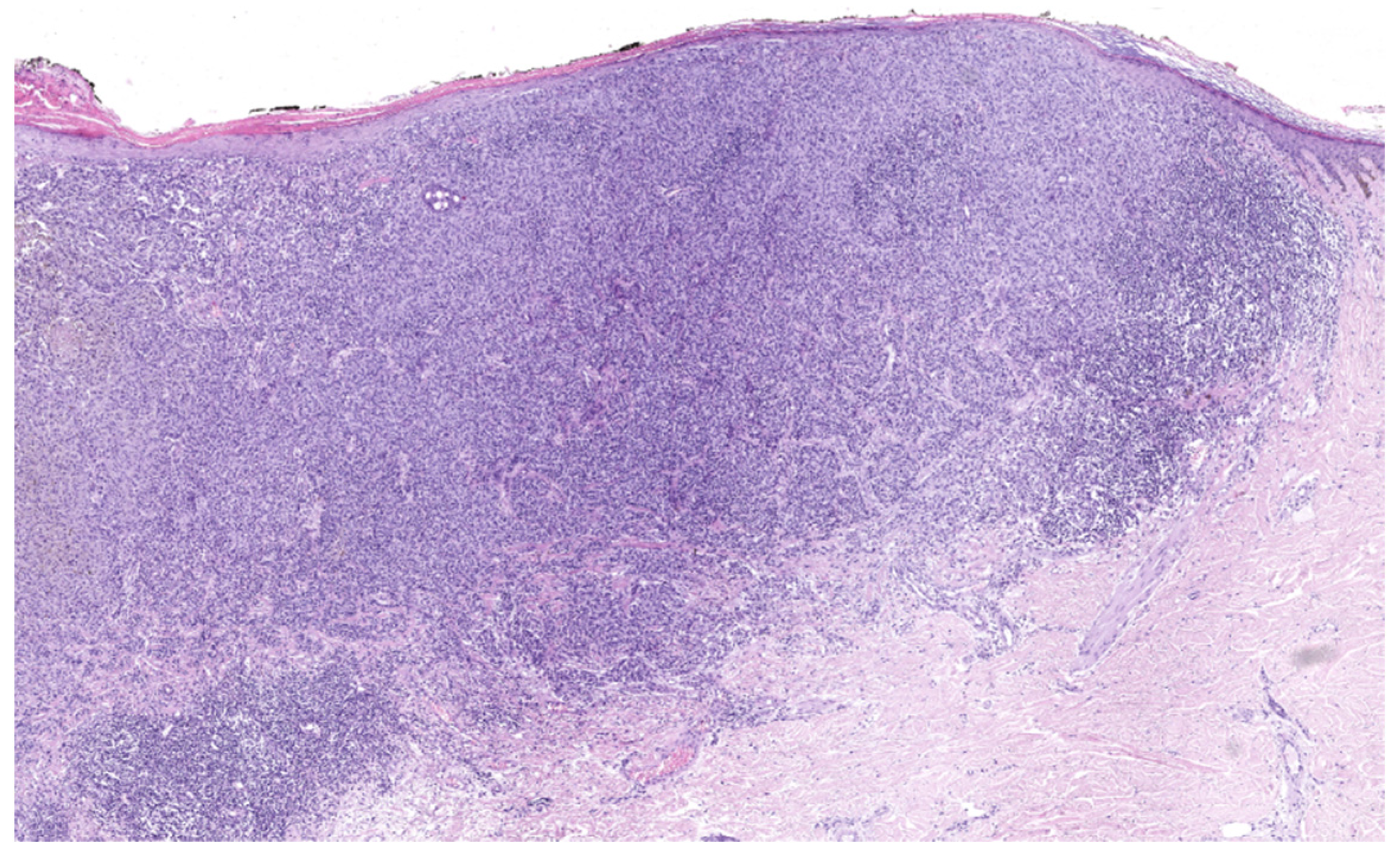
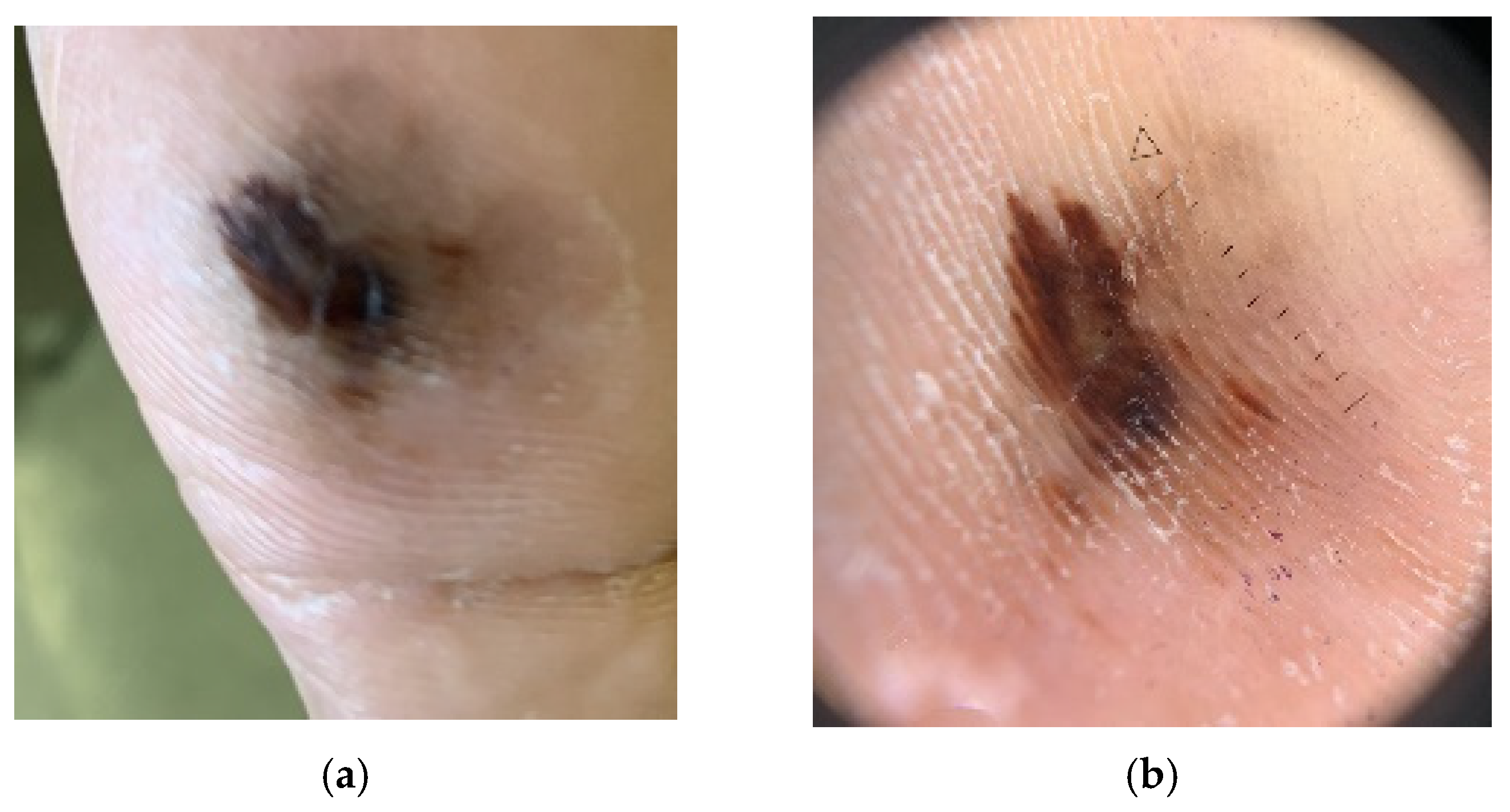
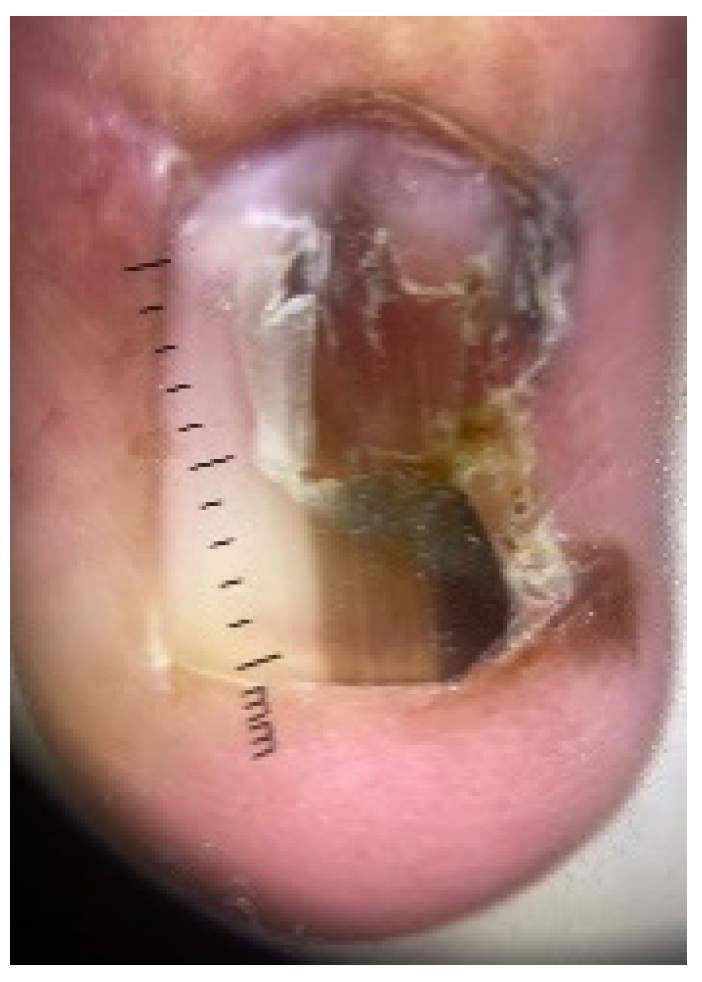
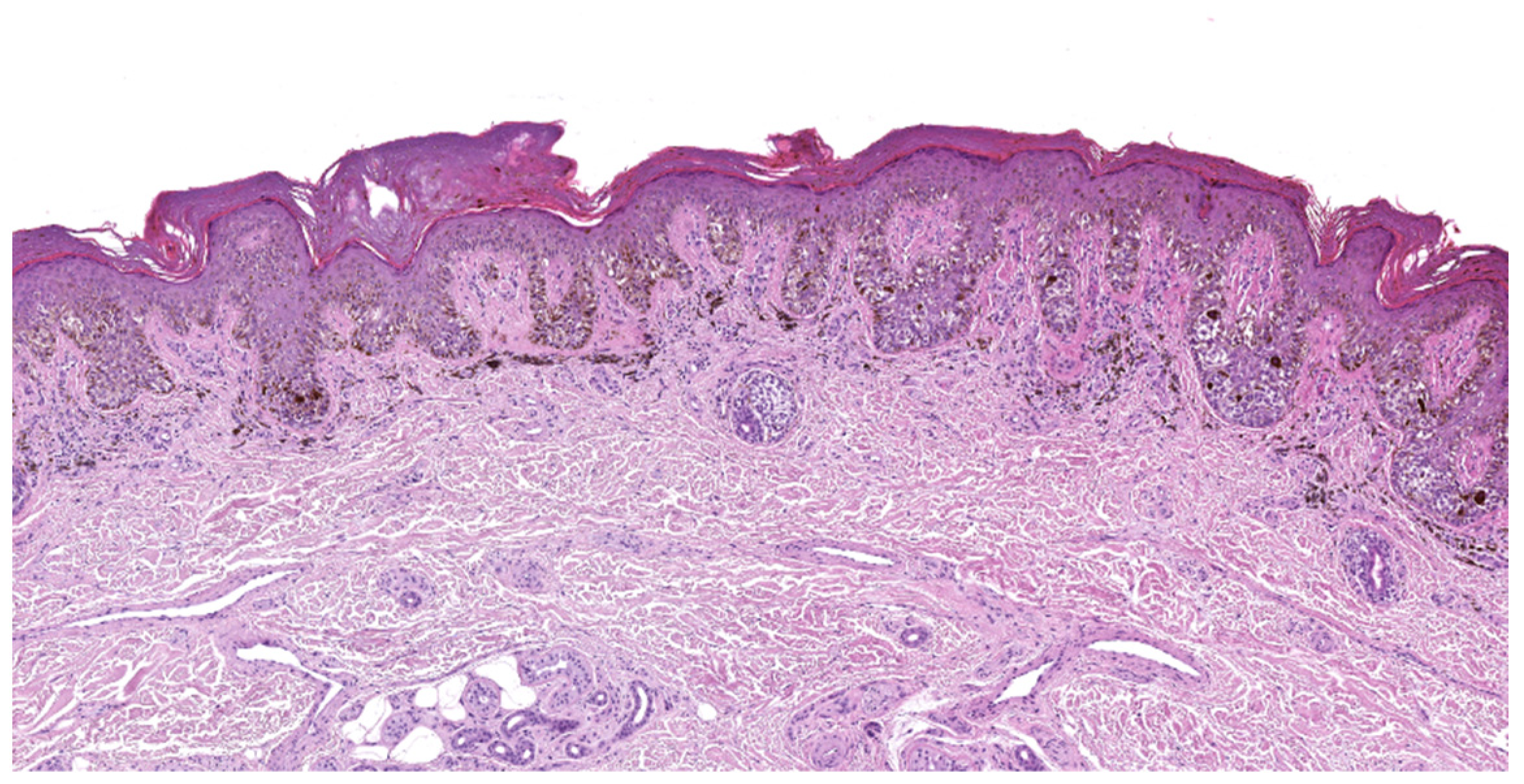
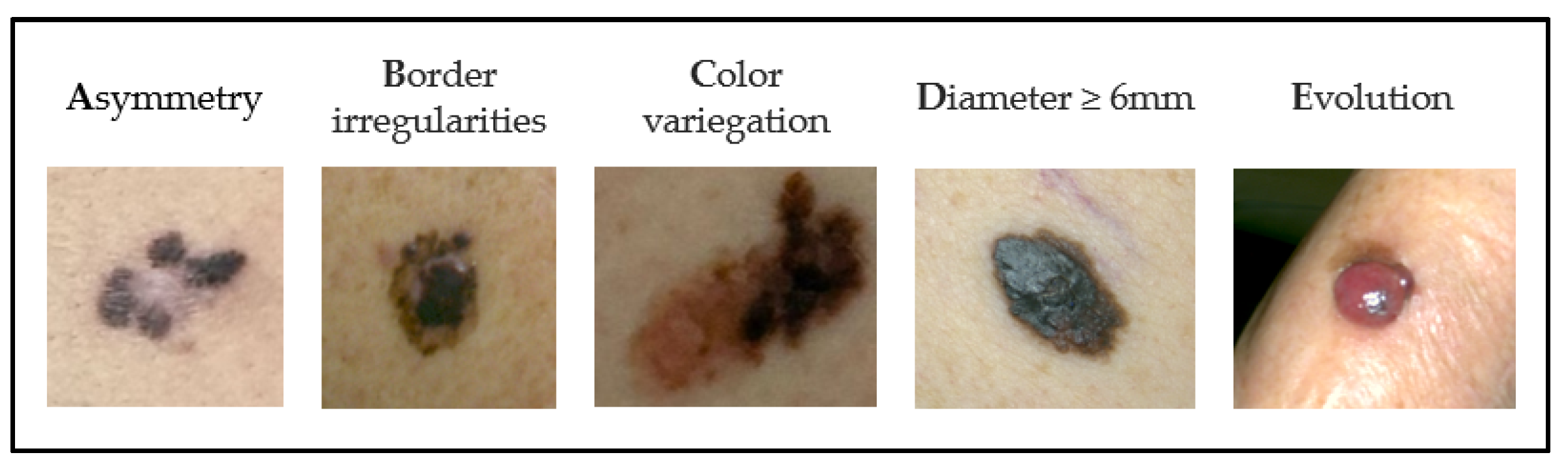
| Low UV Radiation Exposure/ CSD | High UV Radiation Exposure/ CSD | Low to No (or Variable/Incidental) UV Radiation Exposure/CSD | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Pathway | I | II | III | IV | V | VI | VII | VIII | IX |
| Pathway Endpoint | Low-CSD Melanoma/SSM | High-CSD Melanoma/LMM | Desmoplastic Melanoma | Malignant Spitz Tumor/Spitz Melanoma | Acral Melanoma | Mucosal Melanoma | Melanoma Arising in Congenital Naevus | Melanoma in Blue Naevus | Uveal Melanoma |
| Benign Precursor lesion | Nevus | ? IMP | ? IMP | Spitz nevus | ? Acral nevus | ? Melanosis | Congenial nevus | Blue nevus | ? Nevus |
| Intermediate/low-grade dysplasias and melanocytomas | Low-grade dysplasia; BIN; DPN | ? IAMP/dysplasia | ? IAMP/dysplasia | Atypical Spitz tumor (melanocytoma) | IAMPUS/dysplasia | Atypical melanosis/dysplasia/IAMPUS | Nodule in congenial nevus (melanocytoma) | (Atypical) cellular blue naevus (melanocytoma) | ? |
| Intermediate lesions/high-grade dysplasias and melanocytomas | High-grade dysplasia/MIS; BAP1-inactivated melanocytoma, deep penetrating melanocytoma; PEM; MELTUMP | LM (MIS) | MIS | STUMP/ MELTUMP | Acral MIS | Mucosal MIS | MIS in congenital naevus | Atypical cellular blue nevus | ? |
| Malignant neoplasms | Low-CSD melanoma/SSM (VGP); melanoma in BIN; melanoma in DPN; melanoma in PEM | LMM (VGP) | Desmoplastic melanoma | Malignant Spitz tumor/Spitz melanoma (tumorigenic) | Acral melanoma (VGP) | Mucosal lentiginous melanoma (VGP) | Melanoma in congenital naevus (tumorigenic) | Melanoma in blue naevus (tumorigenic) | Uveal melanoma |
| Common Mutations * | BRAF p.V600E, MAP2K1, NRAS + BAP1/CTNNb1/APC, BRAF + PRKAR1A/PRKCA TERT, CDKN2A, TP53, PTEN | NRAS, BRAF (non-p.V600E), KIT, NF1 TERT, CDKN2A, TP53, PTEN, RAC1 | NF1 ERBB2, MAP2K1, MAP3K1, BRAF, EGFR, MET TERT, NFKBIE, NRAS, PIK3CA, PTPN11 | HRAS, ALK, ROS1, RET, NTRK1, NTRK3, BRAF, MET CDKN2A | KIT, NRAS, BRAF, HRAS, KRAS, NTRK3, ALK, NF1, CDKN2A, TERT, CCND1, GAB2 | KIT, NRAS, KRAS, BRAF NF1, CDKN2, SF3B1, CCND1, CDK4, MDM2 | NRAS, BRAF p.V600E (small lesions), BRAF | GNAQ, GNA11, CYSLTR2 BAP1, EIF1AX, SF3B1 | GNA11, GNAQ, CYSLTR2, PLCB4 BAP1, SF3B1, EIF1AX |
| Clinical Features | Score |
|---|---|
| Major Criteria | |
| 1. Change in size/new lesion. | 2 |
| 2. Change in shape/irregular border. | 2 |
| 3. Change in color/irregular pigmentation. | 2 |
| Minor Criteria | |
| 4. Diameter ≥ 7 mm. | 1 |
| 5. Inflammation. | 1 |
| 6. Crusting or bleeding. | 1 |
| 7. Sensory change/itch. | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caraviello, C.; Nazzaro, G.; Tavoletti, G.; Boggio, F.; Denaro, N.; Murgia, G.; Passoni, E.; Benzecry Mancin, V.; Marzano, A.V. Melanoma Skin Cancer: A Comprehensive Review of Current Knowledge. Cancers 2025, 17, 2920. https://doi.org/10.3390/cancers17172920
Caraviello C, Nazzaro G, Tavoletti G, Boggio F, Denaro N, Murgia G, Passoni E, Benzecry Mancin V, Marzano AV. Melanoma Skin Cancer: A Comprehensive Review of Current Knowledge. Cancers. 2025; 17(17):2920. https://doi.org/10.3390/cancers17172920
Chicago/Turabian StyleCaraviello, Camila, Gianluca Nazzaro, Gianluca Tavoletti, Francesca Boggio, Nerina Denaro, Giulia Murgia, Emanuela Passoni, Valentina Benzecry Mancin, and Angelo Valerio Marzano. 2025. "Melanoma Skin Cancer: A Comprehensive Review of Current Knowledge" Cancers 17, no. 17: 2920. https://doi.org/10.3390/cancers17172920
APA StyleCaraviello, C., Nazzaro, G., Tavoletti, G., Boggio, F., Denaro, N., Murgia, G., Passoni, E., Benzecry Mancin, V., & Marzano, A. V. (2025). Melanoma Skin Cancer: A Comprehensive Review of Current Knowledge. Cancers, 17(17), 2920. https://doi.org/10.3390/cancers17172920








