MiRNA Profiling in Premalignant Lesions and Early Glottic Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. Molecular Analysis
2.3. Statistical Analysis
3. Results
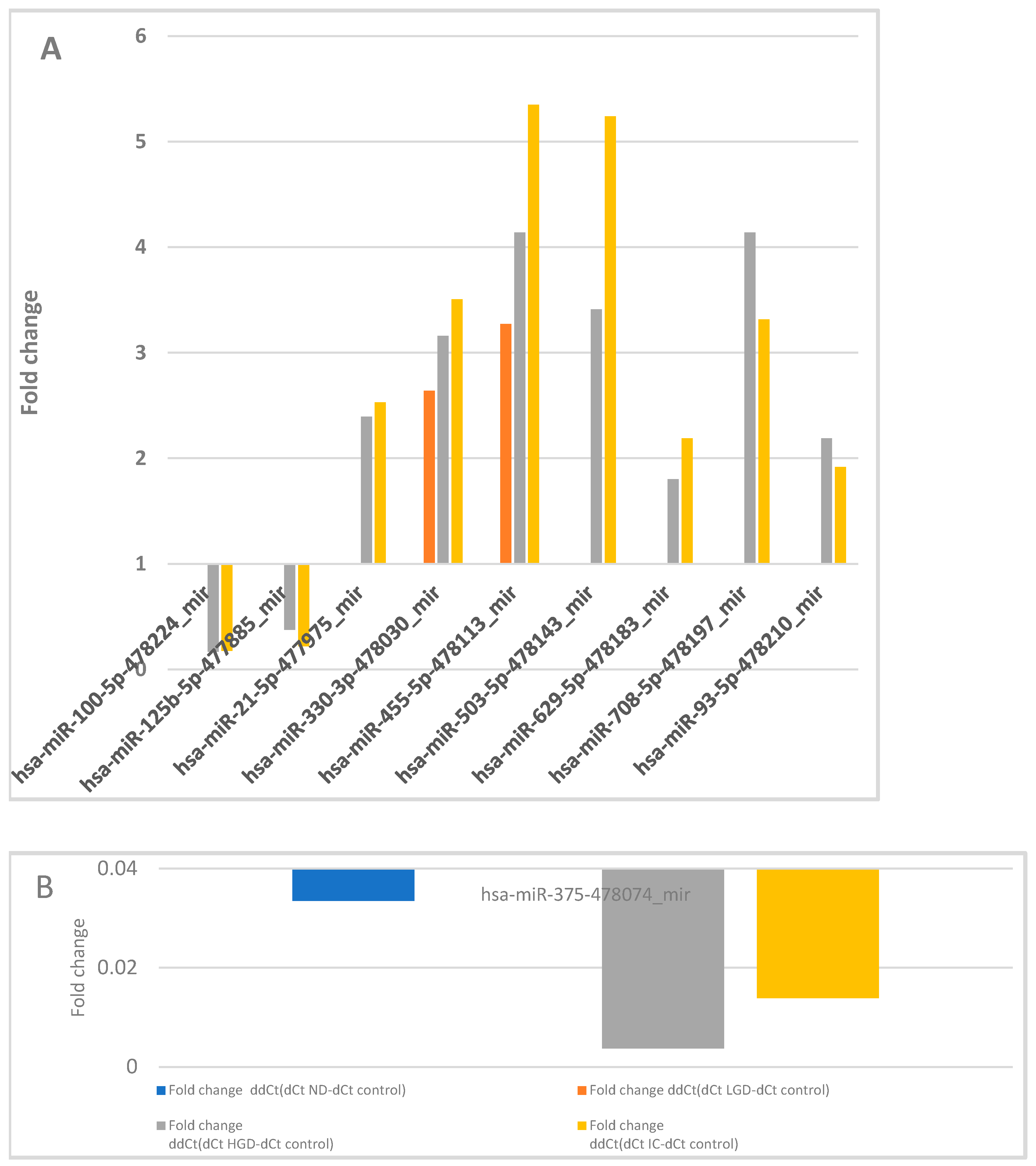
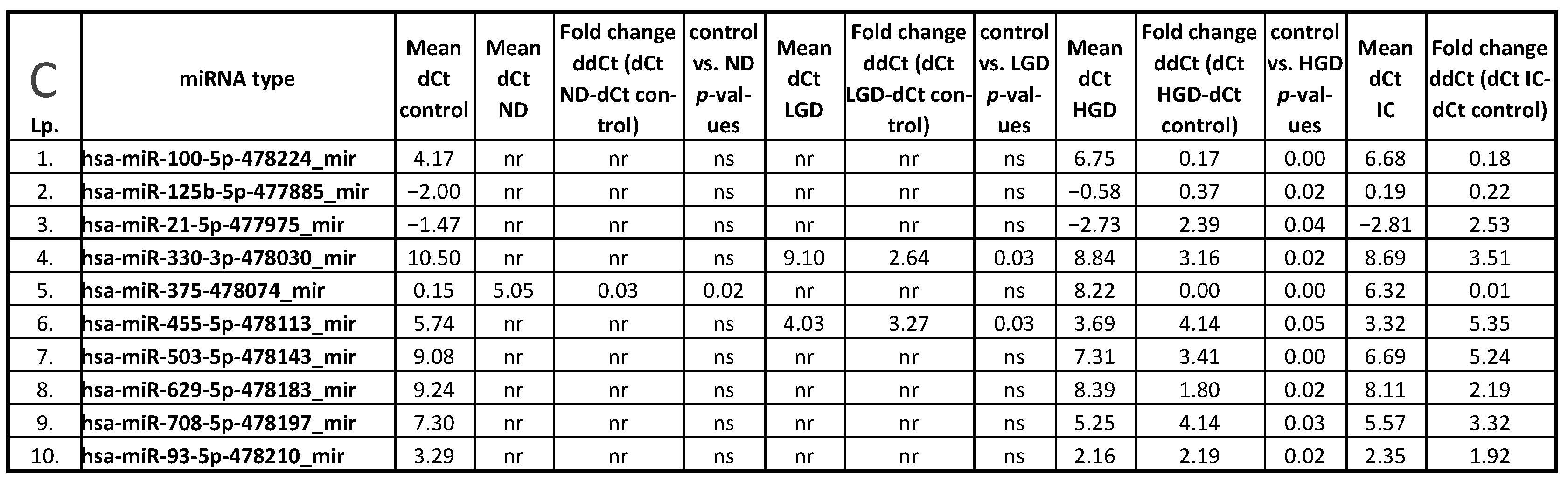
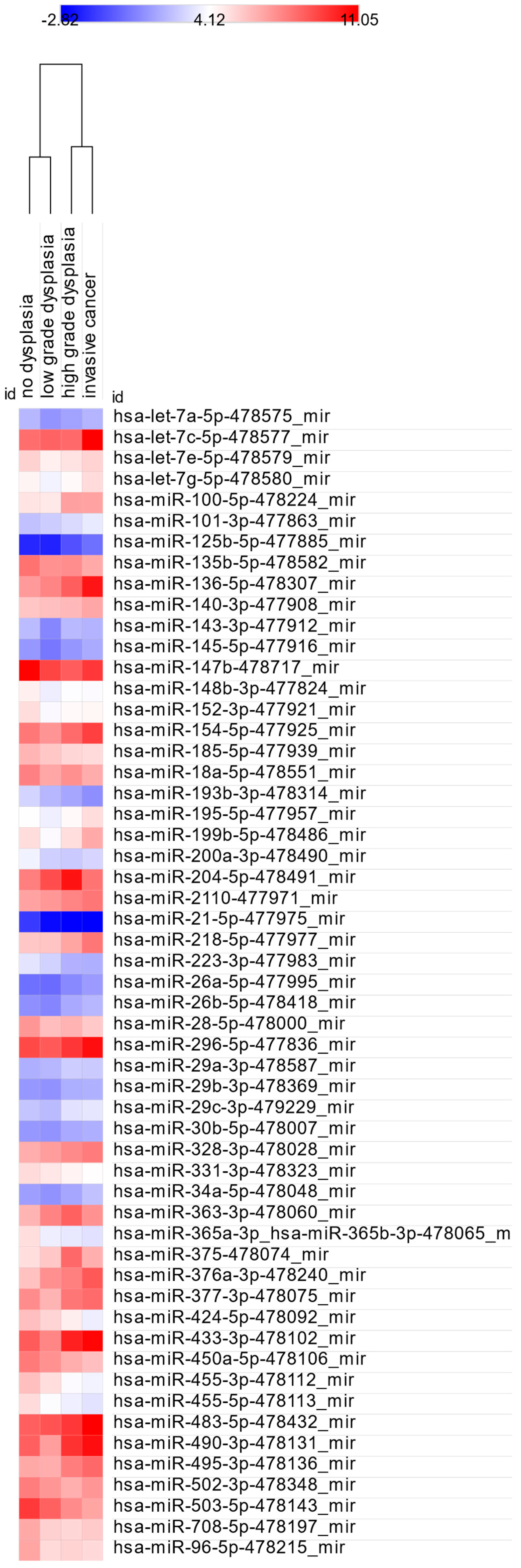
3.1. miRNA Expression: Control vs. ND, LGD, HGD, and IC
3.2. miRNA Expression: ND vs. LGD, HGD, and IC
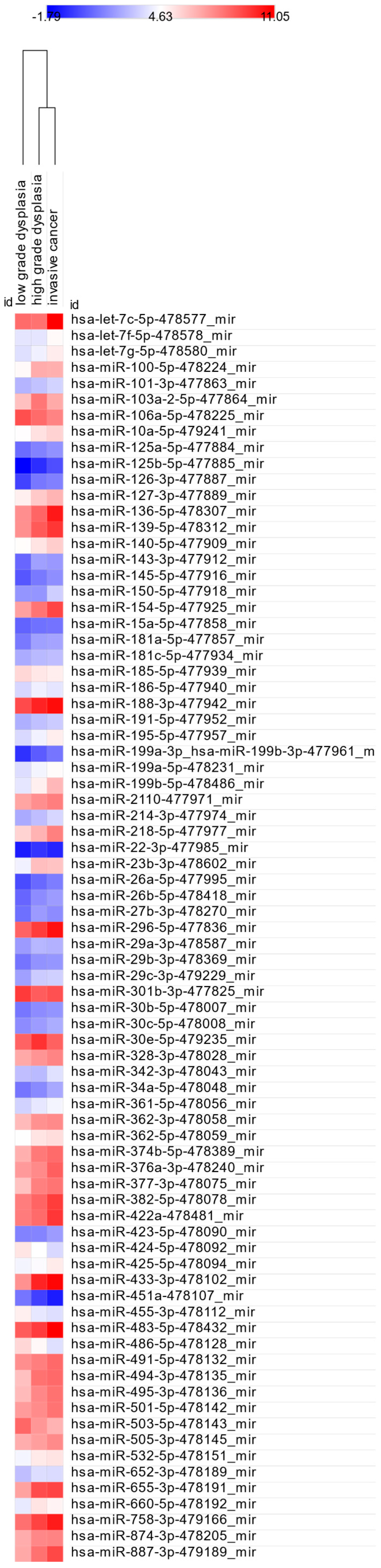
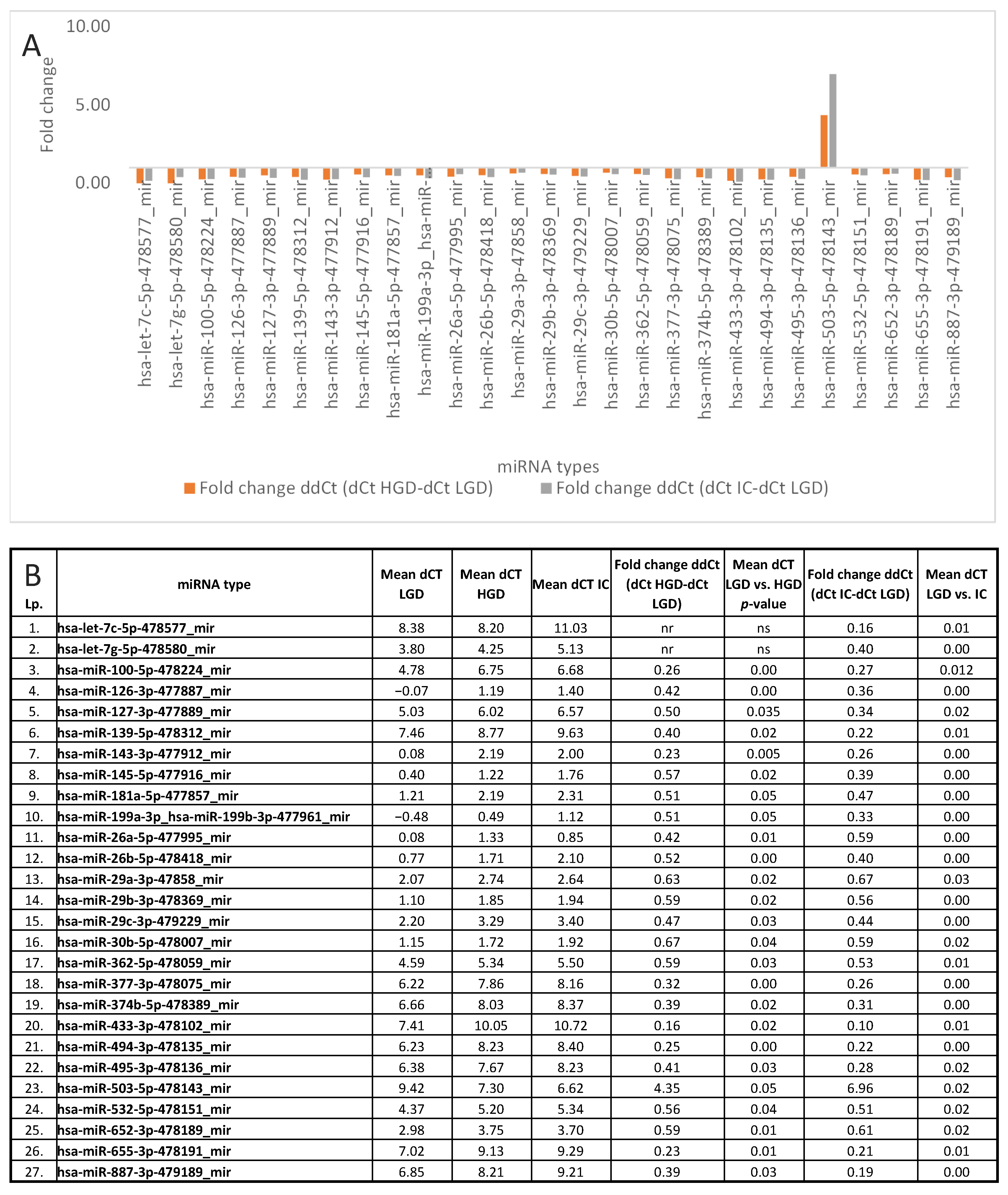
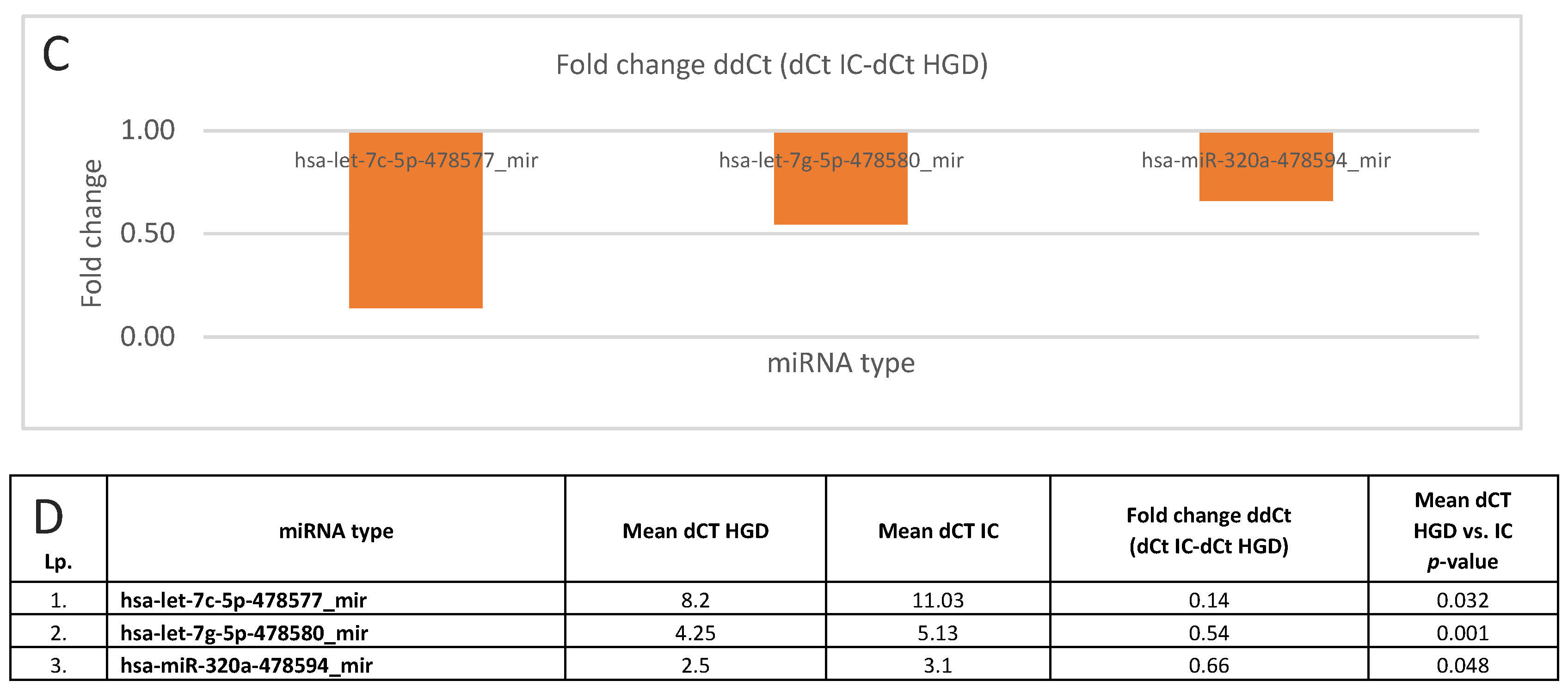
3.3. miRNA Expression: LGD vs. HGD, LGD vs. IC, and HGD vs. IC
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cancer Today. Available online: https://gco.iarc.who.int/today/en/dataviz/bars?types=0_1&mode=population https://gco.iarc.who.int/media/globocan/factsheets/cancers/14-larynx-fact-sheet.pdf (accessed on 20 May 2025).
- Cancer Tomorrow. Available online: https://gco.iarc.who.int/tomorrow/en/dataviz/bars?cancers=14&populations=903_904_905_908_909_935_900 (accessed on 20 May 2025).
- Hegazy, M.; Elkady, M.A.; Yehia, A.M.; Elsakka, E.G.E.; Abulsoud, A.I.; Abdelmaksoud, N.M.; Elshafei, A.; Abdelghany, T.M.; Elkhawaga, S.Y.; Ismail, A.; et al. The role of miRNAs in laryngeal cancer pathogenesis and therapeutic resistance - A focus on signaling pathways interplay. Pathol. Res. Pract. 2023, 246, 154510. [Google Scholar] [CrossRef]
- Zidar, N.; Gale, N. Update from the 5th Edition of the World Health Organization Classification of Head and Neck Tumors: Hypopharynx, Larynx, Trachea and Parapharyngeal Space. Head Neck Pathol. 2022, 16, 31–39. [Google Scholar] [CrossRef]
- van Hulst, A.M.; Kroon, W.; van der Linden, E.S.; Nagtzaam, L.; Ottenhof, S.R.; Wegner, I. Grade of dysplasia and malignant transformation in adults with premalignant laryngeal lesions. Head. Neck. 2016, 38 (Suppl. Sl), E2284–E2290. [Google Scholar] [CrossRef]
- Li, P.; Liu, H.; Wang, Z.; He, F.; Wang, H.; Shi, Z.; Yang, A.; Ye, J. MicroRNAs in laryngeal cancer: Implications for diagnosis, prognosis and therapy. Am. J. Transl.Res. 2016, 8, 1935–1944. [Google Scholar]
- Yu, X.; Li, Z. The role of microRNAs expression in laryngeal cancer. Oncotarget 2015, 6, 23297–23305. [Google Scholar] [CrossRef] [PubMed]
- Shiah, S.G.; Chou, S.T.; Chang, J.Y. MicroRNAs: Their Role in Metabolism, Tumor Microenvironment, and Therapeutic Implications in Head and Neck Squamous Cell Carcinoma. Cancers 2021, 13, 5604. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, S.; Li, K.; Qi, J.; Liu, C.; Zong, L.; Zhang, Y.; Zhao, J.; Zhai, X.; Li, J.; et al. Evaluation of microRNA expression profiling in highly metastatic laryngocarcinoma cells. Acta Otolaryngol. 2018, 138, 1105–1111. [Google Scholar] [CrossRef]
- Wu, Y.; Yu, J.; Ma, Y.; Wang, F.; Liu, H. miR-148a and miR-375 may serve as predictive biomarkers for early diagnosis of laryngeal carcinoma. Oncol. Lett. 2016, 12, 871–878. [Google Scholar] [CrossRef]
- Wei, L.; Mao, M.; Liu, H. Droplet digital PCR and qRT-PCR to detect circulating miR-21 in laryngeal squamous cell carcinoma and pre-malignant laryngeal lesions. Acta Otolaryngol. 2016, 136, 923–932. [Google Scholar] [CrossRef]
- Tuncturk, F.R.; Akalin, I.; Uzun, L.; Zenginkinet, T.J. Comparison of miRNA expressions among benign, premalignant and malignant lesions of the larynx: Could they be transformation biomarkers? Otolaryngol. Head Neck Surg. 2021, 50, 14. [Google Scholar] [CrossRef] [PubMed]
- Dave, V.P.; Ngo, T.A.; Pernestig, A.K.; Tilevik, D.; Kant, K.; Nguyen, T.; Wolff, A.; Bang, D.D. MicroRNA amplification and detection technologies: Opportunities and challenges for point of care diagnostics. Lab. Invest. 2019, 99, 452–469. [Google Scholar] [CrossRef]
- Wang, H. A Review of Nanotechnology in microRNA Detection and Drug Delivery. Cells 2024, 13, 1277. [Google Scholar] [CrossRef]
- Nafari, N.B.; Zamani, M.; Mosayyebi, B. Recent advances in lateral flow assays for MicroRNA detection. Clin. Chim. Acta. 2025, 567, 120096. [Google Scholar] [CrossRef]
- Hwang, J.; Min, B.H.; Jang, J.; Kang, S.Y.; Bae, H.; Jang, S.S.; Kim, J.I.; Kim, K.M. MicroRNA Expression Profiles in Gastric Carcinogenesis. Sci. Rep. 2018, 8, 14393. [Google Scholar] [CrossRef]
- Shirjang, S.; Mansoori, B.; Mohammadi, A.; Shajari, N.; Duijf, P.H.; Najafi, S.; Abedi Gaballu, F.; Nofouzi, K.; Baradaran, B. MiR-330 Regulates Colorectal Cancer Oncogenesis by Targeting BACH1. Adv. Pharm. Bull. 2020, 10, 444–451. [Google Scholar] [CrossRef]
- Sequeira, J.P.; Constâncio, V.; Salta, S.; Lobo, J.; Barros-Silva, D.; Carvalho-Maia, C.; Rodrigues, J.; Braga, I.; Henrique, R.; Jerónimo, C. LiKidMiRs: A ddPCR-Based Panel of 4 Circulating miRNAs for Detection of Renal Cell Carcinoma. Cancers 2022, 14, 858. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Wu, J.; Huang, Q.; Ren, J.; Huang, J.; Yuan, W.; Xuekun, H.; Yuhuan, W.; Congxian, C.; Shengwei, X.; et al. The effects of miR-375 expression in NSCLC via the 14-3-3ζ/ERK/MYC pathway. Oncol. Transl. Med. 2018, 4, 196–202. [Google Scholar] [CrossRef]
- Gan, J.; Liu, S.; Zhang, Y.; He, L.; Bai, L.; Liao, R.; Zhao, J.; Guo, M.; Jiang, W.; Li, J.; et al. MicroRNA-375 is a therapeutic target for castration-resistant prostate cancer through the PTPN4/STAT3 axis. Exp. Mol. Med. 2022, 54, 1290–1305. [Google Scholar] [CrossRef] [PubMed]
- Galuppini, F.; Censi, S.; Moro, M.; Carraro, S.; Sbaraglia, M.; Iacobone, M.; Fassan, M.; Mian, C.; Pennelli, G. MicroRNAs in Medullary Thyroid Carcinoma: A State of the Art Review of the Regulatory Mechanisms and Future Perspectives. Cells 2021, 10, 955. [Google Scholar] [CrossRef]
- Fan, K.; Ritter, C.; Nghiem, P.; Blom, A.; Verhaegen, M.E.; Dlugosz, A.; Ødum, N.; Woetmann, A.; Tothill, R.W.; Hicks, R.J.; et al. Circulating Cell-Free miR-375 as Surrogate Marker of Tumor Burden in Merkel Cell Carcinoma. Clin. Cancer Res. 2018, 24, 5873–5882. [Google Scholar] [CrossRef] [PubMed]
- Zellinger, B.; Bodenhofer, U.; Engländer, I.A.; Kronberger, C.; Strasser, P.; Grambozov, B.; Fastner, G.; Stana, M.; Reitsamer, R.; Sotlar, K.; et al. Hsa-miR-375/RASD1 Signaling May Predict Local Control in Early Breast Cancer. Genes 2020, 11, 1404. [Google Scholar] [CrossRef]
- Chawra, H.S.; Agarwal, M.; Mishra, A.; Chandel, S.S.; Singh, R.P.; Dubey, G.; Kukreti, N.; Singh, M. MicroRNA-21’s role in PTEN suppression and PI3K/AKT activation: Implications for cancer biology. Pathol. Res. Pract. 2024, 254, 155091. [Google Scholar] [CrossRef]
- Liu, C.; Tong, Z.; Tan, J.; Xin, Z.; Wang, Z.; Tian, L. MicroRNA-21-5p targeting PDCD4 suppresses apoptosis via regulating the PI3K/AKT/FOXO1 signaling pathway in tongue squamous cell carcinoma. Exp. Ther. Med. 2019, 18, 3543–3551. [Google Scholar] [CrossRef]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; de La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids. 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Tang, M.; Xu, X.; Jiang, B.; Wei, Y.; Qian, H.; Lu, X. MiR-143-3p suppresses cell proliferation, migration, and invasion by targeting Melanoma-Associated Antigen A9 in laryngeal squamous cell carcinoma. J. Cell Biochem. 2019, 120, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Cao, H. MicroRNA-143-3p suppresses cell growth and invasion in laryngeal squamous cell carcinoma via targeting the k-Ras/Raf/MEK/ERK signaling pathway. Int. J. Oncol. 2019, 54, 689–701. [Google Scholar] [CrossRef]
- Cai, B.; Qu, X.; Kan, D.; Luo, Y. miR-26a-5p suppresses nasopharyngeal carcinoma progression by inhibiting PTGS2 expression. Cell Cycle 2022, 21, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ge, Z.; Chen, S.; Li, S.; Zhang, X.; Hu, J.; Guo, W.; Wang, Y. miR-26a-5p Suppresses Wnt/β-Catenin Signaling Pathway by Inhibiting DNMT3A-Mediated SFRP1 Methylation and Inhibits Cancer Stem Cell-Like Properties of NSCLC. Dis. Markers. 2022, 2022, 7926483. [Google Scholar] [CrossRef]
- Zhu, W.J.; Yan, Y.; Zhang, J.W.; Tang, Y.D.; Han, B. Effect and Mechanism of miR-26a-5p on Proliferation and Apoptosis of Hepatocellular Carcinoma Cells. Cancer Manag. Res. 2020, 12, 3013–3022. [Google Scholar] [CrossRef]
- Yamada, Y.; Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Okato, A.; Idichi, T.; Arai, T.; Sugawara, S.; Katada, K.; Okamoto, Y.; et al. Passenger strand of miR-145-3p acts as a tumor-suppressor by targeting MYO1B in head and neck squamous cell carcinoma. Int. J. Oncol. 2018, 52, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Ghorai, S.; Panda, K.; Santiago, M.J.; Aggarwal, S.; Wang, T.; Rahman, I.; Chinnapaiyan, S.; Unwalla, H.J. Dr. Jekyll or Mr. Hyde: The multifaceted roles of miR-145-5p in human health and disease. Noncoding RNA Res. 2024, 11, 22–37. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sun, X.; Wang, Z.M.; Song, Y.; Tai, X.H.; Ji, W.Y.; Gu, H. MicroRNA-126 modulates the tumor microenvironment by targeting calmodulin-regulated spectrin-associated protein 1 (Camsap1). Int. J. Oncol. 2014, 44, 1678–1684. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Kawasaki, H.; Luce, A.; Cossu, A.M.; Misso, G.; Scrima, M.; Bocchetti, M.; Ricciardiello, F.; Caraglia, M.; Zappavigna, S. Insight toward the MicroRNA Profiling of Laryngeal Cancers: Biological Role and Clinical Impact. Int. J. Mol. Sci. 2020, 21, 3693. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hu, B.; Wang, Y.; Li, Z.; Wu, J.; Yang, Y.; Wei, Y.; Peng, X.; Chen, H.; Chen, R.; et al. miR-216a-5p inhibits malignant progression in small cell lung cancer: Involvement of the Bcl-2 family proteins. Cancer Manag. Res. 2018, 10, 4735–4745. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, S.; Min, Z.; Yu, Z.; Zhang, H.; Jiao, J. Knockdown of circ_0011946 targets miR-216a-5p/BCL2L2 axis to regulate proliferation, migration, invasion and apoptosis of oral squamous cell carcinoma cells. BMC Cancer 2021, 21, 1085. [Google Scholar] [CrossRef]
- Shen, Z.; Zhou, R.; Liu, C.; Wang, Y.; Zhan, W.; Shao, Z.; Liu, J.; Zhang, F.; Xu, L.; Zhou, X.; et al. MicroRNA-105 is involved in TNF-α-related tumor microenvironment enhanced colorectal cancer progression. Cell Death Dis. 2017, 8, 3213. [Google Scholar] [CrossRef]
- Li, M.; Zhang, S.; Ma, Y.; Yang, Y.; An, R. Role of hsa-miR-105 during the pathogenesis of paclitaxel resistance and its clinical implication in ovarian cancer. Oncol. Rep. 2021, 45, 84. [Google Scholar] [CrossRef]
- Shuang, Y.; Zhou, X.; Li, C.; Huang, Y.; Zhang, L. MicroRNA-503 serves an oncogenic role in laryngeal squamous cell carcinoma via targeting programmed cell death protein 4. Mol. Med. Rep. 2017, 16, 5249–5256. [Google Scholar] [CrossRef]
- Caporali, A.; Meloni, M.; Völlenkle, C.; Bonci, D.; Sala-Newby, G.B.; Addis, R.; Spinetti, G.; Losa, S.; Masson, R.; Baker, A.H.; et al. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation 2011, 123, 282–291. [Google Scholar] [CrossRef]



| No | Mean Age (Years) ±SD | Sex (M/F) | |
|---|---|---|---|
| All patients | 31 | 64.16 ± 10.43 | 20/11 |
| Control group | 3 | 48.67 ± 6.55 | 2/1 |
| No dysplasia (ND) | 7 | 70.43 ± 6.3 | 1/6 |
| Low-grade dysplasia (LGD) | 7 | 59.14 ± 8.48 | 5/2 |
| High-grade dysplasia (HGD) | 7 | 66.57 ± 10.2 | 6/1 |
| Invasive cancer (IC) | 7 | 67.14 ± 8.13 | 6/1 |
| 5 | 66.8 ± 5.77 | 4/1 |
| 2 | 70.5 ± 11.5 | 2/0 |
| miRNA Identified only in control group | No of samples with detected miRNA Type | Mean dCt |
| hsa-miR-208b-3p-477806_mir | 1 | 5.82 |
| hsa-miR-216a-5p-477976_mir | 2 | 10.83 |
| hsa-miR-133b-480871_mir | 1 | 4.98 |
| hsa-miR-499a-3p-478948_mir | 1 | 9.26 |
| hsa-miR-876-5p-479187_mir | 1 | 9.70 |
| hsa-miR-208a-3p-477819_mir | 1 | 8.48 |
| hsa-miR-488-3p-478129_mir | 2 | 10.11 |
| miRNA identified only in IC and HGD patients | No of samples with detected miRNA type | Mean dCt |
| hsa-miR-105-5p-477865_mir | 6 | 9.37 |
| hsa-miR-516a-5p-478978_mir | 6 | 10.11 |
| miRNA identified only in IC, HGD, and LGD patients | No of samples with detected miRNA type | Mean dCt |
| hsa-miR-212-3p-478318_mir | 10 | 10.13 |
| hsa-miR-548d-5p-480870_mir | 11 | 10.63 |
| miRNA Type | Control vs. ND p-Value | Control vs. LGD p-Value | Control vs. HGD p-Value | Control vs. IC p-Value |
|---|---|---|---|---|
| hsa-let-7c-5p-478577_mir | 0.029 | |||
| hsa-let-7g-5p-478580_mir | 0.007 | |||
| hsa-let-7i-5p-478375_mir | 0.04 | |||
| hsa-miR-100-5p-478224_mir | 0.002 | 0.012 | ||
| hsa-miR-106a-5p-478225_mir | 0.001 | |||
| hsa-miR-10a-5p-479241_mir | 0.027 | |||
| hsa-miR-125b-5p-477885_mir | 0.015 | 0.003 | ||
| hsa-miR-127-3p-477889_mir | 0.024 | |||
| hsa-miR-134-5p-477901_mir | 0.044 | |||
| hsa-miR-136-5p-478307_mir | 0.014 | |||
| hsa-miR-138-5p-477905_mir | 0.03 | |||
| hsa-miR-140-3p-477908_mir | 0.029 | |||
| hsa-miR-142-3p-477910_mir | 0.023 # | |||
| hsa-miR-142-5p-477911_mir | 0.011 | |||
| hsa-miR-145-5p-477916_mir | 0.046 | |||
| hsa-miR-152-3p-477921_mir | 0.036 # | |||
| hsa-miR-154-5p-477925_mir | 0.016 | |||
| hsa-miR-155-5p-477927_mir | 0.031 | |||
| hsa-miR-181a-5p-477857_mir | 0.028 | |||
| hsa-miR-181b-5p-478583_mir | 0.028 | 0.035 | ||
| hsa-miR-181c-5p-477934_mir | 0.028 | |||
| hsa-miR-18a-5p-478551_mir | 0.03 | 0.027 | ||
| hsa-miR-18b-5p-478584_mir | 0.032 | |||
| hsa-miR-193b-3p-478314_mir | 0.036 # | |||
| hsa-miR-199a-3p_hsa-miR-199b-3p-477961_mir | 0.045 | |||
| hsa-miR-199b-5p-478486_mir | 0.02 | |||
| hsa-miR-204-5p-478491_mir | 0.048 | |||
| hsa-miR-21-5p-477975_mir | 0.037 | 0.034 | ||
| hsa-miR-23a-3p-478532_mir | 0.014 | |||
| hsa-miR-26b-5p-478418_mir | 0.021 | |||
| hsa-miR-296-5p-477836_mir | 0.048 | |||
| hsa-miR-301a-3p-477815_mir | 0.046 | |||
| hsa-miR-301b-3p-477825_mir | 0.002 | |||
| hsa-miR-30e-5p-479235_mir | 0.038 | 0.039 | ||
| hsa-miR-330-3p-478030_mir | 0.033 | 0.016 | 0.019 | |
| hsa-miR-363-3p-478060_mir | 0.013 | 0.004 | ||
| hsa-miR-375-478074_mir | 0.018 | 0.002 | 0.011 | |
| hsa-miR-377-3p-478075_mir | 0.042 | |||
| hsa-miR-381-3p-477816_mir | 0.048 | |||
| hsa-miR-382-5p-478078_mir | 0.038 | |||
| hsa-miR-424-5p-478092_mir | 0.013 | 0.038 | ||
| hsa-miR-433-3p-478102_mir | 0.007 | |||
| hsa-miR-455-3p-478112_mir | 0.040 | |||
| hsa-miR-455-5p-478113_mir | 0.032 | 0.047 | 0.047 | |
| hsa-miR-494-3p-478135_mir | 0.031 | |||
| hsa-miR-495-3p-478136_mir | 0.04 # | |||
| has-miR-499a-5p-478139_mir | 0.041 | |||
| hsa-miR-501-5p-478142_mir | 0.028 | |||
| hsa-miR-502-3p-478348_mir | 0.023 # | |||
| hsa-miR-503-5p-478143_mir | 0.002 | 0.012 | ||
| hsa-miR-504-5p-478144_mir | 0.019 | |||
| hsa-miR-576-5p-478165_mir | 0.029 | |||
| hsa-miR-629-5p-478183_mir | 0.018 | 0.007 | ||
| hsa-miR-653-5p-479134_mir | 0.048 | |||
| hsa-miR-708-5p-478197_mir | 0.030 | 0.042 | ||
| hsa-miR-887-3p-479189_mir | 0.026 | |||
| hsa-miR-93-5p-478210_mir | 0.016 | 0.040 # | ||
| Total number of identified significantly different miRNAs | 4 | 16 | 17 | 38 |
| miRNA Type | ND vs. LGD | ND vs. HGD | ND vs. IC |
|---|---|---|---|
| hsa-let-7a-5p-478575_mir | 0.045 | ||
| hsa-let-7c-5p-478577_mir | 0.006 | ||
| hsa-let-7e-5p-478579_mir | 0.03 # | ||
| hsa-let-7g-5p-478580_mir | 0.029 | 0.016 | |
| hsa-miR-100-5p-478224_mir | 0.003 | 0.012 | |
| hsa-miR-101-3p-477863_mir | 0.013 | ||
| hsa-miR-125b-5p-477885_mir | 0.021 # | 0.005 # | |
| hsa-miR-135b-5p-478582_mir | 0.017 | ||
| hsa-miR-136-5p-478307_mir | 0.007 | ||
| hsa-miR-140-3p-477908_mir | 0.018 | ||
| hsa-miR-143-3p-477912_mir | 0.002 | ||
| hsa-miR-145-5p-477916_mir | 0.045 | ||
| hsa-miR-147b-478717_mir | 0.021 | ||
| hsa-miR-148b-3p-477824_mir | 0.018 | ||
| hsa-miR-152-3p-477921_mir | 0.032 | ||
| hsa-miR-154-5p-477925_mir | 0.014 | ||
| hsa-miR-185-5p-477939_mir | 0.026 | 0.001 | 0.002 # |
| hsa-miR-18a-5p-478551_mir | 0.038 | 0.026 | |
| hsa-miR-193b-3p-478314_mir | 0.045 | ||
| hsa-miR-195-5p-477957_mir | 0.042 | ||
| hsa-miR-199b-5p-478486_mir | 0.003 | ||
| hsa-miR-200a-3p-478490_mir | 0.035 | ||
| hsa-miR-204-5p-478491_mir | 0.026 | ||
| hsa-miR-21-5p-477975_mir | 0.021 | 0.002 | 0.001 |
| hsa-miR-2110-477971_mir | 0.021 | ||
| hsa-miR-218-5p-477977_mir | 0.01 | ||
| hsa-miR-223-3p-477983_mir | 0.049 | 0.041 | |
| hsa-miR-26a-5p-477995_mir | 0.017 | 0.001 | |
| hsa-miR-26b-5p-478418_mir | 0.023 | 0.001 | |
| hsa-miR-28-5p-478000_mir | 0.01 | 0.004 | |
| hsa-miR-296-5p-477836_mir | 0.01 | ||
| hsa-miR-29a-3p498587_mir | 0.021 | 0.026 | |
| hsa-miR-29b-3p-478369_mir | 0.017 | ||
| hsa-miR-29c-3p-479229_mir | 0.018 | ||
| hsa-miR-30b-5p-478007_mir | 0.044 | ||
| hsa-miR-328-3p-478028_mir | 0.025 | ||
| hsa-miR-331-3p-478323_mir | 0.037 | ||
| hsa-miR-34a-5p-478048_mir | 0.03 | ||
| hsa-miR-363-3p-478060_mir | 0.036 | 0.003 | |
| hsa-miR-365a-3p_hsa-miR-365b-3p-478065_mir | 0.15 # | 0.041 # | |
| hsa-miR-375-478074_mir | 0.021 | ||
| has-miR-376a-3p-478240_mir | 0.007 | ||
| hsa-miR-377-3p-478075_mir | 0.041 # | ||
| hsa-miR-424-5p-478092_mir | 0.007 | 0.000. | |
| hsa-miR-433-3p-478102_mir | 0.020 | ||
| hsa-miR-450a-5p-478106_mir | 0.020 | ||
| hsa-miR-455-3p-478112_mir | 0.011 | 0.006 | |
| hsa-miR-455-5p-478113_mir | 0.049 | 0.031 | |
| hsa-miR-483-5p-478432_mir | 0.010 | ||
| hsa-miR-490-3p-478131_mir | 0.037 | ||
| hsa-miR-495-3p-478136_mir | 0.023 # | ||
| hsa-miR-502-3p-478348_mir | 0.049 | ||
| hsa-miR-503-5p-478143_mir | 0.018 | 0.003 | |
| hsa-miR-708-5p-478197_mir | 0.027 | ||
| hsa-miR-96-5p-478215_mir | 0.015 # | 0.03 # | |
| Total number of identified significantly different miRNAs | 16 | 18 | 40 |
| miRNA Type | LGD vs. HGD | LGD vs. IC | HGD vs. IC |
|---|---|---|---|
| hsa-let-7c-5p-478577_mir | 0.009 | 0.032 | |
| hsa-let-7f-5p-478578_mir | 0.04 | ||
| hsa-let-7g-5p-478580_mir | 0.001 | 0.002 # | |
| hsa-miR-100-5p-478224_mir | 0.004 | 0.012 | |
| hsa-miR-101-3p-477863_mir | 0.011 | ||
| hsa-miR-103a-2-5p-477864_mir | 0.026 | ||
| has-miR-106a-5p-478225_mir | 0.021 | ||
| hsa-miR-10a-5p-479241_mir | 0.003 | ||
| hsa-miR-125a-5p-477884_mir | 0.002 | ||
| hsa-miR-125b-5p-477885_mir | 0.047 | ||
| hsa-miR-126-3p-477887_mir | 0.002 | 0.002 | |
| hsa-miR-127-3p-477889_mir | 0.035 | 0.02 | |
| hsa-miR-136-5p-478307_mir | 0.007 | ||
| hsa-miR-139-5p-478312_mir | 0.015 | 0.01 | |
| hsa-miR-140-5p-477909_mir | 0.041 # | ||
| hsa-miR-143-3p-477912_mir | 0.005 | 0.001 | |
| hsa-miR-145-5p-477916_mir | 0.017 | 0.003 | |
| hsa-miR-150-5p-477918_mir | 0.037 | ||
| hsa-miR-154-5p-477925_mir | 0.001 | ||
| hsa-miR-15a-5p-477858_mir | 0.049 | ||
| hsa-miR-181a-5p-477857_mir | 0.046 | 0.010 | |
| hsa-miR-181c-5p-477934_mir | 0.049 | ||
| hsa-miR-185-5p-477939_mir | 0.021 # | ||
| hsa-miR-186-5p-477940_mir | 0.028 | ||
| hsa-miR-188-3p-477942_mir | 0.009 | ||
| hsa-miR-191-5p-477952_mir | 0.007 | ||
| hsa-miR-195-5p-477957_mir | 0.04 | ||
| hsa-miR-199a-5p-478231_mir | 0.019 | ||
| hsa-miR-199a-3p_hsa-miR-199b-3p-477961_mir | 0.045 | 0.004 | |
| hsa-miR-199b-5p-478486_mir | 0.026 | ||
| hsa-miR-214-3p-477974_mir | 0.027 | ||
| hsa-miR-2110-477971_mir | 0.002 | ||
| hsa-miR-218-5p-477977_mir | 0.012 | ||
| hsa-miR-22-3p-477985_mir | 0.046 | ||
| hsa-miR-23b-3p-478602_mir | 0.036 | ||
| hsa-miR-26a-5p-477995_mir | 0.009 | 0.001 | |
| hsa-miR-26b-5p-478418_mir | 0.004 | 0.001 | |
| hsa-miR-27b-3p-478270_mir | 0.006 | ||
| hsa-miR-296-5p-477836_mir | 0.04 | ||
| hsa-miR-29a-3p-47858_mir | 0.022 | 0.025 | |
| hsa-miR-29b-3p-478369_mir | 0.023 | 0.002 | |
| hsa-miR-29c-3p-479229_mir | 0.027 | 0.004 | |
| hsa-miR-301b-3p-477825_mir | 0.020 # | ||
| hsa-miR-30b-5p-478007_mir | 0.036 | 0.015 | |
| hsa-miR-30c-5p-478008_mir | 0.012 | ||
| hsa-miR-30e-5p-479235_mir | 0.032 | ||
| hsa-miR-320a-478594_mir | 0.048 | ||
| hsa-miR-328-3p-478028_mir | 0.037 | ||
| hsa-miR-342-3p-478043_mir | 0.02 | 0.032 | |
| hsa-miR-376a-3p-478240_mir | 0.041 | ||
| hsa-miR-361-5p-478056_mir | 0.033 | ||
| hsa-miR-362-3p-478058_mir | 0.038 | ||
| hsa-miR-362-5p-478059_mir | 0.032 | 0.014 | |
| hsa-miR-374b-5p-478389_mir | 0.015 # | 0.002 # | |
| hsa-miR-377-3p-478075_mir | 0.004 | 0.001 | |
| hsa-miR-382-5p-478078_mir | 0.018 | ||
| hsa-miR-433-3p-478102_mir | 0.02 | 0.005 | |
| hsa-miR-422a-478481_mir | 0.042 | ||
| hsa-miR-34a-5p-478048_mir | 0.04 # | ||
| hsa-miR-423-5p-478090_mir | 0.044 | ||
| hsa-miR-424-5p-478092_mir | 0.043 | ||
| hsa-miR-425-5p-478094_mir | 0.037 | 0.038 | |
| hsa-miR-455-3p-478112_mir | 0.044 | ||
| hsa-miR-451a-478107_mir | 0.042 | ||
| hsa-miR-483-5p-478432_mir | 0.022 | ||
| hsa-miR-486-5p-478128_mir | 0.024 | 0.046 | |
| hsa-miR-491-5p-478132_mir | 0.035 | ||
| hsa-miR-494-3p-478135_mir | 0.004 | 0.001 | |
| hsa-miR-495-3p-478136_mir | 0.026 | 0.015 # | |
| hsa-miR-501-5p-478132_mir | 0.023 | ||
| hsa-miR-503-5p-478143_mir | 0.047 | 0.015 | |
| hsa-miR-505-3p-478145_mir | 0.011 | ||
| hsa-miR-532-5p-478151_mir | 0.040 | 0.016 | |
| hsa-miR-652-3p-478189_mir | 0.005 | 0.020 | |
| hsa-miR-655-3p-478191_mir | 0.008 | 0.005 | |
| hsa-miR-660-5p-478192_mir | 0.041 | ||
| hsa-miR-758-3p-479166_mir | 0.033 | ||
| hsa-miR-874-3p-478205_mir | 0.034 | ||
| hsa-miR-887-3p-479189_mir | 0.026 | 0.001 | |
| Total number of identified significantly different miRNAs | 33 | 65 | 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rzepakowska, A.; Zajkowska, A.; Mękarska, M.; Śladowska, J.; Borowy, A.; Małecki, M. MiRNA Profiling in Premalignant Lesions and Early Glottic Cancer. Cancers 2025, 17, 2883. https://doi.org/10.3390/cancers17172883
Rzepakowska A, Zajkowska A, Mękarska M, Śladowska J, Borowy A, Małecki M. MiRNA Profiling in Premalignant Lesions and Early Glottic Cancer. Cancers. 2025; 17(17):2883. https://doi.org/10.3390/cancers17172883
Chicago/Turabian StyleRzepakowska, Anna, Agnieszka Zajkowska, Marta Mękarska, Julia Śladowska, Aleksandra Borowy, and Maciej Małecki. 2025. "MiRNA Profiling in Premalignant Lesions and Early Glottic Cancer" Cancers 17, no. 17: 2883. https://doi.org/10.3390/cancers17172883
APA StyleRzepakowska, A., Zajkowska, A., Mękarska, M., Śladowska, J., Borowy, A., & Małecki, M. (2025). MiRNA Profiling in Premalignant Lesions and Early Glottic Cancer. Cancers, 17(17), 2883. https://doi.org/10.3390/cancers17172883






