Comment on Bar-Sela et al. Cannabis Consumption Used by Cancer Patients During Immunotherapy Correlates with Poor Clinical Outcome. Cancers 2020, 12, 2447
Simple Summary
Abstract
1. Introduction
- A.
- Table 1 and the associated text is the foundation of this report [1]. If this baseline information is inaccurate and the groups differ significantly on other variables besides cannabis, then it becomes very challenging to meaningfully assess the rest of this study. They reported (page 3, Section 2.1) that liver metastasis of the immunotherapy group (19% of N = 68 is 13) vs. the immunotherapy–cannabis group (67% of N = 34 is 23) had a p = 0.89. Using GraphPad Prism [7] and the two statistics listed in their Methods section (chi-square and Fisher’s exact tests) with the N provided, we were unable to verify this result. We determined that chi-square (1) = 23.375, p < 0.0001 and Fisher’s p < 0.0001.
- B.
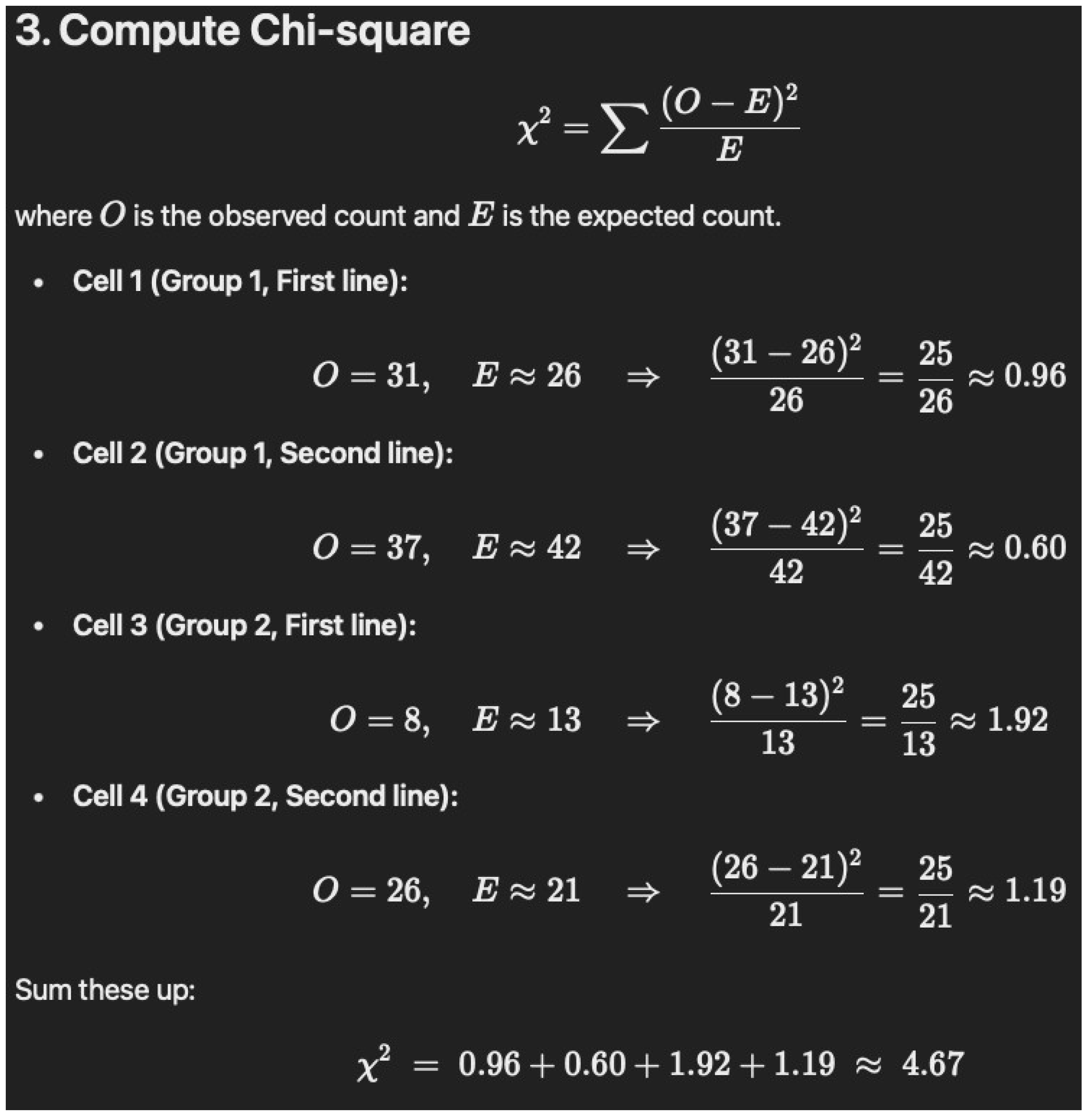
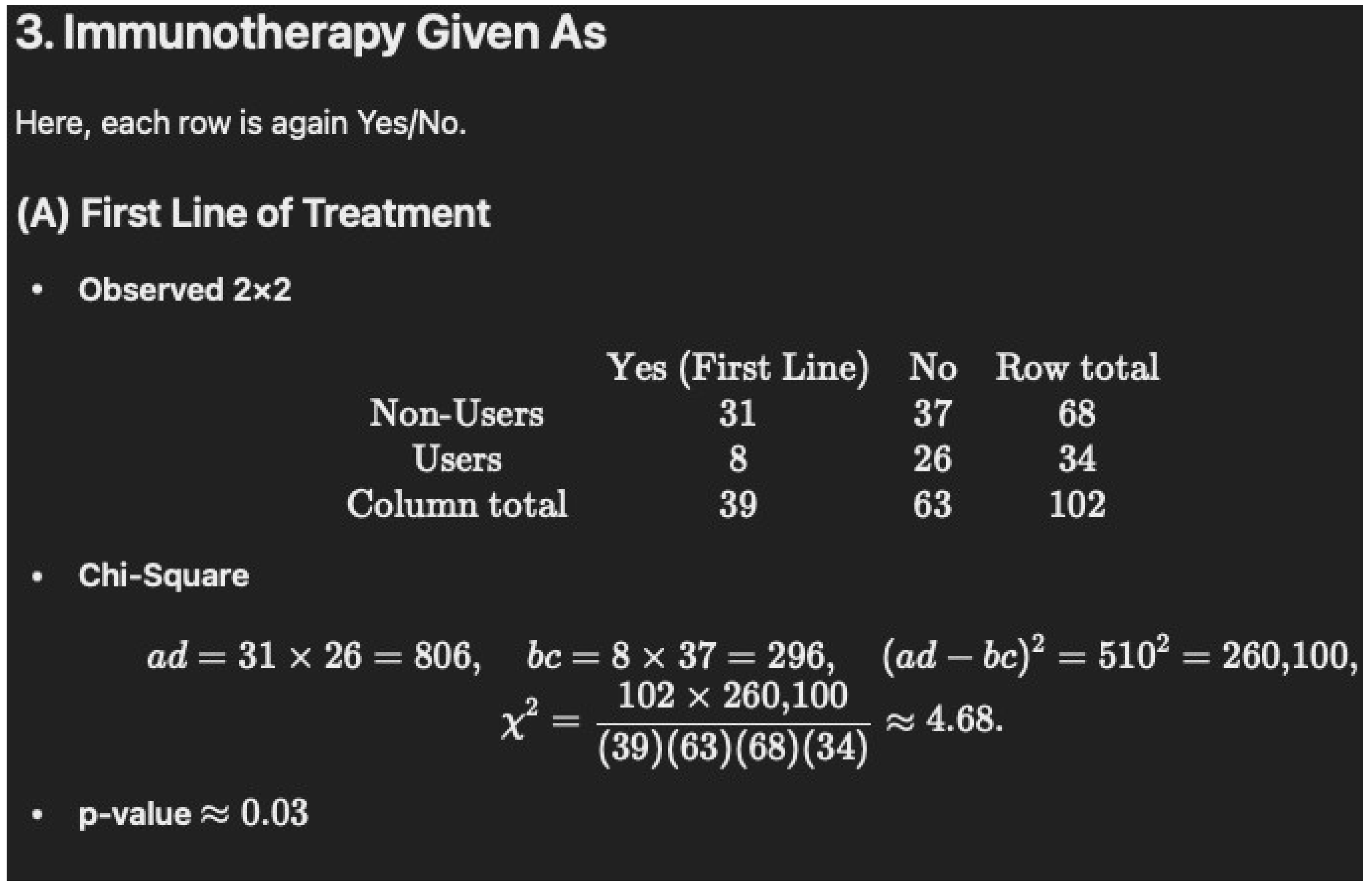
- Table 1 notes that 76% of the immunotherapy–cannabis group (76% of N = 34 is 26) were given immunotherapy as the second line of treatment versus 54% (54% of N = 68 is 37) of immunotherapy only, which had a p-value of “0.05178”. Again, we were unable to verify this with chi-square (1) = 4.67, p = 0.0317 and Fisher’s p = 0.0334.
- C.
- Similarly, only one of the 22 p-values (4.5%) in the corrected Table 1 could be verified using the same statistics, as were reported in the Methods section [1]. Based on p-value agreement to at least three decimal places, the authors appear to have reported a different statistic (Yates) than was in their Methods section (chi-square or Fisher’s) on 18 of 22 (81.8%) of these statistics. This is a deviation from common scientific reporting conventions.
- D.
- Although perhaps less impactful for their core conclusions, there appears to have been some difficulties in calculating percentages (i.e., a difference of 16.2% between the reported 22.0% and our calculated value of 38.2% for chronic diseases = 0 for cannabis users; a difference of 4.1% between the reported 34.1 and calculated 38.2% for high blood pressure for cannabis users; and a 10.3% difference between the reported 13.2% and the calculated 23.5% for bone metastasis for cannabis users) [6], which suggests an inattention to detail in the “corrected” Table 1. Further, the total percentages for chronic diseases for cannabis users (0 = 22.0%, 1 = 20.5%, ≥2 = 41.1%) adds up to 83.6% instead of the usual 100.0%.
- E.
- The original authors also employed a type of “floor rounding” on 19 occasions [6]. For those who are unfamiliar (as we were), a floor function would take an input (e.g., 4.9) and convert it to the next lowest whole integer (4.0) [8]. As applied here, a number like 0.049999 if calculated to two decimal places would be listed as “0.04” with floor rounding. In retrospect, unusual practices like this should be noted in the Methods section.
- F.
- The Methods section includes the standard statement that “We computed 2-tailed p-values, where p < 0.05 was considered a statistically significant result.” The sample size calculation sub-section reinforced the reader’s expectation of two-tailed statistical tests by stating “alpha of 5% for the two-tailed test.” for the complete response and partial response calculations. However, the Results section then notes that there was “(one-tailed p = 0.08) (Table 2).” [1]. Although we found [6,7] the one-tailed Fisher’s was “0.089”, which would commonly be reported as 0.09, this could be attributed to floor-rounding. The alkaline phosphatase level was reported as “0.09”, but we obtained a one-tailed chi-square of 0.0687, one-tailed Fischer’s of 0.1089, and even a one-tailed Yates of 0.1079. Switching the data analysis plan from the Methods to the Results section (i.e., from two-tailed to one-tailed) is incongruent with typical practice in scientific reporting. If the authors made their deidentified raw data and a detailed data dictionary publicly available on GitHub at https://github.com/ (assuming that this were acceptable to the Rambam IRB), this could allow for independent re-analysis of this and other statistics (Table 4, and Figure 3 and Figure 4).
- G.
- Tobacco smoking is a defining theme of oncology that impacts cancer risk, diagnosis, and treatment [9,10]. This includes lung cancer, with a relative risk of seven among smokers [11]. Lung cancer patients that quit smoking may have a 20 to 35% improved survival [10]. Current smokers had a greater (Hazard Ratio = 1.48) likelihood of melanoma-associated death than non-smokers [12]. A meta-analysis revealed that current smokers with renal cell carcinoma had poorer progression-free survival (HR = 2.94) than non-smokers [13]. Note that about half the Bar-Sela sample from Israel consisted of non-small cell lung cancer, one-third melanoma, and the rest were renal cell carcinoma or other malignancies [1]. Among Israeli cancer survivors, significantly more cannabis users also used tobacco [14]. Adding further complexity, about one-third of cancer survivors with an active cannabis permit in Israel reported mixing tobacco with cannabis [14]. The Bar-Sela team should be commended for reporting smoking in Table 2 of their retrospective study [15] as this could be an important confound [9,10,11,12,13,14]. However, this key variable was never mentioned, and therefore, any variance due to this was not removed in their prospective study [1].
- H.
- There is a very limited sample size within the cannabis group (n = 34), who were more likely to receive second line treatment, were more likely to have impaired performance statuses (ECOG ≥ 2) and had higher rates of baseline lymphopenia (all associated with worse clinical outcomes). Although none of these differences reach statistical significance, it does raise questions about important differences that may exist between the groups that may not have been apparent with this limited sample size. A median overall survival of six months seems unusually low in the modern immunotherapy era for these malignancies, which likely suggests additional unmeasured confounds associated with cannabis use and cancer outcomes.
2. Potential Solutions
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
| LLM | Large Language Models |
Appendix A
- -
- We gave the ChatGPT 4o model the paper and instructed it to go to Table 1 and perform a chi-square test for the gender- and number-matched samples. The results were good.
- -
- Then, we instructed the model to now perform the same test for Table 1. Cont., with immunotherapy given as the first-line vs. second-line treatment. The 4o model produced a p-value of 0.0518, which matched that in the paper, likely due to bias. We found this to be inaccurate so we input the data into the o1 model (much better at statistics) and instructed it to perform the same task. See # 2.
- -
- Because the o1 model does not read documents like 4o, it needs to be copy and pasted as text.
- -
- This model found the p-value to be 0.03, not 0.0518 as the paper and 4o reported.
- -
- https://openai.com/index/learning-to-reason-with-llms/ (accessed on 4 January 2025) shows how much more accurate o1 is, especially at math.
- -
- To see how o1 could perform on its own with little guidance, we said “below is a Table 1. Cont. for a paper. please recalculate values and check for accuracy”, and then, we copy and pasted the entire Table 1. Cont.
- -
- The model found an error in the table where it marks the brain as the main site of metastasis, where Users (N = 34): 8/34 = 23.5%. The original table lists 13.2% for 8/34, which is incorrect.
- -
- Then, with a very simple prompt, “thanks, now do chi squared please and thank you”, with very little guidance, it was able to distinguish the following:
- -
- Important Note
- -
- Many of the original p-values in your table appear to come from Fisher’s Exact Tests (common for smaller counts) or from combining multi-category variables into one test.
- -
- Chi--Square results on small or sparse counts (some cells <5) should be interpreted with caution.
- -
- Thus, the p-values below will differ from the original table’s p-values if the original used Fisher’s Exact Test or a multi-level Chi--Square.
- -
- Again, these will differ from your original table if those p-values were based on Fisher’s Exact Test or a multi-category test.
- -
- This shows that intelligent models like o1 can be utilized to readily double-check statistics. It can easily navigate percentages and non-parametric tests with little context.
- •
- Chi-square value (χ2): 0.0057
- •
- p-value: 0.9399
- •
- Degrees of freedom (dof): 1
- •
- Expected frequencies:
- ○
- Female: [21.33, 10.67]
- ○
- Male: [46.67, 23.33]
- •
- Chi-square value (χ2): 3.78
- •
- p-value: 0.0518
- •
- Degrees of freedom (dof): 1
- •
- Expected frequencies:
- ○
- First line: [26, 13]
- ○
- Second line: [42, 21]
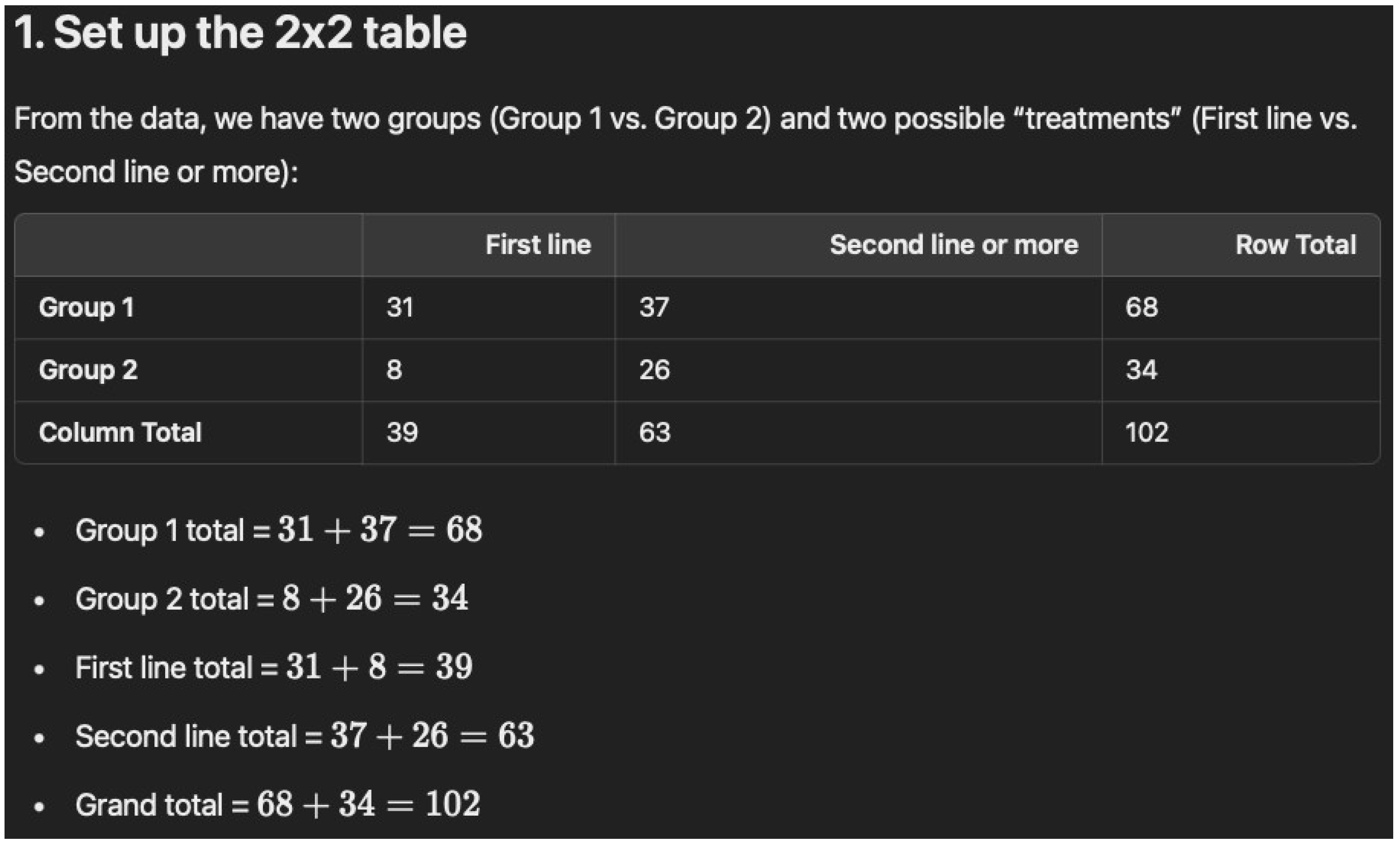
Below is a step-by-step walkthrough of calculating a Chi-square (\(\chi^2\)) test from the table: --- ## 1. Set up the 2 × 2 table From the data, we have two groups (Group 1 vs. Group 2) and two possible “treatments” (First line vs. Second line or more): | | First line | Second line or more | Row Total | |----------------------|-----------:|---------------------:|----------:| | **Group 1** | 31 | 37 | 68 | | **Group 2** | 8 | 26 | 34 | | **Column Total** | 39 | 63 | 102 | - Group 1 total = \(31 + 37 = 68\) - Group 2 total = \(8 + 26 = 34\) - First line total = \(31 + 8 = 39\) - Second line total = \(37 + 26 = 63\) - Grand total = \(68 + 34 = 102\) --- ## 2. Calculate the expected frequen-cies Using the formula for expected counts in each cell of a 2 × 2 table: \[ E_{ij} = \frac{(\text{Row Total}_i) \times (\text{Column Total}_j)}{\text{Grand Total}} \] 1. **Group 1, First line**: \[ E_{11} = \frac{68 \times 39}{102} \approx 26.0 \] 2. **Group 1, Second line**: \[ E_{12} = \frac{68 \times 63}{102} \approx 42.0 \] 3. **Group 2, First line**: \[ E_{21} = \frac{34 \times 39}{102} \approx 13.0 \] 4. **Group 2, Second line**: \[ E_{22} = \frac{34 \times 63}{102} \approx 21.0 \] --- ## 3. Compute Chi-square \[ \chi^2 = \sum \frac{(O - E)^2}{E} \] where \(O\) is the observed count and \(E\) is the expected count. - **Cell 1 (Group 1, First line):** \[ O = 31, \quad E \approx 26 \quad \Rightarrow \quad \frac{(31 - 26)^2}{26} = \frac{25}{26} \approx 0.96 \] - **Cell 2 (Group 1, Second line):** \[ O = 37, \quad E \approx 42 \quad \Rightarrow \quad \frac{(37 - 42)^2}{42} = \frac{25}{42} \approx 0.60 \] - **Cell 3 (Group 2, First line):** \[ O = 8, \quad E \approx 13 \quad \Rightarrow \quad \frac{(8 - 13)^2}{13} = \frac{25}{13} \approx 1.92 \] - **Cell 4 (Group 2, Second line):** \[ O = 26, \quad E \approx 21 \quad \Rightarrow \quad \frac{(26 - 21)^2}{21} = \frac{25}{21} \approx 1.19 \] Sum these up: \[ \chi^2 \;=\; 0.96 + 0.60 + 1.92 + 1.19 \;\approx\; 4.67 \] --- ## 4. Determine degrees of freedom (df) and ap-proximate p-value For a 2 × 2 contingency table, \[ \text{df} = (2–1) \times (2–1) = 1. \] With \(\chi^2 \approx 4.67\) and \( \text{df} = 1\): - The p-value is between 0.025 and 0.05 (since \(\chi^2 = 3.84\) corresponds roughly to p = 0.05 and \(\chi^2 = 5.02\) to p = 0.025). - More precisely, it is around **p ≈ 0.03**. --- ## 5. Conclusion **\(\chi^2 \approx 4.67\) with df = 1, and \(p \approx 0.03\).** Thus, there is a statistically significant (though borderline) difference between the two groups regarding whether immunotherapy is given as first line vs. second line or more. --- *Hope this helps, Dr. Dobbins!*



-
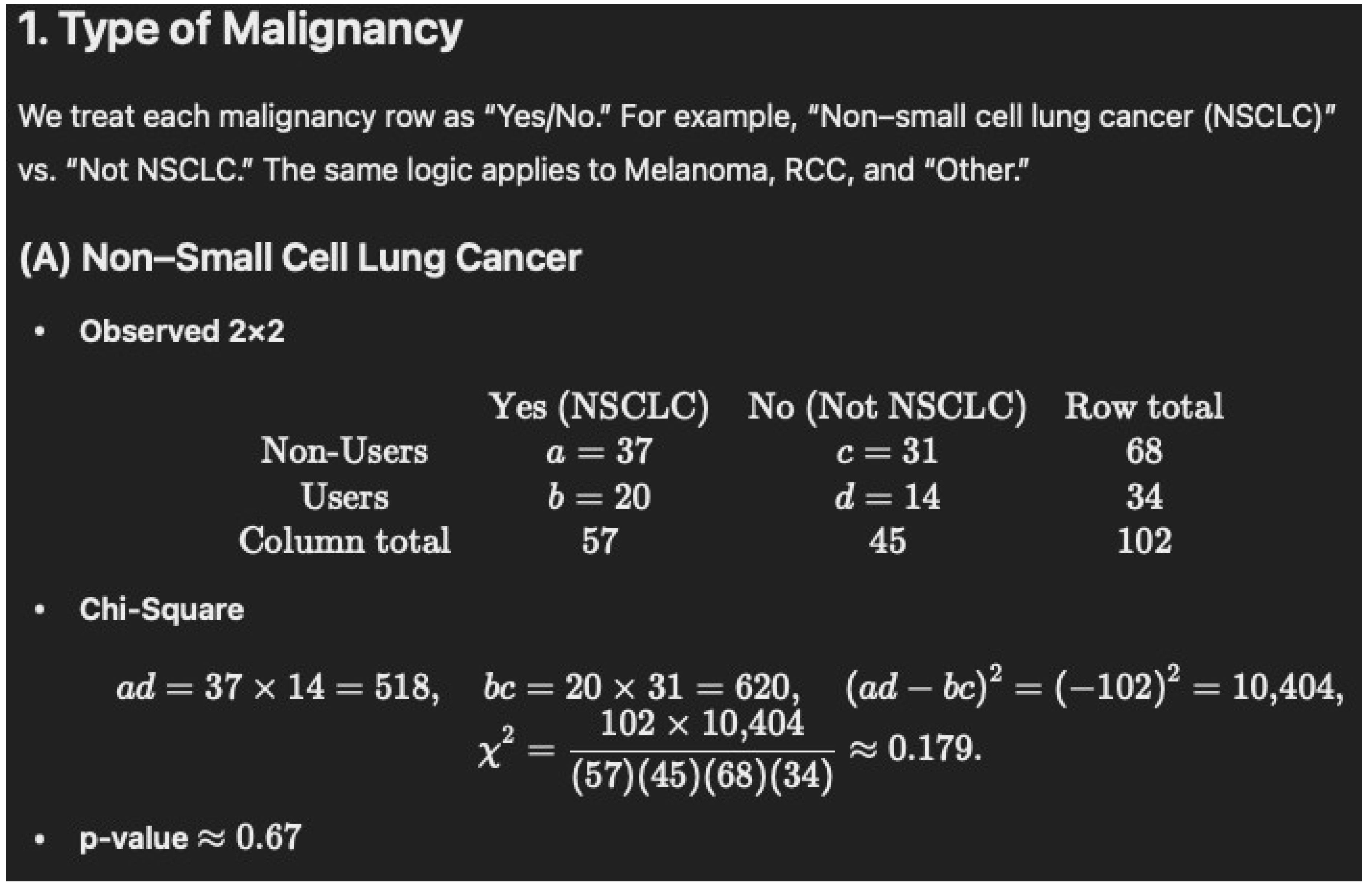
Non-Users (N = 68): 37/68 = 54.4%
-
Users (N = 34): 20/34 = 58.8%
-
p = 0.8325
-
Non-Users (N = 68): 25/68 = 36.8% (rounded to 36.7%)
-
Users (N = 34): 9/34 = 26.5% (rounded to 26.4%)
-
p = 0.414
-
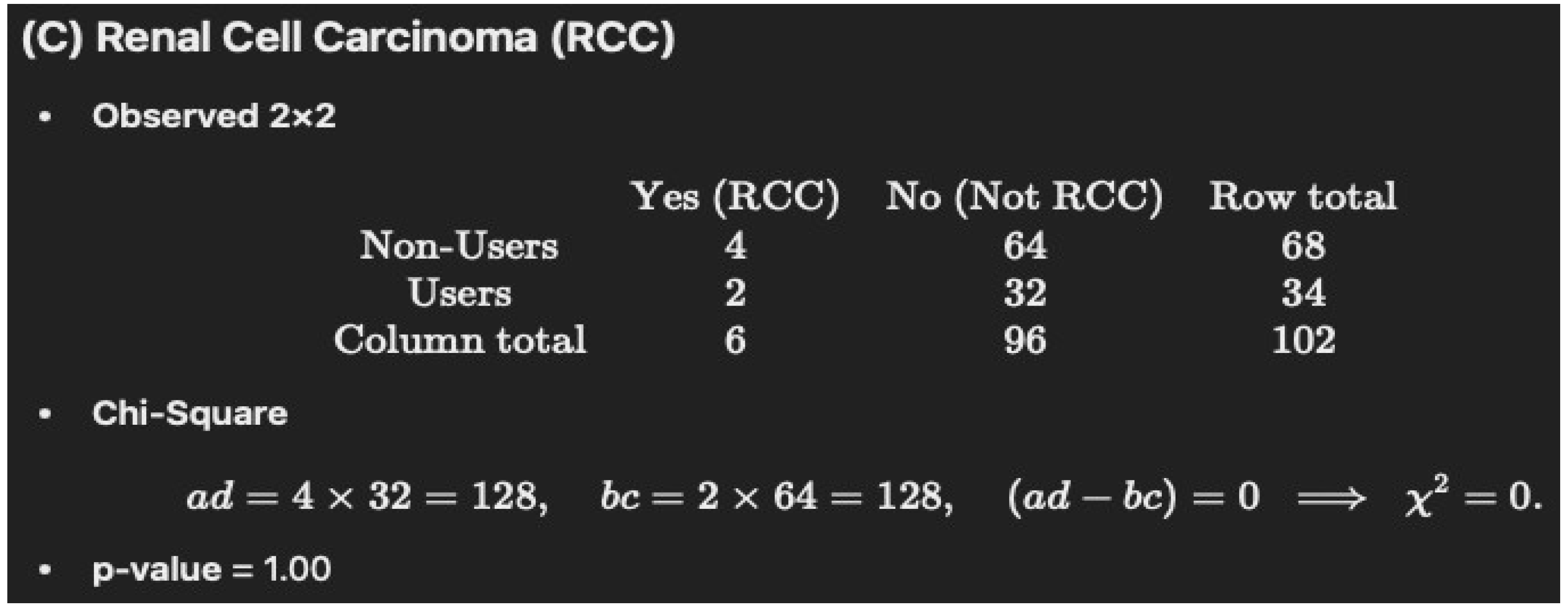
Non-Users (N = 68): 4/68 = 5.88% (rounded to 5.8%)
-
Users (N = 34): 2/34 = 5.88% (rounded to 5.8%)
-
p = 1
-
Non-Users (N = 68): 2/68 = 2.94% (rounded to 2.9%)
-
Users (N = 34): 3/34 = 8.82% (rounded to 8.8%)
-
p = 1
- •
-
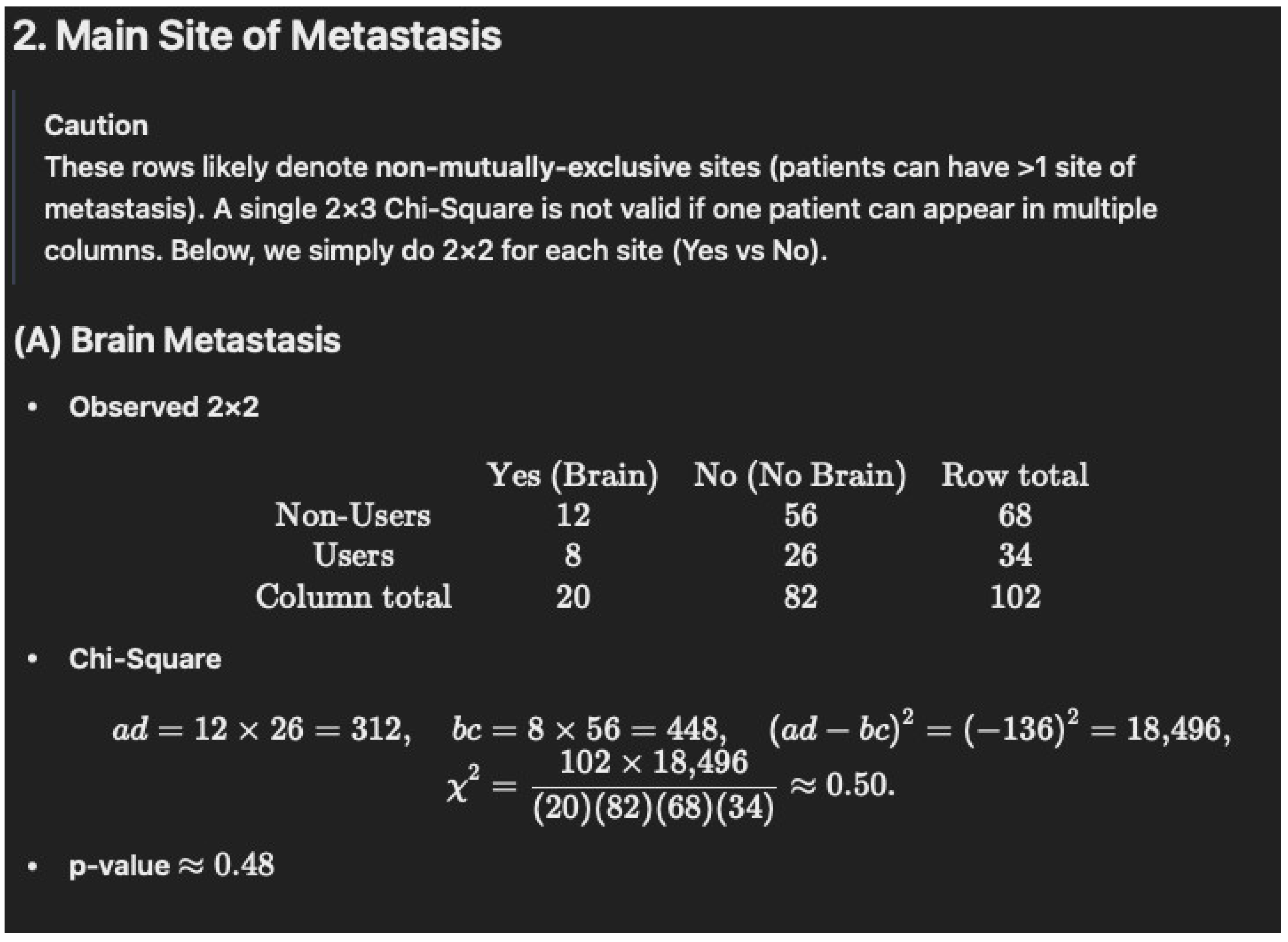
Non-Users (N = 68): 12/68 = 17.6%
- •
-
Users (N = 34): 8/34 = 23.5%
- ○
-
The original table lists 13.2% for 8/34, which is incorrect.
- •
-
p = 0.6593
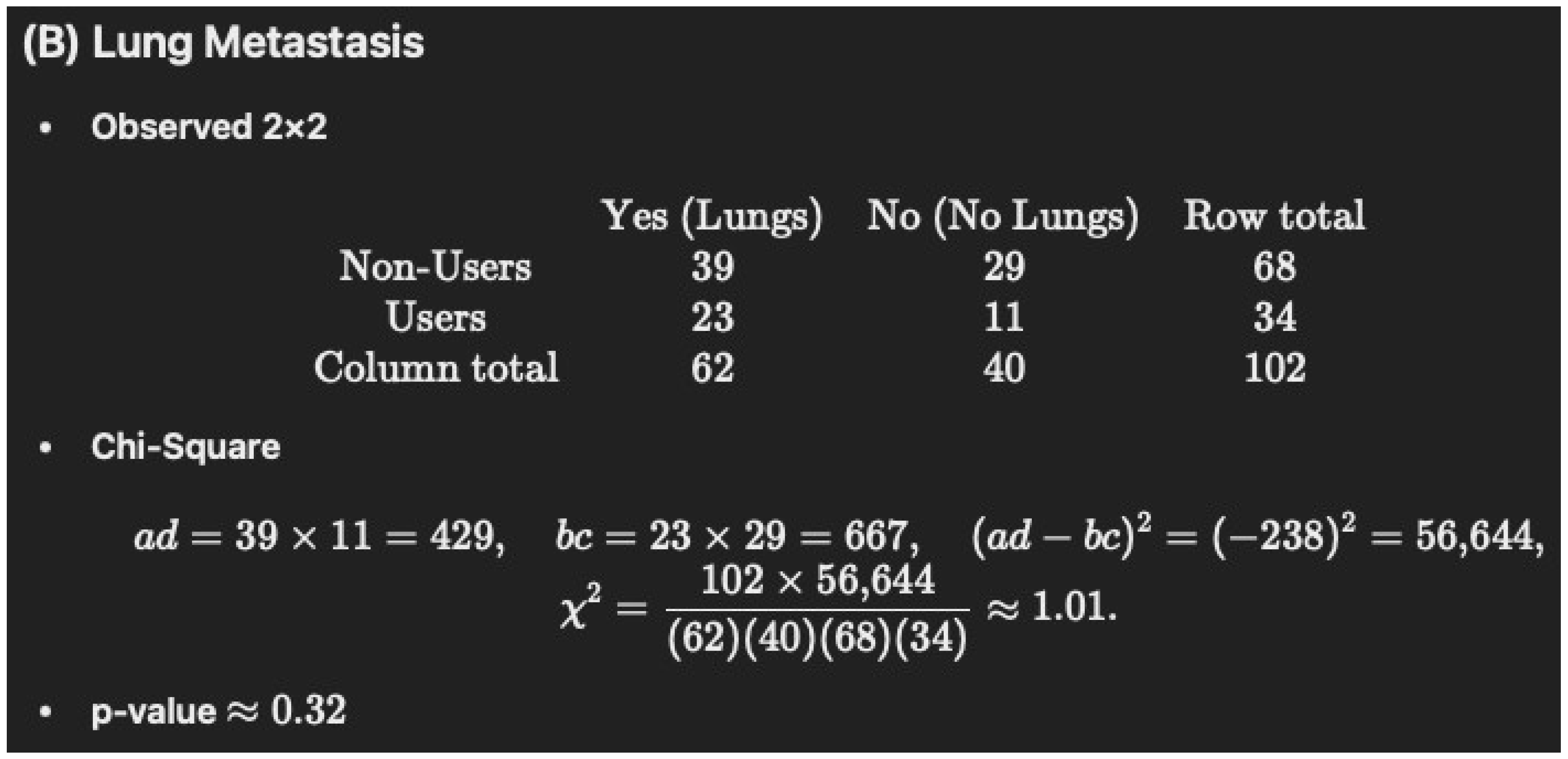
-
Non-Users (N = 68): 39/68 = 57.4% (rounded to 57.3%)
-
Users (N = 34): 23/34 = 67.6%
-
p = 0.4303
-
Non-Users (N = 68): 13/68 = 19.1%
-
Users (N = 34): 11/34 = 32.4% (rounded to 32.3%)
-
p = 0.2157
-
Non-Users (N = 68): 31/68 = 45.6% (rounded to 45.5%)
-
Users (N = 34): 8/34 = 23.5%
-
p = 0.05178
-
Non-Users (N = 68): 37/68 = 54.4%
-
Users (N = 34): 26/34 = 76.5% (rounded to 76.4%)
-
p = 0.05178
-
Non-Users (N = 68): 47/68 = 69.1%
-
Users (N = 34): 29/34 = 85.3% (rounded to 85.2%)
-
p = 0.127
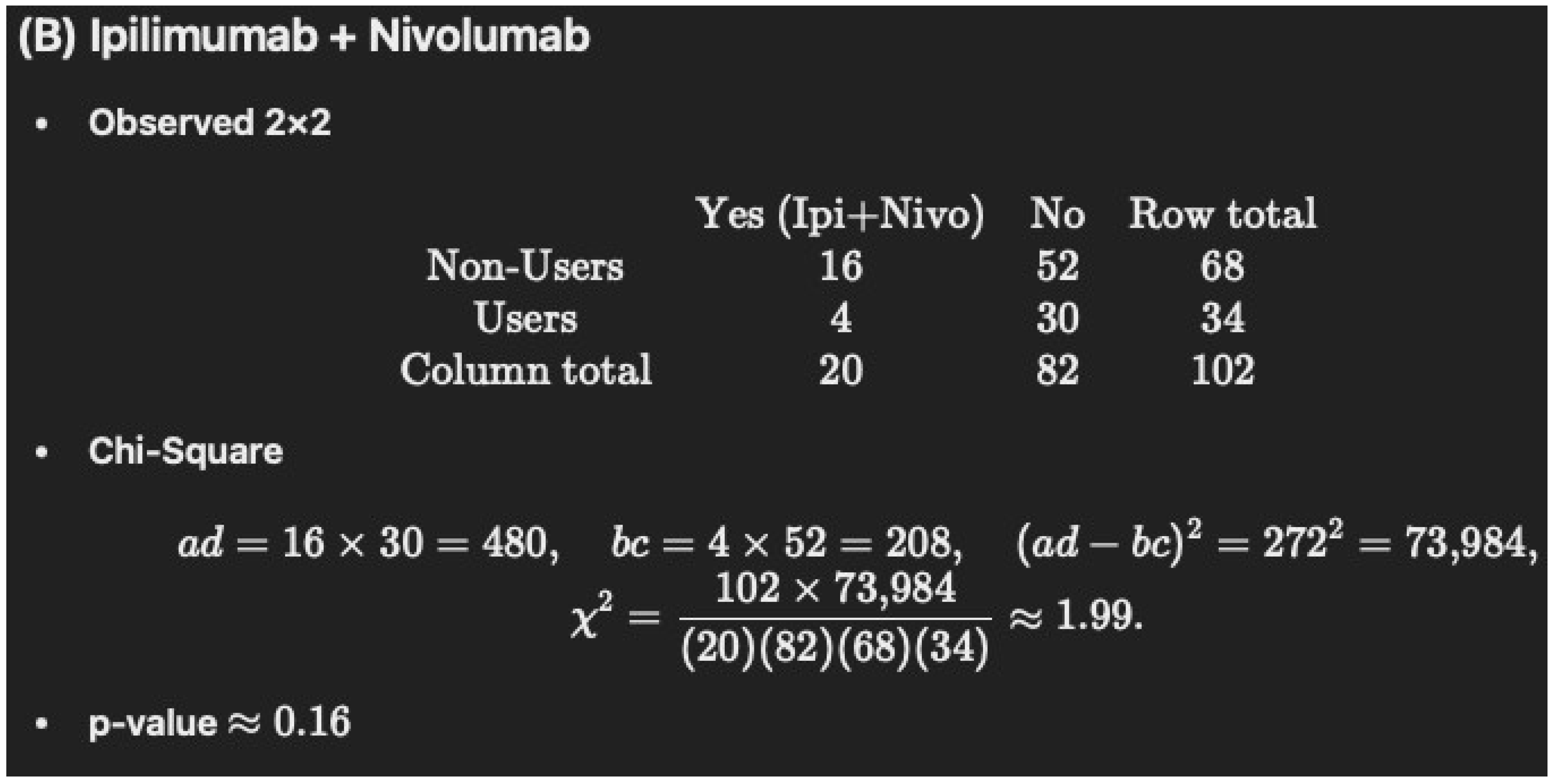
-
Non-Users (N = 68): 16/68 = 23.5%
-
Users (N = 34): 4/34 = 11.8% (rounded to 11.7%)
-
p = 0.2517
-
Non-Users (N = 68): 5/68 = 7.4% (rounded to 7.3%)
-
Users (N = 34): 1/34 = 2.94% (rounded to 2.9%)
-
p = 1
-
Corrected “Table 1 Cont.”
| Characteristics |
Cannabis Non-Users
(n = 68) |
Cannabis Users
(n = 34) |
p-Value |
|---|---|---|---|
| Type of malignancy–n (%) | |||
| Non-small cell lung cancer | 37 (54.4) | 20 (58.8) | 0.8325 |
| Melanoma | 25 (36.7) | 9 (26.4) | 0.414 |
| Renal cell carcinoma | 4 (5.8) | 2 (5.8) | 1 |
| Other | 2 (2.9) | 3 (8.8) | 1 |
| Main site of metastasis–n (%) | |||
| Brain | 12 (17.6) | 8 (23.5) * | 0.6593 |
| Lungs | 39 (57.3) | 23 (67.6) | 0.4303 |
| Liver | 13 (19.1) | 11 (32.3) | 0.2157 |
| Immunotherapy given as–n (%) | |||
| First line of treatment | 31 (45.5) | 8 (23.5) | 0.05178 |
| Second line of treatment or more | 37 (54.4) | 26 (76.4) | 0.05178 |
| Checkpoint therapy–n (%) | |||
| Anti-PD1: Pembrolizumab or Nivolumab | 47 (69.1) | 29 (85.2) | 0.127 |
| Ipilimumab and Nivolumab | 16 (23.5) | 4 (11.7) | 0.2517 |
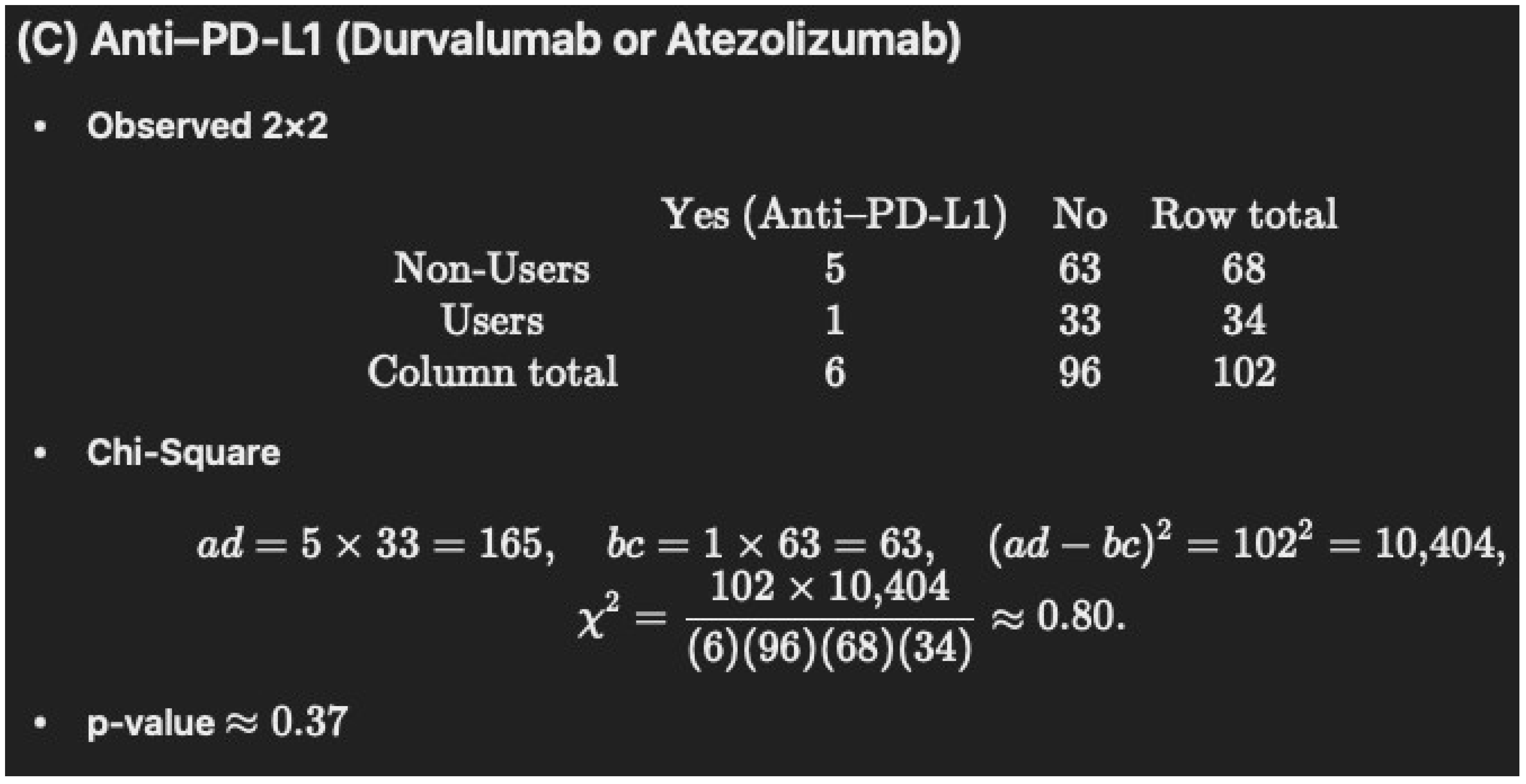
| Anti-PD-L1: Durvalumab or Atezolizumab | 5 (7.3) | 1 (2.9) | 1 |






References
- Bar-Sela, G.; Cohen, I.; Campisi-Pinto, S.; Lewitus, G.M.; Oz-Ari, L.; Jehassi, A.; Peer, A.; Turgeman, I.; Vernicova, O.; Berman, P.; et al. Cannabis consumption used by cancer patients during immunotherapy correlates with poor clinical outcome. Cancers 2020, 12, 2447. [Google Scholar] [CrossRef] [PubMed]
- Ioaniddis, J.P.A. Why most published research findings are false. PLoS Med. 2005, 2, e124. [Google Scholar] [CrossRef] [PubMed]
- To, J.; Davis, M.; Sbrana, A.; Alderman, B.; Alderman, B.; Hui, D.; Mukhopadhyay, S.; Bouleuc, C.; Case, A.A.; Amano, K.; et al. MASCC guideline: Cannabis for cancer-related pain and risk of harms and adverse events. Support. Care Cancer 2023, 31, 202. [Google Scholar] [CrossRef] [PubMed]
- Braun, I.M.; Bohlke, K.; Abrams, D.I.; Anderson, H.; Balnaves, L.G.; Bar-Sela, G.; Bowles, D.W.; Chai, P.R.; Madeni, A.; Gupta, A.; et al. Cannabis and cannabinoids in adults with Cancer: ASCO Guidelines. J. Clin. Oncol. 2023, 42, 1575. [Google Scholar] [CrossRef] [PubMed]
- National Academies of Sciences. The Health Effects of Cannabis and Cannabinoids: The Current State of Evidence and Recommendations for Research; National Academies of Sciences: Washington, DC, USA, 2017. [Google Scholar]
- Piper, B.J.; Tian, M.; Saini, P.; Higazy, A.; Graham, J.; Carbe, C.J.; Bordonaro, M. Immunotherapy and cannabis: A harmful drug interaction or Reefer Madness? Cancers 2024, 16, 1245. [Google Scholar] [CrossRef] [PubMed]
- GraphPad Prism. Analyze a 2x2 Contingency Table. Available online: https://www.graphpad.com/quickcalcs/contingency2/ (accessed on 17 July 2025).
- Wikipedia. Available online: https://en.wikipedia.org/wiki/Floor_and_ceiling_functions (accessed on 21 December 2024).
- National Center for Chronic Disease Prevention and Health Promotion (US) Office on Smoking and Health. The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General; Centers for Disease Control and Prevention (US): Atlanta, GA, USA, 2014.
- Evans, W.K.; Warren, G.W.; Dresler, C. Ignoring the obvious: Smoking cessation improves survival. J. Thorac. Oncol. 2022, 17, 596–598. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, L.M.; Taylor, G.; Huxley, R.R.; Mitchell, P.; Woodward, M.; Peters, S.E. Smoking as a risk factor for lung cancer in women and men: A systematic review and meta-analysis. BMJ Open 2018, 8, e021611. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.M.; Jones, P.C.; Fluke, L.M.; Fischer, T.D.; Thompson, J.F.; Cochan, A.J.; Stern, S.L.; Faries, M.B.; Hoon, D.S.B.; Foshag, L.J. Smoking status and survival in patients with early stage primary cutaneous melanoma. JAMA Netw. Open 2024, 7, e2354751. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Qi, Y.; Zhang, J.; Lu, Y.; Song, J.; Dong, B.; Kong, W.; Xue, W.; Huang, Y. The impact of smoking on survival in renal cell carcinoma: A systematic review and meta-analysis. Tumor Biol. 2014, 35, 6633–6640. [Google Scholar] [CrossRef] [PubMed]
- Halpern, N.; Eshet, L.; Greenhouse, I.; Beller, T.; Hausner, D.; Sella, A. Medical cannabis use in young adults with cancer: A self-reported survey study. BMJ Support. Palliat. Care 2024, 14, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Taha, T.; Meiri, D.; Wolliner, M.; Peer, A.; Bar-Sela, G. Cannabis impacts tumor response rate to nivolumab in patients with advanced malignancies. Oncologist 2019, 24, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Nuijten, M.; Polanin, J.R. “statcheck”: Automatically detect statistical reporting inconsistencies to increase reproducibility of meta-analyses. Res. Synth. Methods 2020, 11, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Waissengrin, B.; Leshem, Y.; Taya, M.; Meiri, D.; Merimsky, O.; Shamai, S.; Wolf, I.; Bubinek, T. The use of medical cannabis concomitantly with immune checkpoint inhibitors in non-small cell lung cancer: A sigh of relief? Eur. J. Cancer 2023, 180, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Tikun. Our Strains. Available online: https://www.tikun-olam.info/our-Strains/ (accessed on 7 August 2025).
- Piper, B.J. Mother of Berries, ACDC, or Chocolope: Examination of the strains used by medical cannabis patients in New England. J. Psychoact. Drugs 2018, 50, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.G.; Kim, B.R.; Kim, D.Y.; Yun, H.; Dayoung, K.; Kim, W.K.; Kang, S.; Lee, S.I.; Oh, S.I. Abstract 5548: Cannabidiol enhances efficacy of atezolizumab via upregulation of PD-L1 expression by cGAS-STING pathway in triple negative breast cancer cells. Cancer Res. 2024, 84 (Suppl. S6), 5548. [Google Scholar] [CrossRef]
- Twelves, C.; Sabel, M.; Checketts, D.; Miller, S.; Tayo, B.; Jove, M.; Brazil, L.; Short, S.C.; GWCA1208 study group; McBain, C.; et al. A phase 1b randomised, placebo-controlled trial of nabiximols cannabinoid oromucosal spray with temozolomide in patients with recurrent glioblastoma. Brit. J. Cancer 2021, 124, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Ionnidis, J.P. Why most discovered true associations are inflated. Epidemiology 2008, 19, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Nahler, G. Phytocannabinoids as chemotherapy adjuncts—A review for users. Onco 2024, 4, 287–321. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piper, B.J.; Dobbins, D.X.; Graham, J.; Churilla, T.M.; Bordonaro, M. Comment on Bar-Sela et al. Cannabis Consumption Used by Cancer Patients During Immunotherapy Correlates with Poor Clinical Outcome. Cancers 2020, 12, 2447. Cancers 2025, 17, 2754. https://doi.org/10.3390/cancers17172754
Piper BJ, Dobbins DX, Graham J, Churilla TM, Bordonaro M. Comment on Bar-Sela et al. Cannabis Consumption Used by Cancer Patients During Immunotherapy Correlates with Poor Clinical Outcome. Cancers 2020, 12, 2447. Cancers. 2025; 17(17):2754. https://doi.org/10.3390/cancers17172754
Chicago/Turabian StylePiper, Brian J., Duncan X. Dobbins, Jason Graham, Thomas M. Churilla, and Michael Bordonaro. 2025. "Comment on Bar-Sela et al. Cannabis Consumption Used by Cancer Patients During Immunotherapy Correlates with Poor Clinical Outcome. Cancers 2020, 12, 2447" Cancers 17, no. 17: 2754. https://doi.org/10.3390/cancers17172754
APA StylePiper, B. J., Dobbins, D. X., Graham, J., Churilla, T. M., & Bordonaro, M. (2025). Comment on Bar-Sela et al. Cannabis Consumption Used by Cancer Patients During Immunotherapy Correlates with Poor Clinical Outcome. Cancers 2020, 12, 2447. Cancers, 17(17), 2754. https://doi.org/10.3390/cancers17172754







