MICA+ Tumor Cells Modulate Macrophage Phenotype and Function via PPAR/EHHADH-Mediated Fatty Acid Metabolism in Hepatocellular Carcinoma (HCC)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Screening for Differentially Expressed Genes (DEGs)
2.3. Screening of Target Genes and Functional Enrichment Analysis
2.4. Expression and Prognostic Analysis of EHHADH
2.5. Immune Cell Infiltration Analysis
2.6. Cancer Genomics Analysis of the cBioPortal Database
2.7. The Acquisition of scRNA-Seq Data
2.8. Preprocessing and Analysis of scRNA-Seq Data
2.9. Patient Sample
2.10. Quantitative Real-Time PCR (qRT-PCR)
2.11. Immunohistochemistry (IHC)
2.12. Cell Culture
2.13. Lentivirus Infection
2.14. Western Blotting
2.15. Co-Culture
2.16. Immunofluorescence (IF) Staining
2.17. Oil Red O Staining
2.18. BODIPY Staining
2.19. FAO Testing
2.20. ELISA Validation
2.21. Cell Proliferation Assay
2.22. Apoptosis Assay
2.23. Statistical Analysis
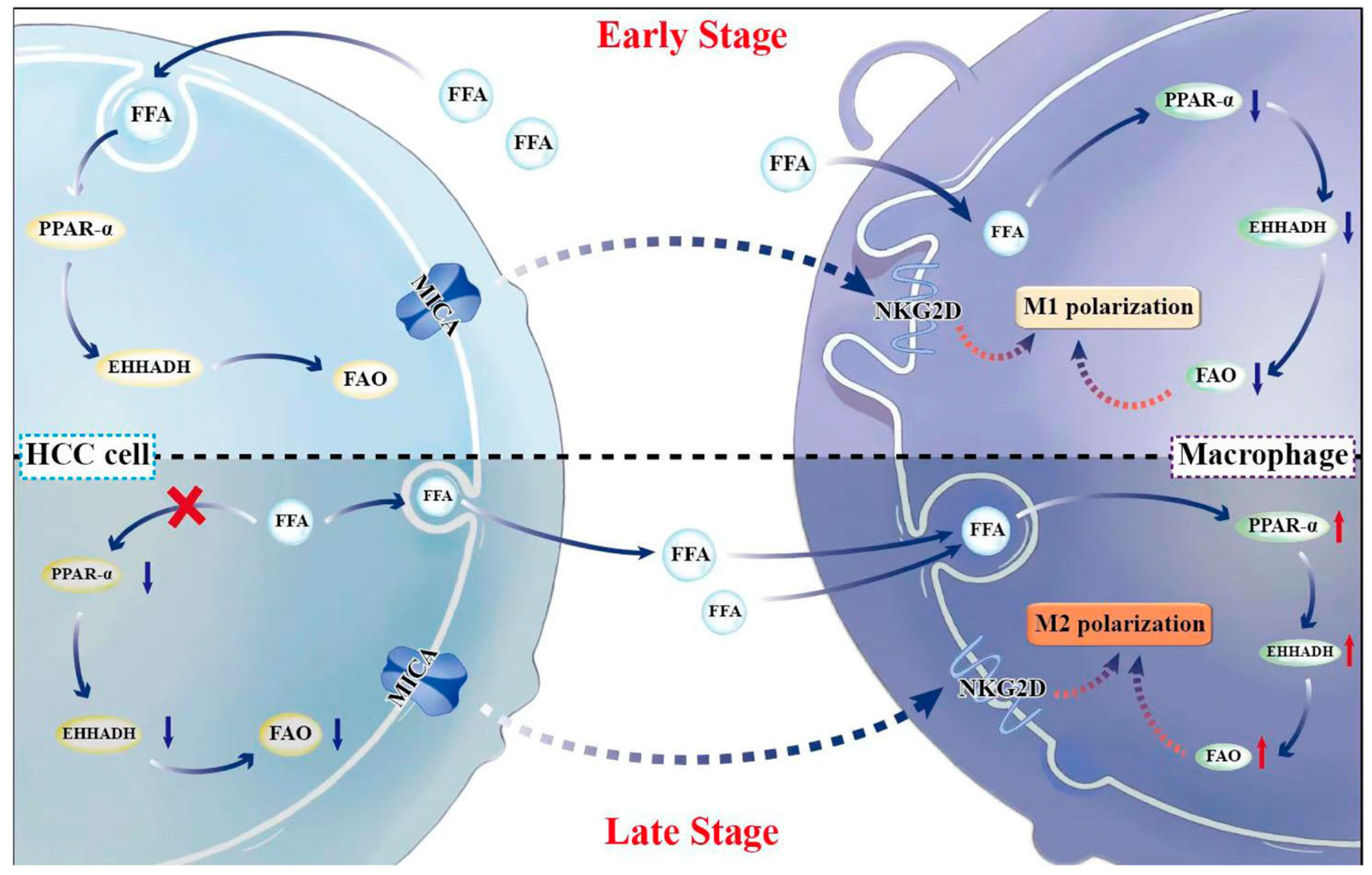
3. Results
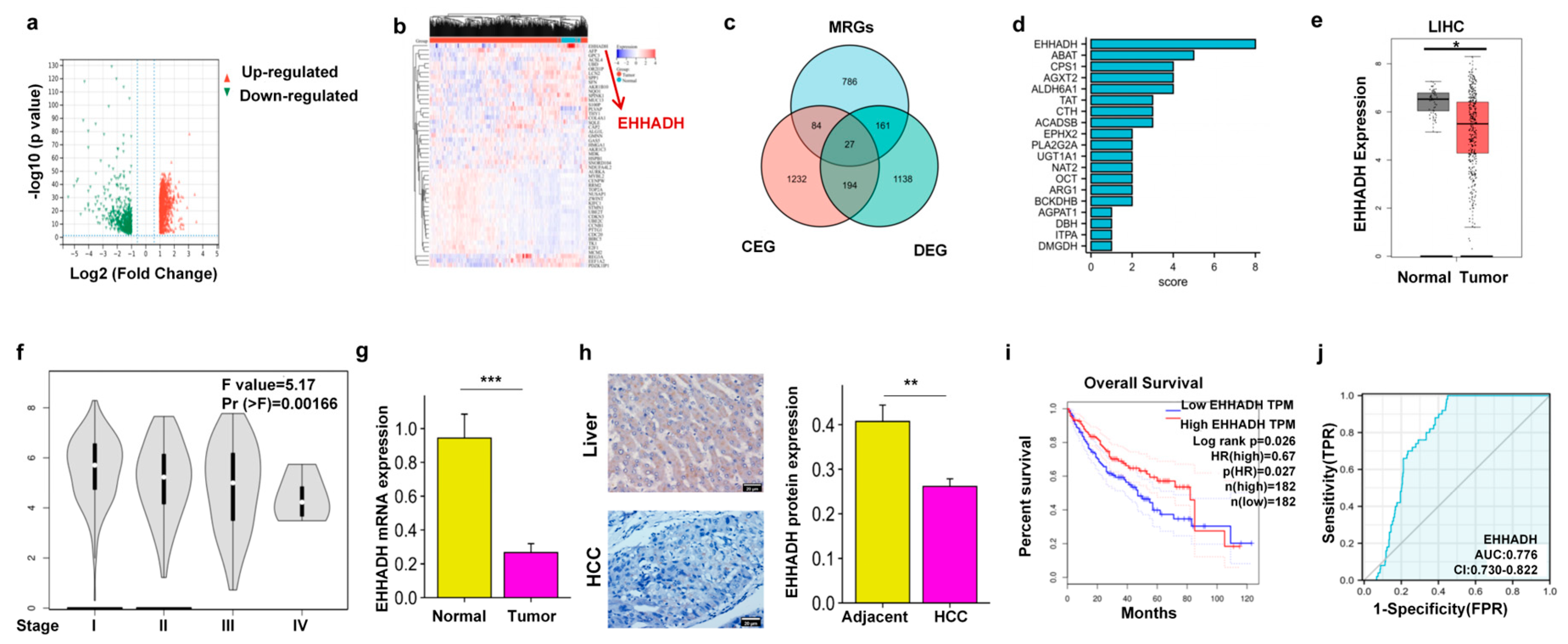
3.1. EHHADH Was Identified as a Candidate Gene Through the Integration of DEGs, MRGs, and CEGs with MICA in HCC
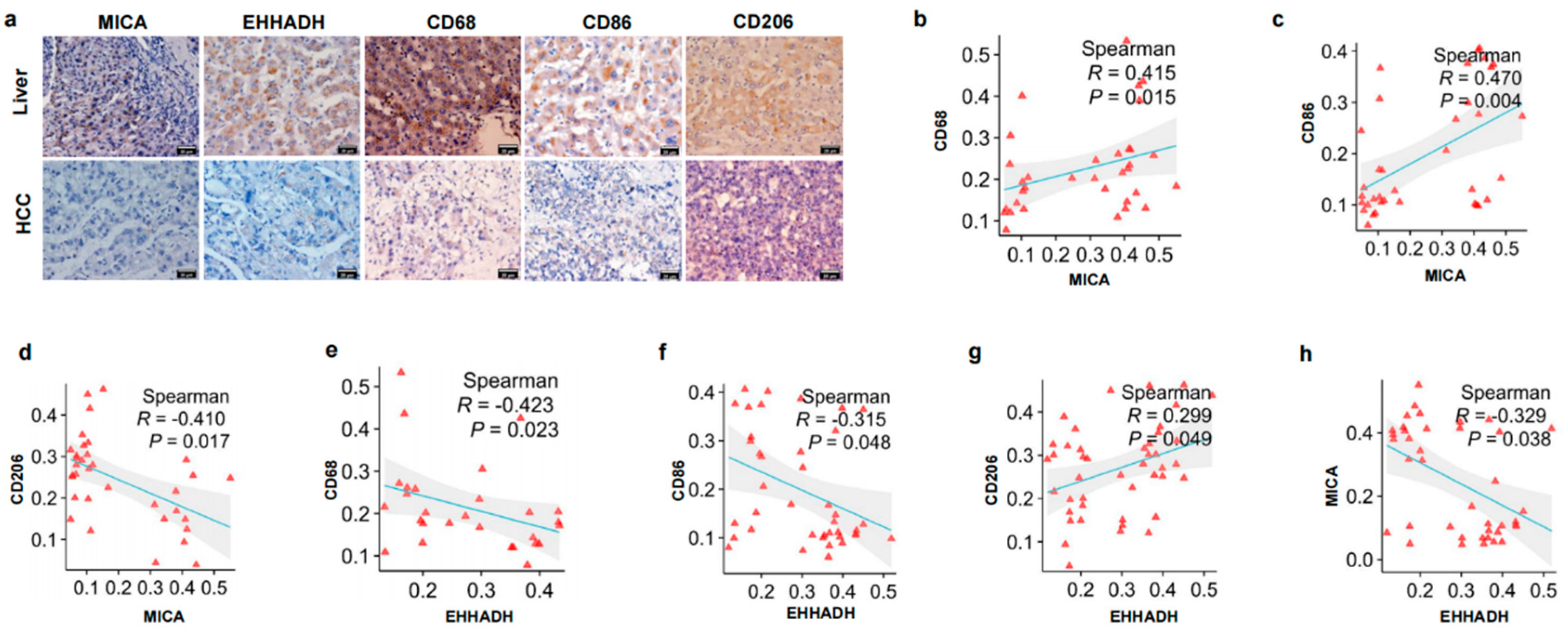
3.2. The Relationship Between MICA and EHHADH Was Associated with Macrophage Infiltration and Its Phenotype in HCC
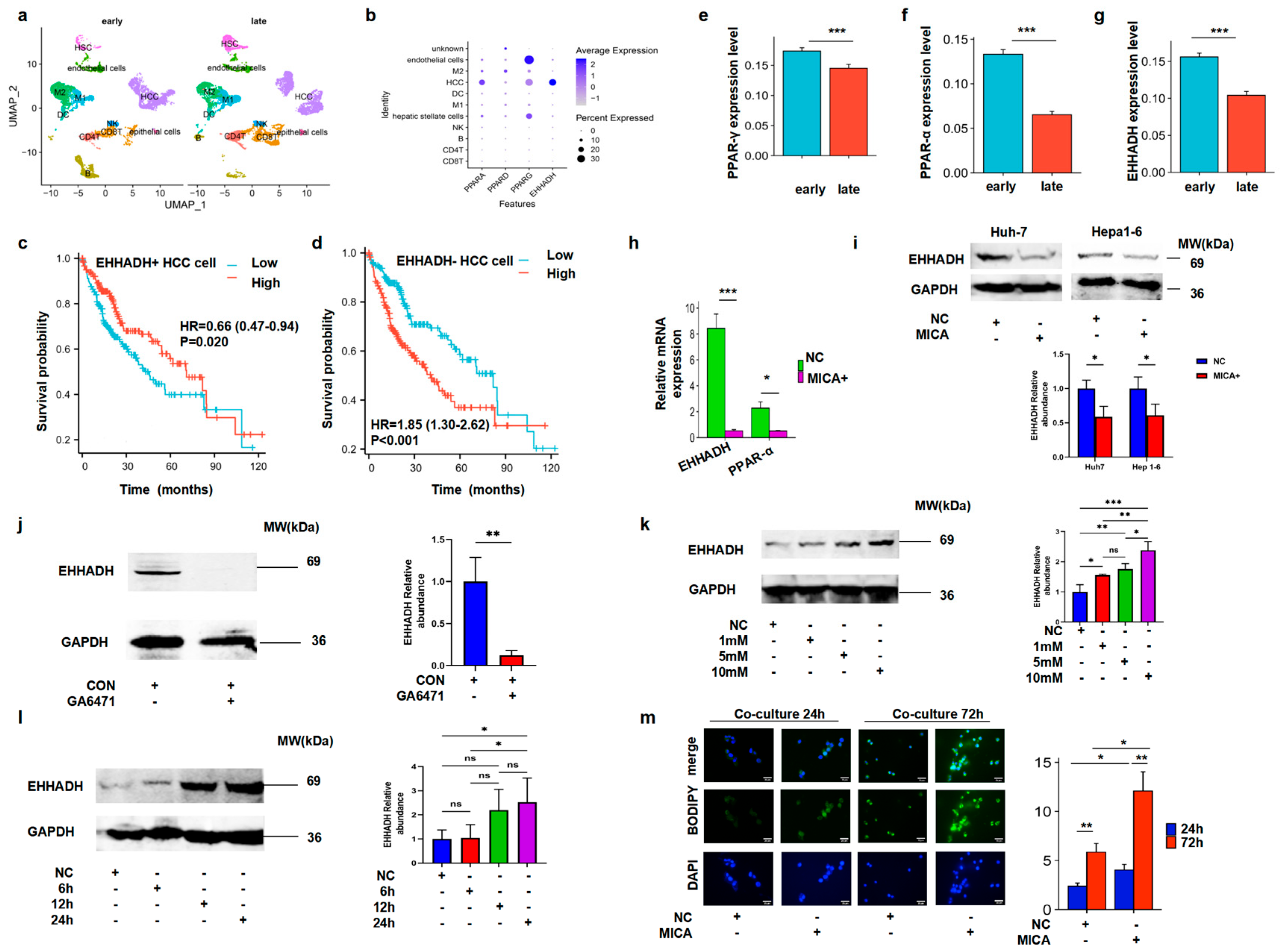
3.3. MICA+ HCC Cells Regulated Macrophage Polarization
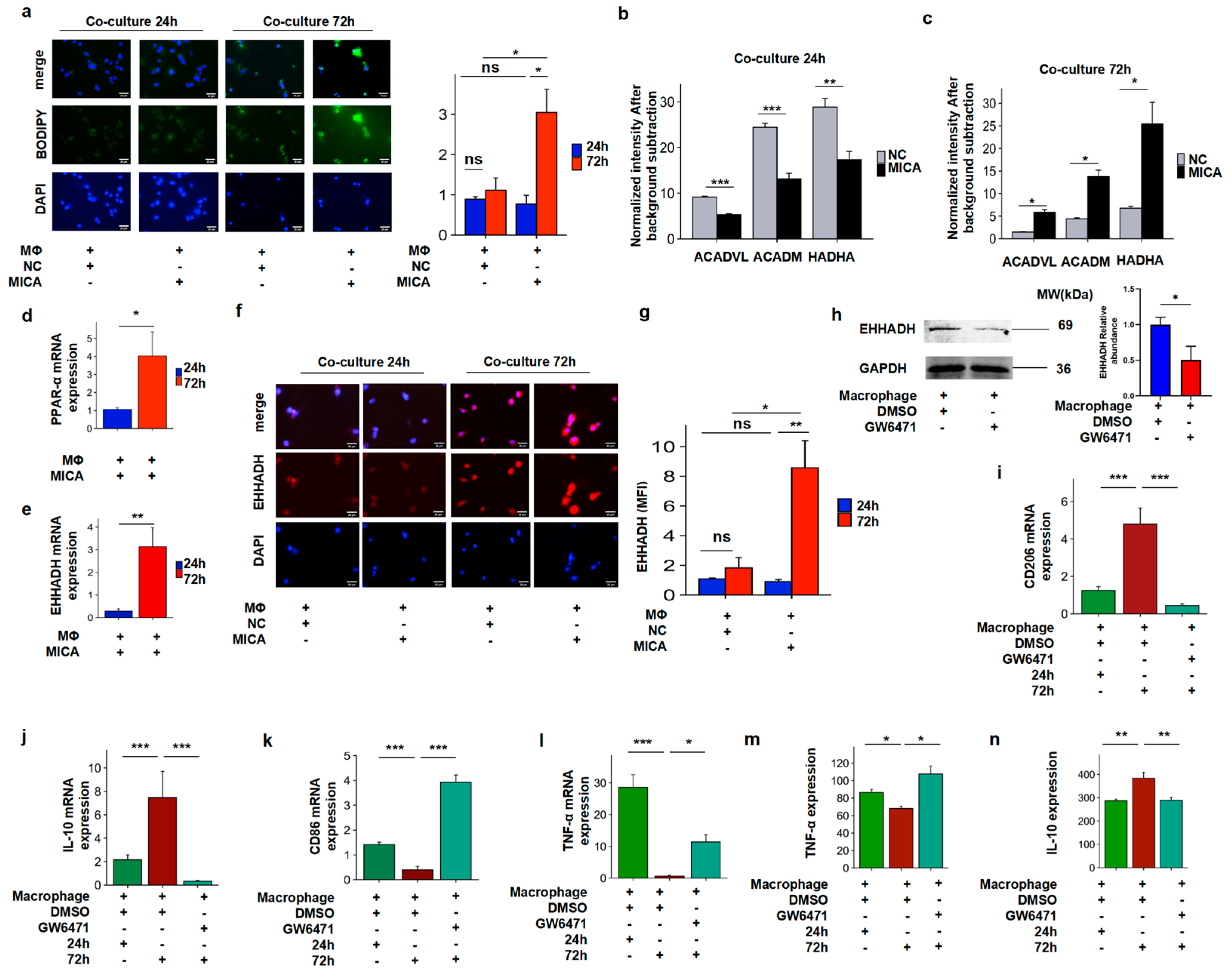
3.4. MICA+ HCC Cells Increased Fatty Acid Accumulation Through the Decreased PPAR-α/EHHADH Signaling Pathway
3.5. The Increased FAO Level Induced Phenotypic Alteration in Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AOD | Average optical density |
| BP | Biological process |
| cBioportal | cBio Cancer Genomics Portal |
| CC | Cellular component |
| CEG | Co-expression genes |
| CNV | Copy number variation |
| DAB | 3,3′-diaminobenzidine |
| DAP10 | DNAX-Activation Protein 10 |
| DEG | Differentially expressed gene |
| EHHADH | Enoyl-CoA Hydratase And 3-Hydroxyacyl CoA Dehydrogenase |
| FAO | Fatty acid oxidation |
| FAS | Fatty acid synthesis |
| FBS | Fetal bovine serum |
| FFA | Free fatty acid |
| GEPIA | Gene Expression Profiling Interaction Analysis |
| GO | Gene Ontology |
| HCC | Hepatocellular carcinoma |
| HPA | The Human Protein Atlas |
| IHC | Immunohistochemistry |
| IL | Interleukin |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LPS | Lipopolysaccharide |
| MF | Molecular function |
| MHC | Major histocompatibility complex |
| MICA | Major histocompatibility complex class I polypeptide-related sequence A |
| MRGs | Metabolism-related genes |
| NC | Negative control |
| NKG2D | Natural killer group 2, member D |
| PCA | Principal component analysis |
| PCs | Principal components |
| PMA | Phorbol 12-myristate 13-acetate |
| PPP | Pentose phosphate pathway |
| qRT-PCR | Quantitative real-time PCR |
| ROC | Receiver operating characteristic |
| scRNA-seq | Single-cell RNA sequence |
| TAM | Tumor-associated macrophages |
| TCA | Tricarboxylic acid cycle |
| TCGA | The Cancer Genome Atlas |
| TIMER | Tumor IMmune Estimation Resource |
| TME | Tumor microenvironment |
References
- Zhou, Z.; Wang, Z.; Gao, J.; Lin, Z.; Wang, Y.; Shan, P.; Li, M.; Zhou, T.; Li, P. Noncoding RNA-mediated macrophage and cancer cell crosstalk in hepatocellular carcinoma. Mol. Ther. Oncolytics 2022, 25, 98–120. [Google Scholar] [CrossRef] [PubMed]
- Colotta, F.; Allavena, P.; Sica, A.; Garlanda, C.; Mantovani, A. Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis 2009, 30, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Zou, W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat. Rev. Cancer 2005, 5, 263–274. [Google Scholar] [CrossRef]
- Rabold, K.; Netea, M.G.; Adema, G.J.; Netea-Maier, R.T. Cellular metabolism of tumor-associated macrophages—Functional impact and consequences. FEBS Lett. 2017, 591, 3022–3041. [Google Scholar] [CrossRef] [PubMed]
- Geiss, C.; Salas, E.; Guevara-Coto, J.; Regnier-Vigouroux, A.; Mora-Rodriguez, R.A. Multistability in Macrophage Activation Pathways and Metabolic Implications. Cells 2022, 11, 404. [Google Scholar] [CrossRef]
- Shi, Y.; Li, S.; Zhang, H.; Zhu, J.; Che, T.; Yan, B.; Li, J.; Liu, C. The effect of macrophage polarization on the expression of the oxytocin signalling system in enteric neurons. J. Neuroinflamm. 2021, 18, 261. [Google Scholar] [CrossRef]
- Kuntzel, T.; Bagnard, D. Manipulating Macrophage/Microglia Polarization to Treat Glioblastoma or Multiple Sclerosis. Pharmaceutics 2022, 14, 344. [Google Scholar] [CrossRef]
- Sarode, A.Y.; Jha, M.K.; Zutshi, S.; Ghosh, S.K.; Mahor, H.; Sarma, U.; Saha, B. Residue-Specific Message Encoding in CD40-Ligand. iScience 2020, 23, 101441. [Google Scholar] [CrossRef]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef]
- Ho, P.C.; Liu, P.S. Metabolic communication in tumors: A new layer of immunoregulation for immune evasion. J. Immunother. Cancer 2016, 4, 4. [Google Scholar] [CrossRef]
- Mills, E.L.; O’Neill, L.A. Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. Eur. J. Immunol. 2016, 46, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Pearce, E.L.; Pearce, E.J. Metabolic pathways in immune cell activation and quiescence. Immunity 2013, 38, 633–643. [Google Scholar] [CrossRef]
- Machuldova, A.; Holubova, M.; Caputo, V.S.; Cedikova, M.; Jindra, P.; Houdova, L.; Pitule, P. Role of Polymorphisms of NKG2D Receptor and Its Ligands in Acute Myeloid Leukemia and Human Stem Cell Transplantation. Front. Immunol. 2021, 12, 651751. [Google Scholar] [CrossRef]
- Delaune, V.; Toso, C.; Kahler-Quesada, A.; Slits, F.; Gex, Q.; Kaya, G.; Lavallard, V.; Orci, L.A.; Peloso, A.; Lacotte, S. Antibody-induced NKG2D blockade in a rat model of intraportal islet transplantation leads to a deleterious reaction. Transpl. Int. 2020, 33, 675–688. [Google Scholar] [CrossRef]
- Li, X.; Huang, J.; Wu, Q.; Du, Q.; Wang, Y.; Huang, Y.; Cai, X.; Geller, D.A.; Yan, Y. Inhibition of Checkpoint Kinase 1 (CHK1) Upregulates Interferon Regulatory Factor 1 (IRF1) to Promote Apoptosis and Activate Anti-Tumor Immunity via MICA in Hepatocellular Carcinoma (HCC). Cancers 2023, 15, 850. [Google Scholar] [CrossRef]
- Navarro, A.; Chen, Y.; Lin, G.; Guo, Z.-q.; Zhou, Z.-f.; He, Z.-y.; Ye, Y.-b. Effects of MICA Expression on the Prognosis of Advanced Non-Small Cell Lung Cancer and the Efficacy of CIK Therapy. PLoS ONE 2013, 8, e69044. [Google Scholar] [CrossRef]
- Yizhou, Z.; Xiaocui, H.; Ming, L.; Shili, L.; Min, X.; Rongjiao, L.; Huiyun, P.; Hongjun, H.; Quan, Z.; Weiguang, L.; et al. Tumor-Derived Soluble MICA Obstructs the NKG2D Pathway to Restrain NK Cytotoxicity. Aging Dis. 2020, 11, 118–128. [Google Scholar] [CrossRef]
- Wu, Q.; Li, X.; Yang, Y.; Huang, J.; Yao, M.; Li, J.; Huang, Y.; Cai, X.; Geller, D.A.; Yan, Y. MICA+ Tumor Cell Upregulated Macrophage-Secreted MMP9 via PROS1-AXL Axis to Induce Tumor Immune Escape in Advanced Hepatocellular Carcinoma (HCC). Cancers 2024, 16, 269. [Google Scholar] [CrossRef]
- Høgh, R.I.; Møller, S.H.; Jepsen, S.D.; Mellergaard, M.; Lund, A.; Pejtersen, M.; Fitzner, E.; Andresen, L.; Skov, S. Metabolism of short-chain fatty acid propionate induces surface expression of NKG2D ligands on cancer cells. FASEB J. 2020, 34, 15531–15546. [Google Scholar] [CrossRef]
- Moller, S.H.; Mellergaard, M.; Madsen, M.; Bermejo, A.V.; Jepsen, S.D.; Hansen, M.H.; Hogh, R.I.; Aldana, B.I.; Desler, C.; Rasmussen, L.J.; et al. Cytoplasmic Citrate Flux Modulates the Immune Stimulatory NKG2D Ligand MICA in Cancer Cells. Front. Immunol. 2020, 11, 1968. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Springer Nature: Berlin/Heidelberg, Germany, 2021; pp. 27–56. [Google Scholar]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 126409. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Lu, Y.; Yang, A.; Quan, C.; Pan, Y.; Zhang, H.; Li, Y.; Gao, C.; Lu, H.; Wang, X.; Cao, P.; et al. A single-cell atlas of the multicellular ecosystem of primary and metastatic hepatocellular carcinoma. Nat. Commun. 2022, 13, 4594. [Google Scholar] [CrossRef]
- Zhang, X.; Lan, Y.; Xu, J.; Quan, F.; Zhao, E.; Deng, C.; Luo, T.; Xu, L.; Liao, G.; Yan, M.; et al. CellMarker: A manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019, 47, D721–D728. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wawryk-Gawda, E.; Chlapek, K.; Zarobkiewicz, M.K.; Lis-Sochocka, M.; Chylinska-Wrzos, P.; Boguszewska-Czubara, A.; Slawinski, M.A.; Franczak, A.; Jodlowska-Jedrych, B.; Biala, G. CB2R agonist prevents nicotine induced lung fibrosis. Exp. Lung Res. 2018, 44, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Jiang, X.M.; Yu, X.N.; Liu, T.T.; Zhu, H.R.; Shi, X.; Bilegsaikhan, E.; Guo, H.Y.; Song, G.Q.; Weng, S.Q.; Huang, X.X.; et al. microRNA-19a-3p promotes tumor metastasis and chemoresistance through the PTEN/Akt pathway in hepatocellular carcinoma. Biomed. Pharmacother. 2018, 105, 1147–1154. [Google Scholar] [CrossRef]
- He, Q.; Yang, C.; Xiang, Z.; Huang, G.; Wu, H.; Chen, T.; Dou, R.; Song, J.; Han, L.; Song, T.; et al. LINC00924-induced fatty acid metabolic reprogramming facilitates gastric cancer peritoneal metastasis via hnRNPC-regulated alternative splicing of Mnk2. Cell Death Dis. 2022, 13, 987. [Google Scholar] [CrossRef]
- Serrano, I.; McDonald, P.C.; Lock, F.; Muller, W.J.; Dedhar, S. Inactivation of the Hippo tumour suppressor pathway by integrin-linked kinase. Nat. Commun. 2013, 4, 2976. [Google Scholar] [CrossRef]
- Collins, T.J. ImageJ for microscopy. Biotechniques 2007, 43, 25–30. [Google Scholar] [CrossRef]
- Jiang, N.; Xie, B.; Xiao, W.; Fan, M.; Xu, S.; Duan, Y.; Hamsafar, Y.; Evans, A.C.; Huang, J.; Zhou, W.; et al. Fatty acid oxidation fuels glioblastoma radioresistance with CD47-mediated immune evasion. Nat. Commun. 2022, 13, 1511. [Google Scholar] [CrossRef]
- Li, L.; Liu, Y.; Liu, X.; Zheng, N.; Gu, Y.; Song, Y.; Wang, X. Regulatory roles of external cholesterol in human airway epithelial mitochondrial function through STARD3 signalling. Clin. Transl. Med. 2022, 12, e902. [Google Scholar] [CrossRef]
- Jung, T.W.; Park, H.S.; Choi, G.H.; Kim, D.; Lee, T. beta-aminoisobutyric acid attenuates LPS-induced inflammation and insulin resistance in adipocytes through AMPK-mediated pathway. J. Biomed. Sci. 2018, 25, 27. [Google Scholar] [CrossRef]
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T.; et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 2022, 1, e36. [Google Scholar] [CrossRef] [PubMed]
- Houten, S.M.; Denis, S.; Argmann, C.A.; Jia, Y.; Ferdinandusse, S.; Reddy, J.K.; Wanders, R.J.A. Peroxisomal L-bifunctional enzyme (Ehhadh) is essential for the production of medium-chain dicarboxylic acids. J. Lipid Res. 2012, 53, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Su, P.; Wang, Q.; Bi, E.; Ma, X.; Liu, L.; Yang, M.; Qian, J.; Yi, Q. Enhanced Lipid Accumulation and Metabolism Are Required for the Differentiation and Activation of Tumor-Associated Macrophages. Cancer Res. 2020, 80, 1438–1450. [Google Scholar] [CrossRef]
- Hachim, D.; LoPresti, S.T.; Rege, R.D.; Umeda, Y.; Iftikhar, A.; Nolfi, A.L.; Skillen, C.D.; Brown, B.N. Distinct macrophage populations and phenotypes associated with IL-4 mediated immunomodulation at the host implant interface. Biomater. Sci. 2020, 8, 5751–5762. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Rőszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef]
- Yang, P.; Qin, H.; Li, Y.; Xiao, A.; Zheng, E.; Zeng, H.; Su, C.; Luo, X.; Lu, Q.; Liao, M.; et al. CD36-mediated metabolic crosstalk between tumor cells and macrophages affects liver metastasis. Nat. Commun. 2022, 13, 5782. [Google Scholar] [CrossRef]
- Ahuja, P.; Ng, C.F.; Pang, B.P.S.; Chan, W.S.; Tse, M.C.L.; Bi, X.; Kwan, H.-L.R.; Brobst, D.; Herlea-Pana, O.; Yang, X.; et al. Muscle-generated BDNF (brain derived neurotrophic factor) maintains mitochondrial quality control in female mice. Autophagy 2021, 18, 1367–1384. [Google Scholar] [CrossRef]
- Ma, A.P.Y.; Yeung, C.L.S.; Tey, S.K.; Mao, X.; Wong, S.W.K.; Ng, T.H.; Ko, F.C.F.; Kwong, E.M.L.; Tang, A.H.N.; Ng, I.O.; et al. Suppression of ACADM-Mediated Fatty Acid Oxidation Promotes Hepatocellular Carcinoma via Aberrant CAV1/SREBP1 Signaling. Cancer Res. 2021, 81, 3679–3692. [Google Scholar] [CrossRef]
- Davuluri, G.V.N.; Chen, C.C.; Chiu, Y.C.; Tsai, H.W.; Chiu, H.C.; Chen, Y.L.; Tsai, P.J.; Kuo, W.T.; Tsao, N.; Lin, Y.S.; et al. Autophagy Drives Galectin-1 Secretion From Tumor-Associated Macrophages Facilitating Hepatocellular Carcinoma Progression. Front. Cell Dev. Biol. 2021, 9, 741820. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, X.; Zheng, L.; Zhao, H.; Yan, G.; Zhang, Q.; Zhou, Y.; Lei, J.; Zhang, J.; Wang, J.; et al. RIPK3 Orchestrates Fatty Acid Metabolism in Tumor-Associated Macrophages and Hepatocarcinogenesis. Cancer Immunol. Res. 2020, 8, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Rui, R.; Zhou, L.; He, S. Cancer immunotherapies: Advances and bottlenecks. Front. Immunol. 2023, 14, 1212476. [Google Scholar] [CrossRef]
- Xie, S.; Li, M.; Jiang, F.; Yi, Q.; Yang, W. EHHADH is a key gene in fatty acid metabolism pathways in hepatocellular carcinoma: A transcriptomic analysis. J. South. Med. Univ. 2023, 43, 680–693. [Google Scholar] [CrossRef]
- Li, H.; Yu, J.; Wu, Y.; Shao, B.; Wei, X. In situ antitumor vaccination: Targeting the tumor microenvironment. J. Cell Physiol. 2020, 235, 5490–5500. [Google Scholar] [CrossRef] [PubMed]
- Galbo, P.M., Jr.; Zang, X.; Zheng, D. Molecular Features of Cancer-associated Fibroblast Subtypes and their Implication on Cancer Pathogenesis, Prognosis, and Immunotherapy Resistance. Clin. Cancer Res. 2021, 27, 2636–2647. [Google Scholar] [CrossRef]
- Zhang, J.; Li, R.; Huang, S. The immunoregulation effect of tumor microenvironment in pancreatic ductal adenocarcinoma. Front. Oncol. 2022, 12, 951019. [Google Scholar] [CrossRef]
- Carracedo, A.; Cantley, L.C.; Pandolfi, P.P. Cancer metabolism: Fatty acid oxidation in the limelight. Nat. Rev. Cancer 2013, 13, 227–232. [Google Scholar] [CrossRef]
- Sun, J.X.; Xu, X.H.; Jin, L. Effects of Metabolism on Macrophage Polarization Under Different Disease Backgrounds. Front. Immunol. 2022, 13, 880286. [Google Scholar] [CrossRef]
- Tian, F.; Chen, H.; Zhang, J.; He, W. Reprogramming Metabolism of Macrophages as a Target for Kidney Dysfunction Treatment in Autoimmune Diseases. Int. J. Mol. Sci. 2022, 23, 8024. [Google Scholar] [CrossRef]
- Jiang, T.; Huang, J.B.; Xu, C.Y.; Lv, Y.L.; Lu, J.; Zhao, Z.Q.; Yang, D.Q.; Lou, Z.H.; Zhang, G.J. Arsenic Trioxide Cooperate Cryptotanshinone Exerts Antitumor Effect by Medicating Macrophage Polarization through Glycolysis. J. Immunol. Res. 2022, 2022, 2619781. [Google Scholar] [CrossRef]

| Gene Name | Sequence of Primer |
|---|---|
| MICA | F: CACAGCGGGAATCACAGCACTC |
| R: ATAGCAGCAGCAGCAACAGCAG | |
| EHHADH | F: GTCAACGCGATCAGTACGAC |
| R: CCTAGGAGCACTGAAGCCAC | |
| PPAR-α | F: TCGGCGAGGATAGTTCTGGAAGC |
| R: ACCACAGGATAAGTCACCGAGGAG | |
| CD68 | F: GTTCATCCAACAAGCAACAGCACTG |
| R: CGGAGAGGGTGGAGGTGGTTC | |
| CD86 | F: GGAACCAACACAATGGAGAG |
| R: AAACACGCTGGGCTTCATC | |
| CD206 | F: TCGGGTTTATGGAGCAGGTG |
| R: TGAACGGGAATGCACAGGTT | |
| GAPDH | F: GCACCGTCAAGGCTGAGAAC |
| R: TGGTGAAGACGCCAGTGGA | |
| TNF-α | F: TGCTCCTCACCCACACCAT |
| R: GGAGGTTGACCTTGGTCTGGTA | |
| IL-10 | F: ATCCAAGACAACACTACTAA |
| R: TAAATATCCTCAAAGTTCC | |
| PPAR-γ | F: CCAGAAGCCTGCATTTCTGC |
| R: GTGTCAACCATGGTCATTTCGTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Teng, Y.; Yan, P.; Yang, Y.; Lin, S.; Wu, Q.; Du, Q.; Li, X.; Yao, M.; Li, J.; et al. MICA+ Tumor Cells Modulate Macrophage Phenotype and Function via PPAR/EHHADH-Mediated Fatty Acid Metabolism in Hepatocellular Carcinoma (HCC). Cancers 2025, 17, 2365. https://doi.org/10.3390/cancers17142365
Huang J, Teng Y, Yan P, Yang Y, Lin S, Wu Q, Du Q, Li X, Yao M, Li J, et al. MICA+ Tumor Cells Modulate Macrophage Phenotype and Function via PPAR/EHHADH-Mediated Fatty Acid Metabolism in Hepatocellular Carcinoma (HCC). Cancers. 2025; 17(14):2365. https://doi.org/10.3390/cancers17142365
Chicago/Turabian StyleHuang, Jingquan, Yumeng Teng, Peng Yan, Yan Yang, Shixun Lin, Qiulin Wu, Qiang Du, Xicai Li, Ming Yao, Jianjun Li, and et al. 2025. "MICA+ Tumor Cells Modulate Macrophage Phenotype and Function via PPAR/EHHADH-Mediated Fatty Acid Metabolism in Hepatocellular Carcinoma (HCC)" Cancers 17, no. 14: 2365. https://doi.org/10.3390/cancers17142365
APA StyleHuang, J., Teng, Y., Yan, P., Yang, Y., Lin, S., Wu, Q., Du, Q., Li, X., Yao, M., Li, J., Huang, Y., Cai, X., Geller, D. A., & Yan, Y. (2025). MICA+ Tumor Cells Modulate Macrophage Phenotype and Function via PPAR/EHHADH-Mediated Fatty Acid Metabolism in Hepatocellular Carcinoma (HCC). Cancers, 17(14), 2365. https://doi.org/10.3390/cancers17142365






