Multi-Cancer Genome Profiling for Neurotrophic Tropomyosin Receptor Kinase (NTRK) Fusion Genes: Analysis of Profiling Database of 88,688 Tumors
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Multi-CGP Tests and C-CAT
2.2. Extraction of Genetic Abnormalities
2.3. Statistical Analysis
3. Results
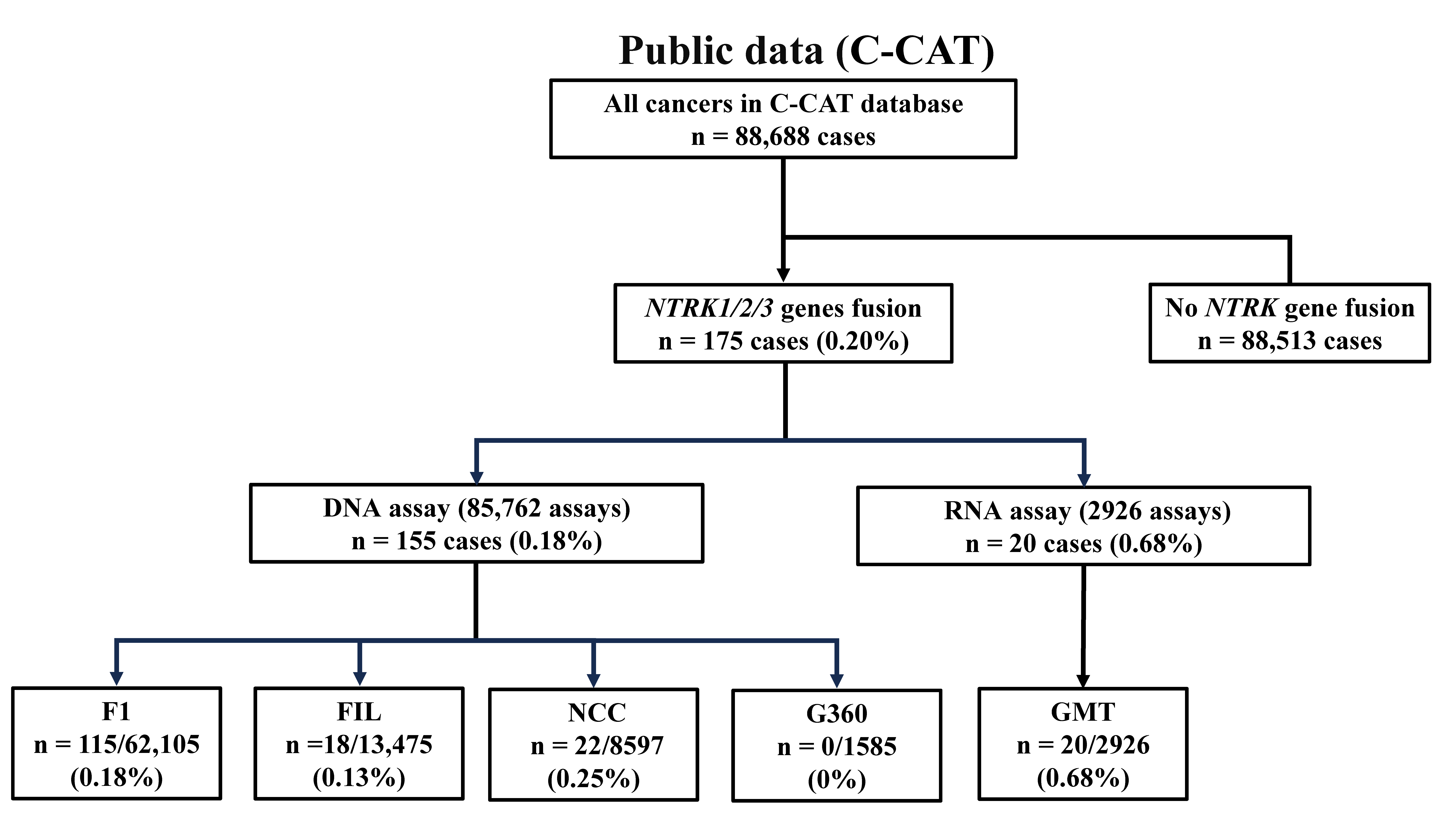
3.1. Patient Background of Cases with NTRK Fusion Genes
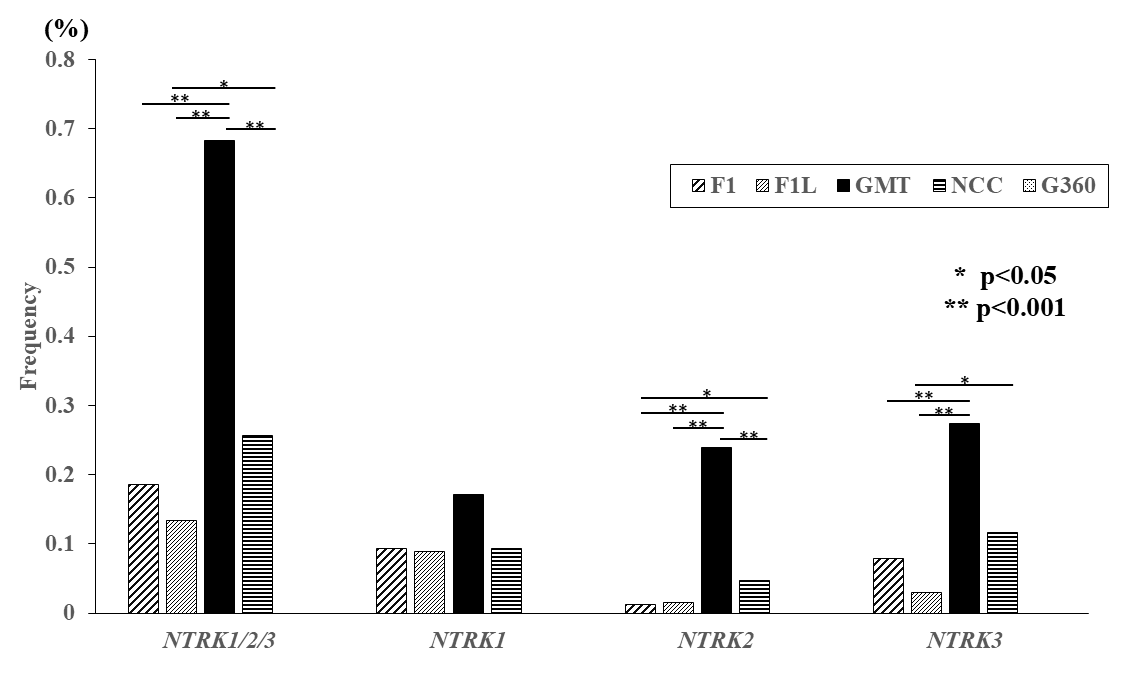
3.2. Detection Rate of NTRK Fusion Genes in Each Multi-CGP Test
3.3. Cases That Reached Treatment After Multi-CGP Tests
3.4. Organs with NTRK Fusion Genes and Their Fusion Partners
3.5. Detection Frequency of Other Fusion Genes, ALK, RET, ROS1, and FGFR1/2/3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cocco, E.; Scaltriti, M.; Drilon, A. NTRK Fusion-Positive Cancers and TRK Inhibitor Therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar] [CrossRef] [PubMed]
- Amatu, A.; Sartore-Bianchi, A.; Siena, S. NTRK Gene Fusions as Novel Targets of Cancer Therapy across Multiple Tumour Types. ESMO Open 2016, 1, 23. [Google Scholar] [CrossRef] [PubMed]
- Thiele, C.J.; Li, Z.; McKee, A.E. On Trk—The TrkB Signal Transduction Pathway Is an Increasingly Important Target in Cancer Biology. Clin. Cancer Res. 2009, 15, 5962–5967. [Google Scholar] [CrossRef] [PubMed]
- Hechtman, J.F. NTRK Insights: Best Practices for Pathologists. Mod. Pathol. 2022, 35, 298–305. [Google Scholar] [CrossRef]
- Penault-Llorca, F.; Rudzinski, E.R.; Sepulveda, A.R. Testing Algorithm for Identification of Patients with TRK Fusion Cancer. J. Clin. Pathol. 2019, 72, 460–467. [Google Scholar] [CrossRef]
- Amatu, A.; Sartore-Bianchi, A.; Bencardino, K.; Pizzutilo, E.G.; Tosi, F.; Siena, S. Tropomyosin Receptor Kinase (TRK) Biology and the Role of NTRK Gene Fusions in Cancer. Ann. Oncol. 2019, 30, VIII5–VIII15. [Google Scholar] [CrossRef]
- Doebele, R.C.; Drilon, A.; Paz-Ares, L.; Siena, S.; Shaw, A.T.; Farago, A.F.; Blakely, C.M.; Seto, T.; Cho, B.C.; Tosi, D.; et al. Entrectinib in Patients with Advanced or Metastatic NTRK Fusion-Positive Solid Tumours: Integrated Analysis of Three Phase 1–2 Trials. Lancet Oncol. 2020, 21, 271–282. [Google Scholar] [CrossRef]
- Blauel, E.R.; Laetsch, T.W. The Promise of TRK Inhibitors in Pediatric Cancers with NTRK Fusions. Cancer Genet. 2022, 262, 71–79. [Google Scholar] [CrossRef]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; DuBois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of Larotrectinib in TRK Fusion–Positive Cancers in Adults and Children. N. Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef]
- Solomon, J.P.; Benayed, R.; Hechtman, J.F.; Ladanyi, M. Identifying Patients with NTRK Fusion Cancer. Ann. Oncol. 2019, 30, VIII16–VIII22. [Google Scholar] [CrossRef]
- Brčić, I.; Godschachner, T.M.; Bergovec, M.; Igrec, J.; Till, H.; Lackner, H.; Scheipl, S.; Kashofer, K.; Brodowicz, T.; Leithner, A.; et al. Broadening the Spectrum of NTRK Rearranged Mesenchymal Tumors and Usefulness of Pan-TRK Immunohistochemistry for Identification of NTRK Fusions. Mod. Pathol. 2021, 34, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Kanai, M. Current Status and Issues of Companion Diagnostics in Cancer Genomic Medicine. Gan Kagaku Ryoho Cancer Chemother. 2024, 51, 388–391. [Google Scholar]
- Mukai, Y.; Ueno, H. Establishment and Implementation of Cancer Genomic Medicine in Japan. Cancer Sci. 2021, 112, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Kato, M.; Kohsaka, S.; Sudo, T.; Tamai, I.; Shiraishi, Y.; Okuma, Y.; Ogasawara, D.; Suzuki, T.; Yoshida, T.; et al. C-CAT: The National Datacenter for Cancer Genomic Medicine in Japan. Cancer Discov. 2022, 12, 2509–2515. [Google Scholar] [CrossRef]
- Sunami, K.; Ichikawa, H.; Kubo, T.; Kato, M.; Fujiwara, Y.; Shimomura, A.; Koyama, T.; Kakishima, H.; Kitami, M.; Matsushita, H.; et al. Feasibility and Utility of a Panel Testing for 114 Cancer-Associated Genes in a Clinical Setting: A Hospital-Based Study. Cancer Sci. 2019, 110, 1480–1490. [Google Scholar] [CrossRef]
- Yatabe, Y.; Sunami, K.; Goto, K.; Nishio, K.; Aragane, N.; Ikeda, S.; Inoue, A.; Kinoshita, I.; Kimura, H.; Sakamoto, T.; et al. Multiplex Gene-Panel Testing for Lung Cancer Patients. Pathol. Int. 2020, 70, 921–931. [Google Scholar] [CrossRef]
- Woodhouse, R.; Li, M.; Hughes, J.; Delfosse, D.; Skoletsky, J.; Ma, P.; Meng, W.; Dewal, N.; Milbury, C.; Clark, T.; et al. Clinical and Analytical Validation of Foundation One Liquid CDx, a Novel 324-Gene CfDNA-Based Comprehensive Genomic Profiling Assay for Cancers of Solid Tumor Origin. PLoS ONE 2020, 15, e0237802. [Google Scholar] [CrossRef]
- Watanabe, K.; Ogawa, M.; Shinozaki-Ushiku, A.; Tsutsumi, S.; Tatsuno, K.; Aburatani, H.; Kage, H.; Oda, K. Real-World Data Analysis of Genomic Alterations Detected by a Dual DNA–RNA Comprehensive Genomic Profiling Test. Cancer Sci. 2025. Online ahead of print. [Google Scholar] [CrossRef]
- Nakata, E.; Osone, T.; Ogawa, T.; Taguchi, T.; Hattori, K.; Kohsaka, S. Prevalence of Neurotrophic Tropomyosin Receptor Kinase (NTRK) Fusion Gene Positivity in Patients with Solid Tumors in Japan. Cancer Med. 2024, 13, e7351. [Google Scholar] [CrossRef]
- O’Haire, S.; Franchini, F.; Kang, Y.J.; Steinberg, J.; Canfell, K.; Desai, J.; Fox, S.; IJzerman, M. Systematic Review of NTRK 1/2/3 Fusion Prevalence Pan-Cancer and across Solid Tumours. Sci. Rep. 2023, 13, 4116. [Google Scholar] [CrossRef]
- Gatalica, Z.; Xiu, J.; Swensen, J.; Vranic, S. Molecular Characterization of Cancers with NTRK Gene Fusions. Mod. Pathol. 2019, 32, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, S.J.; Zehir, A.; Sireci, A.N.; Aisner, D.L. Detection of Tumor NTRK Gene Fusions to Identify Patients Who May Benefit from Tyrosine Kinase (TRK) Inhibitor Therapy. J. Mol. Diagn. 2019, 21, 553–571. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Suurmeijer, A.J.H.; Agaram, N.P.; Antonescu, C.R. Head and Neck Mesenchymal Tumors with Kinase Fusions: A Report of 15 Cases with Emphasis on Wide Anatomic Distribution and Diverse Histologic Appearance. Am. J. Surg. Pathol. 2023, 47, 248–258. [Google Scholar] [CrossRef]
- Okamura, R.; Boichard, A.; Kato, S.; Sicklick, J.K.; Bazhenova, L.; Kurzrock, R. Analysis of NTRK Alterations in Pan-Cancer Adult and Pediatric Malignancies: Implications for NTRK-Targeted Therapeutics. JCO Precis. Oncol. 2018, 2, 1–20. [Google Scholar] [CrossRef]
- Laetsch, T.W.; Hong, D.S. Tropomyosin Receptor Kinase Inhibitors for the Treatment of TRK Fusion Cancer. Clin. Cancer Res. 2021, 27, 4974–4982. [Google Scholar] [CrossRef]
- Solomon, J.P.; Hechtman, J.F. Detection of NTRK Fusions: Merits and Limitations of Current Diagnostic Platforms. Cancer Res. 2019, 79, 3163–3168. [Google Scholar] [CrossRef]
- Benayed, R.; Offin, M.; Mullaney, K.; Sukhadia, P.; Rios, K.; Desmeules, P.; Ptashkin, R.; Won, H.; Chang, J.; Halpenny, D.; et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin. Cancer Res. 2019, 25, 4712–4722. [Google Scholar] [CrossRef]
- Sorokin, M.; Rabushko, E.; Rozenberg, J.M.; Mohammad, T.; Seryakov, A.; Sekacheva, M.; Buzdin, A. Clinically Relevant Fusion Oncogenes: Detection and Practical Implications. Ther. Adv. Med. Oncol. 2022, 14, 17588359221144108. [Google Scholar] [CrossRef]
- Vendrell, J.A.; Taviaux, S.; Béganton, B.; Godreuil, S.; Audran, P.; Grand, D.; Clermont, E.; Serre, I.; Szablewski, V.; Coopman, P.; et al. Detection of Known and Novel ALK Fusion Transcripts in Lung Cancer Patients Using Next-Generation Sequencing Approaches. Sci. Rep. 2017, 7, 12510. [Google Scholar] [CrossRef]
- Mondaca, S.; Lebow, E.S.; Namakydoust, A.; Razavi, P.; Reis-Filho, J.S.; Shen, R.; Offin, M.; Tu, H.Y.; Murciano-Goroff, Y.; Xu, C.; et al. Clinical Utility of Next-Generation Sequencing-Based CtDNA Testing for Common and Novel ALK Fusions. Lung Cancer 2021, 159, 66–73. [Google Scholar] [CrossRef]
- Maansson, C.T.; Andersen, E.R.; Ulhoi, M.P.; Meldgaard, P.; Sorensen, B.S. DNAfusion: An R/Bioconductor Package for Increased Sensitivity of Detecting Gene Fusions in Liquid Biopsies. BMC Bioinform. 2023, 24, 131. [Google Scholar] [CrossRef] [PubMed]
- Li, A.Y.; McCusker, M.G.; Russo, A.; Scilla, K.A.; Gittens, A.; Arensmeyer, K.; Mehra, R.; Adamo, V.; Rolfo, C. RET Fusions in Solid Tumors. Cancer Treat Rev. 2019, 81, 101911. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Wang, W.; Zhang, J.; Li, B.; Lv, D.; Wang, D.; Wang, S.; Cheng, D.; Ma, T. Identification of RET Fusions in a Chinese Multicancer Retrospective Analysis by Next-Generation Sequencing. Cancer Sci. 2022, 113, 308–318. [Google Scholar] [CrossRef] [PubMed]


| Clinical Features | NTRK1/2/3 Fusion | NTRK1 Fusion | NTRK2 Fusion | NTRK3 Fusion | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | |||||
| n = 175 | n = 88,513 | p-Value | n = 83 | n = 88,605 | p-Value | n = 21 | n = 88,667 | p-Value | n = 71 | n = 88,617 | p-Value | |
| Sex | ||||||||||||
| Male (n = 44,337) | 95 (0.21%) | 44,242 (99.79%) | 36 (0.08%) | 44,301 (99.92%) | 14 (0.03%) | 44,323 (99.97%) | 45 (0.10%) | 44,292 (99.90%) | ||||
| Female (n = 44,346) | 80 (0.18%) | 44,266 (99.82%) | n.s. | 47 (0.11%) | 44,299 (99.89%) | n.s. | 7 (0.02%) | 44,339 (99.98%) | n.s. | 26 (0.06%) | 44,320 (99.94%) | 0.02 |
| Age | ||||||||||||
| <20 (n = 1670) | 29 (1.74%) | 1641 (98.26%) | 14 (0.84%) | 1656 (99.16%) | 3 (0.18%) | 1667 (99.82%) | 12 (0.72%) | 1658 (99.28%) | ||||
| ≥20 (n = 87,018) | 146 (0.17%) | 86,872 (99.83%) | <0.001 | 69 (0.08%) | 86,949 (99.92%) | <0.001 | 18 (0.02%) | 87,000 (99.98%) | <0.001 | 59 (0.07%) | 86,959 (99.93%) | <0.001 |
| Multi-CGP tests assay | ||||||||||||
| DNA assay (n = 85,762) | 155 (0.18%) | 85,607 (99.82%) | 78 (0.09%) | 85,684 (99.91%) | 14 (0.02%) | 85,748 (99.98%) | 63 (0.07%) | 85,699 (99.93%) | ||||
| RNA assay (n = 2926) | 20 (0.68%) | 2906 (99.32%) | <0.001 | 5 (0.17%) | 2921 (99.83%) | n.s. | 7 (0.24%) | 2919 (99.76%) | <0.001 | 8 (0.27%) | 2918 (99.73%) | <0.001 |
| Materials | ||||||||||||
| Tissue (n = 73,628) | 157 (0.21%) | 73,471(99.79%) | 71 (0.10%) | 73,557 (99.90%) | 19 (0.03%) | 73,609 (99.97%) | 67 (0.09%) | 73,561 (99.91%) | ||||
| Blood (n = 15,060) | 18 (0.12%) | 15,042 (99.88%) | 0.02 | 12 (0.08%) | 15,048 (99.92%) | n.s. | 2 (0.01%) | 15,058 (99.99%) | n.s. | 4 (0.03%) | 15,056 (99.97%) | 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishikubo, H.; Kawabata, K.; Kanei, S.; Aoyama, R.; Ma, D.; Sano, T.; Imanishi, D.; Sakuma, T.; Maruo, K.; Fan, C.; et al. Multi-Cancer Genome Profiling for Neurotrophic Tropomyosin Receptor Kinase (NTRK) Fusion Genes: Analysis of Profiling Database of 88,688 Tumors. Cancers 2025, 17, 2250. https://doi.org/10.3390/cancers17132250
Nishikubo H, Kawabata K, Kanei S, Aoyama R, Ma D, Sano T, Imanishi D, Sakuma T, Maruo K, Fan C, et al. Multi-Cancer Genome Profiling for Neurotrophic Tropomyosin Receptor Kinase (NTRK) Fusion Genes: Analysis of Profiling Database of 88,688 Tumors. Cancers. 2025; 17(13):2250. https://doi.org/10.3390/cancers17132250
Chicago/Turabian StyleNishikubo, Hinano, Kyoka Kawabata, Saki Kanei, Rika Aoyama, Dongheng Ma, Tomoya Sano, Daiki Imanishi, Takashi Sakuma, Koji Maruo, Canfeng Fan, and et al. 2025. "Multi-Cancer Genome Profiling for Neurotrophic Tropomyosin Receptor Kinase (NTRK) Fusion Genes: Analysis of Profiling Database of 88,688 Tumors" Cancers 17, no. 13: 2250. https://doi.org/10.3390/cancers17132250
APA StyleNishikubo, H., Kawabata, K., Kanei, S., Aoyama, R., Ma, D., Sano, T., Imanishi, D., Sakuma, T., Maruo, K., Fan, C., Yamamoto, Y., & Yashiro, M. (2025). Multi-Cancer Genome Profiling for Neurotrophic Tropomyosin Receptor Kinase (NTRK) Fusion Genes: Analysis of Profiling Database of 88,688 Tumors. Cancers, 17(13), 2250. https://doi.org/10.3390/cancers17132250







