MicroRNAs in Cancer Immunology: Master Regulators of the Tumor Microenvironment and Immune Evasion, with Therapeutic Potential
Simple Summary
Abstract
1. Introduction
2. miRNAs and Immune Surveillance
2.1. T Lymphocytes
2.2. CD8+ Cytotoxic T Lymphocytes (CTLs)
2.3. CD4+ Helper T Lymphocytes
2.4. Regulatory T Cells (Treg)
2.5. Natural Killer (NK) Cells
2.6. Viral miRNAs and NK Cell Evasion
2.7. Dendritic Cells (DCs)
2.8. Tumor-Associated Macrophages (TAMs)
2.9. Tumor-Associated Neutrophils (TANs)
3. miRNAs and Immune Equilibrium
3.1. T Cells
3.2. Regulatory T Cell (Treg)
3.3. Tumor-Associated Macrophages (TAMs)
3.4. Dendritic Cells (DCs)
3.5. Natural Killer (NK) Cells
4. miRNAs and Immune Evasion
4.1. Regulatory T Cells (Tregs)
4.2. Myeloid-Derived Suppressor Cells (MDSCs)
4.3. Tumor-Associated Macrophages (TAMs)
4.4. Cancer-Associated Fibroblast (CAF)
4.5. Endothelial Cells of the Tumor Vasculature
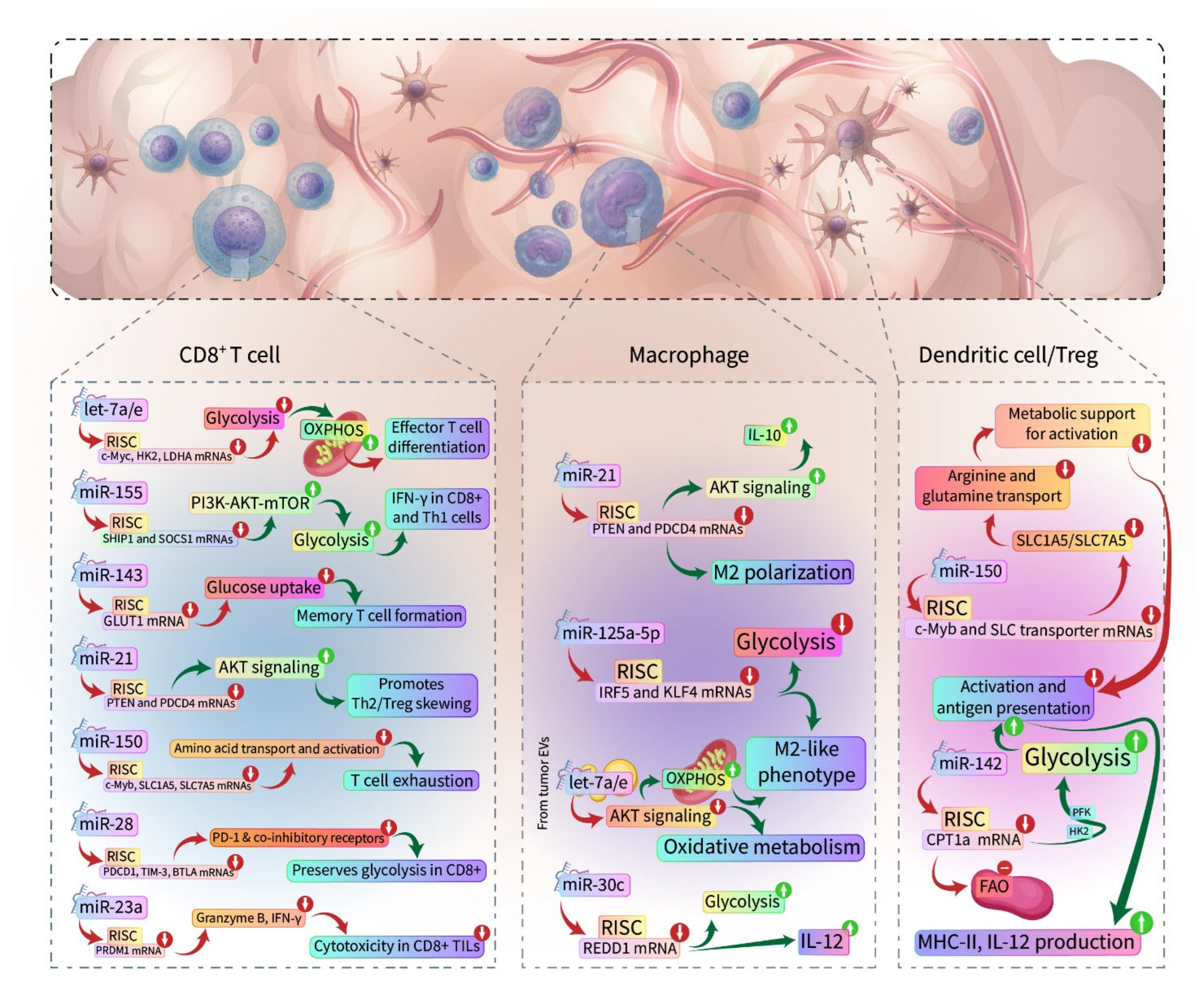
5. miRNAs and Immunometabolism
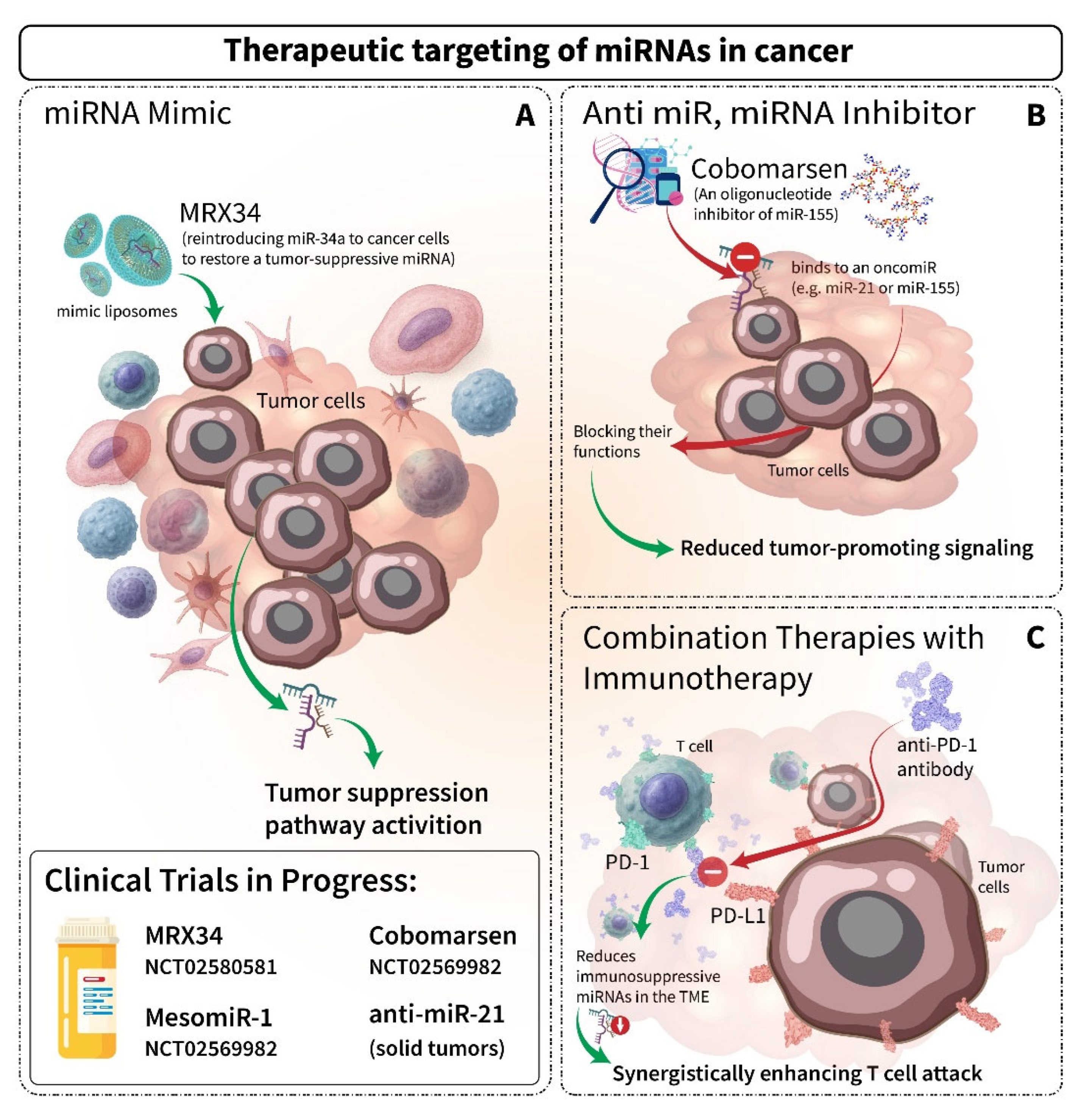
6. Therapeutic Implications and Future Directions of miRNAs
| miR (Drug Name) | RCT ID /Phase | Disease | Delivery Method | Key Findings/Status |
|---|---|---|---|---|
| Mimic | ||||
| miR-34a (MRX34) | NCT01829971 Phase I | Liver, lung, melanoma | Liposomal nanoparticles | The trial was halted due to immune toxicity, but validated miRNA mimic feasibility [204,205] |
| miR-16 (MesomiR-1) | NCT02369198 Phase I | Mesothelioma, NSCLC | EGFR-targeted minicells | Safe, reduced tumor burden in some patients [206] |
| miR-193a-3p | NCT04675996 Phase I | Melanoma | Lipid nanoparticles | Ongoing trial testing safety and efficacy. |
| miR-221 | NCT02716012 Phase I/II | HCC | LNA-modified antisense | Reduced miR-221 levels improved survival in HCC [209] |
| miR-29b (TargomiRs) | Preclinical | Lung cancer | Bacterial minicells | Promising preclinical results; no active trial yet [212] |
| Anti-miR | ||||
| miR-155 (Cobomarsen) | NCT02580552 Phase II | CTCL | LNA-based antisense | Reduced disease activity in CTCL patients [207,208] |
| miR-21 | NCT04556981 Phase I | Solid tumors | LNA antisense | Preclinical data show reduced metastasis; the trial is ongoing [210] |
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
List of Abbreviations
References
- Chen, Y.; Fu, L.L.; Wen, X.; Liu, B.; Huang, J.; Wang, J.H.; Wei, Y.Q. Oncogenic and tumor suppressive roles of microRNAs in apoptosis and autophagy. Apoptosis 2014, 19, 1177–1189. [Google Scholar] [CrossRef] [PubMed]
- Behbahani, G.D.; Ghahhari, N.M.; Javidi, M.A.; Molan, A.F.; Feizi, N.; Babashah, S. MicroRNA-Mediated Post-Transcriptional Regulation of Epithelial to Mesenchymal Transition in Cancer. Pathol. Oncol. Res. 2017, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Altmann, M.; Schmitz, N.; Berset, C.; Trachsel, H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997, 16, 1114–1121. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ostrand-Rosenberg, S. Immune surveillance: A balance between protumor and antitumor immunity. Curr. Opin. Genet. Dev. 2008, 18, 11–18. [Google Scholar] [CrossRef]
- Waddell, N.; Addala, V. Germline variants alter immune surveillance. Science 2024, 384, 961–962. [Google Scholar] [CrossRef]
- Zhang, S.-M.; Cai, W.; Liu, X.; Thakral, D.; Luo, J.; Chan, L.-H.; McGeary, M.; Song, E.; Blenman, K.; Micevic, G.; et al. KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature 2021, 598, 682–687. [Google Scholar] [CrossRef]
- Taghiloo, S.; Asgarian-Omran, H. Immune evasion mechanisms in acute myeloid leukemia: A focus on immune checkpoint pathways. Crit. Rev. Oncol./Hematol. 2020, 157, 103164. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Z.; Ding, Y.; Qin, Y.Y. Tumor microenvironment-mediated immune evasion in hepatocellular carcinoma. Front. Immunol. 2023, 14, 1133308. [Google Scholar] [CrossRef]
- Lu, Y.; Deng, M.; Wang, K.; Peng, Y.; Ouyang, M. The Regulatory Effects of MicroRNAs on Tumor Immunity. BioMed Res. Int. 2022, 2022, 2121993. [Google Scholar] [CrossRef]
- Yi, M.; Xu, L.; Jiao, Y.; Luo, S.; Li, A.; Wu, K. The role of cancer-derived microRNAs in cancer immune escape. J. Hematol. Oncol. 2020, 13, 25. [Google Scholar] [CrossRef]
- Lin, R.; Chen, L.; Chen, G.; Hu, C.; Jiang, S.; Sevilla, J.; Wan, Y.; Sampson, J.H.; Zhu, B.; Li, Q.J. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J. Clin. Investig. 2014, 124, 5352–5367. [Google Scholar] [CrossRef] [PubMed]
- Berchem, G.; Noman, M.Z.; Bosseler, M.; Paggetti, J.; Baconnais, S.; Le Cam, E.; Nanbakhsh, A.; Moussay, E.; Mami-Chouaib, F.; Janji, B.; et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-β and miR23a transfer. Oncoimmunology 2016, 5, e1062968. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Zhang, Y.N.; Li, H.B.; Hu, C.Y.; Wang, N.; Cao, P.P.; Liao, B.; Lu, X.; Cui, Y.H.; Liu, Z. Overexpression of miR-125b, a novel regulator of innate immunity, in eosinophilic chronic rhinosinusitis with nasal polyps. Am. J. Respir. Crit. Care Med. 2012, 185, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Pan, W.; Song, X.; Liu, Y.; Shao, X.; Tang, Y.; Liang, D.; He, D.; Wang, H.; Liu, W.; et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat. Med. 2012, 18, 1077–1086. [Google Scholar] [CrossRef]
- Trifari, S.; Pipkin, M.E.; Bandukwala, H.S.; Äijö, T.; Bassein, J.; Chen, R.; Martinez, G.J.; Rao, A. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, 18608–18613. [Google Scholar] [CrossRef]
- Xiao, C.; Calado, D.P.; Galler, G.; Thai, T.H.; Patterson, H.C.; Wang, J.; Rajewsky, N.; Bender, T.P.; Rajewsky, K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 2007, 131, 146–159. [Google Scholar] [CrossRef]
- Cichocki, F.; Felices, M.; McCullar, V.; Presnell, S.R.; Al-Attar, A.; Lutz, C.T.; Miller, J.S. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J. Immunol. 2011, 187, 6171–6175. [Google Scholar] [CrossRef]
- Bezman, N.A.; Cedars, E.; Steiner, D.F.; Blelloch, R.; Hesslein, D.G.; Lanier, L.L. Distinct requirements of microRNAs in NK cell activation, survival, and function. J. Immunol. 2010, 185, 3835–3846. [Google Scholar] [CrossRef]
- Gracias, D.T.; Stelekati, E.; Hope, J.L.; Boesteanu, A.C.; Doering, T.A.; Norton, J.; Mueller, Y.M.; Fraietta, J.A.; Wherry, E.J.; Turner, M.; et al. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat. Immunol. 2013, 14, 593–602. [Google Scholar] [CrossRef]
- Rodriguez, A.; Vigorito, E.; Clare, S.; Warren, M.V.; Couttet, P.; Soond, D.R.; van Dongen, S.; Grocock, R.J.; Das, P.P.; Miska, E.A.; et al. Requirement of bic/microRNA-155 for normal immune function. Science 2007, 316, 608–611. [Google Scholar] [CrossRef]
- Trotta, R.; Chen, L.; Ciarlariello, D.; Josyula, S.; Mao, C.; Costinean, S.; Yu, L.; Butchar, J.P.; Tridandapani, S.; Croce, C.M.; et al. miR-155 regulates IFN-γ production in natural killer cells. Blood 2012, 119, 3478–3485. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.Q.; Ren, J.P.; Zhao, J.; Wang, J.M.; Zhou, Y.; Li, G.Y.; Moorman, J.P.; Yao, Z.Q. MicroRNA-155 regulates interferon-γ production in natural killer cells via Tim-3 signalling in chronic hepatitis C virus infection. Immunology 2015, 145, 485–497. [Google Scholar] [CrossRef]
- Curtale, G.; Rubino, M.; Locati, M. MicroRNAs as Molecular Switches in Macrophage Activation. Front. Immunol. 2019, 10, 799. [Google Scholar] [CrossRef]
- Hawez, A.; Al-Haidari, A.; Madhi, R.; Rahman, M.; Thorlacius, H. MiR-155 Regulates PAD4-Dependent Formation of Neutrophil Extracellular Traps. Front. Immunol. 2019, 10, 2462. [Google Scholar] [CrossRef]
- Arroyo, A.B.; Fernández-Pérez, M.P.; Del Monte, A.; Águila, S.; Méndez, R.; Hernández-Antolín, R.; García-Barber, N.; de Los Reyes-García, A.M.; González-Jiménez, P.; Arcas, M.I.; et al. miR-146a is a pivotal regulator of neutrophil extracellular trap formation promoting thrombosis. Haematologica 2021, 106, 1636–1646. [Google Scholar] [CrossRef] [PubMed]
- Adel, R.M.; Helal, H.; Ahmed Fouad, M.; Sobhy Abd-Elhalem, S. Regulation of miRNA-155-5p ameliorates NETosis in pulmonary fibrosis rat model via inhibiting its target cytokines IL-1β, TNF-α and TGF-β1. Int. Immunopharmacol. 2024, 127, 111456. [Google Scholar] [CrossRef] [PubMed]
- Hussen, B.M.; Rasul, M.F.; Faraj, G.S.H.; Abdullah, S.R.; Sulaiman, S.H.; Pourmoshtagh, H.; Taheri, M. Role of microRNAs in neutrophil extracellular trap formation and prevention: Systematic narrative review. Mol. Cell. Probes 2024, 78, 101986. [Google Scholar] [CrossRef]
- Zhang, M.; Gao, D.; Shi, Y.; Wang, Y.; Joshi, R.; Yu, Q.; Liu, D.; Alotaibi, F.; Zhang, Y.; Wang, H.; et al. miR-149-3p reverses CD8(+) T-cell exhaustion by reducing inhibitory receptors and promoting cytokine secretion in breast cancer cells. Open Biol. 2019, 9, 190061. [Google Scholar] [CrossRef]
- Yang, J.; Jia, Y.; Wang, B.; Yang, S.; Du, K.; Luo, Y.; Li, Y.; Zhu, B. Circular RNA CHST15 Sponges miR-155-5p and miR-194-5p to Promote the Immune Escape of Lung Cancer Cells Mediated by PD-L1. Front. Oncol. 2021, 11, 595609. [Google Scholar] [CrossRef]
- Matsuyama, H.; Suzuki, H.I.; Nishimori, H.; Noguchi, M.; Yao, T.; Komatsu, N.; Mano, H.; Sugimoto, K.; Miyazono, K. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood 2011, 118, 6881–6892. [Google Scholar] [CrossRef]
- Madrid-Elena, N.; Serrano-Villar, S.; Gutiérrez, C.; Sastre, B.; Morín, M.; Luna, L.; Martín, L.; Santoyo-López, J.; López-Huertas, M.R.; Moreno, E.; et al. Selective miRNA inhibition in CD8(+) cytotoxic T lymphocytes enhances HIV-1 specific cytotoxic responses. Front. Immunol. 2022, 13, 998368. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Liu, C.; Kang, J.; Zhao, G.; Ye, Z.; Huang, S.; Li, Z.; Wu, Z.; Pei, G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 2009, 10, 1252–1259. [Google Scholar] [CrossRef]
- Baumjohann, D.; Kageyama, R.; Clingan, J.M.; Morar, M.M.; Patel, S.; de Kouchkovsky, D.; Bannard, O.; Bluestone, J.A.; Matloubian, M.; Ansel, K.M.; et al. The microRNA cluster miR-17∼92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat. Immunol. 2013, 14, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Regis, S.; Caliendo, F.; Dondero, A.; Casu, B.; Romano, F.; Loiacono, F.; Moretta, A.; Bottino, C.; Castriconi, R. TGF-β1 Downregulates the Expression of CX(3)CR1 by Inducing miR-27a-5p in Primary Human NK Cells. Front. Immunol. 2017, 8, 868. [Google Scholar] [CrossRef]
- Pedersen, C.C.; Refsgaard, J.C.; Østergaard, O.; Jensen, L.J.; Heegaard, N.H.; Borregaard, N.; Cowland, J.B. Impact of microRNA-130a on the neutrophil proteome. BMC Immunol. 2015, 16, 70. [Google Scholar] [CrossRef] [PubMed]
- Cordo, V.; van der Zwet, J.C.G.; Canté-Barrett, K.; Pieters, R.; Meijerink, J.P.P. T-cell Acute Lymphoblastic Leukemia: A Roadmap to Targeted Therapies. Blood Cancer Discov. 2021, 2, 19–31. [Google Scholar] [CrossRef]
- Xue, X.; Cao, A.T.; Cao, X.; Yao, S.; Carlsen, E.D.; Soong, L.; Liu, C.G.; Liu, X.; Liu, Z.; Duck, L.W.; et al. Downregulation of microRNA-107 in intestinal CD11c(+) myeloid cells in response to microbiota and proinflammatory cytokines increases IL-23p19 expression. Eur. J. Immunol. 2014, 44, 673–682. [Google Scholar] [CrossRef]
- Nail, H.M.; Chiu, C.C.; Leung, C.H.; Ahmed, M.M.M.; Wang, H.D. Exosomal miRNA-mediated intercellular communications and immunomodulatory effects in tumor microenvironments. J. Biomed. Sci. 2023, 30, 69. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, F.; Jia, H.; Zhang, Q.; Yin, L.; Liu, W.; Li, H.; Yu, B.; Wu, J. Inhibition of microRNA let-7i depresses maturation and functional state of dendritic cells in response to lipopolysaccharide stimulation via targeting suppressor of cytokine signaling 1. J. Immunol. 2011, 187, 1674–1683. [Google Scholar] [CrossRef]
- Song, J.; Lin, Z.; Liu, Q.; Huang, S.; Han, L.; Fang, Y.; Zhong, P.; Dou, R.; Xiang, Z.; Zheng, J.; et al. MiR-192-5p/RB1/NF-κBp65 signaling axis promotes IL-10 secretion during gastric cancer EMT to induce Treg cell differentiation in the tumour microenvironment. Clin. Transl. Med. 2022, 12, e992. [Google Scholar] [CrossRef]
- Yin, Y.; Cai, X.; Chen, X.; Liang, H.; Zhang, Y.; Li, J.; Wang, Z.; Chen, X.; Zhang, W.; Yokoyama, S.; et al. Tumor-secreted miR-214 induces regulatory T cells: A major link between immune evasion and tumor growth. Cell Res. 2014, 24, 1164–1180. [Google Scholar] [CrossRef]
- Yang, P.; Li, Q.J.; Feng, Y.; Zhang, Y.; Markowitz, G.J.; Ning, S.; Deng, Y.; Zhao, J.; Jiang, S.; Yuan, Y.; et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell 2012, 22, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, A.; Zhao, F.; Pechlivanis, S.; Eberle, J.; Steinle, A.; Diederichs, S.; Schadendorf, D.; Paschen, A. Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012, 72, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Katsura, A.; Matsuyama, H.; Miyazono, K. MicroRNA regulons in tumor microenvironment. Oncogene 2015, 34, 3085–3094. [Google Scholar] [CrossRef] [PubMed]
- Fehniger, T.A.; Wylie, T.; Germino, E.; Leong, J.W.; Magrini, V.J.; Koul, S.; Keppel, C.R.; Schneider, S.E.; Koboldt, D.C.; Sullivan, R.P.; et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 2010, 20, 1590–1604. [Google Scholar] [CrossRef]
- Zhou, H.; Xiao, J.; Wu, N.; Liu, C.; Xu, J.; Liu, F.; Wu, L. MicroRNA-223 Regulates the Differentiation and Function of Intestinal Dendritic Cells and Macrophages by Targeting C/EBPβ. Cell Rep. 2015, 13, 1149–1160. [Google Scholar] [CrossRef]
- Sheedy, F.J.; Palsson-McDermott, E.; Hennessy, E.J.; Martin, C.; O’Leary, J.J.; Ruan, Q.; Johnson, D.S.; Chen, Y.; O’Neill, L.A. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010, 11, 141–147. [Google Scholar] [CrossRef]
- Águila, S.; de Los Reyes-García, A.M.; Fernández-Pérez, M.P.; Reguilón-Gallego, L.; Zapata-Martínez, L.; Ruiz-Lorente, I.; Vicente, V.; González-Conejero, R.; Martínez, C. MicroRNAs as New Regulators of Neutrophil Extracellular Trap Formation. Int. J. Mol. Sci. 2021, 22, 2116. [Google Scholar] [CrossRef]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef]
- Sullivan, R.P.; Leong, J.W.; Schneider, S.E.; Keppel, C.R.; Germino, E.; French, A.R.; Fehniger, T.A. MicroRNA-deficient NK cells exhibit decreased survival but enhanced function. J. Immunol. 2012, 188, 3019–3030. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Komabayashi, Y.; Kishibe, K.; Nagato, T.; Ueda, S.; Takahara, M.; Harabuchi, Y. Downregulation of miR-15a due to LMP1 promotes cell proliferation and predicts poor prognosis in nasal NK/T-cell lymphoma. Am. J. Hematol. 2014, 89, 25–33. [Google Scholar] [CrossRef]
- Yang, J.; Liu, R.; Deng, Y.; Qian, J.; Lu, Z.; Wang, Y.; Zhang, D.; Luo, F.; Chu, Y. MiR-15a/16 deficiency enhances anti-tumor immunity of glioma-infiltrating CD8+ T cells through targeting mTOR. Int. J. Cancer 2017, 141, 2082–2092. [Google Scholar] [CrossRef]
- Xie, J.; Liu, M.; Li, Y.; Nie, Y.; Mi, Q.; Zhao, S. Ovarian tumor-associated microRNA-20a decreases natural killer cell cytotoxicity by downregulating MICA/B expression. Cell. Mol. Immunol. 2014, 11, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Dews, M.; Fox, J.L.; Hultine, S.; Sundaram, P.; Wang, W.; Liu, Y.Y.; Furth, E.; Enders, G.H.; El-Deiry, W.; Schelter, J.M.; et al. The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 2010, 70, 8233–8246. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Qiu, W.; Wang, J.; Liu, Q.; Qian, M.; Wang, S.; Zhang, Z.; Gao, X.; Chen, Z.; Guo, Q.; et al. Glioma exosomes mediate the expansion and function of myeloid-derived suppressor cells through microRNA-29a/Hbp1 and microRNA-92a/Prkar1a pathways. Int. J. Cancer 2019, 144, 3111–3126. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, W.; Zhang, R.; Liu, P.; Ye, Y.; Yu, W.; Guo, X.; Yu, J. Cancer exosome-derived miR-9 and miR-181a promote the development of early-stage MDSCs via interfering with SOCS3 and PIAS3 respectively in breast cancer. Oncogene 2020, 39, 4681–4694. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef]
- Botta, C.; Cucè, M.; Pitari, M.R.; Caracciolo, D.; Gullà, A.; Morelli, E.; Riillo, C.; Biamonte, L.; Gallo Cantafio, M.E.; Prabhala, R.; et al. MiR-29b antagonizes the pro-inflammatory tumor-promoting activity of multiple myeloma-educated dendritic cells. Leukemia 2018, 32, 1003–1015. [Google Scholar] [CrossRef]
- Cooks, T.; Pateras, I.S.; Jenkins, L.M.; Patel, K.M.; Robles, A.I.; Morris, J.; Forshew, T.; Appella, E.; Gorgoulis, V.G.; Harris, C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018, 9, 771. [Google Scholar] [CrossRef]
- Tong, F.; Mao, X.; Zhang, S.; Xie, H.; Yan, B.; Wang, B.; Sun, J.; Wei, L. HPV + HNSCC-derived exosomal miR-9 induces macrophage M1 polarization and increases tumor radiosensitivity. Cancer Lett. 2020, 478, 34–44. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Zhang, D.; Sun, X.; Wu, Y.; Wang, J.; Li, Q.; Jiang, G. The macrophage polarization by miRNAs and its potential role in the treatment of tumor and inflammation (Review). Oncol. Rep. 2023, 50, 190. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cai, B.; Shen, L.; Dong, Y.; Lu, Q.; Sun, S.; Liu, S.; Shu, G.; Chen, J. MiRNA-29b suppresses tumor growth through simultaneously inhibiting angiogenesis and tumorigenesis by targeting Akt3. Cancer Lett. 2017, 397, 111–119. [Google Scholar] [CrossRef]
- Wang, P.; Gu, Y.; Zhang, Q.; Han, Y.; Hou, J.; Lin, L.; Wu, C.; Bao, Y.; Su, X.; Jiang, M.; et al. Identification of resting and type I IFN-activated human NK cell miRNomes reveals microRNA-378 and microRNA-30e as negative regulators of NK cell cytotoxicity. J. Immunol. 2012, 189, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.D.; Lee, S.U.; Yun, S.; Sun, H.N.; Lee, S.H.; Kim, J.W.; Kim, H.M.; Park, S.K.; Lee, C.W.; Yoon, S.R.; et al. Human microRNA-27a* targets Prf1 and GzmB expression to regulate NK-cell cytotoxicity. Blood 2011, 118, 5476–5486. [Google Scholar] [CrossRef]
- Donatelli, S.S.; Zhou, J.M.; Gilvary, D.L.; Eksioglu, E.A.; Chen, X.; Cress, W.D.; Haura, E.B.; Schabath, M.B.; Coppola, D.; Wei, S.; et al. TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc. Natl. Acad. Sci. USA 2014, 111, 4203–4208. [Google Scholar] [CrossRef]
- Zhang, J.; Han, X.; Hu, X.; Jin, F.; Gao, Z.; Yin, L.; Qin, J.; Yin, F.; Li, C.; Wang, Y. IDO1 impairs NK cell cytotoxicity by decreasing NKG2D/NKG2DLs via promoting miR-18a. Mol. Immunol. 2018, 103, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Neviani, P.; Wise, P.M.; Murtadha, M.; Liu, C.W.; Wu, C.H.; Jong, A.Y.; Seeger, R.C.; Fabbri, M. Natural Killer-Derived Exosomal miR-186 Inhibits Neuroblastoma Growth and Immune Escape Mechanisms. Cancer Res. 2019, 79, 1151–1164. [Google Scholar] [CrossRef]
- Nachmani, D.; Stern-Ginossar, N.; Sarid, R.; Mandelboim, O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 2009, 5, 376–385. [Google Scholar] [CrossRef]
- Ueda, R.; Kohanbash, G.; Sasaki, K.; Fujita, M.; Zhu, X.; Kastenhuber, E.R.; McDonald, H.A.; Potter, D.M.; Hamilton, R.L.; Lotze, M.T.; et al. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc. Natl. Acad. Sci. USA 2009, 106, 10746–10751. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Shan, G.; Zhou, X.; Gu, J.; Zhou, D.; Cheng, W.; Wu, H.; Wang, Y.; Tang, T.; Wang, X. Downregulated exosomal microRNA-148b-3p in cancer associated fibroblasts enhance chemosensitivity of bladder cancer cells by downregulating the Wnt/β-catenin pathway and upregulating PTEN. Cell. Oncol. 2021, 44, 45–59. [Google Scholar] [CrossRef]
- Yuan, S.; Zhu, T.; Wang, J.; Jiang, R.; Shu, A.; Zhang, Z.; Zhang, P.; Feng, X.; Zhao, L. miR-22 promotes immunosuppression via activating JAK/STAT3 signaling in cutaneous squamous cell carcinoma. Carcinogenesis 2023, 44, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Zhou, L.; Qian, Y.; Fu, M.; Chen, J.; Chen, J.; Xiang, J.; Wu, Z.; Jiang, G.; Cao, L. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212-3p. Oncotarget 2015, 6, 29877–29888. [Google Scholar] [CrossRef]
- Cui, Z.J.; Xie, X.L.; Qi, W.; Yang, Y.C.; Bai, Y.; Han, J.; Ding, Q.; Jiang, H.Q. Cell-free miR-17-5p as a diagnostic biomarker for gastric cancer inhibits dendritic cell maturation. Onco Targets Ther. 2019, 12, 2661–2675. [Google Scholar] [CrossRef]
- Zheng, G.; Ye, H.; Bai, J.; Zhang, X. Downregulation of lncRNA MIR17HG reduced tumorigenicity and Treg-mediated immune escape of non-small-cell lung cancer cells through targeting the miR-17-5p/RUNX3 axis. J. Biochem. Mol. Toxicol. 2024, 38, e23715. [Google Scholar] [CrossRef]
- Lu, C.; Huang, X.; Zhang, X.; Roensch, K.; Cao, Q.; Nakayama, K.I.; Blazar, B.R.; Zeng, Y.; Zhou, X. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood 2011, 117, 4293–4303. [Google Scholar] [CrossRef]
- Zhang, M.; Shi, Y.; Zhang, Y.; Wang, Y.; Alotaibi, F.; Qiu, L.; Wang, H.; Peng, S.; Liu, Y.; Li, Q.; et al. miRNA-5119 regulates immune checkpoints in dendritic cells to enhance breast cancer immunotherapy. Cancer Immunol. Immunother. 2020, 69, 951–967. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Han, D.; Fan, W. Down-regulation of RBP-J mediated by microRNA-133a suppresses dendritic cells and functions as a potential tumor suppressor in osteosarcoma. Exp. Cell Res. 2016, 349, 264–272. [Google Scholar] [CrossRef]
- Liu, G.; Friggeri, A.; Yang, Y.; Park, Y.J.; Tsuruta, Y.; Abraham, E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc. Natl. Acad. Sci. USA 2009, 106, 15819–15824. [Google Scholar] [CrossRef]
- Zhao, Q.; Huang, L.; Qin, G.; Qiao, Y.; Ren, F.; Shen, C.; Wang, S.; Liu, S.; Lian, J.; Wang, D.; et al. Cancer-associated fibroblasts induce monocytic myeloid-derived suppressor cell generation via IL-6/exosomal miR-21-activated STAT3 signaling to promote cisplatin resistance in esophageal squamous cell carcinoma. Cancer Lett. 2021, 518, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Orso, F.; Quirico, L.; Dettori, D.; Coppo, R.; Virga, F.; Ferreira, L.; Paoletti, C.; Baruffaldi, D.; Penna, E.; Taverna, D. Role of miRNAs in tumor and endothelial cell interactions during tumor progression. Semin. Cancer Biol. 2020, 60, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Guo, X.; Li, B.; Wang, J.; Qi, Y.; Chen, Z.; Zhao, R.; Deng, L.; Qian, M.; Wang, S.; et al. Exosomal miR-1246 from glioma patient body fluids drives the differentiation and activation of myeloid-derived suppressor cells. Mol. Ther. J. Am. Soc. Gene Ther. 2021, 29, 3449–3464. [Google Scholar] [CrossRef]
- Ouimet, M.; Ediriweera, H.N.; Gundra, U.M.; Sheedy, F.J.; Ramkhelawon, B.; Hutchison, S.B.; Rinehold, K.; van Solingen, C.; Fullerton, M.D.; Cecchini, K.; et al. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J. Clin. Investig. 2015, 125, 4334–4348. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Q.; Song, Y.; Lai, L.; Wang, J.; Yu, H.; Cao, X.; Wang, Q. MicroRNA-98 negatively regulates IL-10 production and endotoxin tolerance in macrophages after LPS stimulation. FEBS Lett. 2011, 585, 1963–1968. [Google Scholar] [CrossRef]
- Zhong, Y.; Yi, C. MicroRNA-720 suppresses M2 macrophage polarization by targeting GATA3. Biosci. Rep. 2016, 36, e00363. [Google Scholar] [CrossRef] [PubMed]
- Squadrito, M.L.; Pucci, F.; Magri, L.; Moi, D.; Gilfillan, G.D.; Ranghetti, A.; Casazza, A.; Mazzone, M.; Lyle, R.; Naldini, L.; et al. miR-511-3p modulates genetic programs of tumor-associated macrophages. Cell Rep. 2012, 1, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Wang, P.; Lin, L.; Liu, X.; Ma, F.; An, H.; Wang, Z.; Cao, X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 2009, 183, 2150–2158. [Google Scholar] [CrossRef]
- Ali, S.R.; Humphreys, K.J.; McKinnon, R.A.; Michael, M.Z. Impact of Histone Deacetylase Inhibitors on microRNA Expression and Cancer Therapy: A Review. Drug Dev. Res. 2015, 76, 296–317. [Google Scholar] [CrossRef]
- Huber, V.; Vallacchi, V.; Fleming, V.; Hu, X.; Cova, A.; Dugo, M.; Shahaj, E.; Sulsenti, R.; Vergani, E.; Filipazzi, P.; et al. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J. Clin. Investig. 2018, 128, 5505. [Google Scholar] [CrossRef]
- Zhao, H.; Su, W.; Kang, Q.; Xing, Z.; Lin, X.; Wu, Z. Natural killer cells inhibit oxaliplatin-resistant colorectal cancer by repressing WBSCR22 via upregulating microRNA-146b-5p. Am. J. Cancer Res. 2018, 8, 824–834. [Google Scholar] [PubMed]
- Park, H.; Huang, X.; Lu, C.; Cairo, M.S.; Zhou, X. MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J. Biol. Chem. 2015, 290, 2831–2841. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, S.; Yin, K.; Zhang, Q.; Li, S. MiR-1696/GPx3 axis is involved in oxidative stress mediated neutrophil extracellular traps inhibition in chicken neutrophils. J. Cell Physiol. 2021, 236, 3688–3699. [Google Scholar] [CrossRef]
- Wang, L.; Yang, J.; Huang, J.; Wen, Z.Q.; Xu, N.; Liu, X.; Zhang, J.H.; Li, W.L. miRNA Expression Profile in the N2 Phenotype Neutrophils of Colorectal Cancer and Screen of Putative Key miRNAs. Cancer Manag. Res. 2020, 12, 5491–5503. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.C.; Tzeng, W.F.; Chiou, T.J.; Chu, S.T. MicroRNA-138 suppresses neutrophil gelatinase-associated lipocalin expression and inhibits tumorigenicity. PLoS ONE 2012, 7, e52979. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schwarzenbach, H. Clinical Implementation of MicroRNAs in Cancer Immunology. Int. J. Transl. Med. 2024, 4, 53–71. [Google Scholar] [CrossRef]
- Chen, B.; Han, J.; Chen, S.; Xie, R.; Yang, J.; Zhou, T.; Zhang, Q.; Xia, R. MicroLet-7b Regulates Neutrophil Function and Dampens Neutrophilic Inflammation by Suppressing the Canonical TLR4/NF-κB Pathway. Front. Immunol. 2021, 12, 653344. [Google Scholar] [CrossRef]
- Zhang, L.; Ke, F.; Liu, Z.; Bai, J.; Liu, J.; Yan, S.; Xu, Z.; Lou, F.; Wang, H.; Zhu, H.; et al. MicroRNA-31 negatively regulates peripherally derived regulatory T-cell generation by repressing retinoic acid-inducible protein 3. Nat. Commun. 2015, 6, 7639. [Google Scholar] [CrossRef]
- Xue, F.; Li, H.; Zhang, J.; Lu, J.; Xia, Y.; Xia, Q. miR-31 regulates interleukin 2 and kinase suppressor of ras 2 during T cell activation. Genes. Immun. 2013, 14, 127–131. [Google Scholar] [CrossRef]
- Hsu, A.Y.; Wang, D.; Liu, S.; Lu, J.; Syahirah, R.; Bennin, D.A.; Huttenlocher, A.; Umulis, D.M.; Wan, J.; Deng, Q. Phenotypical microRNA screen reveals a noncanonical role of CDK2 in regulating neutrophil migration. Proc. Natl. Acad. Sci. USA 2019, 116, 18561–18570. [Google Scholar] [CrossRef]
- Hsu, A.Y.; Wang, D.; Gurol, T.; Zhou, W.; Zhu, X.; Lu, H.Y.; Deng, Q. Overexpression of microRNA-722 fine-tunes neutrophilic inflammation by inhibiting Rac2 in zebrafish. Dis. Model. Mech. 2017, 10, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Linhares-Lacerda, L.; Temerozo, J.R.; Ribeiro-Alves, M.; Azevedo, E.P.; Mojoli, A.; Nascimento, M.T.C.; Silva-Oliveira, G.; Savino, W.; Foguel, D.; Bou-Habib, D.C.; et al. Neutrophil extracellular trap-enriched supernatants carry microRNAs able to modulate TNF-α production by macrophages. Sci. Rep. 2020, 10, 2715. [Google Scholar] [CrossRef]
- Lasser, S.; Ozbay Kurt, F.G.; Fritz, L.; Gutzeit, N.; De La Torre, C.; Altevogt, P.; Utikal, J.; Umansky, V. Generation of Myeloid-Derived Suppressor Cells Mediated by MicroRNA-125a-5p in Melanoma. Int. J. Mol. Sci. 2024, 25, 6693. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-L.; Huang, F.; He, F.; Gao, C.-C.; Liang, S.-Q.; Ma, P.-F.; Dong, G.-Y.; Han, H.; Qin, H. Forced Activation of Notch in Macrophages Represses Tumor Growth by Upregulating miR-125a and Disabling Tumor-Associated Macrophages. Cancer Res. 2016, 76, 1403–1415. [Google Scholar] [CrossRef]
- Gerloff, D.; Lützkendorf, J.; Moritz, R.; Wersig, T.; Mäder, K.; Müller, L.; Sunderkötter, C. Melanoma-Derived Exosomal miR-125b-5p Educates Tumor Associated Macrophages (TAMs) by Targeting Lysosomal Acid Lipase A (LIPA). Cancers 2020, 12, 464. [Google Scholar] [CrossRef]
- Li, Q.; Johnston, N.; Zheng, X.; Wang, H.; Zhang, X.; Gao, D.; Min, W. miR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget 2016, 7, 53735–53750. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Lan, P.; Han, Q.; Huang, M.; Zhang, Z.; Xu, G.; Song, J.; Wang, J.; Wei, H.; Zhang, J.; et al. Oncofetal gene SALL4 reactivation by hepatitis B virus counteracts miR-200c in PD-L1-induced T cell exhaustion. Nat. Commun. 2018, 9, 1241. [Google Scholar] [CrossRef]
- Yu, W.Q.; Ji, N.F.; Gu, C.J.; Sun, Z.X.; Wang, Z.X.; Chen, Z.Q.; Ma, Y.; Wu, Z.Z.; Wang, Y.L.; Wu, C.J.; et al. Downregulation of miR-4772-3p promotes enhanced regulatory T cell capacity in malignant pleural effusion by elevating Helios levels. Chin. Med. J. 2019, 132, 2705–2715. [Google Scholar] [CrossRef]
- Lv, M.; Xu, Y.; Tang, R.-J.; Ren, J.; Shen, S.; Chen, Y.; Liu, B.; Hou, Y.; Wang, T. miR141–CXCL1–CXCR2 Signaling–Induced Treg Recruitment Regulates Metastases and Survival of Non–Small Cell Lung Cancer. Mol. Cancer Ther. 2014, 13, 3152–3162. [Google Scholar] [CrossRef]
- Soheilifar, M.H.; Vaseghi, H.; Seif, F.; Ariana, M.; Ghorbanifar, S.; Habibi, N.; Barjasteh, F.P.; Pornour, M. Concomitant overexpression of mir-182-5p and mir-182-3p raises the possibility of IL-17–producing Treg formation in breast cancer by targeting CD3d, ITK, FOXO1, and NFATs: A meta-analysis and experimental study. Cancer Sci. 2020, 112, 589–603. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Z.; Liang, J.; Liu, J.; Hao, J.; Wan, Q.; Liu, J.; Luo, C.; Lu, Z. CircRNA has_circ_0069313 induced OSCC immunity escape by miR-325-3p-Foxp3 axes in both OSCC cells and Treg cells. Aging 2022, 14, 4376–4389. [Google Scholar] [CrossRef]
- Ning, T.; Li, J.; He, Y.; Zhang, H.; Wang, X.; Deng, T.; Liu, R.; Li, H.-L.; Bai, M.; Fan, Q.; et al. Exosomal miR-208b related with oxaliplatin resistance promotes Treg expansion in colorectal cancer. Mol. Ther. 2021, 29, 2723–2736. [Google Scholar] [CrossRef]
- Yu, Z.; Zhao, H.; Feng, X.; Li, H.; Qiu, C.-H.; Yi, X.; Tang, H.; Zhang, J. Long Non-coding RNA FENDRR Acts as a miR-423-5p Sponge to Suppress the Treg-Mediated Immune Escape of Hepatocellular Carcinoma Cells. Mol. Ther. Nucleic Acids 2019, 17, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Fan, X.; Zhang, X.; Wei, P.; Zhou, H.; Liu, D.; Chen, B. miR-429 inhibits the formation of an immunosuppressive microenvironment to counteract hepatocellular carcinoma immune escape by targeting PD-L1. Funct. Integr. Genom. 2023, 23, 312. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Qiu, W.; Liu, Q.; Qian, M.; Wang, S.; Zhang, Z.; Gao, X.; Chen, Z.; Xue, H.; Li, G. Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten Pathways. Oncogene 2018, 37, 4239–4259. [Google Scholar] [CrossRef] [PubMed]
- Noman, M.; Janji, B.; Hu, S.; Wu, J.; Martelli, F.; Bronte, V.; Chouaib, S. Tumor-Promoting Effects of Myeloid-Derived Suppressor Cells Are Potentiated by Hypoxia-Induced Expression of miR-210. Cancer Res. 2015, 75, 3771–3787. [Google Scholar] [CrossRef]
- Qi, Y.; Jin, C.; Qiu, W.; Zhao, R.; Wang, S.; Li, B.; Zhang, Z.; Guo, Q.; Zhang, S.; Gao, Z.; et al. The dual role of glioma exosomal microRNAs: Glioma eliminates tumor suppressor miR-1298-5p via exosomes to promote immunosuppressive effects of MDSCs. Cell Death Dis. 2021, 13, 426. [Google Scholar] [CrossRef]
- Wang, H.; Wei, H.; Wang, J.-S.; Li, L.; Chen, A.; Li, Z. MicroRNA-181d-5p-Containing Exosomes Derived from CAFs Promote EMT by Regulating CDX2/HOXA5 in Breast Cancer. Mol. Ther. Nucleic Acids 2019, 19, 654–667. [Google Scholar] [CrossRef]
- Moaaz, M.; Lotfy, H.; Elsherbini, B.; Motawea, M.; Fadali, G. TGF-β Enhances the Anti-inflammatory Effect of Tumor-Infiltrating CD33+11b+HLA-DR Myeloid-Derived Suppressor Cells in Gastric Cancer: A Possible Relation to MicroRNA-494. Asian Pac. J. Cancer Prev. APJCP 2020, 21, 3393–3403. [Google Scholar] [CrossRef]
- Frank, A.-C.; Ebersberger, S.; Fink, A.; Lampe, S.; Weigert, A.; Schmid, T.; Ebersberger, I.; Syed, S.; Brüne, B. Apoptotic tumor cell-derived microRNA-375 uses CD36 to alter the tumor-associated macrophage phenotype. Nat. Commun. 2019, 10, 1135. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, Z.; Ma, R.; Zhang, Y.; Zhao, L.; Yan, Y.; Lv, X.; Zhang, L.; Su, P.; Bi, J.; et al. lncRNA-Xist/miR-101-3p/KLF6/C/EBPα axis promotes TAM polarization to regulate cancer cell proliferation and migration. Mol. Ther. Nucleic Acids 2020, 23, 536–551. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, Y.; Guo, J.; He, H.; Mi, X.; Chen, C.; Xie, J.; Wang, S.; Wu, P.; Cao, F.; et al. miR-100 maintains phenotype of tumor-associated macrophages by targeting mTOR to promote tumor metastasis via Stat5a/IL-1ra pathway in mouse breast cancer. Oncogenesis 2018, 7, 97. [Google Scholar] [CrossRef] [PubMed]
- Rasoolnezhad, M.; Safaralizadeh, R.; Hosseinpourfeizi, M.A.; Banan-Khojasteh, S.M.; Baradaran, B. MiRNA-138-5p: A strong tumor suppressor targeting PD-L-1 inhibits proliferation and motility of breast cancer cells and induces apoptosis. Eur. J. Pharmacol. 2021, 896, 173933. [Google Scholar] [CrossRef]
- Dastmalchi, N.; Hosseinpourfeizi, M.A.; Khojasteh, S.M.B.; Baradaran, B.; Safaralizadeh, R. Tumor suppressive activity of miR-424-5p in breast cancer cells through targeting PD-L1 and modulating PTEN/PI3K/AKT/mTOR signaling pathway. Life Sci. 2020, 259, 118239. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Shan, J.; Chen, X.-H.; Zhang, D.; Su, L.; Huang, X.; Yu, B.; Zhi, Q.; Li, C.; Wang, Y.-Q.; et al. Epigenetic silencing of microRNA-149 in cancer-associated fibroblasts mediates prostaglandin E2/interleukin-6 signaling in the tumor microenvironment. Cell Res. 2015, 25, 588–603. [Google Scholar] [CrossRef]
- Chen, B.; Sang, Y.; Song, X.; Zhang, D.; Wang, L.; Zhao, W.; Liang, Y.; Zhang, N.; Yang, Q. Exosomal miR-500a-5p derived from cancer-associated fibroblasts promotes breast cancer cell proliferation and metastasis through targeting USP28. Theranostics 2021, 11, 3932–3947. [Google Scholar] [CrossRef]
- Guo, L.-P.; Li, B.; Yang, J.; Shen, J.; Ji, J.; Miao, M. Fibroblast-derived exosomal microRNA-369 potentiates migration and invasion of lung squamous cell carcinoma cells via NF1-mediated MAPK signaling pathway. Int. J. Mol. Med. 2020, 46, 595–608. [Google Scholar] [CrossRef]
- Lee, S.; Hong, J.; Kim, J.S.; Yoon, J.-S.; Chun, S.; Hong, S.; Kim, E.J.; Kang, K.; Kang, J.L.; Ko, Y.; et al. Cancer-associated fibroblasts activated by miR-196a promote the migration and invasion of lung cancer cells. Cancer Lett. 2021, 508, 92–103. [Google Scholar] [CrossRef]
- Plummer, P.; Freeman, R.; Taft, R.; Vider, J.; Sax, M.; Umer, B.; Gao, D.; Johns, C.; Mattick, J.; Wilton, S.; et al. MicroRNAs regulate tumor angiogenesis modulated by endothelial progenitor cells. Cancer Res. 2013, 73, 341–352. [Google Scholar] [CrossRef]
- Stern-Ginossar, N.; Gur, C.; Biton, M.; Horwitz, E.; Elboim, M.; Stanietsky, N.; Mandelboim, M.; Mandelboim, O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat. Immunol. 2008, 9, 1065–1073. [Google Scholar] [CrossRef]
- Xue, X.; Feng, T.; Yao, S.; Wolf, K.J.; Liu, C.G.; Liu, X.; Elson, C.O.; Cong, Y. Microbiota downregulates dendritic cell expression of miR-10a, which targets IL-12/IL-23p40. J. Immunol. 2011, 187, 5879–5886. [Google Scholar] [CrossRef] [PubMed]
- Masoumi-Dehghi, S.; Babashah, S.; Sadeghizadeh, M. microRNA-141-3p-containing small extracellular vesicles derived from epithelial ovarian cancer cells promote endothelial cell angiogenesis through activating the JAK/STAT3 and NF-κB signaling pathways. J. Cell Commun. Signal. 2020, 14, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wei, W.; Ma, J.; Yang, Y.; Liang, L.; Zhang, Y.; Wang, Z.; Chen, X.; Huang, L.; Wang, W.; et al. Cancer-secreted exosomal miR-1468-5p promotes tumor immune escape via the immunosuppressive reprogramming of lymphatic vessels. Mol. Ther. 2020, 29, 1512–1528. [Google Scholar] [CrossRef]
- Hunter, S.; Nault, B.; Ugwuagbo, K.; Maiti, S.; Majumder, M. Mir526b and Mir655 Promote Tumour Associated Angiogenesis and Lymphangiogenesis in Breast Cancer. Cancers 2019, 11, 938. [Google Scholar] [CrossRef]
- Wilson, R.; Espinosa-Diez, C.; Kanner, N.; Chatterjee, N.; Ruhl, R.; Hipfinger, C.; Advani, S.; Li, J.; Khan, O.; Franovic, A.; et al. MicroRNA regulation of endothelial TREX1 reprograms the tumour microenvironment. Nat. Commun. 2016, 7, 13597. [Google Scholar] [CrossRef]
- Yuan, S.; Chen, Y.; Zhang, M.; Wang, Z.; Hu, Z.; Ruan, Y.; Ren, Z.; Shi, F. Overexpression of miR-223 Promotes Tolerogenic Properties of Dendritic Cells Involved in Heart Transplantation Tolerance by Targeting Irak1. Front. Immunol. 2021, 12, 676337. [Google Scholar] [CrossRef] [PubMed]
- Wilczyński, M.; Wilczyński, J.; Nowak, M. MiRNAs as Regulators of Immune Cells in the Tumor Microenvironment of Ovarian Cancer. Cells 2024, 13, 1343. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Z.; Chen, C.; Liu, Y.; Si, Q.; Chuang, T.H.; Li, N.; Gomez-Cabrero, A.; Reisfeld, R.A.; Xiang, R.; et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene 2014, 33, 3014–3023. [Google Scholar] [CrossRef]
- Li, D.; Jia, H.; Zhang, H.; Lv, M.; Liu, J.; Zhang, Y.; Huang, T.; Huang, B. TLR4 signaling induces the release of microparticles by tumor cells that regulate inflammatory cytokine IL-6 of macrophages via microRNA let-7b. Oncoimmunology 2012, 1, 687–693. [Google Scholar] [CrossRef]
- Chen, R.; Coleborn, E.; Bhavsar, C.; Wang, Y.; Alim, L.; Wilkinson, A.N.; Tran, M.A.; Irgam, G.; Atluri, S.; Wong, K.; et al. miR-146a inhibits ovarian tumor growth in vivo via targeting immunosuppressive neutrophils and enhancing CD8(+) T cell infiltration. Mol. Ther. Oncolytics 2023, 31, 100725. [Google Scholar] [CrossRef]
- Palucka, A.K.; Coussens, L.M. The Basis of Oncoimmunology. Cell 2016, 164, 1233–1247. [Google Scholar] [CrossRef] [PubMed]
- Muljo, S.A.; Ansel, K.M.; Kanellopoulou, C.; Livingston, D.M.; Rao, A.; Rajewsky, K. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 2005, 202, 261–269. [Google Scholar] [CrossRef]
- Tagawa, T.; Albanese, M.; Bouvet, M.; Moosmann, A.; Mautner, J.; Heissmeyer, V.; Zielinski, C.; Lutter, D.; Hoser, J.; Hastreiter, M.; et al. Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J. Exp. Med. 2016, 213, 2065–2080. [Google Scholar] [CrossRef]
- Ye, S.B.; Zhang, H.; Cai, T.T.; Liu, Y.N.; Ni, J.J.; He, J.; Peng, J.Y.; Chen, Q.Y.; Mo, H.Y.; Jun, C.; et al. Exosomal miR-24-3p impedes T-cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma. J. Pathol. 2016, 240, 329–340. [Google Scholar] [CrossRef]
- Kobeissy, P.H.; Denève-Larrazet, C.; Marvaud, J.C.; Kansau, I. MicroRNA miR-27a-5p Reduces Intestinal Inflammation Induced by Clostridioides difficile Flagella by Regulating the Nuclear Factor-κB Signaling Pathway. J. Infect. Dis. 2025, 231, e38–e46. [Google Scholar] [CrossRef]
- Wieckowski, E.U.; Visus, C.; Szajnik, M.; Szczepanski, M.J.; Storkus, W.J.; Whiteside, T.L. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J. Immunol. 2009, 183, 3720–3730. [Google Scholar] [CrossRef] [PubMed]
- Portale, F.; Di Mitri, D. NK Cells in Cancer: Mechanisms of Dysfunction and Therapeutic Potential. Int. J. Mol. Sci. 2023, 24, 9521. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; He, S.; Jin, A. MiRNAs and lncRNAs in NK cell biology and NK/T-cell lymphoma. Genes Dis. 2021, 8, 590–602. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Wu, X.; Wang, Y.; Cui, H.; Li, X.; Zhang, J.; Tun, N.; Peng, Y.; Yu, J. Regulation of Human Natural Killer Cell IFN-γ Production by MicroRNA-146a via Targeting the NF-κB Signaling Pathway. Front. Immunol. 2018, 9, 293. [Google Scholar] [CrossRef]
- Xu, H.; Cheung, I.Y.; Guo, H.F.; Cheung, N.K. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: Potential implications for immune based therapy of human solid tumors. Cancer Res. 2009, 69, 6275–6281. [Google Scholar] [CrossRef]
- Wculek, S.K.; Cueto, F.J.; Mujal, A.M.; Melero, I.; Krummel, M.F.; Sancho, D. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2020, 20, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef]
- Xu, W.X.; Wang, D.D.; Zhao, Z.Q.; Zhang, H.D.; Yang, S.J.; Zhang, Q.; Li, L.; Zhang, J. Exosomal microRNAs shuttling between tumor cells and macrophages: Cellular interactions and novel therapeutic strategies. Cancer Cell Int. 2022, 22, 190. [Google Scholar] [CrossRef] [PubMed]
- Shaul, M.E.; Fridlender, Z.G. Tumour-associated neutrophils in patients with cancer. Nat. Rev. Clin. Oncol. 2019, 16, 601–620. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Conklin, D.J.; Li, F.; Dai, Z.; Hua, X.; Li, Y.; Xu-Monette, Z.Y.; Young, K.H.; Xiong, W.; Wysoczynski, M.; et al. The oncogenic microRNA miR-21 promotes regulated necrosis in mice. Nat. Commun. 2015, 6, 7151. [Google Scholar] [CrossRef]
- Dudda, J.C.; Salaun, B.; Ji, Y.; Palmer, D.C.; Monnot, G.C.; Merck, E.; Boudousquie, C.; Utzschneider, D.T.; Escobar, T.M.; Perret, R.; et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity 2013, 38, 742–753. [Google Scholar] [CrossRef]
- Stelekati, E.; Chen, Z.; Manne, S.; Kurachi, M.; Ali, M.A.; Lewy, K.; Cai, Z.; Nzingha, K.; McLane, L.M.; Hope, J.L.; et al. Long-Term Persistence of Exhausted CD8 T Cells in Chronic Infection Is Regulated by MicroRNA-155. Cell Rep. 2018, 23, 2142–2156. [Google Scholar] [CrossRef]
- Ji, Y.; Fioravanti, J.; Zhu, W.; Wang, H.; Wu, T.; Hu, J.; Lacey, N.E.; Gautam, S.; Le Gall, J.B.; Yang, X.; et al. miR-155 harnesses Phf19 to potentiate cancer immunotherapy through epigenetic reprogramming of CD8(+) T cell fate. Nat. Commun. 2019, 10, 2157. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, X.; Wang, Y.; Ren, F.; Sun, D.; Yan, Y.; Kong, X.; Bu, J.; Liu, M.; Xu, S. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020, 11, 32. [Google Scholar] [CrossRef]
- Jiang, M.; Liu, J.; Hu, S.; Yan, X.; Cao, Y.; Wu, Z. Exosomal miR-125b-5p derived from mesenchymal stromal/stem cell enhances anti-PD-1 therapy in mouse colon cancer model. Stem Cell Res. Ther. 2025, 16, 112. [Google Scholar] [CrossRef]
- Vaddi, P.K.; Osborne, D.G.; Nicklawsky, A.; Williams, N.K.; Menon, D.R.; Smith, D.; Mayer, J.; Reid, A.; Domenico, J.; Nguyen, G.H.; et al. CTLA4 mRNA is downregulated by miR-155 in regulatory T cells, and reduced blood CTLA4 levels are associated with poor prognosis in metastatic melanoma patients. Front. Immunol. 2023, 14, 1173035. [Google Scholar] [CrossRef]
- Lu, L.F.; Boldin, M.P.; Chaudhry, A.; Lin, L.L.; Taganov, K.D.; Hanada, T.; Yoshimura, A.; Baltimore, D.; Rudensky, A.Y. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 2010, 142, 914–929. [Google Scholar] [CrossRef] [PubMed]
- Sahraei, M.; Chaube, B.; Liu, Y.; Sun, J.; Kaplan, A.; Price, N.L.; Ding, W.; Oyaghire, S.; García-Milian, R.; Mehta, S.; et al. Suppressing miR-21 activity in tumor-associated macrophages promotes an antitumor immune response. J. Clin. Investig. 2019, 129, 5518–5536. [Google Scholar] [CrossRef]
- Canfrán-Duque, A.; Rotllan, N.; Zhang, X.; Fernández-Fuertes, M.; Ramírez-Hidalgo, C.; Araldi, E.; Daimiel, L.; Busto, R.; Fernández-Hernando, C.; Suárez, Y. Macrophage deficiency of miR-21 promotes apoptosis, plaque necrosis, and vascular inflammation during atherogenesis. EMBO Mol. Med. 2017, 9, 1244–1262. [Google Scholar] [CrossRef]
- Zonari, E.; Pucci, F.; Saini, M.; Mazzieri, R.; Politi, L.S.; Gentner, B.; Naldini, L. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood 2013, 122, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Vadevoo, S.M.P.; Gunassekaran, G.R.; Yoo, J.D.; Kwon, T.H.; Hur, K.; Chae, S.; Lee, B. Epigenetic therapy reprograms M2-type tumor-associated macrophages into an M1-like phenotype by upregulating miR-7083-5p. Front. Immunol. 2022, 13, 976196. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, J.L.; Takami, A.; Yoshioka, K.; Nakata, K.; Sato, T.; Kasahara, Y.; Nakao, S. Human microRNA-1245 down-regulates the NKG2D receptor in natural killer cells and impairs NKG2D-mediated functions. Haematologica 2012, 97, 1295–1303. [Google Scholar] [CrossRef]
- Pesce, S.; Greppi, M.; Ferretti, E.; Obino, V.; Carlomagno, S.; Rutigliani, M.; Thoren, F.B.; Sivori, S.; Castagnola, P.; Candiani, S.; et al. miRNAs in NK Cell-Based Immune Responses and Cancer Immunotherapy. Front. Cell Dev. Biol. 2020, 8, 119. [Google Scholar] [CrossRef]
- Sullivan, R.P.; Fogel, L.A.; Leong, J.W.; Schneider, S.E.; Wong, R.; Romee, R.; Thai, T.H.; Sexl, V.; Matkovich, S.J.; Dorn, G.W., 2nd; et al. MicroRNA-155 tunes both the threshold and extent of NK cell activation via targeting of multiple signaling pathways. J. Immunol. 2013, 191, 5904–5913. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Sun, Q.; Yan, J.; Huang, J.; Zhu, S.; Yu, J. Identification of microRNA transcriptome involved in human natural killer cell activation. Immunol. Lett. 2012, 143, 208–217. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, S.; Liu, J.; Zhang, Z.; Mao, X.; Zhou, H. MicroRNA-130a enhances the killing ability of natural killer cells against non-small cell lung cancer cells by targeting signal transducers and activators of transcription 3. Biochem. Biophys. Res. Commun. 2020, 523, 481–486. [Google Scholar] [CrossRef]
- Di Pace, A.L.; Pelosi, A.; Fiore, P.F.; Tumino, N.; Besi, F.; Quatrini, L.; Santopolo, S.; Vacca, P.; Moretta, L. MicroRNA analysis of Natural Killer cell-derived exosomes: The microRNA let-7b-5p is enriched in exosomes and participates in their anti-tumor effects against pancreatic cancer cells. Oncoimmunology 2023, 12, 2221081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, Q.; Yu, L.; Zhu, D.; Li, Y.; Xue, Z.; Hua, Z.; Luo, X.; Song, Z.-Q.; Lu, C.; et al. The Role of miRNA in Tumor Immune Escape and miRNA-Based Therapeutic Strategies. Front. Immunol. 2022, 12, 807895. [Google Scholar] [CrossRef]
- Nishikawa, H.; Sakaguchi, S. Regulatory T cells in tumor immunity. Int. J. Cancer 2010, 127, 759–767. [Google Scholar] [CrossRef] [PubMed]
- De Cicco, P.; Ercolano, G.; Ianaro, A. The New Era of Cancer Immunotherapy: Targeting Myeloid-Derived Suppressor Cells to Overcome Immune Evasion. Front. Immunol. 2020, 11, 1680. [Google Scholar] [CrossRef]
- Parker, K.; Beury, D.; Ostrand-Rosenberg, S. Myeloid-Derived Suppressor Cells: Critical Cells Driving Immune Suppression in the Tumor Microenvironment. Adv. Cancer Res. 2015, 128, 95–139. [Google Scholar] [CrossRef] [PubMed]
- Ostrand-Rosenberg, S.; Fenselau, C. Myeloid-Derived Suppressor Cells: Immune-Suppressive Cells That Impair Antitumor Immunity and Are Sculpted by Their Environment. J. Immunol. 2018, 200, 422–431. [Google Scholar] [CrossRef]
- Gabrilovich, D. Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 2017, 5, 3–8. [Google Scholar] [CrossRef]
- Marvel, D.; Gabrilovich, D. Myeloid-derived suppressor cells in the tumor microenvironment: Expect the unexpected. J. Clin. Investig. 2015, 125, 3356–3364. [Google Scholar] [CrossRef]
- Jung, M.; Bonavida, B. Immune Evasion in Cancer Is Regulated by Tumor-Asociated Macrophages (TAMs): Targeting TAMs. Crit. Rev. Oncog. 2024, 29, 1–17. [Google Scholar] [CrossRef]
- Qiu, Y.; Chen, T.; Hu, R.; Zhu, R.; Li, C.; Ruan, Y.; Xie, X.-H.; Li, Y. Next frontier in tumor immunotherapy: Macrophage-mediated immune evasion. Biomark. Res. 2021, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Petty, A.; Owen, D.; Yang, Y.; Huang, X. Targeting Tumor-Associated Macrophages in Cancer Immunotherapy. Cancers 2021, 13, 5318. [Google Scholar] [CrossRef] [PubMed]
- Petty, A.; Yang, Y. Tumor-associated macrophages: Implications in cancer immunotherapy. Immunotherapy 2017, 9, 289–302. [Google Scholar] [CrossRef]
- Wang, S.; Gao, Y. Pancreatic cancer cell-derived microRNA-155-5p-containing extracellular vesicles promote immune evasion by triggering EHF-dependent activation of Akt/NF-κB signaling pathway. Int. Immunopharmacol. 2021, 100, 107990. [Google Scholar] [CrossRef]
- Yin, Y.; Liu, B.; Cao, Y.; Yao, S.; Liu, Y.; Jin, G.; Qin, Y.; Chen, Y.; Cui, K.; Zhou, L.; et al. Colorectal Cancer-Derived Small Extracellular Vesicles Promote Tumor Immune Evasion by Upregulating PD-L1 Expression in Tumor-Associated Macrophages. Adv. Sci. 2022, 9, 2102620. [Google Scholar] [CrossRef] [PubMed]
- Gou, Z.; Li, J.; Liu, J.; Yang, N. The hidden messengers: Cancer associated fibroblasts—Derived exosomal miRNAs as key regulators of cancer malignancy. Front. Cell Dev. Biol. 2024, 12, 1378302. [Google Scholar] [CrossRef]
- Shi, L.; Zhu, W.; Huang, Y.; Zhuo, L.; Wang, S.; Chen, S.; Zhang, B.; Ke, B. Cancer-associated fibroblast-derived exosomal microRNA-20a suppresses the PTEN/PI3K-AKT pathway to promote the progression and chemoresistance of non-small cell lung cancer. Clin. Transl. Med. 2022, 12, e989. [Google Scholar] [CrossRef]
- Huijbers, E.; Khan, K.; Kerbel, R.; Griffioen, A. Tumors resurrect an embryonic vascular program to escape immunity. Sci. Immunol. 2022, 7, eabm6388. [Google Scholar] [CrossRef]
- Yang, C.; Gibson, V.P.; Hardy, P. The Role of MiR-181 Family Members in Endothelial Cell Dysfunction and Tumor Angiogenesis. Cells 2022, 11, 1670. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Pearce, E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016, 213, 15–23. [Google Scholar] [CrossRef]
- Biswas, S.K. Metabolic Reprogramming of Immune Cells in Cancer Progression. Immunity 2015, 43, 435–449. [Google Scholar] [CrossRef]
- Pearce, E.J.; Everts, B. Dendritic cell metabolism. Nat. Rev. Immunol. 2015, 15, 18–29. [Google Scholar] [CrossRef]
- Yao, Q.; Song, Z.; Wang, B.; Zhang, J.-a. Emerging roles of microRNAs in the metabolic control of immune cells. Cancer Lett. 2018, 433, 10–17. [Google Scholar] [CrossRef]
- Alshahrani, S.H.; Ibrahim, Y.S.; Jalil, A.T.; Altoum, A.A.; Achmad, H.; Zabibah, R.S.; Gabr, G.A.; Ramírez-Coronel, A.A.; Alameri, A.A.; Qasim, Q.A.; et al. Metabolic reprogramming by miRNAs in the tumor microenvironment: Focused on immunometabolism. Front. Oncol. 2022, 12, 1042196. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Chen, B.; Zhang, Z.; Huang, Y.; Li, J.; Wei, Q.; Cao, D.; Ai, J. Crosstalk between Tumor-Associated Macrophages and MicroRNAs: A Key Role in Tumor Microenvironment. Int. J. Mol. Sci. 2022, 23, 13258. [Google Scholar] [CrossRef]
- Sheedy, F.J. Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front. Immunol. 2015, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Wang, Y.; Zhu, Y.; Li, S.; Liu, Y.; Chen, C.; Wang, X.; Zen, K.; Li, L. The miR-125a/HK2 axis regulates cancer cell energy metabolism reprogramming in hepatocellular carcinoma. Sci. Rep. 2017, 7, 3089. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.W.; Bauer, K.M.; Barba, C.; Ekiz, H.A.; O’Connell, R.M. miR-aculous new avenues for cancer immunotherapy. Front. Immunol. 2022, 13, 929677. [Google Scholar] [CrossRef]
- Yu, T.; Ju, Z.; Luo, M.; Hu, R.; Teng, Y.; Xie, L.; Zhong, C.; Chen, L.; Hou, W.; Xiong, Y. Elevated expression of miR-146a correlates with high levels of immune cell exhaustion markers and suppresses cellular immune function in chronic HIV-1-infected patients. Sci. Rep. 2019, 9, 18829. [Google Scholar] [CrossRef]
- Sun, Y.; Oravecz-Wilson, K.; Bridges, S.; McEachin, R.; Wu, J.; Kim, S.H.; Taylor, A.; Zajac, C.; Fujiwara, H.; Peltier, D.C.; et al. miR-142 controls metabolic reprogramming that regulates dendritic cell activation. J. Clin. Investig. 2019, 129, 2029–2042. [Google Scholar] [CrossRef]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 1–24. [Google Scholar] [CrossRef]
- Ahir, M.; Upadhyay, P.; Ghosh, A.; Sarker, S.; Bhattacharya, S.; Gupta, P.; Ghosh, S.; Chattopadhyay, S.; Adhikary, A. Delivery of dual miRNA through CD44-targeted mesoporous silica nanoparticles for enhanced and effective triple-negative breast cancer therapy. Biomater. Sci. 2020, 8, 2939–2954. [Google Scholar] [CrossRef]
- Miroshnichenko, S.; Patutina, O. Enhanced Inhibition of Tumorigenesis Using Combinations of miRNA-Targeted Therapeutics. Front. Pharmacol. 2019, 10, 488. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.-K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 2018, 183, 428–444. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Seto, A.G.; Beatty, X.; Hermreck, M.; Gilles, M.E.; Stroopinsky, D.; Pinter-Brown, L.C.; Pestano, L.; Marchese, C.; Avigan, D.; et al. Cobomarsen, an Oligonucleotide Inhibitor of miR-155, Slows DLBCL Tumor Cell Growth In Vitro and In Vivo. Clin. Cancer Res. 2021, 27, 1139–1149. [Google Scholar] [CrossRef]
- Fornari, F.; Gramantieri, L.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.; Grazi, G.; Giovannini, C.; Croce, C.M.; Bolondi, L. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 2008, 27, 5651–5661. [Google Scholar] [CrossRef]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef]
- Sulaiman, S.A.; Ab Mutalib, N.S.; Jamal, R. miR-200c Regulation of Metastases in Ovarian Cancer: Potential Role in Epithelial and Mesenchymal Transition. Front. Pharmacol. 2016, 7, 271. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, M.; Jorge, J.; Gonçalves, A.C.; Sarmento-Ribeiro, A.B.; Carvalho, R.; Figueiras, A.; Santos, A.C.; Veiga, F. miR-29b and retinoic acid co-delivery: A promising tool to induce a synergistic antitumoral effect in non-small cell lung cancer cells. Drug Deliv. Transl. Res. 2020, 10, 1367–1380. [Google Scholar] [CrossRef] [PubMed]

| miRNA | Immune Cell/Target | Function/Effect | Context/Mechanism | Ref. |
|---|---|---|---|---|
| miR-23a | CD8+, NK | Suppresses effector functions, Impairs cytotoxicity, upregulated by TGF-β | Inhibits granzyme B, promotes immune evasion, transferred via exosomes under hypoxia | [11,12] |
| miR-23b | DC | Promotes tolerogenic DCs | Suppresses Notch1/NF-κB | [13,14] |
| miR-139 | CD8+ | Inhibits perforin expression, downregulation enhances cytotoxicity | Downregulated during inflammation | [15] |
| miR-150 | CD8+, NK | Regulates differentiation into effector/memory subsets, maturation | Targets c-Myb, represses PI3K-AKT pathway | [15,16,17,18] |
| miR-155 | CD8+, CD4+, DC, NK, TAM | Essential for CTL and Th1 responses, DC maturation, enhances NK cytotoxicity by upregulating NKG2D, IFN-γ, and granzyme B, drives M1 TAM, promotes NETosis by upregulating PAD4, enhances Treg function, modulate PD-L1, drives monocyte conversion to MDSC, and inhibits T cell exhaustion | Regulates interferon signaling, targets SOCS1, SHIP-1, C/EBPβ, c-Fos | [19,20,21,22,23,24,25,26,27,28] |
| miR-194-5p | Treg | Enhance function | Modulate PD-L1 | [29] |
| miR-135b | CD8+, CD4+ | Silencing reduces granzyme B and perforin, enhances IL-17 by repressing STAT6/GATA3 | Silencing impairs cytotoxicity, promotes Th17 | [30,31] |
| miR-17-92 cluster | CD8+, CD4+ | CTL differentiation, supports Tfh function | Regulate CTL differentiation during antiviral and antitumor responses | [15,32,33] |
| miR-326 | CD4+ | Facilitates Th17 development | - | [32,33] |
| miR-27a-5p | CD4+, NK, DC | Inhibit Treg/Th17 and Th1 differentiation, Regulates NK chemotaxis | modulate CX3CR1 and CX3CL1, induced by TGF-β1, effects on chemotactic receptors and immunosuppressive ligands | [34] |
| mir-27a/b | TAM | Promote M1 polarization, pro-inflammatory | increased inflammatory cytokine production and tumoricidal activity | [23] |
| miR-130a/b | TAM, TAN, NK | Promote M1 polarization, pro-inflammatory, Controls neutrophil maturation, targets STAT3, enhance NK cytotoxicity | Regulates MPO, proteinase 3, TGF-β1, controlling cell cycle exit, | [23,35] |
| miR-24 | CD4+, T cells | Promotes Th1/Th17, drives T cell exhaustion | immunosuppressive TME, Upregulates CD39, PD-1, TIM-3, dampens metabolism, targeting Myc and FGF11 | [36] |
| miR-107 | DC | Targets IL-23p19 | alters T cell polarization | [37] |
| let-7i | DC | Suppresses immune response, suppresses SOCS1 and enhances DC activation | Alters cytokine expression, in response to LPS but not other TLR ligands | [38,39] |
| miR-192 | Treg | Promotes Treg differentiation, enhances IL-10 | Regulates via RB1/NF-κBp65 | [40] |
| miR-214 | Treg | Promotes Treg expansion, secretion of IL-10 | Delivered from tumor cells via exosomes, by downregulating PTEN | [41] |
| miR-34 | Treg, NK, Tumor | Suppresses CCL22, limits Treg recruitment, Upregulate ULBP2, enhance NK cytotoxicity | Downregulated by TGF-β, restoration reduces Treg infiltration, Tumor suppressors | [42,43,44] |
| miR-223 | NK, TAN, TAM | Restrains NK cytotoxicity, regulates neutrophil maturation, NETosis, modulates cytokine secretion, promotes pro-metastatic behavior, and shapes cytokine milieu | Targets granzyme B, IGF-R, and influence TAM–tumor interactions | [25,26,27,45,46,47,48,49] |
| miR-15/16 | NK, T cells | Targets IFN-γ, Bcl2, cyclin D1, restrains T cell function, restrains T cell function, and targets mTOR | Knockout enhances T cell responses, Knockout enhances proliferation, cytokine secretion, and T cell hyporesponsiveness | [50,51,52,53] |
| miR-20a | NK, CAF, TAM, TAN | Reduces MICA/B expression, suppresses PTEN/PI3K-AKT pathway, promotes angiogenesis, and neutrophil adhesion/migration | Upregulated in ovarian cancer, CAF-derived exosomes, target HIFs, and upregulate IL-8 | [54,55] |
| miR-29a | NK, DC, TAM, MDSC | Inhibits melanoma, regulates B7-H3, Represses DNA methyltransferases, counters pro-inflammatory DC, enhances MDSC differentiation and function, in TAM creates a Treg/Th17 imbalance, and promotes immune suppression | Inhibits angiogenesis, modulates immune checkpoint, targets Hbp1, Prkar1a in MDSC, and STAT3 in CD4+ | [56,57] |
| miR-29b | DC, TAM, Endothelial | Represses DNA methyltransferases, influencing epigenetic regulation and immune gene expression, regulates B7-H3, counters pro-inflammatory DC phenotype, inhibits angiogenesis, inhibits angiogenesis, and tumor growth | Inhibits melanoma, modulates immune checkpoint, targets Akt3, and inhibits VEGF/C-MYC | [58,59,60,61,62,63] |
| miR-30e | NK | Suppresses granzyme B and perforin in CD56dim NK cells, and is elevated under chronic IFN or IL-15 stimulation | Limits cytotoxicity, suppresses translation of PRF1 | [64,65] |
| miR-183 | NK | elevated in RCC, inhibits DAP12 (TYROBP), and impairs NK function | Upregulated by TGF-β | [66] |
| miR-18a | NK | Downregulates NKG2D and ligands, enhancing IFN-γ secretion and NK cell responses | Induced by IDO1, lncRNA-GAS5 counteracts | [67] |
| miR-561-5p | NK | Regulates NK cell recruitment | modulate CX3CR1 and CX3CL1, affects pulmonary metastasis | [34] |
| miR-186 | NK | Inhibits neuroblastoma growth | Exosomal, suppresses immune escape | [68] |
| miR-BART7 | NK | Downregulates MICA, reduces cytotoxicity | EBV-encoded | [69] |
| miR-222, miR-339 | NK, T cells | Impairs cell interactions | Downregulate ICAM-1 | [70] |
| miR-148a | DC | Inhibition enhances DC function | Targets DNMT1/SOCS1 | [71] |
| miR-148b-3p | CAF | Reduced in CAF exosomes, enhances chemosensitivity | Suppresses Wnt/β-catenin, elevates PTEN | [72] |
| miR-22 | DC, CAF, Treg | Impairs DC function, activates JAK/STAT3 in CAF and Tregs | Targets p38, promotes chemokine secretion (CCL17, CCL20, and CCL22), recruits Tregs via chemokines | [71,73] |
| miR-212-3p | DC | Inhibits MHC II expression and promotes tolerance | Taken up by DCs, inhibits RFXAP, and is delivered by pancreatic cancer exosomes | [74] |
| miR-17-5p | DC, Treg | Suppresses DC maturation and promotes Treg-mediated suppression | Upregulated in gastric cancer, and suppresses RUNX3 | [75,76] |
| miR-221 | DC | Regulates IL-12, apoptosis, and DC maturation | Targets p27kip1, KPC1, and SOCS1 | [77] |
| miR-5119 | DC | Enhances the immunogenicity of DCs by modulating checkpoint molecules | Improves responses in breast cancer | [78] |
| miR-133a | DC | As a tumor suppressor in osteosarcoma, and suppresses DC activation | Targets RBP-J | [79] |
| miR-147 | TAM | Regulates TAM function, Attenuates inflammation | Target components of the TLR/NF-κB pathway | [71,80] |
| miR-21 | TAN, TAM, CAF, MDSC | Modulate cytokine secretion, promote pro-metastatic behavior, shape cytokine milieu, promote expansion, activation, T cell inhibition, shape cytokine milieu, promote M2 TAM, and support the equilibrium, leading to immune evasion and increasing drug resistance | Induced by hypoxia, released by CAFs in exosomes, and stimulates MDSC generation via STAT3 | [46,47,81] |
| miR-21-5p | TAM | Exosomes from TAMs create Treg/Th17 imbalance, and promote immune suppression and metastasis | Target STAT3 in CD4+ | [57] |
| miR-9 | TAM, MDSC, Endothelial | Overexpressed in tumors, skewed toward a tumor-promoting phenotype TAM, promote early development of MDSC via a pathway, and promote migration and angiogenesis | Target SOCS3, PIAS3 in Endothelial and JAK/STAT in MDSC, overexpressed in tumors, transferred from the tumor, and taken up by endothelial cells | [57,60,61,62,82] |
| miR-1246 | TAM, MDSCs | Skewed toward tumor-promoting phenotype, transferred from the tumor, and promotes differentiation and activation | Shape TAM phenotypes via exosomal miRNAs, via DUSP3/ERK pathway | [60,61,62,83] |
| miR-33 | TAM | Regulate the functional polarization of macrophages | Regulates lipid metabolism | [84] |
| miR-17 | DC, Treg, TAM, TAN | Suppresses DC maturation, promotes Treg-mediated suppression, promotes angiogenesis, and neutrophil adhesion/migration | Upregulated in gastric cancer, suppresses RUNX3, targets HIFs, supports vascular remodeling in the TME, and upregulates IL-8 | [17,55,76] |
| miR-98, miR-720 | TAM | Regulate inflammatory cytokine balance and tumor cell migration | Modulate TAM-secreted | [85,86] |
| miR-511-3p | TAM | Limits pro-tumor function | Expressed in CD206+ TAMs | [87] |
| miR-146a | TAN, MDSC, NK, DC, TAN, Treg | Regulates NETosis, promotes immune suppression, suppresses NK proliferation/cytotoxicity, DC maturation, stabilizes Treg, promotes M2 TAM, and monocyte conversion to MDSC | Enhances IL-8, CCL5, induced by NF-κB, targets TRAF6/IRAK1, | [23,88,89,90] |
| miR-146b | NK, MDSC | Enhances NK activity against chemo-resistant cancer, and drives monocyte conversion to MDSC | Targets WBSCR22 | [91,92] |
| miR-1696 | TAN | Promotes NETosis, oxidative stress, and immune cell recruitment | Represses GPx3 | [25,26,27,93] |
| miR-16-5p | TAN | NETosis-associated, regulates autophagy | Neutrophil survival and function in the tumor | [48,49] |
| miR-4780 | TAN | Define N2 pro-tumorigenic phenotype | Upregulated in colon cancer, Target TUSC1 | [94] |
| miR-3938 | TAN | Define N2 pro-tumorigenic phenotype | Downregulated in colon cancer, targets ZNF197 | [94] |
| miR-138 | Tumor, NK, TAN | Modulates PD-L1/PD-1 signaling in tumor and immune cells, and suppresses neutrophil-derived NGAL | Reduces proliferation/metastasis in pancreatic cancer | [95,96] |
| let-7b | TAN, Tumor | Anti-inflammatory, suppresses TLR4, IL-6, IL-8, TNF-α, and suppresses tumor proliferation | Upregulates IL-10, can be delivered in exosomes, secreted via NK exosomes enriched | [97] |
| miR-31 | TAN, CD8+ | Promote neutrophil adhesion, promote CD8+ exhaustion, and upregulate PD-1, LAG3 and IL-10 | Upregulating IL-8, induced by TCR activation, increases the transcription factor c-Maf and the PGE₂ receptor Ptger2 | [17,36,98,99] |
| miR-199, miR-722 | TAN | Limits immune surveillance | Inhibits neutrophil chemotaxis and TAN infiltration | [100,101] |
| miR-142-3p | TAN | Regulates neutrophil–macrophage interaction | Modulates TNF-α and inhibits PKC-α | [38,102] |
| miR-125a | TAM, MDSC | Contribute to PMN-MDSC development, reinforce macrophage-mediated suppression, and enrich in M2 TAMs | Drive monocyte conversion to MDSC. miR-125a redirects TAMs towards an M1 | [23,88,89,90,103,104] |
| miR-125b | TAM, Treg, MDSC | Inhibit Treg proliferation, induce tumor-promoting TAM, increase inflammatory cytokines, promote M1 differentiation, and drive monocyte conversion to MDSC | Targets type I IFN pathway, downregulates TNFR2, FoxP3, targets LIPA, Tilt equilibrium by weakening Treg-mediated immunosuppression | [13,14,23,90,105] |
| miR-28 | T cells | Suppresses checkpoint molecules, revives T cell activity | Downregulated in exhausted CD8+, and suppresses the checkpoint molecules PD-1, TIM-3, and BTLA | [106] |
| miR-200c | Tumor, T cells, Endothelial | Reduce PD-L1, reinvigorate T cells, inhibit migration, and angiogenesis | Suppressed in HBV-driven HCC via TGF-β/STAT3 signaling, | [82,107] |
| miR-4772-3p | Treg | Promotes Treg proliferation | Upregulates Helios (IKZF2) | [108] |
| miR-128-3p | Treg | Influences Treg enrichment in gastric cancer, promoting immune evasion | Interacting with IL16 | [109] |
| miR-182 | Treg | Promotes Treg polarization and formation of IL-17-producing Tregs, facilitating immune evasion | Targets FOXO1, NFATs, Increases FOXP3, TGF-β | [110] |
| miR-325-3p | Treg | Regulates Treg function in OSCC | Stabilizes Foxp3 | [111] |
| miR-208b | Treg | Promotes expansion, chemoresistance | Targets PDCD4 | [112] |
| miR-423-5p | Treg | Supports Treg-mediated immune evasion in HCC | Interacts with FENDRR | [113] |
| miR-429 | Treg | Reduces Treg infiltration in HCC | Targets PD-L1 | [114] |
| miR-210 | MDSC | Promote expansion, MDSC activation, and T cell inhibition | Induced by hypoxia | [115,116] |
| miR-92a | MDSC | Enhance MDSC differentiation, function, and establishment of an immunosuppressive environment | Target Hbp1 and Prkar1a | [56] |
| miR-1298-5p | MDSC | Enhances immunosuppressive effects | Inhibits glioma progression | [117] |
| miR-181a/b | MDSC, NK, TAM | Promote early development via JAK/STAT pathway, enhance MDSC differentiation, immune suppression, tumor progression, promote IFN-γ in NK, and favor M2 polarization | Target SOCS3, PIAS3, NLK | [17,18,57] |
| miR-181d-5p | CAF | Inhibits CDX2, HOXA5, promotes EMT, and invasion in breast cancer | Taken up by cancer cells | [118] |
| miR-494 | MDSC, Endothelial | Reduces immunosuppressive effects, promotes angiogenesis, and evasion | regulated by TGF-β and upregulated in gastric cancer, overexpressed in tumors | [119] |
| miR-375 | TAM | Enhances migration and infiltration | Released from the apoptotic tumor, targets TNS3 and PXN in TAM and CCL2 in the tumor | [120] |
| miR-101 | TAM | Facilitates M1→M2 transition, enhances proliferation/migration | Targets C/EBPα, KLF6, in breast and ovarian cancer | [121] |
| miR-100 | TAM, MDSC | Sustains pro-tumor characteristics, influencing the mTOR pathway, driving monocyte conversion to MDSC | Linked to Stat5a/IL-1ra pathway, Links PGE2 and IL-6 signaling | [90,122] |
| miR-149 | CAF, T cells | Immune evasion and tumor progression, reinvigorates T cells, and reverses exhaustion by targeting PD-1, TIM-3, BTLA, and Foxp1 | Silenced to enhance IL-6 secretion in gastric cancer, links PGE2, and promotes EMT | [123,124,125] |
| miR-500a-5p | CAF | Targets USP28, facilitates proliferation, metastasis, immune evasion, and dissemination | Delivered via exosomes and taken up by cancer cells | [126] |
| miR-369 | CAF | Enhances migration/invasion | Stimulates the MAPK pathway | [127] |
| miR-196a | CAF | Promotes migration/invasion, enhances CCL2 secretion | Targets ANXA1 | [128] |
| miR-196b | Endothelial, NK | Upregulated by VEGF and promote angiogenesis | Targeted to reduce tumor growth | [129] |
| miR-10b | Endothelial | Upregulated by VEGF, promotes angiogenesis in breast tumors, downregulates MICB, a stress ligand for NKG2D | Targeted to reduce tumor growth and facilitate tumor escape | [129,130] |
| miR-10a | CD8+, DC, MDSC | T cell inhibition, influences cytotoxic molecule expression, targets IL-12/IL-23p40 in DC and RORA in glioblastoma, promotes expansion, activation, and alters the MDSCs via NF-κB | Downregulates antigen presentation induced by hypoxia | [115,116,131] |
| miR-141 | Endothelial, Treg | Enhances angiogenesis and migration via JAK/STAT3, NF-Κb in ovarian cancer, recruits Tregs via CXCL1, and enhances angiogenesis | Released in extracellular vesicles, Activates JAK/STAT3, NF-κB | [109,132] |
| miR-1468-5p | Endothelial, T cell | Enhances PD-L1, promotes lymphangiogenesis, and causes immune escape | Activates JAK2/STAT3, secreted by cancer exosomes | [133] |
| miR-526b, miR-655 | Endothelial | Promotes angiogenesis, lymphangiogenesis in breast cancer, upregulates VEGF, and downregulates PTEN | Induced by COX-2 | [134] |
| miR-200 and miR-128 | Endothelial | Inhibits migration and angiogenesis | Downregulated in tumors | [82] |
| miR-103 | Endothelial | Enhances tumor cell death and inhibits angiogenesis | Exacerbates DNA damage | [135] |
| miRNA | Target Immune Cell/Pathway | Mechanism of Action | Ref. |
|---|---|---|---|
| let-7 family | CD8+ (naïve → effector switch); TAMs via exosomes (AKT/mTOR) | In T cells, it suppresses c-Myc and glycolytic enzymes (e.g., HK2 and LDHA), maintaining OXPHOS in naïve cells. In TAMs, tumor exosomal let-7a inhibits AKT/mTOR, shifting macrophages to OXPHOS metabolism. | [194] |
| miR-155 | CD8+ (PI3K/AKT/mTOR); Th1 cells; M1 Macrophages | Targets SHIP1, SOCS1 to unleash PI3K-AKT-mTOR signaling—increases glycolysis, proliferation, and IFN-γ in T cells. Inhibits IL-4/c-Maf to skew TH toward Th1 In macrophages, it promotes M1 polarization via JAK/STAT and NF-κB pathways. | [62,194,198] |
| miR-143 | CD8+ (glucose metabolism via GLUT1) | Directly targets the GLUT1 transporter, reducing glucose uptake and glycolysis in T cells. Favors mitochondrial metabolism, supporting central memory T cell formation. | [194] |
| miR-23a | CD8+ (BLIMP-1, effector TFs) | TGF-β-induced in TILs; targets PRDM1 (BLIMP-1) and other effector genes. Reduces granzyme B, TNF, and IFN-γ in CD8+. Reduces cytotoxicity without affecting proliferation. | [11] |
| miR-146a | T cells (NF-κB/STAT1, checkpoints); Tregs; TAMs (TLR/NF-κB) | In T effector cells: targets IRAK1, TRAF6, STAT1, etc., dampening NF-κB and IFN pathways—leads to increased PD-1, CTLA-4, TIM-3, and LAG-3 on T cells (via indirect c-Fos effects). In Tregs, sustains suppressive function (prevents Th1 conversion). In macrophages, it reduces TLR signaling, curbing pro-IL-12/IL-1β and encouraging IL-10. | [199] |
| miR-21 | Tumor-Associated Macrophages (PTEN/PI3K-Akt); CD4 T cells (indirect); MDSCs | Targets PTEN and PDCD4 in macrophages, activating PI3K/AKT and downregulating IL-12, promoting M2 polarization. TAM-derived exosomal miR-21 delivers these signals to neighboring cells, and also prevents apoptosis in tumor cells (via PTEN→AKT). In CD4+ T (induced by NF-κB), aids Th2 and Treg responses (resolves inflammation). | [195,196] |
| miR-30c | Macrophages (REDD1/mTOR axis in TAMs) | Targets REDD1, a negative regulator of mTOR. miR-30c keeps mTOR active, sustaining glycolysis and M1 phenotype. Hypoxia lowers miR-30c, resulting in REDD1 accumulation and mTOR inhibition (glycolysis ↓). | [194] |
| miR-125a-5p | Macrophages (IRF5, KLF4 pathways); TAMs in late-stage tumors | Likely targets transcripts (e.g., IRF5, a driver of M1; KLF4, influences M2), thereby shifting the balance to M2. (miR-125a also targets HK2 in tumor cells, but in TAMs it is linked to polarization). Induced by tumor cytokines. | [197] |
| miR-142 | Dendritic Cells (CPT1a, metabolic switch); also Tregs (SOCS1) | In DCs, targets CPT1a to suppress FAO, enabling glycolysis and DC activation. Without miR-142, DCs remain oxidative (tolerogenic). In Tregs, targets SOCS1, promoting IL-2 signaling and Treg survival. | [198,200] |
| miR-28 | CD8+ (PD-1 and co-inhibitory receptors) | Directly binds PDCD1 (PD-1) mRNA 3′UTR, preventing its translation. Also reported to downregulate other exhaustion markers (Tim-3 and BTLA). Preserves T cell glycolytic capacity and cytokine secretion by keeping the checkpoint low. | [106] |
| miR-146a/b, miR-155, miR-125b, miR-100, let-7e (tumor EV cluster) | Circulating Monocytes → MDSC differentiation (multiple targets) | These miRNAs are released in melanoma exosomes and taken up by CD14+ monocytes. Collectively, they reprogram transcription to induce an immunosuppressive, metabolically quiescent MDSC phenotype (increased Arg1, IDO, and nitric oxide). let-7e and miR-125b reduce pro-inflammatory mediators; miR-146a/b and miR-155 modulate NF-κB. | [90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zare, E.; Yaghoubi, S.M.; Khoshnazar, M.; Jafari Dargahlou, S.; Machhar, J.S.; Zheng, Z.; Duijf, P.H.G.; Mansoori, B. MicroRNAs in Cancer Immunology: Master Regulators of the Tumor Microenvironment and Immune Evasion, with Therapeutic Potential. Cancers 2025, 17, 2172. https://doi.org/10.3390/cancers17132172
Zare E, Yaghoubi SM, Khoshnazar M, Jafari Dargahlou S, Machhar JS, Zheng Z, Duijf PHG, Mansoori B. MicroRNAs in Cancer Immunology: Master Regulators of the Tumor Microenvironment and Immune Evasion, with Therapeutic Potential. Cancers. 2025; 17(13):2172. https://doi.org/10.3390/cancers17132172
Chicago/Turabian StyleZare, Erfan, Seyyed Mohammad Yaghoubi, Maedeh Khoshnazar, Sina Jafari Dargahlou, Janvhi Suresh Machhar, Zihan Zheng, Pascal H. G. Duijf, and Behzad Mansoori. 2025. "MicroRNAs in Cancer Immunology: Master Regulators of the Tumor Microenvironment and Immune Evasion, with Therapeutic Potential" Cancers 17, no. 13: 2172. https://doi.org/10.3390/cancers17132172
APA StyleZare, E., Yaghoubi, S. M., Khoshnazar, M., Jafari Dargahlou, S., Machhar, J. S., Zheng, Z., Duijf, P. H. G., & Mansoori, B. (2025). MicroRNAs in Cancer Immunology: Master Regulators of the Tumor Microenvironment and Immune Evasion, with Therapeutic Potential. Cancers, 17(13), 2172. https://doi.org/10.3390/cancers17132172







