Glioblastoma: Overview of Proteomic Investigations and Biobank Approaches for the Development of a Multidisciplinary Translational Network
Simple Summary
Abstract
1. Introduction
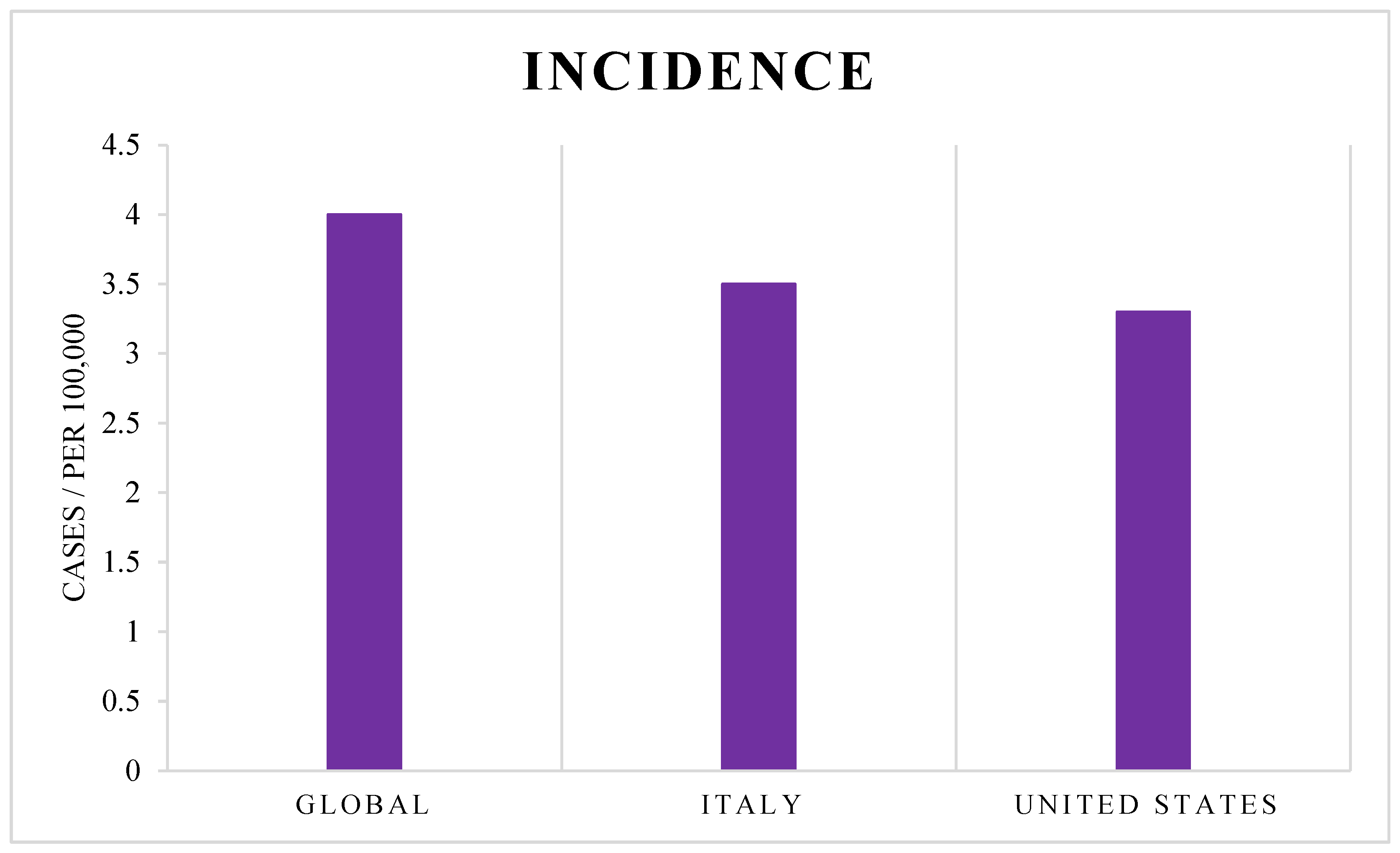
2. GBM Incidence
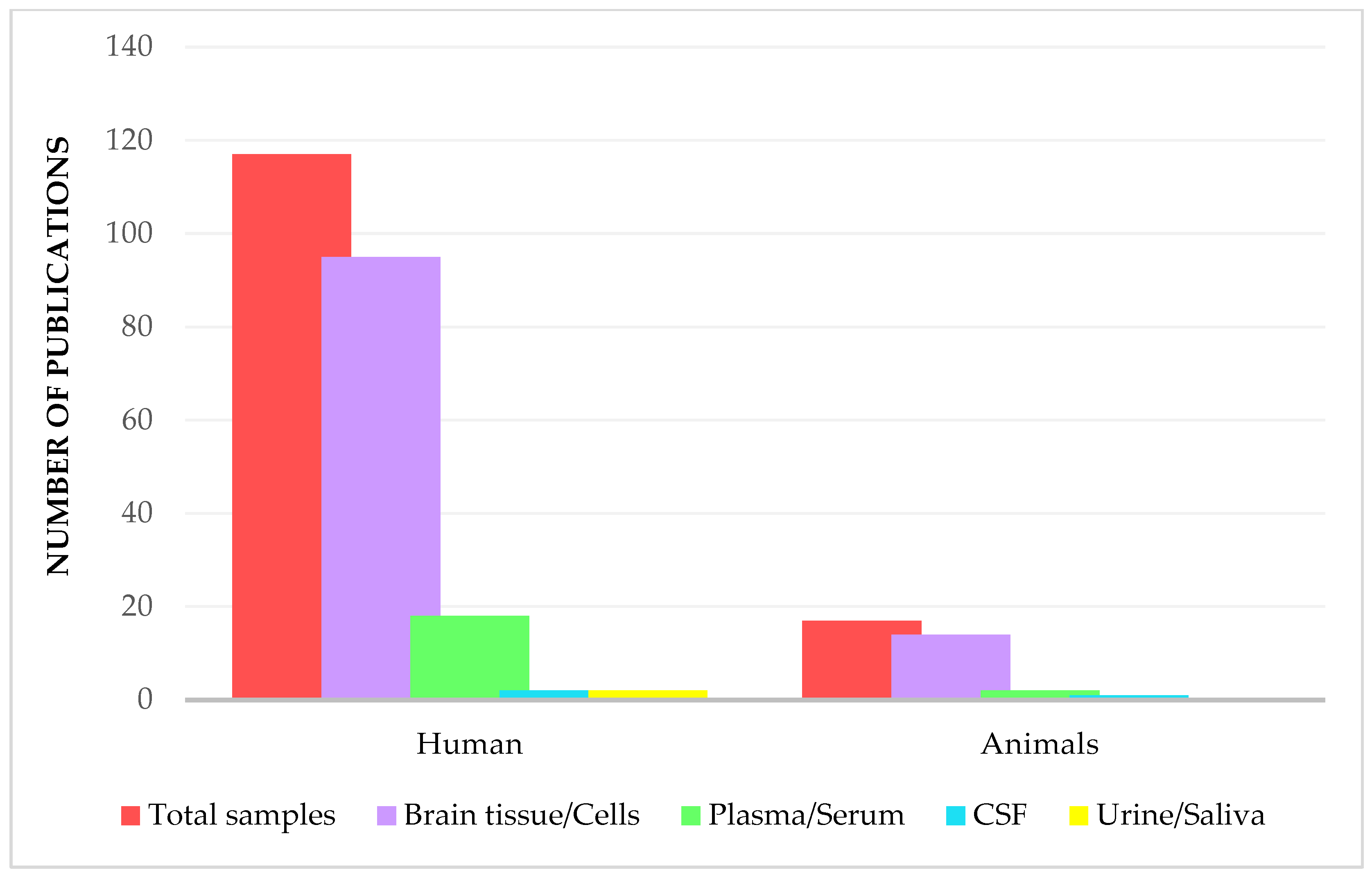
3. Proteomics Analysis in Glioblastoma Disease
3.1. Brain Tissue and Related Cell Lines
3.2. Biofluids
3.2.1. Cerebrospinal Fluid
3.2.2. Plasma
3.2.3. Serum
3.2.4. Urine and Saliva
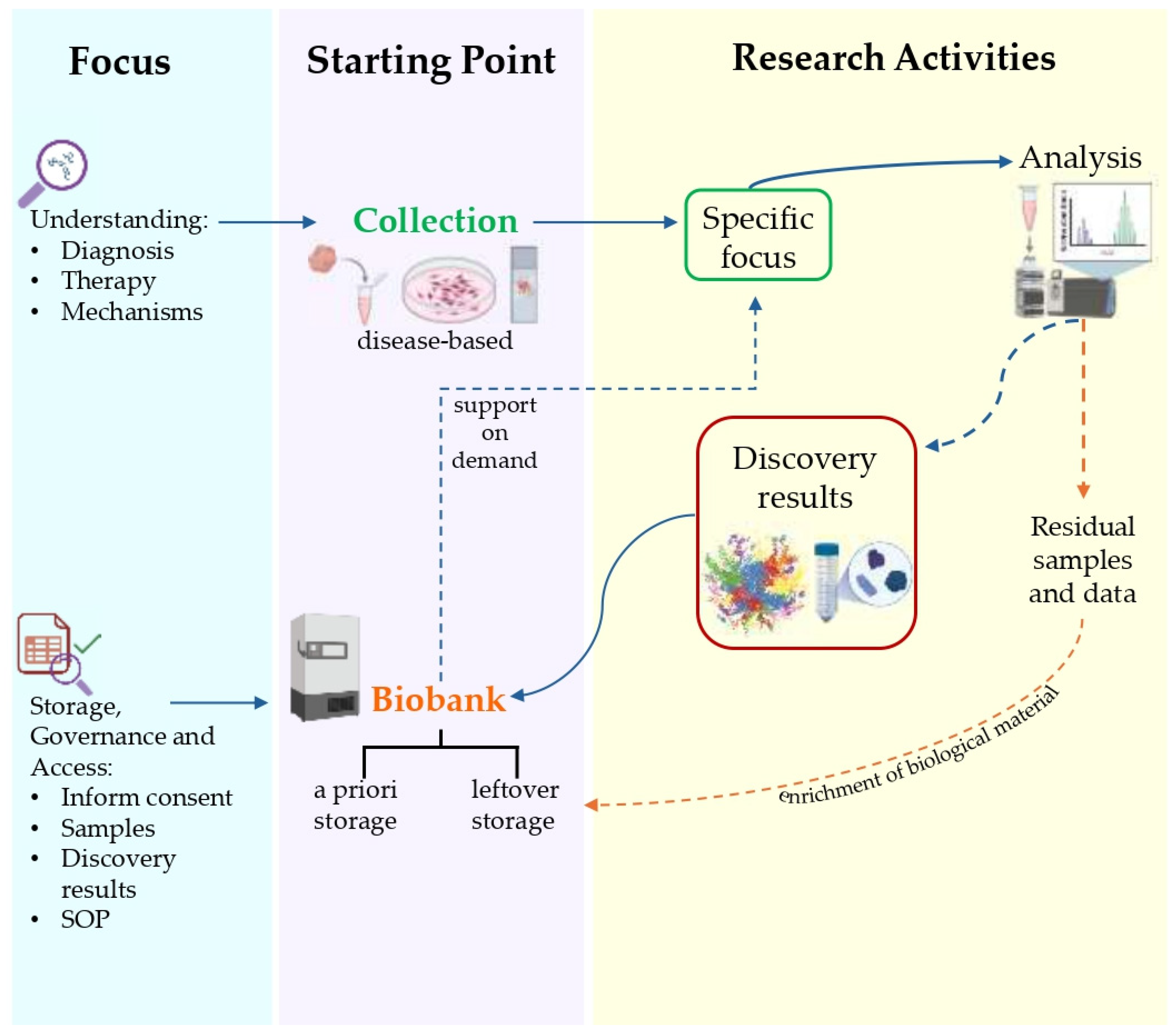
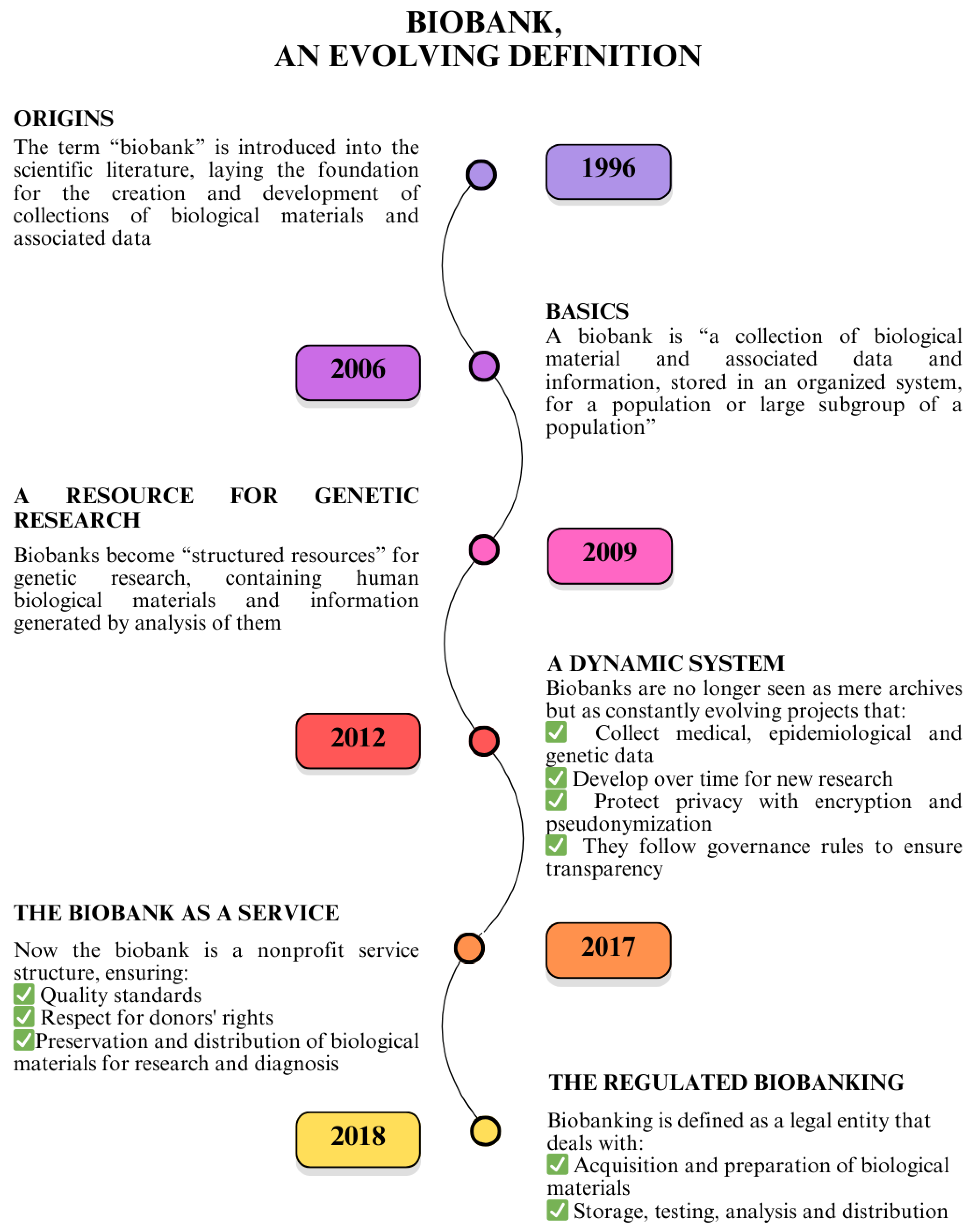
4. Proteomics and Biobanking as Cornerstones of Translational Medicine
- “Study-oriented human biological material Collections”—Collection, storage, and use of human biological materials and related data finalized to a specific project, generally oriented by pathology according to research or clinical protocols and participant’s previous expressed consent. In the protocol as in the specific consent, the start and end of the collection and use are declared, at the end of which the samples must be destroyed or biobanked, based on the further consent to biobanking expressed [163].
- “Biorepository”—A facility that collects, catalogs, and stores samples of biological material, such as urine, blood, tissue, cells, DNA, RNA, and protein, from humans, animals, or plants for laboratory research (https://www.cancer.gov/publications/dictionaries/cancer-terms/def/biorepository, accessed on 3 April 2025). In for-profit contexts, the biorepository is intended for exclusive, corporate use.
- “Research Biobank”—A legal entity, or part of a legal entity, formally established at a public or private institution; a non-profit. The research biobank as a service structure, at the service of the scientific communities, is the guarantor of the principles, rights, and processes that constitute biobanking for future research purposes. In full compliance with the informed consent/assent to research biobanking expressed and the rights of the participants involved, the biobank guarantees and manages, according to proven quality standards, the stable and continuous collection, conservation, use, and access of human biological materials, and/or related and derived data, for research. The sharing of biobanked samples/data, as well as results, is the cornerstone of all the activity of a research biobank (Figure 5) [164,165,166,167].
5. Perspectives for a Glioblastoma Translational Network
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GBM | Glioblastoma Multiforme |
| CNS | Central Nervous System |
| CSF | Cerebrospinal Fluid |
| TCGA | The Cancer Genome Atlas |
| TMZ | Temozolomide |
| BBB | Blood–Brain Barrier |
| GSC | Glioblastoma Stem Cells |
| NCI | National Cancer Institute |
| ISS | Istituto Superiore di Sanità |
| LC-MS/MS | Liquid Chromatography–Tandem Mass Spectrometry |
| DIA | Data-Independent Acquisition |
| ECM | Extracellular Matrix |
| SWATH-MS | Sequential Window Acquisition of All Theoretical Mass Spectra |
| TMT | Tandem Mass Tag |
| CUSA | Cavitron Ultrasonic Surgical Aspirator |
| LFQ | Label-Free Quantification |
| DDA | Data-Dependent Acquisition |
| DEPs | Differentially Expressed Proteins |
| iTRAQ | Isobaric Tags for Relative and Absolute Quantitation |
| EVs | Extracellular Vesicles |
| GASCs | Glioma-Associated Stromal Cells |
| uEV | Urinary Extracellular Vesicles |
| OECD | Organisation for Economic Co-operation and Development |
| BBMRI | Biobanking and Biomolecular Resources Research Infrastructure |
| ERIC | European Research Infrastructure Consortium |
| CIOMS | Council for International Organizations of Medical Sciences |
| ISO | International Organization for Standardization |
| QMS | Quality Management System |
| FAIR | Findability, Accessibility, Interoperability, and Reusability |
| ESFRI | European Strategy Forum on Research Infrastructures |
| SOPs | Standard Operating Procedures |
| TPN | Translational Proteomics Network |
| MTA | Material Transfer Agreement |
| DTA | Data Transfer Agreement |
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2016–2020. Neuro-Oncology 2023, 25, iv1–iv99. [Google Scholar] [CrossRef] [PubMed]
- Taylor, O.G.; Brzozowski, J.S.; Skelding, K.A. Glioblastoma Multiforme: An Overview of Emerging Therapeutic Targets. Front. Oncol. 2019, 9, 963. [Google Scholar] [CrossRef]
- Perez, A.; Huse, J.T. The Evolving Classification of Diffuse Gliomas: World Health Organization Updates for 2021. Curr. Neurol. Neurosci. Rep. 2021, 21, 67. [Google Scholar] [CrossRef]
- Yalamarty, S.S.K.; Filipczak, N.; Li, X.; Subhan, M.A.; Parveen, F.; Ataide, J.A.; Rajmalani, B.A.; Torchilin, V.P. Mechanisms of Resistance and Current Treatment Options for Glioblastoma Multiforme (GBM). Cancers 2023, 15, 2116. [Google Scholar] [CrossRef] [PubMed]
- Onishi, S.; Yamasaki, F.; Amatya, V.J.; Takayasu, T.; Yonezawa, U.; Taguchi, A.; Ohba, S.; Takeshima, Y.; Horie, N.; Sugiyama, K. Characteristics and Therapeutic Strategies of Radiation-Induced Glioma: Case Series and Comprehensive Literature Review. J. Neuro-Oncol. 2022, 159, 531–538. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and Other Malignant Gliomas. JAMA 2013, 310, 1842–1850. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive Genomic Characterization Defines Human Glioblastoma Genes and Core Pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.W.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The Somatic Genomic Landscape of Glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Weller, M. Where Does O6-methylguanine DNA Methyltransferase Promoter Methylation Assessment Place Temozolomide in the Future Standards of Care for Glioblastoma? Cancer 2018, 124, 1316–1318. [Google Scholar] [CrossRef]
- Yu, W.; Zhang, L.; Wei, Q.; Shao, A. O6-Methylguanine-DNA Methyltransferase (MGMT): Challenges and New Opportunities in Glioma Chemotherapy. Front. Oncol. 2020, 9, 1547. [Google Scholar] [CrossRef]
- Lu, X.; Maturi, N.P.; Jarvius, M.; Yildirim, I.; Dang, Y.; Zhao, L.; Xie, Y.; Tan, E.-J.; Xing, P.; Larsson, R.; et al. Cell-Lineage Controlled Epigenetic Regulation in Glioblastoma Stem Cells Determines Functionally Distinct Subgroups and Predicts Patient Survival. Nat. Commun. 2022, 13, 2236. [Google Scholar] [CrossRef] [PubMed]
- Kleihues, P.; Ohgaki, H. Primary and Secondary Glioblastomas: From Concept to Clinical Diagnosis. Neuro-Oncology 1999, 1, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Greco, F.; Anastasi, F.; Pardini, L.F.; Dilillo, M.; Vannini, E.; Baroncelli, L.; Caleo, M.; McDonnell, L.A. Longitudinal Bottom-Up Proteomics of Serum, Serum Extracellular Vesicles, and Cerebrospinal Fluid Reveals Candidate Biomarkers for Early Detection of Glioblastoma in a Murine Model. Molecules 2021, 26, 5992. [Google Scholar] [CrossRef]
- Polisetty, R.V.; Gautam, P.; Sharma, R.; Harsha, H.C.; Nair, S.C.; Gupta, M.K.; Uppin, M.S.; Challa, S.; Puligopu, A.K.; Ankathi, P.; et al. LC-MS/MS Analysis of Differentially Expressed Glioblastoma Membrane Proteome Reveals Altered Calcium Signaling and Other Protein Groups of Regulatory Functions. Mol. Cell. Proteom. 2012, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Frattini, V.; Trifonov, V.; Chan, J.M.; Castano, A.; Lia, M.; Abate, F.; Keir, S.T.; Ji, A.X.; Zoppoli, P.; Niola, F.; et al. The Integrated Landscape of Driver Genomic Alterations in Glioblastoma. Nat. Genet. 2013, 45, 1141–1149. [Google Scholar] [CrossRef]
- Le Rhun, E.; Preusser, M.; Roth, P.; Reardon, D.A.; van den Bent, M.; Wen, P.; Reifenberger, G.; Weller, M. Molecular Targeted Therapy of Glioblastoma. Cancer Treat. Rev. 2019, 80, 101896. [Google Scholar] [CrossRef]
- Kalinina, J.; Peng, J.; Ritchie, J.C.; Van Meir, E.G. Proteomics of Gliomas: Initial Biomarker Discovery and Evolution of Technology. Neuro-Oncology 2011, 13, 926–942. [Google Scholar] [CrossRef]
- Watanabe, K.; Tachibana, O.; Sato, K.; Yonekawa, Y.; Kleihues, P.; Ohgaki, H. Overexpression of the EGF Receptor and P53 Mutations Are Mutually Exclusive in the Evolution of Primary and Secondary Glioblastomas. Brain Pathol. 1996, 6, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Ohgaki, H.; Dessen, P.; Jourde, B.; Horstmann, S.; Nishikawa, T.; Di Patre, P.L.; Burkhard, C.; Schüler, D.; Probst-Hensch, N.M.; Maiorka, P.C.; et al. Genetic Pathways to Glioblastoma: A Population-Based Study. Cancer Res. 2004, 64, 6892–6899. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. Genetic Pathways to Primary and Secondary Glioblastoma. Am. J. Pathol. 2007, 170, 1445–1453. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, H.; Reis, R.M.; Nakamura, M.; Colella, S.; Yonekawa, Y.; Kleihues, P.; Ohgaki, H. Loss of Heterozygosity on Chromosome 10 Is More Extensive in Primary (De Novo) Than in Secondary Glioblastomas. Lab. Investig. 2000, 80, 65–72. [Google Scholar] [CrossRef]
- Nakamura, M.; Yang, F.; Fujisawa, H.; Yonekawa, Y.; Kleihues, P.; Ohgaki, H. Loss of Heterozygosity on Chromosome 19 in Secondary Glioblastomas. J. Neuropathol. Exp. Neurol. 2000, 59, 539–543. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science (1979) 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Nobusawa, S.; Watanabe, T.; Kleihues, P.; Ohgaki, H. IDH1 Mutations as Molecular Signature and Predictive Factor of Secondary Glioblastomas. Clin. Cancer Res. 2009, 15, 6002–6007. [Google Scholar] [CrossRef]
- Dunn, G.P.; Cloughesy, T.F.; Maus, M.V.; Prins, R.M.; Reardon, D.A.; Sonabend, A.M. Emerging Immunotherapies for Malignant Glioma: From Immunogenomics to Cell Therapy. Neuro-Oncology 2020, 22, 1425–1438. [Google Scholar] [CrossRef]
- Fan, Q.-W.; Weiss, W.A. Targeting the RTK-PI3K-MTOR Axis in Malignant Glioma: Overcoming Resistance. Curr. Top. Microbiol. Immunol. 2010, 347, 279–296. [Google Scholar] [CrossRef]
- Koul, D. PTEN Signaling Pathways in Glioblastoma. Cancer Biol. Ther. 2008, 7, 1321–1325. [Google Scholar] [CrossRef]
- Kahlert, U.D.; Suwala, A.K.; Koch, K.; Natsumeda, M.; Orr, B.A.; Hayashi, M.; Maciaczyk, J.; Eberhart, C.G. Pharmacologic Wnt Inhibition Reduces Proliferation, Survival, and Clonogenicity of Glioblastoma Cells. J. Neuropathol. Exp. Neurol. 2015, 74, 889–900. [Google Scholar] [CrossRef]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.L.; Rich, J.N. Cancer Stem Cells in Glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhou, J.; Zou, D.; Hou, D.; Zhang, H.; Zhao, J.; Li, L.; Hu, J.; Zhang, Y.; Jing, Z. Overexpression of Limb-Bud and Heart (LBH) Promotes Angiogenesis in Human Glioma via VEGFA-Mediated ERK Signalling under Hypoxia. EBioMedicine 2019, 48, 36–48. [Google Scholar] [CrossRef]
- Formato, A.; Salbini, M.; Orecchini, E.; Pellegrini, M.; Buccarelli, M.; Vitiani, L.R.; Giannetti, S.; Pallini, R.; D’Alessandris, Q.G.; Lauretti, L.; et al. N-Acetyl-L-Cysteine (NAC) Blunts Axitinib-Related Adverse Effects in Preclinical Models of Glioblastoma. Cancer Med. 2024, 13, e70279. [Google Scholar] [CrossRef]
- Bhat, K.P.L.; Balasubramaniyan, V.; Vaillant, B.; Ezhilarasan, R.; Hummelink, K.; Hollingsworth, F.; Wani, K.; Heathcock, L.; James, J.D.; Goodman, L.D.; et al. Mesenchymal Differentiation Mediated by NF-ΚB Promotes Radiation Resistance in Glioblastoma. Cancer Cell 2013, 24, 331–346. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Zemp, F.J.; Senger, D.; Robbins, S.M.; Yong, V.W. ADAM-9 Is a Novel Mediator of Tenascin-C-Stimulated Invasiveness of Brain Tumor–Initiating Cells. Neuro-Oncology 2015, 17, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Boltman, T.; Meyer, M.; Ekpo, O. Diagnostic and Therapeutic Approaches for Glioblastoma and Neuroblastoma Cancers Using Chlorotoxin Nanoparticles. Cancers 2023, 15, 3388. [Google Scholar] [CrossRef]
- Sweeney, M.D.; Zhao, Z.; Montagne, A.; Nelson, A.R.; Zlokovic, B.V. Blood-Brain Barrier: From Physiology to Disease and Back. Physiol. Rev. 2019, 99, 21–78. [Google Scholar] [CrossRef]
- Shergalis, A.; Bankhead, A.; Luesakul, U.; Muangsin, N.; Neamati, N. Current Challenges and Opportunities in Treating Glioblastomas. Pharmacol. Rev. 2018, 70, 412–445. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Mikolajewicz, N.; Khan, S.; Trifoi, M.; Skakdoub, A.; Ignatchenko, V.; Mansouri, S.; Zuccato, J.; Zacharia, B.E.; Glantz, M.; Zadeh, G.; et al. Leveraging the CSF Proteome toward Minimally-Invasive Diagnostics Surveillance of Brain Malignancies. Neuro-Oncol. Adv. 2022, 4, vdac161. [Google Scholar] [CrossRef]
- Mann, M.; Kumar, C.; Zeng, W.F.; Strauss, M.T. Artificial Intelligence for Proteomics and Biomarker Discovery. Cell Syst. 2021, 12, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Girardi, F.; Matz, M.; Stiller, C.; You, H.; Gragera, R.M.; Valkov, M.Y.; Bulliard, J.L.; De, P.; Morrison, D.; Wanner, M.; et al. Global Survival Trends for Brain Tumors, by Histology: Analysis of Individual Records for 556,237 Adults Diagnosed in 59 Countries during 2000–2014 (CONCORD-3). Neuro-Oncology 2023, 25, 580–592. [Google Scholar] [CrossRef]
- Mousavi, S.E.; Seyedmirzaei, H.; Shahrokhi Nejad, S.; Nejadghaderi, S.A. Epidemiology and Socioeconomic Correlates of Brain and Central Nervous System Cancers in Asia in 2020 and Their Projection to 2040. Sci. Rep. 2024, 14, 21936. [Google Scholar] [CrossRef]
- Price, M.; Ballard, C.; Benedetti, J.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S.; Ostrom, Q.T. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2017–2021. Neuro-Oncology 2024, 26, vi1–vi85. [Google Scholar] [CrossRef] [PubMed]
- PASS. I Numeri Del Cancro in Italia 2024. I Progressi Nelle Aziende Sanitarie per La Salute in Italia, 19 December 2024. [Google Scholar]
- Tamimi, A.F.; Juweid, M. Epidemiology and Outcome of Glioblastoma. In Glioblastoma; Exon Publications: Brisbane, Australia, 2017; pp. 143–153. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Xu, J.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009–2013. Neuro-Oncology 2016, 18, v1–v75. [Google Scholar] [CrossRef]
- Yang, W.; Warrington, N.M.; Taylor, S.J.; Whitmire, P.; Carrasco, E.; Singleton, K.W.; Wu, N.; Lathia, J.D.; Berens, M.E.; Kim, A.H.; et al. Sex Differences in GBM Revealed by Analysis of Patient Imaging, Transcriptome, and Survival Data. Sci. Transl. Med. 2019, 11, eaao5253. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Warrington, N.M.; Luo, J.; Brooks, M.D.; Dahiya, S.; Snyder, S.C.; Sengupta, R.; Rubin, J.B. Sexually Dimorphic RB Inactivation Underlies Mesenchymal Glioblastoma Prevalence in Males. J. Clin. Investig. 2014, 124, 4123–4133. [Google Scholar] [CrossRef]
- Kfoury, N.; Sun, T.; Yu, K.; Rockwell, N.; Tinkum, K.L.; Qi, Z.; Warrington, N.M.; McDonald, P.; Roy, A.; Weir, S.J.; et al. Cooperative P16 and P21 Action Protects Female Astrocytes from Transformation. Acta Neuropathol. Commun. 2018, 6, 12. [Google Scholar] [CrossRef]
- Broestl, L.; Rubin, J.B. Sexual Differentiation Specifies Cellular Responses to DNA Damage. Endocrinology 2021, 162, bqab192. [Google Scholar] [CrossRef]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee, S.U. Glioblastoma Multiforme: A Review of Its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Cordier, S.; Monfort, C.; Filippini, G.; Preston-Martin, S.; Lubin, F.; Mueller, B.A.; Holly, E.A.; Peris-Bonet, R.; McCredie, M.; Choi, W.; et al. Parental Exposure to Polycyclic Aromatic Hydrocarbons and the Risk of Childhood Brain Tumors: The SEARCH International Childhood Brain Tumor Study. Am. J. Epidemiol. 2004, 159, 1109–1116. [Google Scholar] [CrossRef]
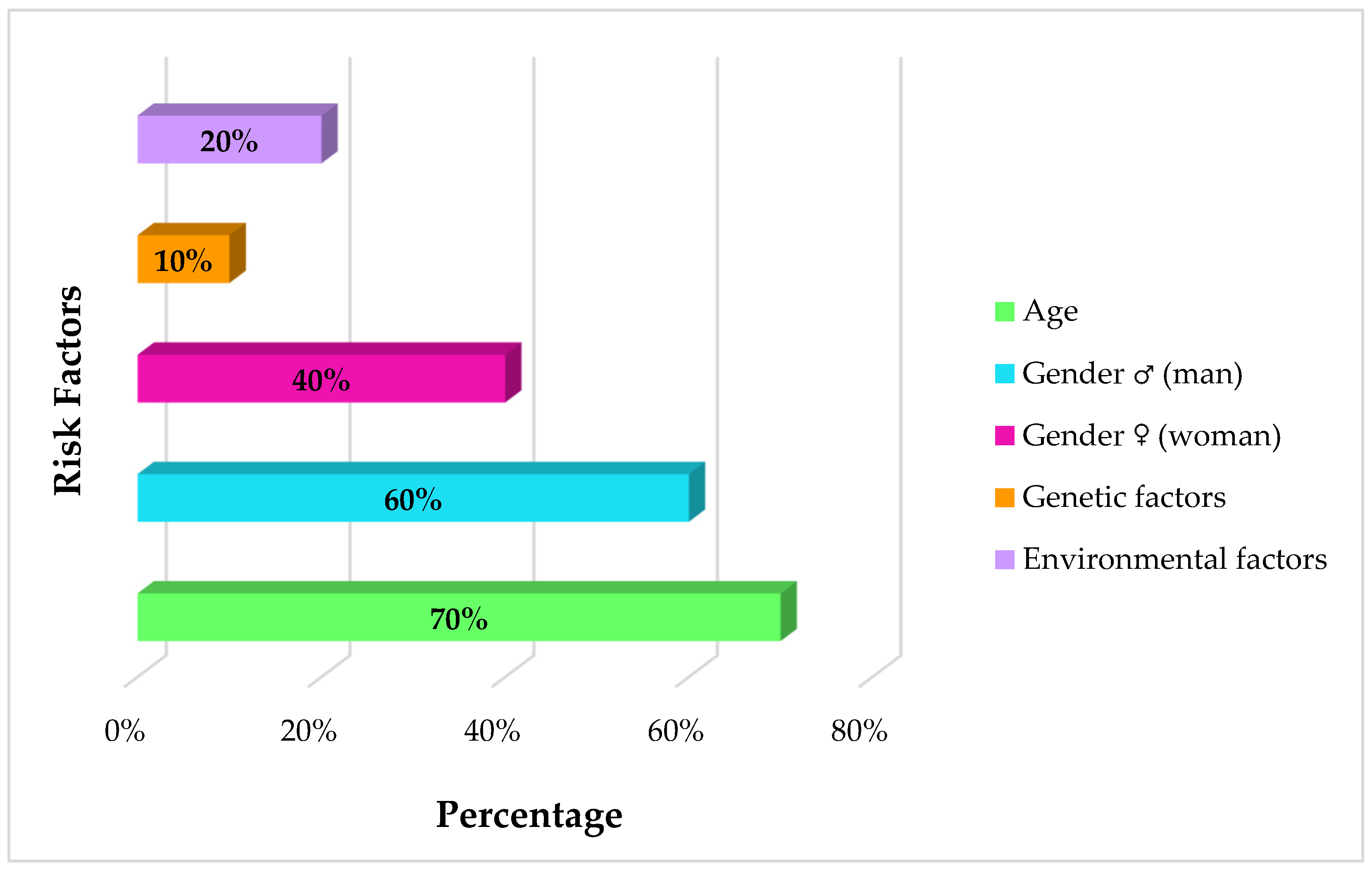
- Colopi, A.; Fuda, S.; Santi, S.; Onorato, A.; Cesarini, V.; Salvati, M.; Balistrieri, C.R.; Dolci, S.; Guida, E. Impact of Age and Gender on Glioblastoma Onset, Progression, and Management. Mech. Ageing Dev. 2023, 211, 111801. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.J.; Armstrong, G.; Lozzi, B.; Vijayaraghavan, P.; Plon, S.E.; Wong, T.C.; Boerwinkle, E.; Muzny, D.M.; Chen, H.C.; Gibbs, R.A.; et al. The Genomic Landscape of Familial Glioma. Sci. Adv. 2023, 9, eade2675. [Google Scholar] [CrossRef]
- Malmer, B.; Henriksson, R.; Grönberg, H. Familial Brain Tumours—Genetics or Environment? A Nationwide Cohort Study of Cancer Risk in Spouses and First-Degree Relatives of Brain Tumour Patients. Int. J. Cancer 2003, 106, 260–263. [Google Scholar] [CrossRef]
- Bailey, H.D.; Rios, P.; Lacour, B.; Guerrini-Rousseau, L.; Bertozzi, A.I.; Leblond, P.; Faure-Conter, C.; Pellier, I.; Freycon, C.; Michon, J.; et al. Factors Related to Pregnancy and Birth and the Risk of Childhood Brain Tumours: The ESTELLE and ESCALE Studies (SFCE, France). Int. J. Cancer 2017, 140, 1757–1769. [Google Scholar] [CrossRef] [PubMed]
- Fisher, P.G.; Reynolds, P.; Von Behren, J.; Carmichael, S.L.; Rasmussen, S.A.; Shaw, G.M. Cancer in Children with Nonchromosomal Birth Defects. J. Pediatr. 2012, 160, 978–983. [Google Scholar] [CrossRef]
- Cosenza-Contreras, M.; Schäfer, A.; Sing, J.; Cook, L.; Stillger, M.N.; Chen, C.Y.; Hidalgo, J.V.; Pinter, N.; Meyer, L.; Werner, T.; et al. Proteometabolomics of Initial and Recurrent Glioblastoma Highlights an Increased Immune Cell Signature with Altered Lipid Metabolism. Neuro-Oncology 2024, 26, 488–502. [Google Scholar] [CrossRef]
- Zhang, P.; Guo, Z.; Zhang, Y.; Gao, Z.; Ji, N.; Wang, D.; Zou, L.; Sun, W.; Zhang, L. A Preliminary Quantitative Proteomic Analysis of Glioblastoma Pseudoprogression. Proteome Sci. 2015, 13, 12. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Zhang, M.; Zhang, X.; Mao, W.; Gao, M. Proteogenomic Characterization of Ferroptosis Regulators Reveals Therapeutic Potential in Glioblastoma. BMC Cancer 2023, 23, 415. [Google Scholar] [CrossRef]
- Oh, S.; Yeom, J.; Cho, H.J.; Kim, J.H.; Yoon, S.J.; Kim, H.; Sa, J.K.; Ju, S.; Lee, H.; Oh, M.J.; et al. Integrated Pharmaco-Proteogenomics Defines Two Subgroups in Isocitrate Dehydrogenase Wild-Type Glioblastoma with Prognostic and Therapeutic Opportunities. Nat. Commun. 2020, 11, 3288. [Google Scholar] [CrossRef] [PubMed]
- Simeone, P.; Trerotola, M.; Urbanella, A.; Lattanzio, R.; Ciavardelli, D.; Di Giuseppe, F.; Eleuterio, E.; Sulpizio, M.; Eusebi, V.; Pession, A.; et al. A Unique Four-Hub Protein Cluster Associates to Glioblastoma Progression. PLoS ONE 2014, 9, e103030. [Google Scholar] [CrossRef]
- Zheng, W.; Chen, Q.; Liu, H.; Zeng, L.; Zhou, Y.; Liu, X.; Bai, Y.; Zhang, J.; Pan, Y.; Shao, C. SDC1-Dependent TGM2 Determines Radiosensitivity in Glioblastoma by Coordinating EPG5-Mediated Fusion of Autophagosomes with Lysosomes. Autophagy 2023, 19, 839–857. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Lin, S.; Wang, Z.; Shang, A. Differential Proteomics Analysis of Low- and High-Grade of Astrocytoma Using ITRAQ Quantification. Onco Targets Ther. 2016, 9, 5883–5895. [Google Scholar] [CrossRef]
- Jovčevska, I.; Zupanec, N.; Kočevar, N.; Cesselli, D.; Podergajs, N.; Stokin, C.L.; Myers, M.P.; Muyldermans, S.; Ghassabeh, G.H.; Motaln, H.; et al. TRIM28 and β-Actin Identified via Nanobody-Based Reverse Proteomics Approach as Possible Human Glioblastoma Biomarkers. PLoS ONE 2014, 9, e113688. [Google Scholar] [CrossRef]
- Heroux, M.S.; Chesnik, M.A.; Halligan, B.D.; Al-Gizawiy, M.; Connelly, J.M.; Mueller, W.M.; Rand, S.D.; Cochran, E.J.; LaViolette, P.S.; Malkin, M.G.; et al. Comprehensive Characterization of Glioblastoma Tumor Tissues for Biomarker Identification Using Mass Spectrometry-Based Label-Free Quantitative Proteomics. Physiol. Genom. 2014, 46, 467–481. [Google Scholar] [CrossRef]
- Jang, B.; Yoon, D.; Lee, J.Y.; Kim, J.; Hong, J.; Koo, H.; Sa, J.K. Integrative Multi-Omics Characterization Reveals Sex Differences in Glioblastoma. Biol. Sex Differ. 2024, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Sang, W.; Su, L.P.; Gao, H.X.; Cui, W.L.; Abulajiang, G.; Wang, Q.; Zhang, J.; Zhang, W. Proteomics Reveals Protein Phosphatase 1γ as a Biomarker Associated with Hippo Signal Pathway in Glioma. Pathol. Res. Pract. 2020, 216, 153187. [Google Scholar] [CrossRef]
- Jeon, H.; Byun, J.; Kang, H.; Kim, K.; Lee, E.; Kim, J.H.; Hong, C.K.; Song, S.W.; Kim, Y.H.; Chong, S.; et al. Proteomic Analysis Predicts Anti-Angiogenic Resistance in Recurred Glioblastoma. J. Transl. Med. 2023, 21, 69. [Google Scholar] [CrossRef]
- Rapp, C.; Warta, R.; Stamova, S.; Nowrouzi, A.; Geisenberger, C.; Gal, Z.; Roesch, S.; Dettling, S.; Juenger, S.; Bucur, M.; et al. Identification of T Cell Target Antigens in Glioblastoma Stem-like Cells Using an Integrated Proteomics-Based Approach in Patient Specimens. Acta Neuropathol. 2017, 134, 297–316. [Google Scholar] [CrossRef]
- Sethi, M.K.; Downs, M.; Shao, C.; Hackett, W.E.; Phillips, J.J.; Zaia, J. In-Depth Matrisome and Glycoproteomic Analysis of Human Brain Glioblastoma Versus Control Tissue. Mol. Cell. Proteom. 2022, 21, 100216. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-C.; Lu, G.-X.; Zhang, H.-W.; Zhong, X.-M.; Cong, X.-L.; Xue, S.-B.; Kong, R.; Li, D.; Chang, Z.-Y.; Wang, X.-F.; et al. Proteogenomic Characterization and Integrative Analysis of Glioblastoma Multiforme. Oncotarget 2017, 8, 97304–97312. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yan, S.; Chen, X.; Wang, A.; Han, Z.; Liu, B.; Shen, H. Identification of Prognostic Biomarkers for Glioblastoma Based on Transcriptome and Proteome Association Analysis. Technol. Cancer Res. Treat. 2022, 21. [Google Scholar] [CrossRef] [PubMed]
- Gularyan, S.K.; Gulin, A.A.; Anufrieva, K.S.; Shender, V.O.; Shakhparonov, M.I.; Bastola, S.; Antipova, N.V.; Kovalenko, T.F.; Rubtsov, Y.P.; Latyshev, Y.A.; et al. Investigation of Inter- And Intratumoral Heterogeneity of Glioblastoma Using TOF-SIMS. Mol. Cell. Proteom. 2020, 19, 960–970. [Google Scholar] [CrossRef]
- Ait-Belkacem, R.; Berenguer, C.; Villard, C.; Ouafik, L.; Figarella-Branger, D.; Chinot, O.; Lafitte, D. MALDI Imaging and In-Source Decay for Top-down Characterization of Glioblastoma. Proteomics 2014, 14, 1290–1301. [Google Scholar] [CrossRef]
- Zhao, R.; Pan, Z.; Li, B.; Zhao, S.; Zhang, S.; Qi, Y.; Qiu, J.; Gao, Z.; Fan, Y.; Guo, Q.; et al. Comprehensive Analysis of the Tumor Immune Microenvironment Landscape in Glioblastoma Reveals Tumor Heterogeneity and Implications for Prognosis and Immunotherapy. Front. Immunol. 2022, 13, 820673. [Google Scholar] [CrossRef]
- Bi, B.; Li, F.; Guo, J.; Li, C.; Jing, R.; Lv, X.; Chen, X.; Wang, F.; Azadzoi, K.M.; Wang, L.; et al. Label-Free Quantitative Proteomics Unravels the Importance of RNA Processing in Glioma Malignancy. Neuroscience 2017, 351, 84–95. [Google Scholar] [CrossRef]
- Doan, N.B.; Alhajala, H.; Al-Gizawiy, M.M.; Mueller, W.M.; Rand, S.D.; Connelly, J.M.; Cochran, E.J.; Chitambar, C.R.; Clark, P.; Kuo, J.; et al. Acid Ceramidase and Its Inhibitors: A de Novo Drug Target and a New Class of Drugs for Killing Glioblastoma Cancer Stem Cells with High Efficiency. Oncotarget 2017, 8, 112662–112674. [Google Scholar] [CrossRef]
- Djuric, U.; Lam, K.H.B.; Kao, J.; Batruch, I.; Jevtic, S.; Papaioannou, M.D.; Diamandis, P. Defining Protein Pattern Differences among Molecular Subtypes of Diffuse Gliomas Using Mass Spectrometry. Mol. Cell. Proteom. 2019, 18, 2029–2043. [Google Scholar] [CrossRef]
- Maire, C.L.; Fuh, M.M.; Kaulich, K.; Fita, K.D.; Stevic, I.; Heiland, D.H.; Welsh, J.A.; Jones, J.C.; Görgens, A.; Ricklefs, T.; et al. Genome-Wide Methylation Profiling of Glioblastoma Cell-Derived Extracellular Vesicle DNA Allows Tumor Classification. Neuro-Oncology 2021, 23, 1087–1099. [Google Scholar] [CrossRef]
- El-Baba, C.; Ayache, Z.; Goli, M.; Hayar, B.; Kawtharani, Z.; Pisano, C.; Kobeissy, F.; Mechref, Y.; Darwiche, N. The Antitumor Effect of the DNA Polymerase Alpha Inhibitor ST1926 in Glioblastoma: A Proteomics Approach. Int. J. Mol. Sci. 2023, 24, 14069. [Google Scholar] [CrossRef] [PubMed]
- Auzmendi-Iriarte, J.; Otaegi-Ugartemendia, M.; Carrasco-Garcia, E.; Azkargorta, M.; Diaz, A.; Saenz-Antoñanzas, A.; Andermatten, J.A.; Garcia-Puga, M.; Garcia, I.; Elua-Pinin, A.; et al. Chaperone-Mediated Autophagy Controls Proteomic and Transcriptomic Pathways to Maintain Glioma Stem Cell Activity. Cancer Res. 2022, 82, 1283–1297. [Google Scholar] [CrossRef]
- Naryzhny, S.; Volnitskiy, A.; Kopylov, A.; Zorina, E.; Kamyshinsky, R.; Bairamukov, V.; Garaeva, L.; Shlikht, A.; Shtam, T. Proteome of Glioblastoma-Derived Exosomes as a Source of Biomarkers. Biomedicines 2020, 8, 216. [Google Scholar] [CrossRef]
- Hu, Y.; Ye, S.; Li, Q.; Yin, T.; Wu, J.; He, J. Quantitative Proteomics Analysis Indicates That Upregulation of LncRNA HULC Promotes Pathogenesis of Glioblastoma Cells. Onco Targets Ther. 2020, 13, 5927–5938. [Google Scholar] [CrossRef]
- Menezes, A.; Julião, G.; Mariath, F.; Ferreira, A.L.; Oliveira-Nunes, M.C.; Gallucci, L.; Evaristo, J.A.M.; Nogueira, F.C.S.; De Abreu Pereira, D.; Carneiro, K. Epigenetic Mechanisms Histone Deacetylase-Dependent Regulate the Glioblastoma Angiogenic Matrisome and Disrupt Endothelial Cell Behavior In Vitro. Mol. Cell. Proteom. 2024, 23, 100722. [Google Scholar] [CrossRef]
- Ghosh, D.; Funk, C.C.; Caballero, J.; Shah, N.; Rouleau, K.; Earls, J.C.; Soroceanu, L.; Foltz, G.; Cobbs, C.S.; Price, N.D.; et al. A Cell-Surface Membrane Protein Signature for Glioblastoma. Cell Syst. 2017, 4, 516–529.e7. [Google Scholar] [CrossRef] [PubMed]
- Schulze, M.; Violonchi, C.; Swoboda, S.; Welz, T.; Kerkhoff, E.; Hoja, S.; Brüggemann, S.; Simbürger, J.; Reinders, J.; Riemenschneider, M.J. RELN Signaling Modulates Glioblastoma Growth and Substrate-Dependent Migration. Brain Pathol. 2018, 28, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Jovčevska, I.; Zupanec, N.; Urlep, Ž.; Vranič, A.; Matos, B.; Stokin, C.L.; Muyldermans, S.; Myers, M.P.; Buzdin, A.A.; Petrov, I.; et al. Differentially Expressed Proteins in Glioblastoma Multiforme Identified with a Nanobody-Based Anti-Proteome Approach and Confirmed by OncoFinder as Possible Tumor-Class Predictive Biomarker Candidates. Oncotarget 2017, 8, 44141–44158. [Google Scholar] [CrossRef]
- Yi, G.Z.; Xiang, W.; Feng, W.Y.; Chen, Z.Y.; Li, Y.M.; Deng, S.Z.; Guo, M.L.; Zhao, L.; Sun, X.G.; He, M.Y.; et al. Identification of Key Candidate Proteins and Pathways Associated with Temozolomide Resistance in Glioblastoma Based on Subcellular Proteomics and Bioinformatical Analysis. Biomed. Res. Int. 2018, 2018, 5238760. [Google Scholar] [CrossRef]
- Kang, N.; Oh, H.J.; Hong, J.H.; Moon, H.E.; Kim, Y.; Lee, H.J.; Min, H.; Park, H.; Lee, S.H.; Peak, S.H.; et al. Glial Cell Proteome Using Targeted Quantitative Methods for Potential Multi-Diagnostic Biomarkers. Clin. Proteom. 2023, 20, 45. [Google Scholar] [CrossRef]
- Haas, T.L.; Sciuto, M.R.; Brunetto, L.; Valvo, C.; Signore, M.; Fiori, M.E.; di Martino, S.; Giannetti, S.; Morgante, L.; Boe, A.; et al. Integrin A7 Is a Functional Marker and Potential Therapeutic Target in Glioblastoma. Cell Stem Cell 2017, 21, 35–50.e9. [Google Scholar] [CrossRef] [PubMed]
- Gyuris, A.; Navarrete-Perea, J.; Jo, A.; Cristea, S.; Zhou, S.; Fraser, K.; Wei, Z.; Krichevsky, A.M.; Weissleder, R.; Lee, H.; et al. Physical and Molecular Landscapes of Mouse Glioma Extracellular Vesicles Define Heterogeneity. Cell Rep. 2019, 27, 3972–3987.e6. [Google Scholar] [CrossRef] [PubMed]
- Ahmadov, U.; Picard, D.; Bartl, J.; Silginer, M.; Trajkovic-Arsic, M.; Qin, N.; Blümel, L.; Wolter, M.; Lim, J.K.M.; Pauck, D.; et al. The Long Non-Coding RNA HOTAIRM1 Promotes Tumor Aggressiveness and Radiotherapy Resistance in Glioblastoma. Cell Death Dis. 2021, 12, 885. [Google Scholar] [CrossRef]
- Mallawaaratchy, D.M.; Buckland, M.E.; McDonald, K.L.; Li, C.C.Y.; Ly, L.; Sykes, E.K.; Christopherson, R.I.; Kaufman, K.L. Membrane Proteome Analysis of Glioblastoma Cell Invasion. J. Neuropathol. Exp. Neurol. 2015, 74, 425–441. [Google Scholar] [CrossRef]
- Mallawaaratchy, D.M.; Hallal, S.; Russell, B.; Ly, L.; Ebrahimkhani, S.; Wei, H.; Christopherson, R.I.; Buckland, M.E.; Kaufman, K.L. Comprehensive Proteome Profiling of Glioblastoma-Derived Extracellular Vesicles Identifies Markers for More Aggressive Disease. J. Neuro-Oncol. 2017, 131, 233–244. [Google Scholar] [CrossRef]
- Tarasova, I.A.; Tereshkova, A.V.; Lobas, A.A.; Solovyeva, E.M.; Sidorenko, A.S.; Gorshkov, V.; Kjeldsen, F.; Bubis, J.A.; Ivanov, M.V.; Ilina, I.Y.; et al. Comparative Proteomics as a Tool for Identifying Specific Alterations within Interferon Response Pathways in Human Glioblastoma Multiforme Cells. Oncotarget 2017, 9, 1785–1802. [Google Scholar] [CrossRef]
- Guffens, L.; Derua, R.; Janssens, V. PME-1 Sensitizes Glioblastoma Cells to Oxidative Stress-Induced Cell Death by Attenuating PP2A-B55α-Mediated Inactivation of MAPKAPK2-RIPK1 Signaling. Cell Death Discov. 2023, 9, 265. [Google Scholar] [CrossRef]
- Hvinden, I.C.; Berg, H.E.; Sachse, D.; Skaga, E.; Skottvoll, F.S.; Lundanes, E.; Sandberg, C.J.; Vik-Mo, E.O.; Rise, F.; Wilson, S.R. Nuclear Magnetic Resonance Spectroscopy to Identify Metabolite Biomarkers of Nonresponsiveness to Targeted Therapy in Glioblastoma Tumor Stem Cells. J. Proteome Res. 2019, 18, 2012–2020. [Google Scholar] [CrossRef] [PubMed]
- Bijnsdorp, I.V.; Schelfhorst, T.; Luinenburg, M.; Rolfs, F.; Piersma, S.R.; de Haas, R.R.; Pham, T.V.; Jimenez, C.R. Feasibility of Phosphoproteomics to Uncover Oncogenic Signalling in Secreted Extracellular Vesicles Using Glioblastoma-EGFRVIII Cells as a Model. J. Proteom. 2021, 232, 104076. [Google Scholar] [CrossRef]
- González-Morales, A.; Zabaleta, A.; Guruceaga, E.; Alonso, M.M.; García-Moure, M.; Fernández-Irigoyen, J.; Santamaría, E. Spatial and Temporal Proteome Dynamics of Glioma Cells during Oncolytic Adenovirus Delta-24-RGD Infection. Oncotarget 2018, 9, 31045–31065. [Google Scholar] [CrossRef]
- Nagashima, S.; Maruyama, J.; Honda, K.; Kondoh, Y.; Osada, H.; Nawa, M.; Nakahama, K.I.; Ishigami-Yuasa, M.; Kagechika, H.; Sugimura, H.; et al. CSE1L Promotes Nuclear Accumulation of Transcriptional Coactivator TAZ and Enhances Invasiveness of Human Cancer Cells. J. Biol. Chem. 2021, 297, 100803. [Google Scholar] [CrossRef] [PubMed]
- Kohata, T.; Ito, S.; Masuda, T.; Furuta, T.; Nakada, M.; Ohtsuki, S. Laminin Subunit Alpha-4 and Osteopontin Are Glioblastoma-Selective Secreted Proteins That Are Increased in the Cerebrospinal Fluid of Glioblastoma Patients. J. Proteome Res. 2020, 19, 3542–3553. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Montermini, L.; Kim, D.K.; Meehan, B.; Roth, F.P.; Rak, J. The Impact of Oncogenic Egfrviii on the Proteome of Extracellular Vesicles Released from Glioblastoma Cells. Mol. Cell. Proteom. 2018, 17, 1948–1964. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ren, T.; Lin, M.; Wang, Z.; Zhang, J. Integrated Proteomic and Metabolomic Profiling the Global Response of Rat Glioma Model by Temozolomide Treatment. J. Proteom. 2020, 211, 103578. [Google Scholar] [CrossRef]
- Sangar, V.; Funk, C.C.; Kusebauch, U.; Campbell, D.S.; Moritz, R.L.; Price, N.D. Quantitative Proteomic Analysis Reveals Effects of Epidermal Growth Factor Receptor (EGFR) on Invasion-Promoting Proteins Secreted by Glioblastoma Cells. Mol. Cell. Proteom. 2014, 13, 2618–2631. [Google Scholar] [CrossRef]
- Spinelli, C.; Montermini, L.; Meehan, B.; Brisson, A.R.; Tan, S.; Choi, D.; Nakano, I.; Rak, J. Molecular Subtypes and Differentiation Programmes of Glioma Stem Cells as Determinants of Extracellular Vesicle Profiles and Endothelial Cell-Stimulating Activities. J. Extracell. Vesicles 2018, 7, 1490144. [Google Scholar] [CrossRef]
- Autelitano, F.; Loyaux, D.; Roudières, S.; Déon, C.; Guette, F.; Fabre, P.; Ping, Q.; Wang, S.; Auvergne, R.; Badarinarayana, V.; et al. Identification of Novel Tumor-Associated Cell Surface Sialoglycoproteins in Human Glioblastoma Tumors Using Quantitative Proteomics. PLoS ONE 2014, 9, e110316. [Google Scholar] [CrossRef] [PubMed]
- Turtoi, A.; Blomme, A.; Bianchi, E.; Maris, P.; Vannozzi, R.; Naccarato, A.G.; Delvenne, P.; De Pauw, E.; Bevilacqua, G.; Castronovo, V. Accessibilome of Human Glioblastoma: Collagen-VI-Alpha-1 Is a New Target and a Marker of Poor Outcome. J. Proteome Res. 2014, 13, 5660–5669. [Google Scholar] [CrossRef]
- Clavreul, A.; Guette, C.; Faguer, R.; Tétaud, C.; Boissard, A.; Lemaire, L.; Rousseau, A.; Avril, T.; Henry, C.; Coqueret, O.; et al. Glioblastoma-Associated Stromal Cells (GASCs) from Histologically Normal Surgical Margins Have a Myofibroblast Phenotype and Angiogenic Properties. J. Pathol. 2014, 233, 74–88. [Google Scholar] [CrossRef]
- Yu, X.; Feng, L.; Liu, D.; Zhang, L.; Wu, B.; Jiang, W.; Han, Z.; Cheng, S. Quantitative Proteomics Reveals the Novel Co-Expression Signatures in Early Brain Development for Prognosis of Glioblastoma Multiforme. Oncotarget 2016, 7, 14161–14171. [Google Scholar] [CrossRef]
- Buehler, M.; Yi, X.; Ge, W.; Blattmann, P.; Rushing, E.; Reifenberger, G.; Felsberg, J.; Yeh, C.; Corn, J.E.; Regli, L.; et al. Quantitative Proteomic Landscapes of Primary and Recurrent Glioblastoma Reveal a Protumorigeneic Role for FBXO2-Dependent Glioma-Microenvironment Interactions. Neuro-Oncology 2023, 25, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, A.; Brambilla, F.; Arbustini, E.; Basello, K.; Speciani, A.; Mauri, P.; Bezzi, P.; Magrassi, L. A New Pathway Promotes Adaptation of Human Glioblastoma Cells to Glucose Starvation. Cells 2020, 9, 1249. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Migliozzi, S.; Koo, H.; Hong, J.H.; Park, S.M.; Kim, S.; Kwon, H.J.; Ha, S.; Garofano, L.; Oh, Y.T.; et al. Integrated Proteogenomic Characterization of Glioblastoma Evolution. Cancer Cell 2024, 42, 358–377.e8. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, A.S.; Lipatova, A.V.; Goncharov, A.O.; Kliuchnikova, A.A.; Pyatnitskiy, M.A.; Kuznetsova, K.G.; Hamad, A.; Vorobyev, P.O.; Alekseeva, O.N.; Mahmoud, M.; et al. Multiomic Profiling Identified EGF Receptor Signaling as a Potential Inhibitor of Type I Interferon Response in Models of Oncolytic Therapy by Vesicular Stomatitis Virus. Int. J. Mol. Sci. 2022, 23, 5244. [Google Scholar] [CrossRef]
- Hallal, S.; Khani, S.E.; Wei, H.; Lee, M.Y.T.; Sim, H.W.; Sy, J.; Shivalingam, B.; Buckland, M.E.; Alexander-Kaufman, K.L. Deep Sequencing of Small RNAs from Neurosurgical Extracellular Vesicles Substantiates MiR-486-3p as a Circulating Biomarker That Distinguishes Glioblastoma from Lower-Grade Astrocytoma Patients. Int. J. Mol. Sci. 2020, 21, 4954. [Google Scholar] [CrossRef]
- Hallal, S.; Azimi, A.; Wei, H.; Ho, N.; Lee, M.Y.T.; Sim, H.W.; Sy, J.; Shivalingam, B.; Buckland, M.E.; Alexander-Kaufman, K.L. A Comprehensive Proteomic SWATH-MS Workflow for Profiling Blood Extracellular Vesicles: A New Avenue for Glioma Tumour Surveillance. Int. J. Mol. Sci. 2020, 21, 4754. [Google Scholar] [CrossRef]
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Skog, J.; Nakano, I.; Pingle, S.; Kalinina, J.; Hua, W.; Kesari, S.; Mao, Y.; et al. MiR-21 in the Extracellular Vesicles (EVs) of Cerebrospinal Fluid (CSF): A Platform for Glioblastoma Biomarker Development. PLoS ONE 2013, 8, e78115. [Google Scholar] [CrossRef]
- Ter-Ovanesyan, D.; Norman, M.; Lazarovits, R.; Trieu, W.; Lee, J.H.; Church, G.M.; Walt, D.R. Framework for Rapid Comparison of Extracellular Vesicle Isolation Methods. Elife 2021, 10, e70725. [Google Scholar] [CrossRef]
- Schmid, D.; Warnken, U.; Latzer, P.; Hoffmann, D.C.; Roth, J.; Kutschmann, S.; Jaschonek, H.; Rübmann, P.; Foltyn, M.; Vollmuth, P.; et al. Diagnostic Biomarkers from Proteomic Characterization of Cerebrospinal Fluid in Patients with Brain Malignancies. J. Neurochem. 2021, 158, 522–538. [Google Scholar] [CrossRef]
- Magrassi, L.; Brambilla, F.; Viganò, R.; Di Silvestre, D.; Benazzi, L.; Bellantoni, G.; Danesino, G.M.; Comincini, S.; Mauri, P. Proteomic Analysis on Sequential Samples of Cystic Fluid Obtained from Human Brain Tumors. Cancers 2023, 15, 4070. [Google Scholar] [CrossRef]
- Naryzhny, S.; Ronzhina, N.; Zorina, E.; Kabachenko, F.; Zavialova, M.; Zgoda, V.; Klopov, N.; Legina, O.; Pantina, R. Evaluation of Haptoglobin and Its Proteoforms as Glioblastoma Markers. Int. J. Mol. Sci. 2021, 22, 6533. [Google Scholar] [CrossRef]
- Sabbagh, Q.; André-Grégoire, G.; Alves-Nicolau, C.; Dupont, A.; Bidère, N.; Jouglar, E.; Guével, L.; Frénel, J.S.; Gavard, J. The von Willebrand Factor Stamps Plasmatic Extracellular Vesicles from Glioblastoma Patients. Sci. Rep. 2021, 11, 22792. [Google Scholar] [CrossRef] [PubMed]
- Popescu, I.D.; Codrici, E.; Albulescu, L.; Mihai, S.; Enciu, A.M.; Albulescu, R.; Tanase, C.P. Potential Serum Biomarkers for Glioblastoma Diagnostic Assessed by Proteomic Approaches. Proteome Sci. 2014, 12, 47. [Google Scholar] [CrossRef] [PubMed]
- Clavreul, A.; Guette, C.; Lasla, H.; Rousseau, A.; Blanchet, O.; Henry, C.; Boissard, A.; Cherel, M.; Jézéquel, P.; Guillonneau, F.; et al. Proteomics of Tumor and Serum Samples from Isocitrate Dehydrogenase-Wildtype Glioblastoma Patients: Is the Detoxification of Reactive Oxygen Species Associated with Shorter Survival? Mol. Oncol. 2024, 18, 2783–2800. [Google Scholar] [CrossRef]
- Kun, S.; Duan, Q.; Liu, G.; Lu, J.-M. Prognostic Value of DNA Repair Genes Based on Stratification of Glioblastomas. Oncotarget 2017, 8, 58222–58230. [Google Scholar] [CrossRef] [PubMed]
- Hallal, S.M.; Tűzesi, Á.; Sida, L.A.; Xian, E.; Madani, D.; Muralidharan, K.; Shivalingam, B.; Buckland, M.E.; Satgunaseelan, L.; Alexander, K.L. Glioblastoma Biomarkers in Urinary Extracellular Vesicles Reveal the Potential for a ‘Liquid Gold’ Biopsy. Br. J. Cancer 2024, 130, 836–851. [Google Scholar] [CrossRef]
- Bark, J.M.; Trevisan França de Lima, L.; Zhang, X.; Broszczak, D.; Leo, P.J.; Jeffree, R.L.; Chua, B.; Day, B.W.; Punyadeera, C. Proteome Profiling of Salivary Small Extracellular Vesicles in Glioblastoma Patients. Cancer 2023, 129, 2836–2847. [Google Scholar] [CrossRef] [PubMed]
- Sastry, R.A.; Shankar, G.M.; Gerstner, E.R.; Curry, W.T. The Impact of Surgery on Survival after Progression of Glioblastoma: A Retrospective Cohort Analysis of a Contemporary Patient Population. J. Clin. Neurosci. 2018, 53, 41–47. [Google Scholar] [CrossRef]
- Craig-Schapiro, R.; Perrin, R.J.; Roe, C.M.; Xiong, C.; Carter, D.; Cairns, N.J.; Mintun, M.A.; Peskind, E.R.; Li, G.; Galasko, D.R.; et al. YKL-40: A Novel Prognostic Fluid Biomarker for Preclinical Alzheimer’s Disease. Biol. Psychiatry 2010, 68, 903–912. [Google Scholar] [CrossRef]
- Kušnierová, P.; Zeman, D.; Hradílek, P.; Zapletalová, O.; Stejskal, D. Determination of Chitinase 3-like 1 in Cerebrospinal Fluid in Multiple Sclerosis and Other Neurological Diseases. PLoS ONE 2020, 15, e0233519. [Google Scholar] [CrossRef]
- Ku, B.M.; Lee, Y.K.; Ryu, J.; Jeong, J.Y.; Choi, J.; Eun, K.M.; Shin, H.Y.; Kim, D.G.; Hwang, E.M.; Yoo, J.C.; et al. CHI3L1 (YKL-40) Is Expressed in Human Gliomas and Regulates the Invasion, Growth and Survival of Glioma Cells. Int. J. Cancer 2011, 128, 1316–1326. [Google Scholar] [CrossRef] [PubMed]
- Tichy, J.; Spechtmeyer, S.; Mittelbronn, M.; Hattingen, E.; Rieger, J.; Senft, C.; Foerch, C. Prospective Evaluation of Serum Glial Fibrillary Acidic Protein (GFAP) as a Diagnostic Marker for Glioblastoma. J. Neuro-Oncol. 2015, 126, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Larraya, J.G.; Paris, S.; Idbaih, A.; Dehais, C.; Laigle-Donadey, F.; Navarro, S.; Capelle, L.; Mokhtari, K.; Marie, Y.; Sanson, M.; et al. Diagnostic and Prognostic Value of Preoperative Combined GFAP, IGFBP-2, and YKL-40 Plasma Levels in Patients with Glioblastoma. Cancer 2014, 120, 3972–3980. [Google Scholar] [CrossRef]
- Kiviniemi, A.; Gardberg, M.; Frantzén, J.; Parkkola, R.; Vuorinen, V.; Pesola, M.; Minn, H. Serum Levels of GFAP and EGFR in Primary and Recurrent High-Grade Gliomas: Correlation to Tumor Volume, Molecular Markers, and Progression-Free Survival. J. Neuro-Oncol. 2015, 124, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Piehowski, P.D.; Petyuk, V.A.; Orton, D.J.; Xie, F.; Moore, R.J.; Ramirez-Restrepo, M.; Engel, A.; Lieberman, A.P.; Albin, R.L.; Camp, D.G.; et al. Sources of Technical Variability in Quantitative LC-MS Proteomics: Human Brain Tissue Sample Analysis. J. Proteome Res. 2013, 12, 2128–2137. [Google Scholar] [CrossRef]
- Mehta, A.I.; Ross, S.; Lowenthal, M.S.; Fusaro, V.; Fishman, D.A.; Petricoin, E.F.; Liotta, L.A. Biomarker Amplification by Serum Carrier Protein Binding. Dis. Markers 2003, 19, 104879. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, Z.; Zou, L.; Yang, Y.; Zhang, L.; Ji, N.; Shao, C.; Sun, W.; Wang, Y. A Comprehensive Map and Functional Annotation of the Normal Human Cerebrospinal Fluid Proteome. J. Proteom. 2015, 119, 90–99. [Google Scholar] [CrossRef]
- Oh, M.K.; Park, H.J.; Lee, J.H.; Bae, H.M.; Kim, I.S. Single Chain Precursor Prohaptoglobin Promotes Angiogenesis by Upregulating Expression of Vascular Endothelial Growth Factor (VEGF) and VEGF Receptor2. FEBS Lett. 2015, 589, 1009–1017. [Google Scholar] [CrossRef]
- Skardelly, M.; Armbruster, F.P.; Meixensberger, J.; Hilbig, H. Expression of Zonulin, c-Kit, and Glial Fibrillary Acidic Protein in Human Gliomas. Transl. Oncol. 2009, 2, 117–120. [Google Scholar] [CrossRef][Green Version]
- Díaz-Coránguez, M.; Segovia, J.; López-Ornelas, A.; Puerta-Guardo, H.; Ludert, J.; Chávez, B.; Meraz-Cruz, N.; González-Mariscal, L. Transmigration of Neural Stem Cells across the Blood Brain Barrier Induced by Glioma Cells. PLoS ONE 2013, 8, e60655. [Google Scholar] [CrossRef]
- Sandset, P.M. CXCL4-Platelet Factor 4, Heparin-Induced Thrombocytopenia and Cancer. Thromb. Res. 2012, 129 (Suppl. 1), S97–S100. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Rothenberg, M.L.; Tabernero, J.; Seoane, J.; Daly, T.; Cleverly, A.; Berry, B.; Rhoades, S.K.; Ray, C.A.; Fill, J.; et al. TGF-β Signalling-Related Markers in Cancer Patients with Bone Metastasis. Biomarkers 2008, 13, 217–236. [Google Scholar] [CrossRef] [PubMed]
- Gautam, P.; Nair, S.C.; Gupta, M.K.; Sharma, R.; Polisetty, R.V.; Uppin, M.S.; Sundaram, C.; Puligopu, A.K.; Ankathi, P.; Purohit, A.K.; et al. Proteins with Altered Levels in Plasma from Glioblastoma Patients as Revealed by ITRAQ-Based Quantitative Proteomic Analysis. PLoS ONE 2012, 7, e46153. [Google Scholar] [CrossRef] [PubMed]
- Clavreul, A.; Menei, P. Mesenchymal Stromal-Like Cells in the Glioma Microenvironment: What Are These Cells? Cancers 2020, 12, 2628. [Google Scholar] [CrossRef]
- Krasny, L.; Huang, P.H. Data-Independent Acquisition Mass Spectrometry (DIA-MS) for Proteomic Applications in Oncology. Mol. Omics 2021, 17, 29–42. [Google Scholar] [CrossRef]
- Li, K.W.; Gonzalez-Lozano, M.A.; Koopmans, F.; Smit, A.B. Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome. Front. Mol. Neurosci. 2020, 13. [Google Scholar] [CrossRef]
- Weke, K.; Kote, S.; Faktor, J.; Al Shboul, S.; Uwugiaren, N.; Brennan, P.M.; Goodlett, D.R.; Hupp, T.R.; Dapic, I. DIA-MS Proteome Analysis of Formalin-Fixed Paraffin-Embedded Glioblastoma Tissues. Anal. Chim. Acta 2022, 1204, 339695. [Google Scholar] [CrossRef]
- Bikfalvi, A.; da Costa, C.A.; Avril, T.; Barnier, J.V.; Bauchet, L.; Brisson, L.; Cartron, P.F.; Castel, H.; Chevet, E.; Chneiweiss, H.; et al. Challenges in Glioblastoma Research: Focus on the Tumor Microenvironment. Trends Cancer 2023, 9, 9–27. [Google Scholar] [CrossRef]
- Lozada-Delgado, E.L.; Grafals-Ruiz, N.; Miranda-Román, M.A.; Santana-Rivera, Y.; Valiyeva, F.; Rivera-Díaz, M.; Marcos-Martínez, M.J.; Vivas-Mejía, P.E. Targeting MicroRNA-143 Leads to Inhibition of Glioblastoma Tumor Progression. Cancers 2018, 10, 382. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, T.; Wong, D.T.W. Saliva-Exosomics in Cancer: Molecular Characterization of Cancer-Derived Exosomes in Saliva. Enzymes 2017, 42, 125–151. [Google Scholar] [CrossRef]
- Trevisan França de Lima, L.; Müller Bark, J.; Rasheduzzaman, M.; Ekanayake Weeramange, C.; Punyadeera, C.; Ekanayake Weeramange, C. Saliva as a Matrix for Measurement of Cancer Biomarkers. Cancer Biomark. Clin. Asp. Lab. Determ. 2022, 297–351. [Google Scholar] [CrossRef]
- Suma, H.; Prabhu, K.; Shenoy, R.; Annaswamy, R.; Rao, S.; Rao, A. Estimation of Salivary Protein Thiols and Total Antioxidant Power of Saliva in Brain Tumor Patients. J. Cancer Res. Ther. 2010, 6, 278–281. [Google Scholar] [CrossRef]
- García-Villaescusa, A.; Morales-Tatay, J.M.; Monleón-Salvadó, D.; González-Darder, J.M.; Bellot-Arcis, C.; Montiel-Company, J.M.; Almerich-Silla, J.M. Using NMR in Saliva to Identify Possible Biomarkers of Glioblastoma and Chronic Periodontitis. PLoS ONE 2018, 13, e0188710. [Google Scholar] [CrossRef] [PubMed]
- Sanzey, M.; Abdul Rahim, S.A.; Oudin, A.; Dirkse, A.; Kaoma, T.; Vallar, L.; Herold-Mende, C.; Bjerkvig, R.; Golebiewska, A.; Niclou, S.P. Comprehensive Analysis of Glycolytic Enzymes as Therapeutic Targets in the Treatment of Glioblastoma. PLoS ONE 2015, 10, e0123544. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; He, D.; Fu, Y.; Zhang, R.; Guo, H.; Wang, Z.; Wang, Y.; Gao, T.; Wei, Y.; Guo, Y.; et al. A Novel LncRNA ARST Represses Glioma Progression by Inhibiting ALDOA-Mediated Actin Cytoskeleton Integrity. J. Exp. Clin. Cancer Res. 2021, 40, 187. [Google Scholar] [CrossRef]
- Comprehensive Analysis of a Long Non-Coding RNA-Associated Competing Endogenous RNA Network in Glioma. Available online: https://www.spandidos-publications.com/10.3892/ol.2020.11924 (accessed on 9 December 2024).
- Huo, J.F.; Chen, X.B. Knockdown of TMPRSS3 Inhibits Cell Proliferation, Migration/Invasion and Induces Apoptosis of Glioma Cells. J. Cell. Biochem. 2019, 120, 7794–7801. [Google Scholar] [CrossRef]
- Dunkelberger, J.R.; Song, W.C. Complement and Its Role in Innate and Adaptive Immune Responses. Cell Res. 2009, 20, 34–50. [Google Scholar] [CrossRef]
- Bouwens, T.A.M.; Trouw, L.A.; Veerhuis, R.; Dirven, C.M.F.; Lamfers, M.L.M.; Al-Khawaja, H. Complement Activation in Glioblastoma Multiforme Pathophysiology: Evidence from Serum Levels and Presence of Complement Activation Products in Tumor Tissue. J. Neuroimmunol. 2015, 278, 271–276. [Google Scholar] [CrossRef]
- Nigro, P.; Pompilio, G.; Capogrossi, M.C. Cyclophilin A: A Key Player for Human Disease. Cell Death Dis. 2013, 4, e888. [Google Scholar] [CrossRef]
- World Health Organization (WHO). International Ethical Guidelines for Health-Related Research Involving Humans; Prepared by the Council for International Organizations of Medical Sciences (CIOMS) in Collaboration with the World Health Organization (WHO); Council for International Organizations of Medical Sciences: Geneva, Switzerland, 2016. [Google Scholar]
- WMA Declaration of Taipei on Ethical Considerations Regarding Health Databases and Biobanks—WMA—The World Medical Association. Available online: https://www.wma.net/policies-post/wma-declaration-of-taipei-on-ethical-considerations-regarding-health-databases-and-biobanks/ (accessed on 5 May 2025).
- Research on Biological Materials of Human Origin (Recommendation CM/Rec(2016)6 and Explanatory Memorandum 2016)—European Sources Online. Available online: https://www.europeansources.info/record/research-on-biological-materials-of-human-origin-recommendation-cm-rec20166-and-explanatory-memorandum-2016/ (accessed on 5 May 2025).
- Per Una Buona Pratica Del Biobanking Di Ricerca. Available online: https://www.senato.it/application/xmanager/projects/leg18/attachments/documento_evento_procedura_commissione/files/000/001/414/IORNO_2.pdf (accessed on 5 May 2025).
- 2020 Ministero Della Salute Direzione Generale Della Ricerca e Dell’innovazione in Sanità. Available online: http://www.bibliosan.it/bussole_IRCCS/il_materiale_biologico_IRCCS_n_1.pdf (accessed on 5 May 2025).
- Quinn, C.M.; Porwal, M.; Meagher, N.S.; Hettiaratchi, A.; Power, C.; Jonnaggadala, J.; McCullough, S.; Macmillan, S.; Tang, K.; Liauw, W.; et al. Moving with the Times: The Health Science Alliance (HSA) Biobank, Pathway to Sustainability. Biomark. Insights 2021, 16, 1. [Google Scholar] [CrossRef]
- Cohrs, R.J.; Bidaut, L.; Shahzad, A.; Martin, T.; Ghahramani, P.; Higgins, P.J. VZV Infection of Human Neurons View Project Posterior Cruciate and Meniscofemoral Ligaments View Project Translational Medicine Definition by the European Society for Translational Medicine. New Horiz. Transl. Med. 2015, 2, 86–88. [Google Scholar] [CrossRef]
- Annaratone, L.; De Palma, G.; Bonizzi, G.; Sapino, A.; Botti, G.; Berrino, E.; Mannelli, C.; Arcella, P.; Di Martino, S.; Steffan, A.; et al. Basic Principles of Biobanking: From Biological Samples to Precision Medicine for Patients. Virchows Arch. 2021, 479, 233–246. [Google Scholar] [CrossRef]
- Grizzle, W.E.; Gunter, E.W.; Sexton, K.C.; Bell, W.C. Quality Management of Biorepositories. Biopreserv. Biobank. 2015, 13, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Riegman, P.H.J.; Becker, K.F.; Zatloukal, K.; Pazzagli, M.; Schröder, U.; Oelmuller, U. How Standardization of the Pre-Analytical Phase of Both Research and Diagnostic Biomaterials Can Increase Reproducibility of Biomedical Research and Diagnostics. N. Biotechnol. 2019, 53, 35–40. [Google Scholar] [CrossRef]
- Loft, S.; Poulsen, H.E. Cancer Risk and Oxidative DNA Damage in Man. J. Mol. Med. 1996, 74, 297–312. [Google Scholar] [CrossRef] [PubMed]
- OECD Legal Instruments. Available online: https://legalinstruments.oecd.org/en/instruments/OECD-LEGAL-0375 (accessed on 8 April 2025).
- Dagher, G. Quality Matters: International Standards for Biobanking. Cell Prolif. 2022, 55, e13282. [Google Scholar] [CrossRef]
- BBMRI.It. Available online: https://repository.bbmri.it/s/stC8Lc4kPDn2qQt (accessed on 8 April 2025).
- ISO 20387:2018(En); Biotechnology—Biobanking—General Requirements for Biobanking. International Organization for Standardization: Geneva, Switzerland, 2018. Available online: https://www.iso.org/obp/ui/en/#iso:std:iso:20387:ed-1:v1:en (accessed on 6 May 2025).
- Bledsoe, M.J. Ethical Legal and Social Issues of Biobanking: Past, Present, and Future. Biopreserv. Biobank. 2017, 15, 142–147. [Google Scholar] [CrossRef]
- Staunton, C.; Slokenberga, S.; Mascalzoni, D. The GDPR and the Research Exemption: Considerations on the Necessary Safeguards for Research Biobanks. Eur. J. Hum. Genet. 2019, 27, 1159–1167. [Google Scholar] [CrossRef]
- Halley, M.C.; Olson, N.W.; Ashley, E.A.; Goldenberg, A.J.; Tabor, H.K. A Just Genomics Needs an ELSI of Translation. Hastings Cent. Rep. 2024, 54, S126–S135. [Google Scholar] [CrossRef]
- Shabihkhani, M.; Lucey, G.M.; Wei, B.; Mareninov, S.; Lou, J.J.; Vinters, H.V.; Singer, E.J.; Cloughesy, T.F.; Yong, W.H. The Procurement, Storage, and Quality Assurance of Frozen Blood and Tissue Biospecimens in Pathology, Biorepository, and Biobank Settings. Clin. Biochem. 2014, 47, 258–266. [Google Scholar] [CrossRef]
- Müller, H.; Dagher, G.; Loibner, M.; Stumptner, C.; Kungl, P.; Zatloukal, K. Biobanks for Life Sciences and Personalized Medicine: Importance of Standardization, Biosafety, Biosecurity, and Data Management. Curr. Opin. Biotechnol. 2020, 65, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Rush, A.; Byrne, J.A.; Watson, P.H. Applying Findable, Accessible, Interoperable, and Reusable Principles to Biospecimens and Biobanks. Biopreservation Biobanking 2024, 22, 550–556. [Google Scholar] [CrossRef]
- Carneiro, R.L. The Transition from Quantity to Quality: A Neglected Causal Mechanism in Accounting for Social Evolution. Proc. Natl. Acad. Sci. USA 2000, 97, 12926–12931. [Google Scholar] [CrossRef] [PubMed]
- Gutland, C. The Shift from Quantitative to Qualitative Thinking—Problems and Prospects as Viewed from Husserl’s and Hegel’s Philosophy. Front. Psychol. 2023, 14, 1232420. [Google Scholar] [CrossRef] [PubMed]
- Hood, L.; Flores, M. A Personal View on Systems Medicine and the Emergence of Proactive P4 Medicine: Predictive, Preventive, Personalized and Participatory. N. Biotechnol. 2012, 29, 613–624. [Google Scholar] [CrossRef]
- Pitt, S.J.; Gunn, A. The One Health Concept. Br. J. Biomed. Sci. 2024, 81, 12366. [Google Scholar] [CrossRef]
- Gorini, A.; Pravettoni, G. P5 Medicine: A plus for a Personalized Approach to Oncology. Nat. Rev. Clin. Oncol. 2011, 8, 444. [Google Scholar] [CrossRef]

| Species | Sample Type | Proteomic Approach | Biomarkers Identified | Functional Relevance | Ref. |
|---|---|---|---|---|---|
| Human | Tissue | Labeling (TMT; iTRAQ) | ASAH1, GPNMB MMP9, TIMP1, Fibulins EGFR, NPM1, RKIP HNRNPK, ELAVL1, NOVA1 | Sphingolipid metabolism and ferroptosis Immune microenvironment Tumor progression, migration and angiogenesis Signaling growth and resistance to therapy Controlling gene expression in GBM | [60,61,62,63,64,65,66] |
| No labeling: LC-MS/MS (LFQ; DDA; DIA) | YAP1, SOX2, PP1γ EGFR, FN1, PTEN, BRAF FN1, TNC, ICAM1, GAGs HIF1α, IDH1, OXPHOS, Cholesterol, HSPD1, Granzyme A, STAT3, CHI3L1 RPS5, SF3B2, HMGB2 ASAH1, p21-p53-RB, ERCC2, POLD1 | Proliferation and survival Tumor growth and migration ECM regulation and cell adhesion Tumor metabolism and hypoxia Immune response and immunosuppression RNA processing and splicing Cell survival, apoptosis, DNA damage | [67,68,69,70,71,72,73,74,75,76,77,78,79,80,81] | ||
| Cells | No labeling: LC-MS/MS (LFQ; DDA; DIA) | ADAM10, ADAM15, COL6A1, COL1A2, COL6A3, TIMPs, Fibulin-2/-5/-7 STAT1, STAT2, OAS, IFIT, TRIM25, PME-1, PP2A-B55α, MAPKAPK2, RIPK1 CSE1L, TAZ, Importin α5, WWTR1, RAD51 | ECM regulation and tumor progression IFN signaling Sensitivity to oxidative stress Apoptosis, DNA damage | [82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111] | |
| Animal | Tissue | Labeling: iTRAQ | ILF2, CCT7, CCT4, RPL10A, MSN, PRPS1, TFRC, APEX1 | Early brain development Primary formation of the neural tube, Regulation of neuronal differentiation, Synaptic transmission, Regulation of the nervous system Regulation of cell survival and tumor proliferation Mechanisms of drug resistance and chemosensitivity | [112] |
| Cells | No labeling: LC-MS/MS (DDA; DIA) | CaMK2, BCAS1, FBXO2, INF2, PRPS2 CD9, CD81, Nono, Gja1 | Tumor growth, microenvironment Response to hypoxia, glycosaminoglycan biosynthesis Integrin-mediated signaling pathways, regulation of TGFβ pathways | [94,113] |
| Sample Type | Proteomic Approach | Biomarkers Identified | Functional Relevance | Ref. |
|---|---|---|---|---|
| Cerebrospinal Fluid | LC-MS/MS (DDA) | CHI3L1 GFAP, GAP43, SERPIN3, APOE, FGA, FGB, FGG, F2 | Tumor aggressiveness BBB disruption Synaptogenesis Coagulation Angiogenesis LXR/RXR Activation pathway Stemness, Immune modulation | [41,121] |
| Cystic Fluid | LC-MS/MS (DDA) | Albumin, Haptoglobin, Fibrinogen, Transferrin, Prostaglandin D2 synthase, IgG, IgA, IgM, S100B, GFAP | Cell adhesion, angiogenesis and cytoskeleton Acute Inflammatory Response Immunomodulation | [122] |
| Plasma | LC-MS/MS (DDA) | ASAH1, SYNM, GPNMB, VWF, Hp (α, β chains), zonulin | Tumor progression, invasiveness and vascularization Neutrophil involvement Pro-angiogenic processes and pro-thrombotic response Oxidative stress protection Inflammation regulation and homeostasis | [60,123,124] |
| Serum | LC-MS/MS (DIA) | CXCL4 (PF4), S100A8, S100A9, MDH1, RNH1, FABP7, TJAP1, AHSP | Inflammation Ros metabolism Nucleotide metabolism Metabolic reprogramming Cellular homeostasis Lipid metabolism and transport VEGF and IL-18 signaling | [125,126,127] |
| Urine | LC-MS/MS (DIA) | GRN, PSAP, ALDOA, S100A11, ITM2B, TCP1, CCT2, CCT3, CCT4, CCT6A, CCT7, CCT8 | Proteostasis and protein folding Metabolic reprogramming Tumor progression Stress response | [128] |
| Saliva | LC-MS/MS (DIA) | ALDOA, 14-3-3ε (YWHAE), TM11B, C3, PPIA, TGF-β-related proteins | Cellular proliferation Cell cycle Signaling regulation Complement system activation Protein folding and trafficking TGF-β signaling Immune response Iron metabolism | [129] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciuffreda, G.; Casati, S.; Brambilla, F.; Campello, M.; De Falco, V.; Di Silvestre, D.; Frigeri, A.; Locatelli, M.; Magrassi, L.; Salmaggi, A.; et al. Glioblastoma: Overview of Proteomic Investigations and Biobank Approaches for the Development of a Multidisciplinary Translational Network. Cancers 2025, 17, 2151. https://doi.org/10.3390/cancers17132151
Ciuffreda G, Casati S, Brambilla F, Campello M, De Falco V, Di Silvestre D, Frigeri A, Locatelli M, Magrassi L, Salmaggi A, et al. Glioblastoma: Overview of Proteomic Investigations and Biobank Approaches for the Development of a Multidisciplinary Translational Network. Cancers. 2025; 17(13):2151. https://doi.org/10.3390/cancers17132151
Chicago/Turabian StyleCiuffreda, Giusy, Sara Casati, Francesca Brambilla, Mauro Campello, Valentina De Falco, Dario Di Silvestre, Antonio Frigeri, Marco Locatelli, Lorenzo Magrassi, Andrea Salmaggi, and et al. 2025. "Glioblastoma: Overview of Proteomic Investigations and Biobank Approaches for the Development of a Multidisciplinary Translational Network" Cancers 17, no. 13: 2151. https://doi.org/10.3390/cancers17132151
APA StyleCiuffreda, G., Casati, S., Brambilla, F., Campello, M., De Falco, V., Di Silvestre, D., Frigeri, A., Locatelli, M., Magrassi, L., Salmaggi, A., Salvetti, M., Signorelli, F., Torrente, Y., Umana, G. E., Viganò, R., & Mauri, P. L. (2025). Glioblastoma: Overview of Proteomic Investigations and Biobank Approaches for the Development of a Multidisciplinary Translational Network. Cancers, 17(13), 2151. https://doi.org/10.3390/cancers17132151









