Simple Summary
Acute myeloid leukemia (AML) is an aggressive, heterogeneous blood malignancy harboring various gene mutations. The nucleophosmin 1 (NPM1) gene is a common mutated gene in AML, with almost 30% of AML patients having NPM1 mutations. Despite our increasing knowledge of the pathogenesis and treatment options of NPM1-mutated AML, there are many unanswered questions concerning this subtype of leukemia. This study is a detailed review of what is known about NPM1-mutated AML. An overview of its definition, subtypes and classification, along with the mutational spectrum that frequently co-exists with NPM1 mutation in AML and affects prognosis, is provided. The clinical and laboratory characteristics of NPM1-mutated AML are also described, with emphasis on disease biology. All important information concerning measurable residual disease (MRD) in patients with NPM1-mutated AML is also highlighted. New drugs under development are also presented, as well as how treatment based on precision medicine might improve outcomes. This review attempts to categorize conflicting study results and provide guidance for the future study and optimal treatment of patients with this type of leukemia.
Abstract
The aberrant localization of the mutated nucleophosmin (NPM1) protein in the cytoplasm is the hallmark of the development of acute myeloid leukemia (AML); the gene, located in the nucleolus, codes for a protein that normally shuttles between the nucleus and the cytoplasm of the normal hematopoietic cells. Patients harboring NPM1 mutations are diagnosed as having NPM1-mutated AMLs, which are types of leukemia with distinct clinical and laboratory characteristics. The essential diagnostics for investigating NPM1-mutated AMLs, the interactions with concomitant mutations affecting prognosis and the therapeutic interventions that the treatment of such patients requires are discussed in this review. Novel investigational agents in current clinical trials are also highlighted, along with the roles of exportin 1 (XPO1), menin-KMT2A inhibitors and immunotherapy in NPM1-mutated AMLs. This review focuses on critically evaluating the available data and aims to reveal the secrets of NPM1-mutated AMLs.
Keywords:
acute myeloid leukemia (AML); nucleophosmin (NPM1) gene; driver mutation; leukemogenesis; nuclear–cytoplasmic localization; concomitant mutations; measurable residual disease (MRD); menin inhibitors; ziftomenib; revumenib; exportin 1; knockout and knock-in phenotype; clinical trials; NPM1-mutated AML; treatment 1. NPM1-Mutated AML: An Overview of Its Definition, Subtypes and Classification
1.1. Background
The link between the nucleophosmin (NPM1) protein and the initiation of acute myeloid leukemia (AML) has been a long-term scientific project. The mutated NPM1 gene in the nucleus of the leukemic cell, located on chromosome 5q35, was discovered in 2005 by Falini B et al. [1], following the impressive finding of the aberrant localization of the mutated NPM1 protein in the cytoplasm of leukemic cells with a specific immunohistochemistry (IHC) stain [1,2]. The NPM1 gene consists of 12 exons and encodes a multifunctional protein comprising 294 amino acids (35–40 kDa) [3,4,5,6]. Normally, the NPM1 wild type (NPM1wt) is located inside the nucleolus of the hematopoietic cells and encodes a multifunctional protein, which shuttles between the nucleolus, the nucleus and the cytoplasm. The normal functions of NPM1wt in the nucleolus include controlling centrosome duplication to ensure effective cell division, regulating ribosome biogenesis, forming heterochromatin to control standard gene expression, ensuring the stability of DNA repair and enhancing TP53 transcriptional activity, among others. It must be mentioned that most of this functional workload is mediated by its potential to chaperone RNA molecules from the nucleus to the cytoplasm. This nucleocytoplasmic transport is essential for the cellular regulatory and maintenance processes [3,7,8,9]. Hence, the location of the NPM1 gene (either in mutated or wild-type form) is the nucleus of the hematopoietic cells, whereas the aberrant localization of the NPM1-mutated protein, which can be identified via IHC staining on a bone marrow (BM) biopsy, occurs in the cytoplasm. IHC was the initial method used for identifying the NPM1-mutated protein, long before the advent of next-generation sequencing (NGS) data [10,11].
1.2. NPM1 Mutational Landscape
As highlighted, the hallmark of NPM1-mutated AML is the cytoplasmic localization of the mutant protein, primarily due to specific genetic alterations, as opposed to the natural nucleolar habitation of the NPM1-mutated and NPM1wt genes. The latter is attributed to the NPM1wt C terminus amino acid positions W288 and W290, which are highly conserved and indispensable for forming a globular structure, responsible for the nucleolar localization of the gene [7]. Conversely, NPM1 mutations delete or alter key amino acids. Thus, the C terminus of the mutated NPM1 loses the amino acid tryptophan in positions W288 and W290 (or W290 alone), causing the disruption of the folded helix structure, the loss of the nucleolar localization signal and the aberrant cytoplasmic localization of NPM1 mutants, as observed in AML [7,11,12,13,14]. As a result of these mutations, the mutant NPM1 protein loses its nuclear localization signal. The latter acquired characteristic leads to the cytoplasmic export and retention of the NPM1-mutated protein, a defining hallmark of NPM1-mutated AML. Despite its high prevalence, the mechanisms of action of the cytoplasmic mutated NPM1 protein in AML remain poorly understood [4,12].
Normally, nuclear import predominates over export, so the normal NPM1 protein mainly remains in the nucleus. However, when NPM1 mutants are present, nuclear export predominates over import and the NPM1 proteinic mutants are relocated in the cytoplasm. In other words, NPM1 mutants are ‘born to be exported’ [7,8,13].
NPM1 mutations are categorized based on variations in their mutational spectrum. Type A mutations harbor the combination of TCTG between nucleotides 860 and 863 and type B mutations insert CATG between nucleotides 863 and 864, whereas type D mutations add CCTG between nucleotides 863 and 864 [3,15]. Almost 70% of the total NPM1 mutations are type A, while 11% are type B. Type A mutations are very often observed with concomitant DNMT3A mutations, which are responsible for worse prognoses in type A and type D NPM1-mutated AML [15]. Intriguingly, the mutated NPM1 allele exerts a dominant biological behavior over the wild-type NPM1 allele due to the preferential transcription of the mutated NPM1 allele [16]. As a result, the mutant/normal NPM1 heterodimers are relocated to the cytoplasm of the leukemic cells [17].
NPM1 mutations in AML are further classified based on the number of the lost tryptophan residues: the mutations causing the loss of two tryptophan residues are characterized as A-like, in contrast to those inducing the loss of one tryptophan residue, which are characterized as non-A-like, more extensively studied in pediatric AML [18]. Patients with non-A-like NPM1-mutated AML have a better prognosis and increased sensitivity to chemotherapy, probably because NPM1 partially remains in the nucleolus. Contrarily, patients with A-like NPM1-mutated AML have a more dismal prognosis and a disease with a more aggressive clinical course. They exhibit a decreased response to chemotherapy, mainly because of HOXA5, HOXA10 and HOXB5 up-regulation and p14ARF/p21/p53 pathway deregulation [18].
1.3. Geographic and Ethnic Variations in NPM1 Mutation Prevalence
Several studies have yielded geographical and ethnic differences in the prevalence of NPM1 mutations among AML patients. These differences are particularly age-dependent and vary significantly between Western and Asian populations. For example, a higher prevalence of this mutation in older AML patients has been observed in Europe compared with Asian countries, such as China, Japan and Korea [19,20,21,22,23]. This mutation is quite common in newly diagnosed (ND) AML patients: it is found in around 30% of cases and its prevalence shows minor variations across different European registries, ranging from 27% [24] to 33% [25]. However, Asian AML patients show lower percentages of the NPM1 mutation, ranging from 13.3% to 28%. The latter could indicate a potential ethnic effect on the probability of NPM1 prevalence [20,21,22,23].
The observed age variations in the mutational landscape between younger and older adults in AML [26] do not influence the aforementioned ethnic differences. Thus, NPM1 occurs in 23.6% of patients older than 40 years old in the Chinese data, significantly lower than in the German AML2003 data, even after age adjustment [20]. Interestingly, lower frequencies of NPM1 mutations have also been reported in Black AML patients [27]. Indeed, Black ethnicity is an independent prognosticator of poor survival in AML patients, probably because fewer NPM1 and more IDH2 mutations are observed in younger Black patients [28]. Intriguingly, the OS of younger Black AML patients in contrast to White patients was not improved by NPM1 mutations, even though this measure was adversely affected by FLT3-ITD and IDH2 mutations [28].
The reasons for these genetic discrepancies among international cohorts and the biology of the disease behind these differences are largely unknown. It could be hypothesized that environmental factors affect the epigenetic modification of the genome, whereas the complex interplay between biological (genetic predisposition) and environmental factors causes genetic variations. Due to the disease heterogeneity, particular AML subtypes may be biologically distinct across different ethnic backgrounds, potentially affecting mutation rates. All these ethnic differences suggest a need for different treatment intensities and modalities in specific AML subgroups, which will be the intended outcomes of well-designed clinical trials comparing different ethnic populations. The latter might lead to different prognostic risk grouping and variations in AML treatment in different parts of the world.
1.4. Classification of NPM1-Mutated AML
In every recent AML classification system, ranging from the one comprising 13 subgroups proposed by Papaemmanuil E et al. in 2016 [24] to the newest involving 16 distinct subgroups suggested by Tazi Y et al. in 2022 [29], AML with the NPM1 mutation is classified as a separate category. The International Consensus Classification (ICC) of 2022 classifies AML with mutated NPM1 as a separate category requiring 10% blasts or more in peripheral blood (PB) or BM for establishing an AML diagnosis [30]. Likewise, the NPM1 mutation is considered to be an AML-defining genetic abnormality, also forming the entity of AML with mutated NPM1 in the European Leukemia Net (ELN) 2022 Guidelines [31]. This classification has also incorporated NPM1 mutations as AML-differentiating genetic lesions, thereby confirming these mutations’ unique position within the subtypes of AML [31]. Similarly, in the 5th World Health Organization (WHO) 2022 classification, AML with NPM1 mutations belongs to the diagnostic group known as AML with defining genetic abnormalities [32]. The only important difference compared to the ICC 2022 is that in the WHO 2022 classification, no blast enumeration threshold exists as a prerequisite for diagnosis, as AML with NPM1 mutation is diagnosed irrespective of the blast count [32,33,34].
1.5. NPM1 as a Genetic Driver Mutation in AML Initiation: The Never-Alone, Usual Suspect
The nature of a ‘driver’ mutation must be justified based on its ability to independently initiate a neoplasm without requiring additional genetic alterations. One way to test whether a mutation meets this criterion is by replicating it in a mouse model and observing whether leukemia develops.
It was reported that the activation of a humanized NPM1 cytoplasmic knock-in allele in mouse hematopoietic stem cells caused delayed-onset AML in one third of the mice, along with HOX gene overexpression, suggesting that additional mutations are needed in this model to initiate leukemogenesis [35]. To determine which additional mutations could accelerate this process, scientists used a transposon system that randomly inserts itself into the genome, disrupting various genes. In 2011, these results provided initial insights showing that mutant NPM1, in co-operation with other secondary mutations, drives leukemia initiation and progression in mice. Furthermore, possible therapeutic targets, such as HOX, were identified in NPM1-mutated AML [35].
Recently, a well-designed study showed that NPM1 haploinsufficiency in collaboration with MEIS1 overexpression is sufficient to induce complete AML that transcriptionally resembles human NPM1-mutated AML in mice [36]. Moreover, the MEIS1-SMC4 axis is a potential therapeutic target in this type of AML. The latter experiment showed that NPM1 mutations, in collaboration with DNA-binding transcription activators, such as MEIS1, can drive AML. Until today, the downstream targets or effectors of the NPM1-mutated protein, such as HOX, exportin-1 (XPO1) or the menin-KMT2A axis, have been inhibited in clinical trials. These treatment approaches are highlighted in the section of this manuscript describing the treatment of NPM1-mutated AML. All the above notions are demonstrated in Figure 1.
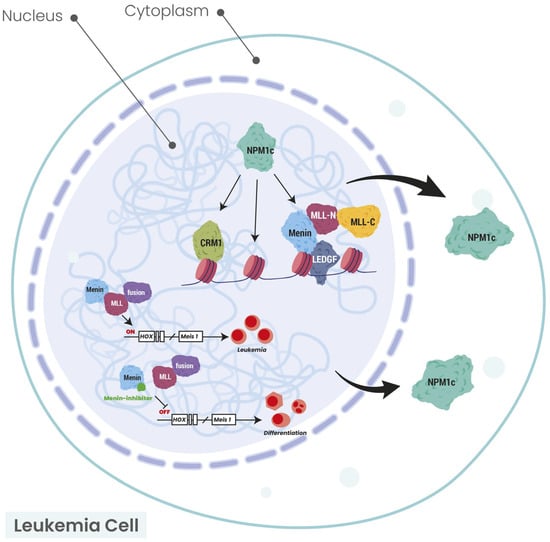
Figure 1.
Schematic illustration of the connection between NPM1 and HOX/MEIS1/XPO1/menin axis. The arrows show the cytoplasmic localization of NPM1 protein in NPM1-mutated AML. Modified from Uckelmann HJ et al. [37] and Krivtsov AV et al. [38]. CRM1 chromosomal region maintenance 1 (XPO1: exportin 1); HOX: homeobox; LEDGF: lens epithelium-derived growth factor; MEIS1: MEIS homeobox 1; MLL: mixed lineage leukemia; MLL-N: mixed lineage leukemia—nuclear; MLL-C: mixed lineage leukemia—cytoplasmic; NPM1: nucleophosmin 1; NPM1c: nucleophosmin 1 cytoplasmic.
As more NGS data became available, it became evident that NPM1 mutations rarely occur in isolation, instead co-existing with other genetic alterations, reinforcing the idea that they function within a broader leukemogenic framework. However, NPM1 mutation is considered to be a driver mutation and a major gene causing leukemogenesis by the scientific community, probably due to its leading role and tight connection with AML pathogenesis [3,4,7,12], even though it does not completely fulfill the strict definition of initiating AML alone in the mouse models, having a strong influence on and collaborating with MEIS1 overexpression to induce leukemia [36].
1.6. The Role of Genomic Instability and Additional Mutations in AML Development
Genomic instability, defined as an increased tendency of the genome to acquire mutations, is a common mechanism in cancer. Nevertheless, in AML, genomic instability is uncommon. Most of the mutations in AML are random and occur in hematopoietic stem cells or progenitors before the acquisition of the initiating mutation. Only one or two additional mutations in combination are required to generate the initial neoplastic clone [39].
It is important to state that many genes are recurrently mutated in AML; moreover, individual leukemias harbor multiple mutations (molecular aberrations or defects) and are potentially composed of subclones with different mutational compositions, genetic profiles and leukemogenic capacities, rendering each patient’s AML genetically unique. Furthermore, the biology of AML significantly differs between younger and elderly patients. Driver mutations such as DNMT3A, TET2, IDH2, TP53, RUNX1, NPM1, NRAS, FLT3-ITD, FLT3-TKD, ASXL1 and STAG2 are often enriched in elderly AML patients (>65–70 years old), thereby differing in type and prevalence from those encountered in younger patients, who exhibit a comparatively different mutational landscape [19,26,40]. DNMT3A, TET2, ASXL1 and other genes are also involved in the PRC1/PRC2 regulation circuit, offering another point of view on gene regulation in AML.
There are emerging data suggesting that cases previously classified as Myelodysplastic syndromes (MDSs) or MDS/myeloproliferative neoplasms (MPNs) with NPM1 progress to AML shortly. The latter means that NPM1 mutation is linked with AML development, as a driver mutation, in a small period from the time of acquisition of the mutation, causing either de novo or secondary AML. Similar findings also exist from patients with clonal hematopoiesis (CH) who acquire an NPM1 mutation; AML evolution occurs within a short period of time [32]. Thus, NPM1 mutation is strongly correlated with the presence of AML and the emergence of AML, shortly after the diagnosis of MDS and MDS/MPN. For all the above reasons, the acquisition of an NPM1 mutation is considered a driver mutation at the clinical level [3,7,12] and a later event in leukemogenesis [24]. NPM1 mutations act as ‘gatekeepers’ for AML, mainly de novo AML [7]. These mutations are found in the entire leukemic population by IHC; they are stable over time, also being detected at relapse; and they are related to the mutated protein, causing its production. This is why NPM1 gene transcripts are an excellent marker for evaluating measurable residual disease (MRD) in AML [25,41,42].
1.7. NPM1 as an MRD Marker in AML
According to 2022 ELN guidelines, MRD monitoring in patients with NPM1-mutated AML is critical in evaluating disease response to drugs, guiding post-remission therapy and, in parallel, predicting relapse [31]. There are established guidelines for monitoring AML patients with NPM1 mutations under intensive induction chemotherapy regimens, whereas there are no current guidelines for MRD testing for patients receiving non-intensive venetoclax (VEN)-based regimens.
Real-time quantitative PCR (RT-qPCR) is the recommended method for detecting NPM1 mutations because of its high sensitivity and specificity [31,42]. Thus, MRD should be assessed with NPM1 transcripts (PCR) in the PB after two cycles of intensive chemotherapy, in the BM at the end of treatment, and either in the BM every 3 months or in the PB every 4–6 weeks for 24 months after treatment completion [31,42,43]. These guidelines were derived from patients with NPM1-mutated AML who achieved CR after two cycles of intensive induction chemotherapy.
MRD positivity is defined as ≥2% in the BM or failure to achieve a 3 to 4 log reduction in either the BM or PB at completion of consolidation. Molecular MRD detectable at a low level (MRD-LL) is defined as <2% and designated as negative, because when measured at the end of consolidation treatment, it is associated with a very low relapse rate. MRD relapse is defined as conversion from MRD negativity to MRD positivity or an increase in MRD ≥ 1 log10 between any two positive samples for patients with MRD-LL [42,43,44].
Othman et al. evaluated patients with ND NPM1-mutated AML regarding their MRD status. These patients achieved CR after therapy with VEN plus HMAs or low-dose cytarabine. MRD was assessed via RT-qPCR. Achieving BM MRD negativity after the end of four cycles of therapy was linked with the greatest improvement in OS; in particular, it predicted a better 2-year OS (58% exhibited BM MRD negativity) [45]. Several issues arose because of these important results. It is not clear if MRD-negative patients at the end of cycle 4 should continue treatment with VEN plus HMAs, decrease the dosages or the days exposed to the administered drugs or even stop therapy [46]. The latter is extremely important, as long exposure to VEN might lead to resistance to the drug due to the appearance of novel KRAS or NRAS mutations [47]. Interestingly, for MRD-positive patients at the end of cycle 4, preemptive therapy is proposed by some clinicians, with significant advantages and disadvantages. Moreover, other issues regarding MRD in AML patients who received VEN/HMAs include the role of MRD in earlier cycles, defining the most clinically actionable MRD level and selecting either the BM or the PB as a better strategy for MRD monitoring [46].
1.8. The Order of Mutations in AML Development
It has long been known that the primary leukemogenic mutations are usually epigenetic modifiers (TET2, DNMT3A, IDH1/2, ASXL1, because they are part of the PRC1/PRC2 circuit), followed by NPM1 or RAS mutations that eventually lead to AML progression [24]. Thus, the AML mutations that can be found together or at different time intervals and the reasons why this occurs are known in individual AML patients. For example, DNMT3A mutations and type A NPM1 mutations co-operate to initiate leukemogenesis. Further molecular lesions in FLT3-ITD drive proliferation, setting the stage for expanding and establishing a predominant clone [3,7]. The order of the mutations in time can be indirectly inferred by the Varied Allele Frequency (VAF), defined as the number of alleles harboring the respective mutation compared to the normal alleles. A higher VAF is indicative of an earlier mutation in the complex mutational leukemogenic landscape in an individual AML patient [24]. These insights highlight the sequential nature of AML progression, where certain mutations establish a foundation for leukemogenesis, whereas others contribute to disease aggressiveness.
1.9. The Functional Impact of NPM1 Mutations on Leukemia Biology
At the molecular level, the NPM1 mutant protein interacts with other oncogenic proteins, engaging with various oncogenic pathways within the leukemic cell cytoplasm. A significant interaction occurs with FOXM1, a transcription factor trapped in the cytoplasm by mutated NPM1, hindering its normal transcriptional function. NPM1 mutations are also associated with increased up-regulation of the MLL/MEIS1/HOX axis, a critical effect for maintaining the leukemic stem cell fate [48]. Another oncogenic mechanism of the NPM1 mutation involves the nuclear export of the tumor suppressor p14 (ARF), causing a poor p53 response, with the cytoplasmic export of PU.1 resulting in the suppression of more than 300 myeloid differentiation genes or the overexpression of the oncogenic long-noncoding RNA LONA, which becomes nuclear as the mutant NPM1 protein relocalizes into the cytoplasm [48].
Additionally, while NPM, FLT3-ITD and DNMT3A mutations rarely co-occur in AML, they interact to drive the increased expression of the transcription factor hepatic leukemia factor (HLF) [49]. The result is an adverse phenotype with an increased leukemic stem cell burden with an aberrant immunophenotype [CD34-low, G protein-coupled receptor 56 (GPR56)-high] harboring a poor prognosis [49]. Furthermore, the aforementioned leukemic phenotype is significantly related to HOX and histone H1 signal transduction pathways, associated with transcriptional misregulation and clinically with older age, lower complete remission (CR) rates and poor clinical outcome [50].
2. Clinical and Laboratory Characteristics of NPM1-Mutated AML
2.1. Chromosomal Aberrations of NPM1-Mutated AML
Most AML patients harboring NPM1 mutations have normal karyotypes (85% of total patients). The remaining 15% of patients carry chromosomal aberrations, the most frequent being +8 (trisomy), del(9q), +4 (trisomy), −Y, +21 (trisomy) and monosomies 5 and 7 [7,14,51,52]. These aberrations are considered secondary late events during the clonal evolution of NPM1-mutated AML [11,52]. Such leukemias show overlapping biologic, pathologic and immunophenotypic features. Prognosis is variable in all the above-mentioned abnormalities, also depending on the presence of other mutations, along with the NPM1 mutation and the cytogenetic lesions [53,54,55,56]. Intriguingly, all AML patients with del(9q) as a sole abnormality consistently display a distinct, exclusive combination of DNMT3A and NPM1 mutations [56].
It is well established that NPM1-ALK fusions are very common in anaplastic large-cell lymphoma (ALCL) patients [57]. ALCL is a type of T-cell non-Hodgkin lymphoma (NHL). A few cases harboring NPM1-ALK fusions have been documented in B-cell acute lymphoblastic leukemia (ALL) [58]. Interestingly, oncogenic ALK point mutations (A348D and F856S) have also been discovered in a B-ALL and an AML patient, respectively [59]. ALK fusions have very rarely been observed in AML patients [60,61].
2.2. Demographics, Clinical Presentation and Immunophenotypic Features
NPM1-mutated AMLs show female predominance, with increased White Blood Cells (WBCs) and platelets and a lower incidence in children (2–8%) [17,51,62]. Moreover, there is a frequent association with extramedullary involvement, particularly in the skin, which is easily diagnosed via IHC. The bone marrow is highly hypercellular, with high blast percentages in most cases, and cup-like nuclei are observed [63], whereas multilineage dysplasia is evident in almost 20% of patients [51]. Even though all FAB categories can be represented, myelomonocytic (M4) and monocytic (M5) types are the most often encountered [10,17,62,64]. The high mutation incidence of NPM1 (38%) may partly explain the relatively benign clinical manifestations of FAB M4 and M5 AML [65]. The majority of these patients have favorable prognosis [31] and do not undergo hematopoietic stem cell transplantation (HSCT). However, a recent study highlighted that the combination of NPM1 and FLT3-ITD mutations (intermediate prognosis) correlated with a significantly higher incidence of M5 morphology (67.4%) compared to NPM1-positive and FLT3-ITD-negative mutations (54.5%) [66]. Thus, due to the known limitations of the retrospective nature, the small sample size and the lack of evaluation of co-existing mutations of the conducted study [65], larger prospective trials incorporating different age subgroups and various applied treatments, along with the presence of concomitant mutations, are necessary to assess the effects of possible allogeneic HSCT in NPM1-positive, FAB M4 and M5 AML patients.
The number of WBCs in NPM1-mutated AMLs increases progressively from concomitant FLT3-ITD wild-type to FLT3-ITD mutated cases, being much higher comparatively in the latter type. There is no or low expression of CD34, while CD34 positivity has been linked with adverse outcomes; on the contrary, the blastic cells of NPM1-mutated AMLs exhibit increased CD33 expression [67]. The response of NPM1-mutated AMLs to the induction of chemotherapy is excellent [10]. It is important to emphasize that NPM1 mutants are restricted to myeloid cells and are never found either in B or T cells in BM or PB [68].
2.3. Diagnostic Challenges of NPM1-Mutated AML
Another important element diagnostically is that even though the majority of cases harbor NPM1 mutations in exon 12 (99%) [10], there are extremely rare cases (<1%) involving exons 11 [69], 5 [70] and 9 [71]. Diagnosing such cases can be challenging, as these very rare mutations may be missed. That is why IHC should always be conducted in BM biopsies in AML patients for detecting the cytoplasmic expression of NPM1, which is a surrogate for NPM1 mutations [1]. Thus, when there is discrepancy between a positive IHC (cytoplasmic NPM1) and a negative conventional molecular analysis of exon 12 (absence of exon 12 NPM1 mutation), there is an NPM1 mutation in another exon, suggesting that the entire coding sequence of NPM1 should be investigated by properly modifying the NGS panels, as the commercially available NGS panels are designed to recognize only NPM1 mutations of exon 12 [11]. Thus, such patients are properly classified prognostically as belonging to the ELN-favorable risk group (mutated NPM1 in exon 11 without FLT3-ITD), instead of being wrongly classified in the ELN-intermediate risk group (NPM1 wild-type without FLT3-ITD) [31]. MRD monitoring in these rare NPM1-mutated AMLs requires designing a patient-specific real-time q-polymerase chain reaction (RT-qPCR) assay [41].
Intriguingly, the principle that NPM1 gene transcripts are an excellent marker for evaluating MRD in AML has some even more extreme rare exceptions. Indeed, NPM1-mutated, fit, older AML patients (≥65 years) at diagnosis have been reported to relapse and acquire resistance with negative NPM1-mutated transcripts after the application of VEN plus modified intensive chemotherapy (CAVEAT study) [72,73]. In these AML patients, the NPM1 gene could not be used as a marker for evaluating MRD status, since it was negative at relapse and not positive, despite initial positivity at diagnosis [72]. The anti-apoptotic proteins MCL-1 and BCL-XL exhibited an elevated expression at the relapse of the NPM1-mutated AML, and this up-regulation was established via flow cytometry [72]. In contrast to younger populations, NPM1-mutated AML confers a favorable prognosis in older AML patients ≥ 65 years old, who experience durable treatment-free remission with the above-mentioned time-limited regimen (CAVEAT study) [73].
2.4. Essential Diagnostics for Investigating and Treating NPM1-Mutated AML
The tests and procedures used at diagnosis for a patient with AML were proposed by the ELN 2022 recommendations [31]. Beyond the routine tests to establish the diagnosis and necessary cytogenetics within 5–7 days, the following screening process for gene mutations required for diagnosis, risk stratification and the identification of actionable therapeutic targets is necessary: FLT3-ITD, FLT3-TKD, IDH1, IDH2, NPM1 (within 3–5 days), CEBPA, DDX41, TP53, ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2 (within the first treatment cycle). The screening process for routine gene rearrangements should also be performed within 3–5 days. The following additional genes are recommended to be tested for at diagnosis: ANKRD26, BCORL1, BRAF, CBL, CSF3R, DNMT3A, ETV6, GATA2, JAK2, KIT, KRAS, NRAS, NF1, PHF6, PPM1D, PTPN11, RAD21, SETBP1, TET2, and WT1 [31]. This is also significant, as mutations co-existing with an NPM1 mutation affect prognosis and alter the risk stratification of the patient (analyzed in the risk stratification part of this manuscript). In cases of delayed screening results not only for NPM1 but also for the other above-mentioned key mutations, important information regarding the prognostic classification of the AML patient is lacking at the critical time point for making a treatment decision on whether to transplant the patient or not.
Moreover, biobanking and the proper registry of family medical history are extremely important for both ongoing research and future therapeutic developments. Advances in genetic testing, in combination with a detailed family history, have enabled identifying germline predisposition syndromes [74]. The HSCT donors carrying deleterious germline variants, such as RUNX1 and/or CEBPA, should be excluded from transplantation [31]. Furthermore, AML patients harboring a germline mutation should be under close surveillance concerning their donor selection, optimal timing for HSCT, conditioning regimen, and comorbidities, aiming to minimize toxicities [74]. Ensuring the proper biobanking of biologic tissue is of the utmost importance for enhancing research and therapeutic strategies. Recent studies have investigated the predisposition potential of several germline variants in large cohorts of AML patients [75] or evaluated the clonal evolution and early detection of DDX41-mutant AML by defining the risk associated with DDX41 variants [76]. The latter method enables closer AML monitoring and disease management for patients and their families.
3. The Prognostic Complexity of NPM1-Mutated AML
NPM1-mutated AML is considered a separate entity in most classifications. However, it is contradictory and peculiar that this mutation is never observed alone, but always with other mutations, affecting and modifying the clinical course of the individual AML patient. It is, therefore, essential to note that NPM1-mutated AML should always be evaluated in the constellation of mutations in the specific individual with AML. In addition, such an evaluation requires NGS data for proper risk assignment.
Large patient studies have revealed the mutations most frequently co-existing with NPM1 mutations. In a cohort of 418 patients harboring NPM1 mutations, the most common co-mutated genes were as follows: DNMT3A (54%), FLT3-ITD (39%), NRAS (19%), TET2 (16%) and PTPN11 (15%) [24]. Almost identical results for DNMTRA, FLT3-ITD and PTPN11 with the addition of cohesin Complex mutations (20%), IDH1 (15%) and IDH2R140 (15%) were reported in ND AML patients [25]. A more recent NGS analysis of 71 patients with NPM1-mutated AML yielded the following results: DNMT3A (62%), FLT3-ITD (38%), FLT3-TKD (14%), TET2 (27%), IDH2 (23%), IDH1 (18%), PTPN11 (18%) and NRAS (17%) [77].
Interestingly, the combinations of NPM1 mutations with IDH2R172 for all AML patients and the mutational combinations NPM1/TP53, NPM1/RUNX1 and NPM1/ASXL1 for elderly patients are mutually exclusive [19]. The data highlight that an IDH2 mutation usually remains in CR and precedes an NPM1 mutation in leukemogenesis, whereas an IDH1 mutation does not remain in complete remission (CR) and is usually subclonal [77]. The remains of an IDH2 mutation and the hierarchy of the IDH clone in CR with the concomitant clearance of an NPM1 mutation do not affect overall survival (OS) [77].
Regarding prognosis, the presence of other molecular defects, along with NPM1, influence the outcome of the individual AML patient. In general, AML patients with an NPM1 mutation in the absence of FLT3-ITD belong to the favorable prognostic risk category (ELN 2022 risk stratification for patients receiving intensive treatments) [31]. These fit patients are treated with chemotherapy without HSCT, while the unfit patients or those >75 years old are treated with a combination of VEN plus a hypomethylating agent (VEN-HMA). However, when the NPM1 mutation co-exists with the FLT3-ITD mutation [29,31], the prognostic risk category is intermediate, and HSCT after chemotherapy is necessary for fit patients. Moreover, patients with the NPM1 mutation and adverse-risk cytogenetic abnormalities, even in the absence of FLT3-ITD mutation, are categorized as adverse-risk patients [31], indicating the inadequate tumor suppressor potential of the NPM1 mutation and the predominance of cytogenetic risk over molecular risk [14].
For patients receiving less intensive treatments, an important factor affecting the prognosis and the response to VEN-azacitidine (AZA) in NPM1-mutated AML is the co-presence of activating signaling mutations (FLT3-ITD, KRAS, NRAS). NPM1-mutated AML when these mutations are absent has a median OS of 39 months; in contrast, this figure decreases to 9.9 months when these kinase-signaling mutations are present in parallel with the NPM1 mutation [78,79].
Moreover, for patients receiving less intensive treatments, TP53, KRAS, NRAS and FLT3-ITD guide prognosis and classify AML patients who received VEN-AZA into three different prognostic groups. A favorable-risk group with all involves four genes being negative (median OS: 26.5 months), an intermediate-risk group has positive FLT3-ITD, mutated KRAS and/or mutated NRAS (median OS: 12.1 months) and an adverse-risk group has mutated TP53 (median OS: 5.5 months) (2024 ELN recommendations) [78]. Thus, a patient with NPM1-mutated AML who received VEN-AZA will have a favorable prognosis if TP53, KRAS, NRAS and FLT3-ITD are negative, while their prognosis will be intermediate in case of positivity of FLT3-ITD, KRAS and/or NRAS (Table 1) [78,79,80].

Table 1.
Optimal treatment for an NPM1-mutated AML in 2025 based on the mutational patient profile. The reader should see the detailed text for dosing regimens and number of cycles for induction, consolidation and the use of gemtuzumab ozogamicin (GO). The question mark (?) refers to combinations of mutations where there is no consensus regarding the prognostic classification.
NGS data play a crucial role in stratifying AML risk. Although the clinical application of these data can sometimes be contentious, they are instrumental in identifying prognostically significant mutations. As an example, AML patients with an NPM1 mutation and no WT1 mutation are stratified as favorable-risk individuals, whereas the concomitant presence of both mutations significantly affects prognosis, so such patients are categorized as adverse-risk individuals [88]. Patients stratified as adverse-risk have a reduced survival rate, and HSCT is the optimal treatment for those who are eligible.
In general, the combination of mutations in NPM1 and IDH (either IDH1 or IDH2) is considered a favorable profile, especially for those AML patients destined to be treated with VEN plus HMA [77,89]. A better OS after CR has been observed for patients harboring both mutations compared to mutated NPM1 with wild-type IDH1-2. Intriguingly, R140 IDH2 mutations are highly associated with NPM1 mutations [90]. The association of NPM1 with R140 IDH2 might justify the favorable prognosis of R140 IDH2-mutated AML, especially in patients receiving VEN-AZA [91,92]. Furthermore, the triple combination of mutated NPM1/DNMT3A/NRASG12/13 has been linked with a favorable prognosis, whereas mutated NPM1/DNMT3A/FLT3-ITD has been associated with an intermediate [77] or adverse [49,50] prognosis, as expected from the presence of FLT3-ITD.
Interestingly, genomic and clinical data from 1961 AML patients analyzed via a machine learning process have been categorized as follows: combinations of mutated NPM1/wild-type IDH1-2, mutated NPM1/mutated IDH2, mutated NPM1/wild-type cohesin/mutated NRAS and mutated NPM1/mutated cohesin/mutated FLT3-ITD were categorized as good risk with a 60–80% 4-year OS, while the combination of mutated NPM1/mutated IDH1 was classified as intermediate risk with a 40–60% 4-year OS [93]. The triple combination of mutated NPM1/mutated cohesin/mutated NRAS is extremely favorable, with a more than 80% 4-year OS, while the combination of mutated NPM1/DNMT3A/FLT3-ITD is classified as poor risk (20–40% 4-year OS) [93].
Finally, the prognosis of AML when NPM1 mutations (in the absence of FLT3-ITD mutations) co-exist with myelodysplasia-related gene (MRG) mutations (ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1 or ZRSR2) remains controversial. One recent German study found that these secondary-type mutations do not impact the favorable outcome of NPM1-mutated AML (no HSCT) [94,95], whereas another synchronous American study showed that AML patients with co-mutations in MRGs and NPM1 harbor a survival similar rate to ELN 2022 intermediate risk when treated with intensive chemotherapy without VEN and that, thus, fit patients should undergo HSCT [96].
Other large studies have also confirmed that the presence of MRG and NPM1 mutations, in the absence of FLT3-ITD, does not alter the favorable prognoses and treatment outcomes of patients with NPM1-mutated AML [97,98]. Intriguingly, Wang Y et al. found a statistically significant worse progression-free survival (PFS) and non-OS for AML patients with NPM1 and MRG mutations, even though patients with concomitant FLT3-ITD mutations exhibited OS and PFS comparable to those with MRGs [97]. Zhao D et al. also demonstrated that NPM1-mutated AML, after a previous diagnosis of MDS or MDS/MPN, is enriched in MRGs and has an inferior prognosis compared to de novo NPM1-mutated AML [99].
However, there are studies highlighting that AML patients with MRG and NPM1 mutations should be classified as intermediate-risk individuals, not as favorable-risk individuals, due to the negative impact of MRGs in the prognosis of NPM1-mutated patients [100]. In a recent series of 568 patients with NPM1-mutated AML treated with intensive chemotherapy, those harboring MRGs had significantly decreased event-free survival (EFS) and a higher probability of MRD-positive disease at the end of induction [101].
More precisely, age, DNMT3A (R882 variant), IDH1 and MRGs have been identified as unfavorable factors for EFS, while mutations in the cohesin complex and treatment with gemtuzumab ozogamicin (GO) have been described as favorable EFS factors in NPM1-mutated AML [101]. The inferior EFS associated with MRGs in NPM1-mutated patients does not remain when data from NPM1-mutated MRD analysis post-cycle 2 are included. Nevertheless, DNMT3A (R882 variant) and MYC mutations retain their adverse effect and cohesin complex mutations have a favorable effect on EFS, regardless of the NPM1-mutated MRD status [101].
The reasons for these discrepancies are unknown. A different selection of the population of patients, a different type of MRG mutation for each patient or a varied order of acquisition of these mutations might explain the conflicting results of these studies, which were derived at recognized institutions.
4. Targeted Agents
4.1. NPM1-Mutated AML and Exportin 1 (XPO1)
XPO1 is a nuclear transporter, acting as a direct carrier of NPM1 mutants from the nucleus to the cytoplasm, favoring leukemogenesis [102]. The latter causes the aberrant cytoplasmic localization of NPM1 proteins and in parallel promotes the high expression of homeobox (HOX) genes [103,104], along with the relevant co-factors MEIS1 and PBX3 [105,106,107], critical for oncogenesis [103]. Thus, XPO1 inhibition might serve as a promising targeted strategy for NPM1-mutated AMLs [103]. Novel drugs, such as selinexor or eltanexor, known as selective inhibitors of nuclear export (SINEs), are under evaluation in several clinical trials for AML patients.
Selinexor (KPT-330) administered once or twice weekly in a phase I clinical trial (NCT01607892) did not show significant clinical efficacy in NPM1-mutated AMLs. On the contrary, grade ≥ 3 toxicities, such as neutropenia, thrombocytopenia, fatigue and anorexia, were observed [108]. The above-mentioned dosing strategy for selinexor, resulting in a temporary disruption of the XPO1-NPM1 oncogenic interaction and limiting drug efficacy, received much criticism. Since it has been reported that prolonged (in contrast to intermittent) XPO1 inhibition down-regulates the HOX/MEIS axis, inducing the differentiation of the NPM1-mutated AML cells, the more frequent administration of the second-generation XPO1 inhibitor eltanexor (KPT-8602) has been proposed. Eltanexor prolonged the survival of leukemic mice, caused HOX down-regulation and induced AML differentiation in [103]. Moreover, eltanexor has the advantage of lower central nervous system (CNS) penetration with less anorexia, permitting frequent dosing and higher drug concentration, resulting in the stable inhibition of the NPM1–XPO1 interaction and promising activity preclinically [109].
Interestingly, the combination of the known B-cell lymphoma 2 (BCL2) inhibitor VEN with SINEs has shown important efficacy in preclinical models [110] and is being evaluated currently in clinical trials (NCT03955783). Even though the full mechanism of MCL1-mediated synergy between VEN and selinexor or eltanexor remains unknown, it is more likely that VEN enhances the selinexor- and eltanexor-induced DNA damage of leukemic cells, mainly through inhibiting DNA damage repair [111]. Furthermore, VEN exerts its anti-leukemic action against AML cells in synergy with selinexor in vitro by inhibiting the glycolytic leukemic cell function, along with down-regulating the DNA replication-related genes [112]. The relevant synergistic clinical trials are eagerly anticipated.
4.2. NPM1-Mutated AML and Menin-KMT2A Inhibitors
NPM1-mutated AMLs overexpress the HOX gene (HOXA9) and activate the oncogenic axis HOXA9/MEIS/CCAAT enhancer-binding protein alpha (CEBPA)/lysine methyl transferase 2 (KMT2A) [104,106,107]. KMT2A is a transcriptional regulator, binding to menin and forming the menin–KMT2A oncogenic complex, which alters HOXA9 expression [104,113,114]. As NPM1-mutated AMLs overexpress the menin-KMT2A-HOX axis, the inhibition of menin-KMT2A could potentially serve as a therapeutic target for this leukemic subset. The latter inhibition has shown efficacy in animal models [115]. In conclusion, cytoplasmic-mutated NPM1 is highly dependent on the menin–MLL interaction. Pre-leukemic and leukemic NPM1-mutated cells can also be eradicated in vivo by menin inhibitors. The links between cytoplasmic mutated NPM1 and chromatin and between cytoplasmic mutated NPM1 and MLL1 remain undetermined.
Revumenib (SNDX-5613), a menin inhibitor, has shown a 60% overall response rate (ORR) in KMT2A- or NPM1-mutated AML. The early phase I/II study (AUGMENT-101, NCT04065399) involved heavily pretreated AML patients [relapsed/refractory (R/R) AML] [116]. The NPM1-mutated AMLs were 21% of the whole study cohort, whereas 68% of the patients were diagnosed with a KMT2A-r AML. SNDX-5613 was administered orally, Q12h, in 28-day cycles. Grade 2 acute differentiation syndrome was observed in 16% of the patients and resolved with steroids and/or hydroxyurea. A grade 3 QTc prolongation was also evident in a subset of the patients [116]. These promising results were also confirmed by a longer follow-up in the AUGMENT-101 phase 2 study of revumenib in patients with R/R KMT2Ar acute leukemias [117].
Based on the results of the AUGMENT-101 clinical trial, the Food and Drug Administration (FDA) approved revumenib on 15 November 2024 in the United States for treating relapsed/refractory acute leukemia with a KMT2A translocation in adult and pediatric patients aged 1 year old and older [118]. This oral drug has not yet been approved in Europe. KMT2A abnormalities are observed in 5–15% of the cases of acute lymphoblastic leukemia (ALL) and in 3% of adult patients with AML [119]. Revumenib addresses a challenging subset of leukemias, often resistant to standard treatment. The drug achieves deep responses, maintains remission after HSCT and has a manageable safety profile (Table 2) [118].

Table 2.
Clinical trials involving AML patients with NPM1-mutated AML with promising clinical responses. Regarding toxicity, grade ≥ 3 is provided, unless otherwise stated for any-grade side effects. Abbreviations: AE: adverse events; ALL: acute lymphoblastic leukemia; allo-HCT/alloSCT: allogeneic hematopoietic cell/stem cell transplantation; AML: acute myeloid leukemia; ATRA: all-trans retinoic acid; AZA: azacitidine; BID: bis in die (twice daily); CI: confidence interval; CR: complete remission; CRc: composite complete remission; CRh: complete remission with partial hematologic recovery; CRi: complete remission with incomplete hematologic recovery; CTCAE: Common Terminology Criteria for Adverse Events; DA: Daunorubicin + Cytarabine; DL: dose level; DS: differentiation syndrome; ETO: Etoposide; FLA: Fludarabine + Cytarabine; FLT3-ITD: FMS-related tyrosine kinase 3—internal tandem duplication; GO: Gemtuzumab Ozogamicin; ITT: intention to treat; IV: intravenously; MRD: measurable residual disease; ND: newly diagnosed; NR: not reached; ORR: overall response rate; OS: overall survival; PO: per os; PS: performance status; pts: patients; q12h: every 12 h (twice daily); QD: quaque die (once daily); RP2D: recommended phase 2 dose; RR: response rate; R/R: relapsed/refractory; SC: subcutaneously; SOC: standard of care; VEN: Venetoclax.
The safety and activity of revumenib in combination with fludarabine/cytarabine (FLA) in patients with relapsed/refractory acute leukemias has been evaluated recently with encouraging results (NCT05326516) (Table 2) [120]. Furthermore, the phase I/II study of the oral combination of revumenib with Decitabine/Cedazuridine (ASTX727) and VEN (SAVE) in R/R AML is underway (NCT05360160) (Table 2) [127]. Finally, the following interesting, randomized phase III clinical trial will compare revumenib plus VEN-AZA with placebo plus VEN-AZA in NPM1-mutated or KMT2A-rearranged AML patients (NCT06652438).
Ziftomenib (KO-539) is another menin inhibitor under evaluation in the phase I/II clinical trial KOMET-001 (NCT04067336) for treating R/R AML patients [130]. Ziftomenib was dosed orally, once daily, in 28-day cycles, and the optimal dose was found to be 600 mg. A CR of 30% and an ORR of 40% were demonstrated in 20 patients with R/R NPM1-mutated AML [130]. No QTc prolongation related to the drug was observed. Acute differentiation syndrome and lung inflammation were manageable adverse events and well tolerated. Ziftomenib seems to be a promising agent for KMT2A- or NPM1-mutated AML, and ongoing evaluation is necessary in future clinical trials [130]. The interim phase 1a results from the KOMET-007 study regarding the application of Ziftomenib combined with VEN/AZA in R/R KMT2A or NPM1-mutated AML (NCT05735184) are shown in Table 2 [121]. Ziftomenib can also be combined with intensive induction (7 + 3) in ND KMT2A- or NPM1-mutated AML (NCT05735184) [131].
Other menin inhibitors such as enzomenib (DSP-5336) or bleximenib are currently being tested in KMT2A or NPM1-mutated AML. The phase 1 results of the use of enzomenib in patients with R/R acute leukemia are presented in Table 2 (NCT04988555) [125]. The specific chemical properties of enzomenib compared to other menin inhibitors minimize off-target toxicity. Bleximenib combined with intensive chemotherapy in ND AML with KMT2Ar or NPM1 alterations is evaluated in a phase 1b study (NCT05453903) (Table 2) [126]. A bleximenib RP2D dose of 100 mg twice daily with optimal safety has been recently demonstrated [132]. A phase II study of bleximenib monotherapy (NCT04811560, namely a cAMeLot-1 study) in R/R AML patients with KMT2Ar or NPM1 alterations is ongoing [132]. Moreover, a phase III, randomized, double blind, placebo-controlled clinical trial, cAMeLot-2, will evaluate the combination of bleximenib, VEN and AZA in AML patients (NCT06852222, Table 3). In general, the toxicity of menin inhibitors involves differentiation syndrome and QTc prolongation in all clinical trials and all tested lines of therapy, and they are well-known complications treated by physicians [118,125,126,130].

Table 3.
Clinical trials in NPM1-mutated AML (Clinicaltrials.gov, accessed on 1 June 2025).
Finally, in an important work, Uckelmann HJ et al. described the crucial functional role of mutant NPM1 as a direct driver of oncogenic gene expression in AML [133]. They highlighted that cytoplasmic NPM1 binds to chromatin and co-operates with the MLL complex. This provides functional insight into the mechanism of menin–MLL inhibition, which disrupts the binding between NPM1 and chromatin, in NPM-1-mutated AMLs [37,38,133].
4.3. NPM1-Mutated AML, Knockout and Knock-In Phenotype
The complete pathophysiological linkage between menin or exportin-1 inhibitors and NPM1 mutants remains largely unknown. The knockout (KO) of mutant NPM1 from AML cells, using CRISPR-Cas9 gene editing, significantly eliminates the sensitivity of the leukemic cells to menin, exportin-1 inhibitors and cytarabine [113], indicating the presence of a therapeutic window for these substances. On the other hand, the knock-in of a copy of mutant NPM1 sensitizes AML cells to treatment with cytarabine or menin inhibitors (revumenib, ziftomenib). Furthermore, treatment with pan-HDAC inhibitors (panobinostat) or WEE1 tyrosine kinase inhibitors (adavosertib) exhibits synergistic in vitro action with menin inhibitors in NPM1-mutated AMLs. The latter combination also shows important efficacy in AML xenograft models [113].
4.4. NPM1-Mutated AML, All-Trans Retinoic Acid (ATRA) and Arsenic Trioxide (ATO)
The anti-leukemic activity of all-trans retinoic acid (ATRA) in combination with chemotherapy in non-acute promyelocytic leukemia (APL) has been initially demonstrated in vitro. ATRA enhances the cytotoxic effect of cytarabine or idarubicin when administered sequentially, after the cytotoxic drug [134,135]. This sequence-dependent synergy may partly be attributed to the reduction in BCL2’s half-life, a mechanism also implicated in drug resistance in AML [134,135]. Notably, in NPM1-mutated AML, ATRA decreases NPM1 protein levels by selectively inducing apoptosis and sensitizing leukemic cells to cytarabine [136]. Furthermore, the combination of ATRA and arsenic trioxide (ATO) synergistically promotes the proteasomal degradation of the mutant NPM1 protein with an unclear mechanism, resulting in leukemic cell differentiation, growth inhibition and apoptosis in NPM1-mutated AML treated with daunorubicin [137,138,139].
It has been proposed that the synergy of ATRA and ATO reverses the characteristic disorganization of promyelocytic leukemia (PML) bodies, as also observed in all types of NPM1-mutated AML, apart from APL. ATRA and ATO induce oxidative stress in the leukemic cells, in parallel with the non-specific disruption of the stress-related pathways required for oncoprotein maintenance in NPM1-mutated AML [139].
Nevertheless, data from large, randomized trials exploring the administration of ATRA combined with intensive or non-intensive chemotherapy in NPM1-mutated AML yielded conflicting results. Some studies demonstrated clinical benefit, whereas others reported no significant impact of the drug. In the UK MRC AML12 trial, adding ATRA to induction chemotherapy in young patients (15–60 years) with NPM1-mutated AML showed no significant survival benefit [140]. Moreover, the results of the randomized AMLSG 15–10 trial exhibited an inferior outcome and decreased OS for the ATRA arm in combination with low-dose cytarabine plus etoposide in older patients (>60 years) with ND NPM1-mutated AML, making it unfit for intensive chemotherapy [123]. Contrarily, in the AMLSG 07-04 trial, adding ATRA to standard induction therapy in younger adults (18–60 years) with NPM1-mutated/FLT3-ITD-negative AML led to improved EFS and OS [141]. Similarly, adding ATRA to induction and consolidation therapy in elderly AML patients (>60 years old) improved CR rates, EFS and OS [142]. Despite the heterogeneous results, these studies highlight the biological and therapeutic rationale of ATRA in NPM1-mutated AML, supporting the continued investigation of ATRA/ATO-based strategies, possibly in combination with other targeted therapies, in this AML molecular subtype.
4.5. NPM1-Mutated AML and Vitamin C and D Supplements
The role of vitamin C and D supplementation in AML has recently been investigated. Patients benefiting the most from vitamin C and D supplementation were those diagnosed with NPM1-mutated AML in a recent study [143]. These vitamins are usually administered as supportive care from day 10 of AML induction chemotherapy until hematologic recovery from induction and consolidation treatments. Interestingly, during AML induction, a lower incidence of complications including macrophage activation syndrome or hemorrhage and a significantly lower rate of bacterial and fungal infections were observed in the vitamin C and D arms for all AML patients.
Moreover, while the aforementioned study showed no difference in OS between those who received supplementation and those who did not receive it, a subgroup analysis yielded that the risk of death was, surprisingly, nearly 50% lower among those who were positive for the NPM1 mutation and receiving vitamin C and D supplements during treatment [143]. Based on the above finding, vitamin C and D supplementation is recommended in AML patients harboring NPM1 mutations for improving their OS and in all AML patients for lowering the rates of grade 3 and 4 adverse events [143]. The possible anti-leukemic mechanisms of vitamin C and D still need to be fully elucidated.
5. Optimal Treatment Approach for NPM1-Mutated AML in 2025
5.1. General Treatment Decisions and Standard Chemotherapy Regimens
Patients with an NPM1 mutation in the absence of FLT3-ITD are classified in the favorable prognostic risk category. Those who are fit for intensive chemotherapy will receive induction treatment intravenously, usually a combination of cytarabine in a continuous 7-day infusion (100–200 mg/m2/day) with an anthracycline (either daunorubicin 60 mg/m2 or idarubicin 12 mg/m2) for 3 days (7 + 3 regimen), depending on the protocol applied at each center. In this subset of patients, HSCT in CR is not recommended due to the favorable prognosis [81]. After CR, these patients receive consolidation therapy. Maintenance therapy with oral AZA (CC-486) can be administered as monotherapy after induction treatment (with or without consolidation) for AML patients who will not be transplanted [144]. The unfit patients of the above prognostic category or those >75 years old will receive a combination of VEN plus HMA [85,86].
On the contrary, patients with an NPM1 mutation in the presence of FLT3-ITD are classified in the intermediate prognostic risk category [31], and eligible patients will undergo allogeneic HSCT after induction and/or consolidation treatment. Interestingly, AML patients with an NPM1 and a WT1 mutation belong to the adverse prognostic group [88]. Thus, a patient with an NPM1 mutation can even belong to the adverse group. This is also the case when the NPM1 mutation and adverse-risk cytogenetic abnormalities co-exist. NPM1-mutated AMLs with an FLT3-ITD mutation should also receive midostaurin (50 mg of q12h per OS) during days 8–21 of induction and in all cycles of consolidation [87]. Unfit patients at intermediate or adverse risk with NPM1-mutated AML or those >75 years old should also receive VEN plus HMA [85,86]. VEN-based combination therapies are a promising targeted approach for NPM1-mutated AML because they have been linked with durable molecular remission and increased OS [91,145].
MRD assessment should be performed after two cycles of chemotherapy and at the end of treatment [31]. There is a correlation between MRD positivity and the risk of relapse. AML patients who are MRD-positive at the end of consolidation benefit from allogeneic HSCT regarding OS. Without HSCT, they will relapse. Before transplantation, MRD evaluation should also be conducted, because positivity has been associated with poor outcomes even after HSCT [146].
Because increased CD33 expression is observed in blastic cells of NPM1-mutated AML, the anti-CD33 monoclonal antibody GO (3 mg/m2) is used in younger fit adults with this AML subtype [82,147]. This antibody is chemically linked to a calicheamicin-based cytotoxic warhead. A meta-analysis showed a clinical benefit for GO in frontline AML therapy, especially in patients with core binding factor (CBF) AML [84]. Furthermore, a reduction in the probability of relapse and greater molecular clearance of the NPM1 mutants, albeit with EFS difference, has been observed in NPM1-mutated AML [83,148]. According to these findings, based on the National Comprehensive Cancer Network (NCCN) guidelines, GO is a category 2B recommendation option for favorable-risk AMLs, such as NPM1-mutated AML. The use of GO is, therefore, recommended on days 1, 4 and 7 of induction and on day 1 of consolidation, but a single dose on day 1 of induction may also be effective [84,149,150].
The optimal number of consolidation cycles is unknown. There is a lack of prospective data supporting the four-cycle standard of care. The current opinion is that the number of cycles is linked to the dose of cytarabine. The European Leukemia Net (ELN) 2022 guidelines recommend administering three to four cycles of intermediate-dose cytarabine (IDAC, 1–1.5 g/m2 every 12 h on days 1, 2 and 3), which can be reduced to 0.5–1 g/m2 in AML patients older than 60 years old [31]. The number of cycles required for applying IDAC (1–1.5 g/m2) is a matter of debate, as insufficient evidence-based data exist. A decrease to two IDAC cycles is recommended for elderly patients. Therefore, either a dose reduction (0.5–1 g/m2, 3–4 cycles) or a cycle reduction (1–1.5 g/m2, 2 cycles) is usually applied for older AML patients [151].
The Hellenic AML National protocol (AML-HSH-2019) for AML patients aged 15–65 years old, eligible for intensive chemotherapy, recommends three consolidation cycles of IDAC (1–1.5 g/m2) in most patients after two cycles of induction treatment. An exception is the application of HiDAC (3 g/m2) in all patients < 45 years old with core binding factor (CBF) leukemias [152]. Additional dose modifications for cytarabine for the second and third consolidation cycles are applied based on the age of the patient for CBF (3 g/m2 < 45 years old, 1.5 g/m2 45–60 years old, 1 g/m2 > 60 years old) and NPM1-mutant (1.5 g/m2 < 60 years old, 1 g/m2 > 60 years old) AMLs. Moreover, a cycle of GO (3 g/m2), along with cytarabine (IDAC, 1–1.5 g/m2) for only the second induction and the first consolidation cycle, is recommended for patients with CBF or NPM1-mutant AML (the protocol is a close variation of the ELN recommendations of that time [153]).
The optimal treatment for an NPM1-mutated AML patient according to the mutational pattern is shown in Table 1.
5.2. Emerging Clinical Trials for NPM1-Mutated AML
A list of 35 clinical trials (8 randomized) is shown in Table 3 (updated 1 June 2025). Interestingly, two thirds of these clinical trials (23/35) involve menin inhibitors, either alone or in combination with other drugs, which is a rapidly evolving field in NPM1-mutated AML. The clinical trials with the most promising clinical responses in patients with NPM1-mutated AML are shown in Table 2.
The phase III, randomized, two-arm, open-label study of chemotherapy in combination with ATRA with or without GO in patients with NPM1-mutated AML (NCT00893399) yielded a statistically significant relapse reduction with GO (Table 2) [122]. Another phase III, randomized, two-arm study investigated the use of low-dose cytarabine and etoposide with or without ATRA in older patients not eligible for intensive chemotherapy with AML and NPM1 mutation (NCT01237808, Table 2) [123]. The retrospective observational clinical study conducted by Orvain C et al. evaluated the outcomes of patients with CBF leukemias and/or NPM1-mutated AML after first-line intensive chemotherapy in their first molecular relapses (NCT04931992, Table 2) [124]. Promising results that render the combination of VEN-AZA as a bridge to transplant therapy have been documented in the single-arm, phase II, non-randomized study of VEN and AZA for managing molecular relapse/progression in adult NPM1-mutated AML (NCT04867928, Table 2) [129,154]. Finally, the phase II results of the randomized study comparing VEN-AZA and intensive chemotherapy in fit patients with ND NPM1-mutated AML (VINCENT study, NCT05904106, Table 2) are eagerly anticipated [128].
6. Immunotherapies—Immunomodulatory Drugs
6.1. Checkpoint Inhibitors
Immune checkpoint molecules, such as PD-1 and its ligand PD-L1, play a pivotal role in leukemogenesis by fostering an immunosuppressive tumor microenvironment. The signal transducer and activator of transcription 5 (STAT5) is overexpressed in AML and activates the promoter of glycolytic genes to enhance glycolysis in leukemic cells. Thus, increased lactate accumulation promotes histone lactylation, thereby inducing PD-L1 transcription, another leukemogenic mechanism [155]. The presence of NPM1 mutations in AML appears to enhance responsiveness to PD-1-based immunotherapies, suggesting that the underlying genetic alterations of the disease might critically influence the effectiveness of checkpoint blockade [156,157].
Nivolumab, a monoclonal antibody targeting PD-1, has shown promising results in preclinical and early-phase clinical trials by boosting the cytotoxic activity of T cells against leukemic cells harboring NPM1 mutations. These findings suggest that the inhibition of the PD-1 signaling pathway can restore the function of exhausted T cells, thereby enhancing the immune system’s ability to eliminate leukemic progenitor cells [156,158].
Although PD-1 inhibition alone has shown limited clinical efficacy in AML, combining PD-1 blockade with other agents may enhance both anti-leukemic and immunostimulatory responses. Notably, AZA induces the up-regulation of PD-1 and PD-L1 expression in AML cells, a phenomenon linked to the development of resistance to AZA treatment. This mechanism of immune escape might be overcome by targeting PD-1 with checkpoint inhibitors, such as nivolumab and pembrolizumab [157,159].
Pembrolizumab has been evaluated mainly in combination with HMAs, such as AZA. This combination can induce molecular remission in a subset of patients with relapsed NPM1-mutated AML (NCT02845297). Furthermore, the combination of nivolumab and AZA with or without the monoclonal antibody Ipilimumab has been tested in patients with R/R or ND AML, including patients harboring NPM1 mutations, with variable responses and outcomes, along with significant toxicity (NCT02397720).
The first randomized study of an anti-PD1 antibody (pembrolizumab) added to AZA+VEN in ND AML patients ineligible for induction therapy highlighted significant toxicity. The addition of pembrolizumab to AZA plus VEN did not improve MRD-negative CR/CRi and was associated with a trend for worse outcomes (NCT04284787) [160].
In conclusion, checkpoint blockade offers a novel therapeutic avenue in NPM1-mutated AML, but further clinical trials are essential for optimizing treatment protocols, identifying predictive biomarkers and establishing long-term benefits for patients.
6.2. Lenalidomide
Lenalidomide exerts its anti-leukemic actions by activating T and natural killer (NK) cells and through inhibiting proinflammatory cytokines, such as interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNF-α). In general, the drug has anti-proliferative, anti-angiogenetic, anti-inflammatory and pro-apoptotic capacities. It also enhances anti-tumor immunity [161]. A 30% response rate has been reported for lenalidomide as a first-line treatment in older ND AML patients [162]. However, the HOVON-SAKK-132 clinical trial found limited therapeutic benefit, especially in younger and middle-aged adults [163].
Lenalidomide could be used as a potential treatment for NPM1-mutated AML, especially when combined with checkpoint inhibitors to improve leukemia-associated antigen-specific response [164]. When both these drugs are used alone as standard induction therapy, they do not have a significant effect, unless other requirements are met; the SRSF2 genotype is used for lenalidomide- [163] or biomarker-driven approaches for checkpoint inhibitors [164]. Despite the promising effect of this treatment combination, limitations and questions still arise, requiring further preclinical and clinical evaluation, along with new promising treatment combination testing. Interestingly, the immunomodulatory agent mezigdomide has shown efficacy alone and in combination with menin inhibition in preclinical models of KMT2Ar- and NPM1-mutated AML [165].
6.3. Imiquimod Analogs
Imiquimod analogs, particularly imiqualine, have emerged as promising agents in treating NPM1-mutated AML. The imiqualine EAPB0503 has shown selective potency, inducing apoptosis. The molecule promotes the proteasomal-mediated degradation of mutant NPM1, restoring the nuclear localization of normal NPM1 and suppressing leukemic cell proliferation [166].
Recently, a second-generation imiqualine, EAPB02303, has shown 200-fold greater potency and broader activity across AML subtypes, exhibiting selective efficacy against NPM1-mutated AML, making it a promising candidate for further clinical evaluation [167].
7. Unmet Needs—CAR-T Cells: Proposed Future Directions
There is an unmet need for novel investigational agents or combinations of novel agents with already well-established drugs in NPM1-mutated AMLs. The detailed description of chimeric antigen receptor (CAR)-T and T-cell receptor (TCR) therapies are investigational approaches that are beyond the scope of this review.
Due to the lack of selective target antigen overexpression on AML blasts, along with the required absence of the same antigen on normal tissues, there are hurdles limiting the clinical application of CAR-T cell therapies in AML patients, as non-specific targeting causes severe toxicities [168,169]. The cytoplasmic NPM1–HLA complex can be targeted by anti-NPM1c/HLA-A2 CAR-T or memory NK cells [170,171,172]. An important limitation of this strategy is the difficulties in triggering full T-cell stimulation. Constructing dual CAR-T cells that co-express an anti-NPM1c/HLA-A2 CAR and an anti-CD123 co-stimulatory receptor (CCR) without activating the signaling domain might offer a possible solution to the problem. CD123 is highly expressed in NPM1-mutated AML [173,174]. Moreover, Van der Lee et al. successfully transduced CD8+ and CD4+ T cells with specificity for NPM1-mutated peptides, targeting HLA A2-restricted primary leukemic blasts [172,175].
While CAR-T cell therapy is still in its early stages in AML, cytotoxic T cells may help in maintaining remission and achieving prolonged survival. They could also play a role in eradicating residual disease, especially for patients ineligible for allogeneic HSCT. In conclusion, ongoing research is essential for deciphering the complex pathophysiology of the NPM1 mutation in AML, which will lead to novel targeted therapies.
Author Contributions
M.D.D. designed the concept of the article, performed the literature review and wrote and corrected the article; M.S.V. performed the literature review, wrote parts of the manuscript and assisted in the formation of tables; A.K., S.K., V.B. and G.I. performed the literature review and assisted in writing parts of the manuscript; G.I. assisted in the formation of the figure. All authors assisted in answering questions raised by the reviewers. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| A | |
| AZA | Azacitidine |
| ALCL | Anaplastic large cell lymphoma |
| ALL | Acute Lymphoblastic Leukemia |
| AML | Acute Myeloid Leukemia |
| ANKRD26 | Ankyring Repeat Domain Containing protein 26 |
| APL | Acute promyelocytic leukemia |
| ARF | Alternative Reading Frame |
| ASXL1 | Additional Sex Combs-like 1 |
| ATO | Arsenic trioxide |
| ATRA | All-trans retinoic acid |
| B | |
| BCL-2 | B-cell lymphoma-2 |
| BCL-XL | B-cell lymphoma extra large |
| BCOR | BCL6 corepressor |
| BCORL1 | BCL6 corepressor-like 1 |
| BM | Bone Marrow |
| BRAF | V-Raf Murine Sarcoma Viral Oncogene Homolog B1 |
| C | |
| CAR | Chimeric Antigen Receptor |
| CBF | Core Binding Factor |
| CBL | Casitas B-lineage Lymphoma |
| CD | Cluster of Differentiation |
| CEBPA | CCAAT enhancer binding protein alpha |
| CH | Clonal Hematopoiesis |
| CNS | Central Nervous System |
| CR | Complete Remission |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CSF3R | Colony Stimulating Factor 3 receptor |
| D | |
| DDX41 | DEAD Box helicase 41 |
| DNA | Deoxyribonucleic acid |
| DNMT3A | DNA (cytosine-5)-methyltransferase 3A |
| E | |
| EFS | Event-Free Survival |
| ELN | European Leukemia Net |
| ETV6 | ETS variant transcription factor 6 |
| EZH2 | Enhancer of Zeste Homolog 2 |
| F | |
| FAB | French–American–British Classification |
| FDA | Food and Drug Administration |
| FLT3-ITD | FMS-like tyrosine kinase 3 internal tandem duplication |
| FLT3-TKD | FMS-like tyrosine kinase 3 tyrosine kinase domain |
| FOXM1 | Forkhead box M1 |
| G | |
| GATA2 | GATA Binding protein 2 |
| GPR56 | G protein coupled receptor 56 |
| GO | Gemtuzumab ozogamicin |
| H | |
| HDAC | Histone Deacetylase |
| HiDAC | High-Dose Cytarabine |
| HLA | Human Leukocyte Antigen |
| HLF | Hepatic Leukemia Factor |
| HMA | Hypomethylating agent |
| HSCT | Hematopoietic Stem Cell Transplantation |
| HOX | Homeobox |
| I | |
| ICC | International Consensus Classification |
| IDAC | Intermediate Dose Cytarabine |
| IDH1 | Isocitrate Dehydrogenase 1 |
| IDH 2 | Isocitrate Dehydrogenase 2 |
| IHC | Immunohistochemistry |
| J | |
| JAK2 | Janus Kinase 2 |
| K | |
| KMT2A | Lysine Methyltransferase 2A |
| KIT | KIT protooncogene, receptor tyrosine kinase |
| KPT-330 | Selinexor |
| KPT-8602 | Eltaxenor |
| KO | Knockout |
| KO-539 | Ziftomenib |
| KRAS | Kirsten rat sarcoma viral oncogene homolog |
| M | |
| MDS | Myelodysplastic Syndrome |
| MCL1 | Myeloid Cell Leukemia sequence 1 |
| MEIS1 | Myeloid Ecotropic viral Integration site 1 |
| MEIS-SMC4 | Myeloid Ecotropic viral Integration site 1-Structural Maintenance of Chromosome 4 |
| MLL | Mixed Lineage Leukemia |
| MPNs | Myeloproliferative Neoplasms |
| MRGs | Myelodysplasia related gene mutations |
| MRD | Measurable Residual Disease |
| MRD-LL | Measurable Residual Disease at Low Level |
| N | |
| NCCN | National Comprehensive Cancer Network |
| ND | Newly Diagnosed |
| NF1 | Neurofibromatosis type 1 |
| NGS | Next-Generation Sequencing |
| NHL | Non-Hodgkin lymphoma |
| NK | Natural Killer |
| NPM1 | Nucleophosmin 1 |
| NPM1wt | NPM1 wild type |
| NRAS | Neuroblastoma RAS viral oncogene homolog |
| O | |
| ORR | Overall Response Rate |
| OS | Overall Survival |
| P | |
| PB | Peripheral Blood |
| PBX3 | Pre-B-cell leukemia transcription factor 3 |
| PD-1 | Programmed (Cell) Death 1 |
| PD-L1 | Programmed (Cell) Death Ligand 1 |
| PFS | Progression-free survival |
| PHF6 | Plant Homeodomain Finger Protein 6 |
| PPM1D | Protein phosphatase magnesium-dependent 1 delta |
| PRC1 | Polycomb repressive complex 1 |
| PRC2 | Polycomb repressive complex 2 |
| PTPN11 | Protein Tyrosine Phosphatase Non-Receptor Type 11 |
| R | |
| RNA | Ribonucleic acid |
| RP2D | Recommended phase 2 dose |
| R/R AML | Relapsed or refractory acute myeloid leukemia |
| RT-qPCR | Quantitative Reverse Transcription Polymerase Chain Reaction |
| RUNX1 | Runt-related transcription factor 1 |
| S | |
| SETBP1 | Set Binding Protein 1 |
| SF3B1 | Splicing Factor 3B Subunit 1 |
| SINE | Selective Inhibitors of Nuclear Export |
| SNDX-5613 | Revumenib |
| SRSF2 | Serine/Arginine-rich Splicing Factor 2 |
| STAG2 | Stromal Antigen 2 |
| STAT5 | Signal transducer and activator of transcription 5 |
| T | |
| TCR | T-cell receptor |
| TET2 | TET methylcytosine dioxygenase 2 |
| TP53 | Tumor Protein p53 |
| U | |
| U2AF1 | U2 small nuclear RNA auxiliary factor 1 |
| V | |
| VAF | Varied Allele Frequency |
| VEN | Venetoclax |
| W | |
| WHO | World Health Organization |
| WT1 | Wilms Tumor 1 |
| X | |
| XPO1 | Exportin 1 |
| Z | |
| ZRSR2 | Zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
References
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. New Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Martelli, M.P.; Bolli, N.; Bonasso, R.; Ghia, E.; Pallotta, M.T.; Diverio, D.; Nicoletti, I.; Pacini, R.; Tabarrini, A.; et al. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood 2006, 108, 1999–2005. [Google Scholar] [CrossRef]
- Hindley, A.; Catherwood, M.A.; McMullin, M.F.; Mills, K.I. Significance of NPM1 Gene Mutations in AML. Int. J. Mol. Sci. 2021, 22, 10040. [Google Scholar] [CrossRef] [PubMed]
- Falini, B. NPM1-mutated acute myeloid leukemia: New pathogenetic and therapeutic insights and open questions. Am. J. Hematol. 2023, 98, 1452–1464. [Google Scholar] [CrossRef]
- Box, J.K.; Paquet, N.; Adams, M.N.; Boucher, D.; Bolderson, E.; O’Byrne, K.J.; Richard, D.J. Nucleophosmin: From structure and function to disease development. BMC Mol. Biol. 2016, 17, 19. [Google Scholar] [CrossRef]
- Zarka, J.; Short, N.J.; Kanagal-Shamanna, R.; Issa, G.C. Nucleophosmin 1 Mutations in Acute Myeloid Leukemia. Genes 2020, 11, 649. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Brunetti, L.; Sportoletti, P.; Martelli, M.P. NPM1-mutated acute myeloid leukemia: From bench to bedside. Blood 2020, 136, 1707–1721. [Google Scholar] [CrossRef]
- Brunetti, L.; Gundry, M.C.; Goodell, M.A. New insights into the biology of acute myeloid leukemia with mutated NPM1. Int. J. Hematol. 2019, 110, 150–160. [Google Scholar] [CrossRef]
- Cela, I.; Di Matteo, A.; Federici, L. Nucleophosmin in Its Interaction with Ligands. Int. J. Mol. Sci. 2020, 21, 4885. [Google Scholar] [CrossRef]
- Falini, B.; Brunetti, L.; Martelli, M.P. How I diagnose and treat NPM1-mutated AML. Blood 2021, 137, 589–599. [Google Scholar] [CrossRef]
- Falini, B.; Sciabolacci, S.; Falini, L.; Brunetti, L.; Martelli, M.P. Diagnostic and therapeutic pitfalls in NPM1-mutated AML: Notes from the field. Leukemia 2021, 35, 3113–3126. [Google Scholar] [CrossRef]
- Falini, B.; Martelli, M.P.; Brunetti, L. Mutant NPM1: Nuclear export and the mechanism of leukemogenesis. Am. J. Hematol. 2023, 98, 550–552. [Google Scholar] [CrossRef]
- Kunchala, P.; Kuravi, S.; Jensen, R.; McGuirk, J.; Balusu, R. When the good go bad: Mutant NPM1 in acute myeloid leukemia. Blood Rev. 2018, 32, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Angenendt, L.; Röllig, C.; Montesinos, P.; Martínez-Cuadrón, D.; Barragan, E.; García, R.; Botella, C.; Martínez, P.; Ravandi, F.; Kadia, T.; et al. Chromosomal Abnormalities and Prognosis in NPM1-Mutated Acute Myeloid Leukemia: A Pooled Analysis of Individual Patient Data From Nine International Cohorts. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 2632–2642. [Google Scholar] [CrossRef] [PubMed]
- Alpermann, T.; Schnittger, S.; Eder, C.; Dicker, F.; Meggendorfer, M.; Kern, W.; Schmid, C.; Aul, C.; Staib, P.; Wendtner, C.M.; et al. Molecular subtypes of NPM1 mutations have different clinical profiles, specific patterns of accompanying molecular mutations and varying outcomes in intermediate risk acute myeloid leukemia. Haematologica 2016, 101, e55–e58. [Google Scholar] [CrossRef]
- Bailey, G.D.; Doolan, L.; Baskar, A.; Smith, L.C.; Seedhouse, C.H. Preferential transcription of the mutated allele in NPM1 mutated acute myeloid leukaemia. Sci. Rep. 2020, 10, 17695. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Pelosi, E.; Castelli, G. Genetic, Phenotypic, and Clinical Heterogeneity of NPM1-Mutant Acute Myeloid Leukemias. Biomedicines 2023, 11, 1805. [Google Scholar] [CrossRef]
- Tregnago, C.; Benetton, M.; Padrin, D.; Polato, K.; Borella, G.; Da Ros, A.; Marchetti, A.; Porcù, E.; Del Bufalo, F.; Mecucci, C.; et al. NPM1 Mutational Status Underlines Different Biological Features in Pediatric AML. Cancers 2021, 13, 3457. [Google Scholar] [CrossRef]
- Döhner, H.; Dolnik, A.; Tang, L.; Seymour, J.F.; Minden, M.D.; Stone, R.M.; Del Castillo, T.B.; Al-Ali, H.K.; Santini, V.; Vyas, P.; et al. Cytogenetics and gene mutations influence survival in older patients with acute myeloid leukemia treated with azacitidine or conventional care. Leukemia 2018, 32, 2546–2557. [Google Scholar] [CrossRef]
- Wei, H.; Wang, Y.; Zhou, C.; Lin, D.; Liu, B.; Liu, K.; Qiu, S.; Gong, B.; Li, Y.; Zhang, G.; et al. Distinct genetic alteration profiles of acute myeloid leukemia between Caucasian and Eastern Asian population. J. Hematol. Oncol. 2018, 11, 18. [Google Scholar] [CrossRef]
- Suzuki, T.; Kiyoi, H.; Ozeki, K.; Tomita, A.; Yamaji, S.; Suzuki, R.; Kodera, Y.; Miyawaki, S.; Asou, N.; Kuriyama, K.; et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood 2005, 106, 2854–2861. [Google Scholar] [CrossRef]
- Yan, L.; Chen, S.; Liang, J.; Feng, Y.; Cen, J.; He, J.; Chang, W.; Zhu, Z.; Pan, J.; Wu, Y.; et al. Analysis of NPM1 gene mutations in Chinese adults with acute myeloid leukemia. Int. J. Hematol. 2007, 86, 143–146. [Google Scholar] [CrossRef]
- Jin, J.; Wang, J.X.; Chen, F.F.; Wu, D.P.; Hu, J.; Zhou, J.F.; Hu, J.D.; Wang, J.M.; Li, J.Y.; Huang, X.J.; et al. Homoharringtonine-based induction regimens for patients with de-novo acute myeloid leukaemia: A multicentre, open-label, randomised, controlled phase 3 trial. Lancet. Oncol. 2013, 14, 599–608. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. New Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef] [PubMed]
- Grimwade, D.; Ivey, A.; Huntly, B.J. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood 2016, 127, 29–41. [Google Scholar] [CrossRef]
- Bullinger, L.; Döhner, K.; Döhner, H. Genomics of Acute Myeloid Leukemia Diagnosis and Pathways. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Larkin, K.T.; Nicolet, D.; Kelly, B.J.; Mrózek, K.; LaHaye, S.; Miller, K.E.; Wijeratne, S.; Wheeler, G.; Kohlschmidt, J.; Blachly, J.S.; et al. High early death rates, treatment resistance, and short survival of Black adolescents and young adults with AML. Blood Adv. 2022, 6, 5570–5581. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, B.; Kohlschmidt, J.; Mrózek, K.; Zhao, Q.; Fisher, J.L.; Nicolet, D.; Walker, C.J.; Mims, A.S.; Oakes, C.; Giacopelli, B.; et al. Poor Survival and Differential Impact of Genetic Features of Black Patients with Acute Myeloid Leukemia. Cancer Discov. 2021, 11, 626–637. [Google Scholar] [CrossRef]
- Tazi, Y.; Arango-Ossa, J.E.; Zhou, Y.; Bernard, E.; Thomas, I.; Gilkes, A.; Freeman, S.; Pradat, Y.; Johnson, S.J.; Hills, R.; et al. Unified classification and risk-stratification in Acute Myeloid Leukemia. Nat. Commun. 2022, 13, 4622. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating morphologic, clinical, and genomic data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef] [PubMed]
- Chandra, D.J.; Lachowiez, C.A.; Loghavi, S. Practical considerations in clinical application of WHO 5th and ICC classification schemes for acute myeloid leukemia. Blood Rev. 2023, 64, 101156. [Google Scholar] [CrossRef] [PubMed]
- Shimony, S.; Stahl, M.; Stone, R.M. Acute myeloid leukemia: 2023 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2023, 98, 502–526. [Google Scholar] [CrossRef]
- Vassiliou, G.S.; Cooper, J.L.; Rad, R.; Li, J.; Rice, S.; Uren, A.; Rad, L.; Ellis, P.; Andrews, R.; Banerjee, R.; et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat. Genet. 2011, 43, 470–475. [Google Scholar] [CrossRef]
- Muranyi, A.; Ammer, T.; Kechter, A.; Rawat, V.P.S.; Sinha, A.; Gonzalez-Menendez, I.; Quintanilla-Martinez, L.; Azoitei, A.; Günes, C.; Mupo, A.; et al. Npm1 haploinsufficiency in collaboration with MEIS1 is sufficient to induce AML in mice. Blood Adv. 2023, 7, 351–364. [Google Scholar] [CrossRef]
- Uckelmann, H.J.; Haarer, E.L.; Takeda, R.; Wong, E.M.; Hatton, C.; Marinaccio, C.; Perner, F.; Rajput, M.; Antonissen, N.J.C.; Wen, Y.; et al. Mutant NPM1 Directly Regulates Oncogenic Transcription in Acute Myeloid Leukemia. Cancer Discov. 2023, 13, 746–765. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Evans, K.; Gadrey, J.Y.; Eschle, B.K.; Hatton, C.; Uckelmann, H.J.; Ross, K.N.; Perner, F.; Olsen, S.N.; Pritchard, T.; et al. A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL-Rearranged Leukemia. Cancer Cell 2019, 36, 660–673.e611. [Google Scholar] [CrossRef]
- Welch, J.S.; Ley, T.J.; Link, D.C.; Miller, C.A.; Larson, D.E.; Koboldt, D.C.; Wartman, L.D.; Lamprecht, T.L.; Liu, F.; Xia, J.; et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012, 150, 264–278. [Google Scholar] [CrossRef]
- Zuckerman, T.; Rowe, J.M. Shifting therapeutic paradigms in induction and consolidation for older adults with acute myeloid leukemia. Curr. Opin. Hematol. 2019, 26, 51–57. [Google Scholar] [CrossRef]
- Dillon, R.; Potter, N.; Freeman, S.; Russell, N. How we use molecular minimal residual disease (MRD) testing in acute myeloid leukaemia (AML). Br. J. Haematol. 2021, 193, 231–244. [Google Scholar] [CrossRef]
- Heuser, M.; Freeman, S.D.; Ossenkoppele, G.J.; Buccisano, F.; Hourigan, C.S.; Ngai, L.L.; Tettero, J.M.; Bachas, C.; Baer, C.; Béné, M.C.; et al. 2021 Update on MRD in acute myeloid leukemia: A consensus document from the European LeukemiaNet MRD Working Party. Blood 2021, 138, 2753–2767. [Google Scholar] [CrossRef] [PubMed]
- Heuser, M.; Shahswar, R. Mutation- and MRD-informed treatment decisions for the transplant-eligible AML patient. Hematol. Am. Soc. Hematol. Educ. Program 2024, 2024, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Cloos, J.; Ngai, L.L.; Heuser, M. Understanding differential technologies for detection of MRD and how to incorporate into clinical practice. Hematol. Am. Soc. Hematol. Educ. Program 2023, 2023, 682–690. [Google Scholar] [CrossRef]
- Othman, J.; Tiong, I.S.; O’Nions, J.; Dennis, M.; Mokretar, K.; Ivey, A.; Austin, M.; Latif, A.L.; Amer, M.; Chan, W.Y.; et al. Molecular MRD is strongly prognostic in patients with NPM1-mutated AML receiving venetoclax-based nonintensive therapy. Blood 2024, 143, 336–341. [Google Scholar] [CrossRef]
- Sahasrabudhe, K.D.; Mims, A.S. MRD in AML: Who, what, when, where, and how? Blood 2024, 143, 296–298. [Google Scholar] [CrossRef]
- Sango, J.; Carcamo, S.; Sirenko, M.; Maiti, A.; Mansour, H.; Ulukaya, G.; Tomalin, L.E.; Cruz-Rodriguez, N.; Wang, T.; Olszewska, M.; et al. RAS-mutant leukaemia stem cells drive clinical resistance to venetoclax. Nature 2024, 636, 241–250. [Google Scholar] [CrossRef]
- Khan, I.; Gartel, A.L. The antagonistic duality of NPM1 mutations in AML. Blood Adv. 2022, 6, 4028–4030. [Google Scholar] [CrossRef]
- Garg, S.; Reyes-Palomares, A.; He, L.; Bergeron, A.; Lavallée, V.P.; Lemieux, S.; Gendron, P.; Rohde, C.; Xia, J.; Jagdhane, P.; et al. Hepatic leukemia factor is a novel leukemic stem cell regulator in DNMT3A, NPM1, and FLT3-ITD triple-mutated AML. Blood 2019, 134, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zeng, Z.; Li, X.; Qin, W.; Cai, X.; Chen, S.; Lu, X. Clinical features and prognostic significance of DNMT3A, FLT3, and NPM1 mutations in de novo acute myeloid leukemia patients. Int. J. Lab. Hematol. 2023, 45, 899–907. [Google Scholar] [CrossRef]
- Thiede, C.; Koch, S.; Creutzig, E.; Steudel, C.; Illmer, T.; Schaich, M.; Ehninger, G. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood 2006, 107, 4011–4020. [Google Scholar] [CrossRef] [PubMed]
- Haferlach, C.; Mecucci, C.; Schnittger, S.; Kohlmann, A.; Mancini, M.; Cuneo, A.; Testoni, N.; Rege-Cambrin, G.; Santucci, A.; Vignetti, M.; et al. AML with mutated NPM1 carrying a normal or aberrant karyotype show overlapping biologic, pathologic, immunophenotypic, and prognostic features. Blood 2009, 114, 3024–3032. [Google Scholar] [CrossRef]
- Liu, J.; Han, W.; Cai, X.; Wang, Z.; Cao, L.; Hua, H.; Jia, Z.; Chao, H.; Lu, X.; Shen, H. Molecular genetic and clinical characterization of acute myeloid leukemia with trisomy 8 as the sole chromosome abnormality. Hematology 2022, 27, 565–574. [Google Scholar] [CrossRef]
- Larsson, N.; Lilljebjörn, H.; Lassen, C.; Johansson, B.; Fioretos, T. Myeloid malignancies with acquired trisomy 21 as the sole cytogenetic change are clinically highly variable and display a heterogeneous pattern of copy number alterations and mutations. Eur. J. Haematol. 2012, 88, 136–143. [Google Scholar] [CrossRef]
- Kayser, S.; Martínez-Cuadrón, D.; Hanoun, M.; Stölzel, F.; Gil, C.; Reinhardt, H.C.; Aguiar, E.; Schäfer-Eckart, K.; Burgues, J.M.B.; Steffen, B.; et al. Characteristics and outcome of patients with acute myeloid leukemia and trisomy 4. Haematologica 2023, 108, 34–41. [Google Scholar] [CrossRef]
- Herold, T.; Metzeler, K.H.; Vosberg, S.; Hartmann, L.; Jurinovic, V.; Opatz, S.; Konstandin, N.P.; Schneider, S.; Zellmeier, E.; Ksienzyk, B.; et al. Acute myeloid leukemia with del(9q) is characterized by frequent mutations of NPM1, DNMT3A, WT1 and low expression of TLE4. Genes Chromosomes Cancer 2017, 56, 75–86. [Google Scholar] [CrossRef]
- Shreenivas, A.; Janku, F.; Gouda, M.A.; Chen, H.Z.; George, B.; Kato, S.; Kurzrock, R. ALK fusions in the pan-cancer setting: Another tumor-agnostic target? NPJ Precis. Oncol. 2023, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.S.; Ali, S.M.; Fasan, O.; Block, J.; Pal, S.; Elvin, J.A.; Schrock, A.B.; Suh, J.; Nozad, S.; Kim, S.; et al. ALK Fusions in a Wide Variety of Tumor Types Respond to Anti-ALK Targeted Therapy. Oncologist 2017, 22, 1444–1450. [Google Scholar] [CrossRef] [PubMed]
- Maxson, J.E.; Davare, M.A.; Luty, S.B.; Eide, C.A.; Chang, B.H.; Loriaux, M.M.; Tognon, C.E.; Bottomly, D.; Wilmot, B.; McWeeney, S.K.; et al. Therapeutically Targetable ALK Mutations in Leukemia. Cancer Res. 2015, 75, 2146–2150. [Google Scholar] [CrossRef]
- Shekar, M.; Llaurador Caraballo, G.; Punia, J.N.; Curry, C.V.; Fisher, K.E.; Redell, M.S. ALK Fusion in an Adolescent with Acute Myeloid Leukemia: A Case Report and Review of the Literature. Biomedicines 2023, 11, 1842. [Google Scholar] [CrossRef]
- Manselle, M.K.; Ries, R.E.; Hylkema, T.; Leonti, A.; Kirkey, D.C.; Furlan, S.N.; Meshinchi, S. Functional consequence and therapeutic targeting of cryptic ALK fusions in monosomy 7 acute myeloid leukemia. Pediatr. Blood Cancer 2023, 70, e30180. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Liesveld, J.L. NPM 1 Mutations in AML-The Landscape in 2023. Cancers 2023, 15, 1177. [Google Scholar] [CrossRef]
- Chen, W.; Konoplev, S.; Medeiros, L.J.; Koeppen, H.; Leventaki, V.; Vadhan-Raj, S.; Jones, D.; Kantarjian, H.M.; Falini, B.; Bueso-Ramos, C.E. Cuplike nuclei (prominent nuclear invaginations) in acute myeloid leukemia are highly associated with FLT3 internal tandem duplication and NPM1 mutation. Cancer 2009, 115, 5481–5489. [Google Scholar] [CrossRef] [PubMed]
- Naseem, S.; Binota, J.; Varma, N.; Virk, H.; Varma, S.; Malhotra, P. NPM1 and FLT3-ITD/TKD Gene Mutations in Acute Myeloid Leukemia. Int. J. Hematol.-Oncol. Stem Cell Res. 2021, 15, 15–26. [Google Scholar] [CrossRef]
- Cheng, Z.; Hu, K.; Tian, L.; Dai, Y.; Pang, Y.; Cui, W.; Zhao, H.; Qin, T.; Han, Y.; Hu, N.; et al. Clinical and biological implications of mutational spectrum in acute myeloid leukemia of FAB subtypes M4 and M5. Cancer Gene Ther. 2018, 25, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, B.; Wu, W.; Liu, X.; Li, H. The correlation of next-generation sequencing-based genotypic profiles with clinicopathologic characteristics in NPM1-mutated acute myeloid leukemia. BMC Cancer 2021, 21, 788. [Google Scholar] [CrossRef]
- De Propris, M.S.; Raponi, S.; Diverio, D.; Milani, M.L.; Meloni, G.; Falini, B.; Foà, R.; Guarini, A. High CD33 expression levels in acute myeloid leukemia cells carrying the nucleophosmin (NPM1) mutation. Haematologica 2011, 96, 1548–1551. [Google Scholar] [CrossRef]
- Martelli, M.P.; Manes, N.; Pettirossi, V.; Liso, A.; Pacini, R.; Mannucci, R.; Zei, T.; Bolli, N.; di Raimondo, F.; Specchia, G.; et al. Absence of nucleophosmin leukaemic mutants in B and T cells from AML with NPM1 mutations: Implications for the cell of origin of NPMc+ AML. Leukemia 2008, 22, 195–198. [Google Scholar] [CrossRef]
- Albiero, E.; Madeo, D.; Bolli, N.; Giaretta, I.; Bona, E.D.; Martelli, M.F.; Nicoletti, I.; Rodeghiero, F.; Falini, B. Identification and functional characterization of a cytoplasmic nucleophosmin leukaemic mutant generated by a novel exon-11 NPM1 mutation. Leukemia 2007, 21, 1099–1103. [Google Scholar] [CrossRef]
- Martelli, M.P.; Rossi, R.; Venanzi, A.; Meggendorfer, M.; Perriello, V.M.; Martino, G.; Spinelli, O.; Ciurnelli, R.; Varasano, E.; Brunetti, L.; et al. Novel NPM1 exon 5 mutations and gene fusions leading to aberrant cytoplasmic nucleophosmin in AML. Blood 2021, 138, 2696–2701. [Google Scholar] [CrossRef]
- Mariano, A.R.; Colombo, E.; Luzi, L.; Martinelli, P.; Volorio, S.; Bernard, L.; Meani, N.; Bergomas, R.; Alcalay, M.; Pelicci, P.G. Cytoplasmic localization of NPM in myeloid leukemias is dictated by gain-of-function mutations that create a functional nuclear export signal. Oncogene 2006, 25, 4376–4380. [Google Scholar] [CrossRef] [PubMed]
- Chua, C.C.; Anstee, N.; Flensburg, C.; Teh, C.; Ivey, A.; Amin, N.; Thijssen, R.; Bohlander, S.K.; Kakadia, P.M.; Xu, Z.; et al. Venetoclax has potent efficacy in NPM1 mutated AML with acquired resistance associated with either perturbed pro-survival signalling or NPM1 wild-type populations. Blood 2023, 142 (Suppl. S1), 423–425. [Google Scholar] [CrossRef]
- Chua, C.C.; Loo, S.; Fong, C.Y.; Ting, S.B.; Tiong, I.S.; Fleming, S.; Anstee, N.S.; Ivey, A.; Ashby, M.; Teh, T.C.; et al. Final analysis of the phase 1b Chemotherapy and Venetoclax in Elderly Acute Myeloid Leukemia Trial (CAVEAT). Blood Adv. 2025, 9, 1827–1835. [Google Scholar] [CrossRef]
- Fabozzi, F.; Mastronuzzi, A. Genetic Predisposition to Hematologic Malignancies in Childhood and Adolescence. Mediterr. J. Hematol. Infect. Dis. 2023, 15, e2023032. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Long, N.; Anekpuritanang, T.; Bottomly, D.; Savage, J.C.; Lee, T.; Solis-Ruiz, J.; Borate, U.; Wilmot, B.; Tognon, C.; et al. Identification and prioritization of myeloid malignancy germline variants in a large cohort of adult patients with AML. Blood 2022, 139, 1208–1221. [Google Scholar] [CrossRef]
- Cheloor Kovilakam, S.; Gu, M.; Dunn, W.G.; Marando, L.; Barcena, C.; Nik-Zainal, S.; Mohorianu, I.; Kar, S.P.; Fabre, M.A.; Quiros, P.M.; et al. Prevalence and significance of DDX41 gene variants in the general population. Blood 2023, 142, 1185–1192. [Google Scholar] [CrossRef]
- Lachowiez, C.A.; DiNardo, C.D.; Morita, K.; Furudate, K.; Wang, F.; Tanaka, T.; Wang, S.A.; Kadia, T.M.; Daver, N.; Short, N.J.; et al. Longitudinal Next Generation Sequencing Reveals the Clonal Hierarchy of IDH Mutated Clones and Impact on Survival in NPM1 Mutated AML. Blood ASH Annu. Meet. Abstr. 2021, 138 (Suppl. S1), 607. [Google Scholar] [CrossRef]
- Döhner, H.; DiNardo, C.D.; Appelbaum, F.R.; Craddock, C.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; Larson, R.A.; et al. Genetic risk classification for adults with AML receiving less-intensive therapies: The 2024 ELN recommendations. Blood 2024, 144, 2169–2173. [Google Scholar] [CrossRef]
- Döhner, H.; Pratz, K.W.; DiNardo, C.D.; Wei, A.H.; Jonas, B.A.; Pullarkat, V.A.; Thirman, M.J.; Récher, C.; Schuh, A.C.; Babu, S.; et al. Genetic risk stratification and outcomes among treatment-naive patients with AML treated with venetoclax and azacitidine. Blood 2024, 144, 2211–2222. [Google Scholar] [CrossRef]
- Lachowiez, C.A.; Ravikumar, V.I.; Othman, J.; O’Nions, J.; Peters, D.T.; McMahon, C.; Swords, R.; Cook, R.; Saultz, J.N.; Tyner, J.W.; et al. Refined ELN 2024 risk stratification improves survival prognostication following venetoclax-based therapy in AML. Blood 2024, 144, 2788–2792. [Google Scholar] [CrossRef]
- Pratcorona, M.; Brunet, S.; Nomdedéu, J.; Ribera, J.M.; Tormo, M.; Duarte, R.; Escoda, L.; Guàrdia, R.; Queipo de Llano, M.P.; Salamero, O.; et al. Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: Relevance to post-remission therapy. Blood 2013, 121, 2734–2738. [Google Scholar] [CrossRef] [PubMed]
- Castaigne, S.; Pautas, C.; Terré, C.; Raffoux, E.; Bordessoule, D.; Bastie, J.N.; Legrand, O.; Thomas, X.; Turlure, P.; Reman, O.; et al. Effect of gemtuzumab ozogamicin on survival of adult patients with de-novo acute myeloid leukaemia (ALFA-0701): A randomised, open-label, phase 3 study. Lancet 2012, 379, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Schlenk, R.F.; Paschka, P.; Krzykalla, J.; Weber, D.; Kapp-Schwoerer, S.; Gaidzik, V.I.; Leis, C.; Fiedler, W.; Kindler, T.; Schroeder, T.; et al. Gemtuzumab Ozogamicin in NPM1-Mutated Acute Myeloid Leukemia: Early Results From the Prospective Randomized AMLSG 09-09 Phase III Study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 623–632. [Google Scholar] [CrossRef]
- Hills, R.K.; Castaigne, S.; Appelbaum, F.R.; Delaunay, J.; Petersdorf, S.; Othus, M.; Estey, E.H.; Dombret, H.; Chevret, S.; Ifrah, N.; et al. Addition of gemtuzumab ozogamicin to induction chemotherapy in adult patients with acute myeloid leukaemia: A meta-analysis of individual patient data from randomised controlled trials. Lancet Oncol. 2014, 15, 986–996. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Jonas, B.A.; Pullarkat, V.; Thirman, M.J.; Garcia, J.S.; Wei, A.H.; Konopleva, M.; Döhner, H.; Letai, A.; Fenaux, P.; et al. Azacitidine and Venetoclax in Previously Untreated Acute Myeloid Leukemia. New Engl. J. Med. 2020, 383, 617–629. [Google Scholar] [CrossRef]
- Pratz, K.W.; Jonas, B.A.; Pullarkat, V.A.; Thirman, M.J.; Garcia, J.S.; Fiedler, W.; Yamamoto, K.; Wang, J.; Yoon, S.S.; Wolach, O.; et al. Long-Term Follow-up of the Phase 3 Viale-a Clinical Trial of Venetoclax Plus Azacitidine for Patients with Untreated Acute Myeloid Leukemia Ineligible for Intensive Chemotherapy. Blood ASH Annu. Meet. Abstr. 2022, 140 (Suppl. S1), 529–531. [Google Scholar] [CrossRef]
- Stone, R.M.; Mandrekar, S.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D.; Thiede, C.; Prior, T.W.; Döhner, K.; Marcucci, G.; et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. New Engl. J. Med. 2017, 377, 454–464. [Google Scholar] [CrossRef]
- Eisfeld, A.K.; Kohlschmidt, J.; Mims, A.; Nicolet, D.; Walker, C.J.; Blachly, J.S.; Carroll, A.J.; Papaioannou, D.; Kolitz, J.E.; Powell, B.E.; et al. Additional gene mutations may refine the 2017 European LeukemiaNet classification in adult patients with de novo acute myeloid leukemia aged <60 years. Leukemia 2020, 34, 3215–3227. [Google Scholar] [CrossRef]
- Lachowiez, C.A.; Reville, P.K.; Kantarjian, H.; Jabbour, E.; Borthakur, G.; Daver, N.; Issa, G.; Furudate, K.; Tanaka, T.; Pierce, S.; et al. Contemporary outcomes in IDH-mutated acute myeloid leukemia: The impact of co-occurring NPM1 mutations and venetoclax-based treatment. Am. J. Hematol. 2022, 97, 1443–1452. [Google Scholar] [CrossRef]
- Green, C.L.; Evans, C.M.; Zhao, L.; Hills, R.K.; Burnett, A.K.; Linch, D.C.; Gale, R.E. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood 2011, 118, 409–412. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Tiong, I.S.; Quaglieri, A.; MacRaild, S.; Loghavi, S.; Brown, F.C.; Thijssen, R.; Pomilio, G.; Ivey, A.; Salmon, J.M.; et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood 2020, 135, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Fruchtman, H.; Avigan, Z.M.; Waksal, J.A.; Brennan, N.; Mascarenhas, J.O. Management of isocitrate dehydrogenase 1/2 mutated acute myeloid leukemia. Leukemia 2024, 38, 927–935. [Google Scholar] [CrossRef]
- Fleming, S.; Tsai, C.H.; Döhner, H.; Döhner, K.; Papaemmanuil, E.; Tien, H.F.; Reynolds, J.; Wei, A.H.; Hou, H.A. Machine Learning of Genomic Factors in 1,961 Patients with Acute Myeloid Leukemia Identifies Patients with Very Good or Very Poor Prognosis Who Do Not Benefit from Allogeneic Hematopoietic Cell Transplant in First Remission. Blood ASH Annu. Meet. Abstr. 2021, 138 (Suppl. S1), 225. [Google Scholar] [CrossRef]
- Bill, M.; Eckardt, J.N.; Rausch, C.; Metzeler, K.H.; Spiekermann, K.; Stasik, S.; Sauer, T.; Scholl, S.; Hochhaus, A.; Crysandt, M.; et al. Secondary-type mutations do not impact the favorable outome of NPM1-mutated acute myeloid leukemia patients-results from a large cohort of intensively treated patients. Blood 2023, 142 (Suppl. S1), 721–724. [Google Scholar] [CrossRef]
- Eckardt, J.N.; Bill, M.; Rausch, C.; Metzeler, K.; Spiekermann, K.; Stasik, S.; Sauer, T.; Scholl, S.; Hochhaus, A.; Crysandt, M.; et al. Secondary-type mutations do not impact outcome in NPM1-mutated acute myeloid leukemia-implications for the European LeukemiaNet risk classification. Leukemia 2023, 37, 2282–2285. [Google Scholar] [CrossRef]
- Lachowiez, C.A.; Asimonitis, G.; Bernard, E.; Tang, I.; Tazi, Y.; Gilkes, A.; Thomas, I.; Bullinger, L.; Dohner, K.; Dohner, H.; et al. Influence of bone marrow blast enumeration and co-occuring myelodysplasia related gene mutations in NPM1-mutated myeloid malignancies. Blood 2023, 142 (Suppl. S1), 818–820. [Google Scholar] [CrossRef]
- Wang, Y.; Quesada, A.E.; Zuo, Z.; Medeiros, L.J.; Yin, C.C.; Li, S.; Xu, J.; Borthakur, G.; Li, Y.; Yang, C.; et al. The Impact of Mutation of Myelodysplasia-Related Genes in De Novo Acute Myeloid Leukemia Carrying NPM1 Mutation. Cancers 2022, 15, 198. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.F.; Pozdnyakova, O.; Hasserjian, R.P.; Aggarwal, N.; Shaver, A.C.; Weinberg, O.K.; Irlmeier, R.; Koyama, T.; Seegmiller, A.C.; Strickland, S.A.; et al. Secondary-type mutations do not impact prognosis in acute myelogenous leukemia AML with mutated NPM1. Am. J. Hematol. 2022, 97, E462–E465. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zarif, M.; Eladl, E.; Capo-Chichi, J.M.; Smith, A.C.; Atenafu, E.G.; Tierens, A.; Minden, M.D.; Schuh, A.; Chang, H. NPM1-mutated AML-MRC diagnosed on the basis of history of MDS or MDS/MPN frequently harbours secondary-type mutations and confers inferior outcome compared to AML with mutated NPM1. Leuk. Res. 2022, 118, 106869. [Google Scholar] [CrossRef]
- Mrózek, K.; Kohlschmidt, J.; Blachly, J.S.; Nicolet, D.; Carroll, A.J.; Archer, K.J.; Mims, A.S.; Larkin, K.T.; Orwick, S.; Oakes, C.C.; et al. Outcome prediction by the 2022 European LeukemiaNet genetic-risk classification for adults with acute myeloid leukemia: An Alliance study. Leukemia 2023, 37, 788–798. [Google Scholar] [CrossRef]
- Cocciardi, S.; Saadati, M.; Weiß, N.; Späth, D.; Kapp-Schwoerer, S.; Schneider, I.; Meid, A.; Gaidzik, V.I.; Skambraks, S.; Fiedler, W.; et al. Impact of myelodysplasia-related and additional gene mutations in intensively treated patients with NPM1-mutated AML. Hemasphere 2025, 9, e70060. [Google Scholar] [CrossRef] [PubMed]
- Stade, K.; Ford, C.S.; Guthrie, C.; Weis, K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 1997, 90, 1041–1050. [Google Scholar] [CrossRef]
- Pianigiani, G.; Gagliardi, A.; Mezzasoma, F.; Rocchio, F.; Tini, V.; Bigerna, B.; Sportoletti, P.; Caruso, S.; Marra, A.; Peruzzi, S.; et al. Prolonged XPO1 inhibition is essential for optimal antileukemic activity in NPM1-mutated AML. Blood Adv. 2022, 6, 5938–5949. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, R.; Pianigiani, G.; Sciabolacci, S.; Perriello, V.M.; Marra, A.; Cardinali, V.; Pierangeli, S.; Milano, F.; Gionfriddo, I.; Brunetti, L.; et al. Current status and future perspectives in targeted therapy of NPM1-mutated AML. Leukemia 2022, 36, 2351–2367. [Google Scholar] [CrossRef] [PubMed]
- Alcalay, M.; Tiacci, E.; Bergomas, R.; Bigerna, B.; Venturini, E.; Minardi, S.P.; Meani, N.; Diverio, D.; Bernard, L.; Tizzoni, L.; et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood 2005, 106, 899–902. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, B.; Mao, F.; Xu, J.; Miao, H.; Zou, Z.; Phuc Khoa, L.T.; Jang, Y.; Cai, S.; Witkin, M.; et al. HOXA9 Reprograms the Enhancer Landscape to Promote Leukemogenesis. Cancer Cell 2018, 34, 643–658.e645. [Google Scholar] [CrossRef]
- Chen, S.L.; Qin, Z.Y.; Hu, F.; Wang, Y.; Dai, Y.J.; Liang, Y. The Role of the HOXA Gene Family in Acute Myeloid Leukemia. Genes 2019, 10, 621. [Google Scholar] [CrossRef]
- Garzon, R.; Savona, M.; Baz, R.; Andreeff, M.; Gabrail, N.; Gutierrez, M.; Savoie, L.; Mau-Sorensen, P.M.; Wagner-Johnston, N.; Yee, K.; et al. A phase 1 clinical trial of single-agent selinexor in acute myeloid leukemia. Blood 2017, 129, 3165–3174. [Google Scholar] [CrossRef]
- Etchin, J.; Montero, J.; Berezovskaya, A.; Le, B.T.; Kentsis, A.; Christie, A.L.; Conway, A.S.; Chen, W.C.; Reed, C.; Mansour, M.R.; et al. Activity of a selective inhibitor of nuclear export, selinexor (KPT-330), against AML-initiating cells engrafted into immunosuppressed NSG mice. Leukemia 2016, 30, 190–199. [Google Scholar] [CrossRef]
- Fischer, M.A.; Friedlander, S.Y.; Arrate, M.P.; Chang, H.; Gorska, A.E.; Fuller, L.D.; Ramsey, H.E.; Kashyap, T.; Argueta, C.; Debler, S.; et al. Venetoclax response is enhanced by selective inhibitor of nuclear export compounds in hematologic malignancies. Blood Adv. 2020, 4, 586–598. [Google Scholar] [CrossRef]
- Yu, H.; Wu, S.; Liu, S.; Li, X.; Gai, Y.; Lin, H.; Wang, Y.; Edwards, H.; Ge, Y.; Wang, G. Venetoclax enhances DNA damage induced by XPO1 inhibitors: A novel mechanism underlying the synergistic antileukaemic effect in acute myeloid leukaemia. J. Cell Mol. Med. 2022, 26, 2646–2657. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, Y.; Liu, D.; Wang, X.; Zhu, Y.; Tong, J.; Chen, E.; Xue, L.; Zhao, N.; Liang, T.; et al. Selinexor Synergistically Promotes the Antileukemia Activity of Venetoclax in Acute Myeloid Leukemia by Inhibiting Glycolytic Function and Downregulating the Expression of DNA Replication Genes. Immunotargets Ther. 2023, 12, 135–147. [Google Scholar] [CrossRef]
- Mill, C.P.; Fiskus, W.; Das, K.; Davis, J.A.; Birdwell, C.E.; Kadia, T.M.; DiNardo, C.D.; Daver, N.; Takahashi, K.; Sasaki, K.; et al. Causal linkage of presence of mutant NPM1 to efficacy of novel therapeutic agents against AML cells with mutant NPM1. Leukemia 2023, 37, 1336–1348. [Google Scholar] [CrossRef]
- Brunetti, L.; Gundry, M.C.; Sorcini, D.; Guzman, A.G.; Huang, Y.H.; Ramabadran, R.; Gionfriddo, I.; Mezzasoma, F.; Milano, F.; Nabet, B.; et al. Mutant NPM1 Maintains the Leukemic State through HOX Expression. Cancer Cell 2018, 34, 499–512.e499. [Google Scholar] [CrossRef] [PubMed]
- Klossowski, S.; Miao, H.; Kempinska, K.; Wu, T.; Purohit, T.; Kim, E.; Linhares, B.M.; Chen, D.; Jih, G.; Perkey, E.; et al. Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia. J. Clin. Investig. 2020, 130, 981–997. [Google Scholar] [CrossRef] [PubMed]
- Issa, G.C.; Aldoss, I.; DiPersio, J.F.; Cuglievan, B.; Stone, R.M.; Arellano, M.L.; Thirman, M.J.; Patel, M.R.; Dickens, D.; Shenoy, S.; et al. The Menin Inhibitor SNDX-5613 (revumenib) Leads to Durable Responses in Patients (Pts) with KMT2A-Rearranged or NPM1 Mutant AML: Updated Results of a Phase (Ph) 1 Study. Blood ASH Annu. Meet. Abstr. 2022, 140 (Suppl. S1), 150–152. [Google Scholar] [CrossRef]
- Aldoss, I.; Issa, G.C.; Blachly, J.S.; Thirman, M.J.; Mannis, G.N.; Arellano, M.L.; DiPersio, J.F.; Traer, E.; Zwaan, C.M.; Shukla, N.; et al. Updated Results and Longer Follow-up from the AUGMENT-101 Phase 2 Study of Revumenib in All Patients with Relapsed or Refractory (R/R) KMT2Ar Acute Leukemia. Blood 2024, 144 (Suppl. S1), 211–214. [Google Scholar] [CrossRef]
- Issa, G.C.; Aldoss, I.; Thirman, M.J.; DiPersio, J.; Arellano, M.; Blachly, J.S.; Mannis, G.N.; Perl, A.; Dickens, D.S.; McMahon, C.M.; et al. Menin Inhibition With Revumenib for KMT2A-Rearranged Relapsed or Refractory Acute Leukemia (AUGMENT-101). J. Clin. Oncol. Off. J. Am. Soc. of. Clinical. Oncology 2025, 43, 75–84. [Google Scholar] [CrossRef]
- Bataller, A.; Guijarro, F.; Caye-Eude, A.; Strullu, M.; Sterin, A.; Molina, O.; Chevallier, P.; Zaliova, M.; Zuna, J.; Mozas, P.; et al. KMT2A-CBL rearrangements in acute leukemias: Clinical characteristics and genetic breakpoints. Blood Adv. 2021, 5, 5617–5620. [Google Scholar] [CrossRef]
- Shukla, N.; Guest, E.; Tasian, S.K.; Breese, E.; Schafer, E.; DiPersio, J.; Issa, G.C.; Silverman, L.B.; Stieglitz, E.; Pollard, J.; et al. Safety and activity of Revumenib in combination with fludarabine/cytarabine (FLA) in patients with relapsed/refractory acute leukemias (P540). HemaSphere 2024, 8, 891–893. [Google Scholar]
- Fathi, A.T.; Issa, G.C.; Wang, E.S.; Erba, H.; Kaplan Altman, J.; Balasubramanian, S.K.; Roboz, G.J.; Schiller, G.J.; McMahon, C.M.; Palmisiano, N.D.; et al. Ziftomenib Combined with Venetoclax/Azacitidine in Relapsed/Refractory NPM1-m or KMT2A-r Acute Myeloid Leukemia: Interim Phase 1a Results from KOMET-007. Blood 2024, 144 (Suppl. S1), 2880–2882. [Google Scholar] [CrossRef]
- Döhner, H.; Weber, D.; Krzykalla, J.; Fiedler, W.; Kühn, M.W.M.; Schroeder, T.; Mayer, K.; Lübbert, M.; Wattad, M.; Götze, K.; et al. Intensive chemotherapy with or without gemtuzumab ozogamicin in patients with NPM1-mutated acute myeloid leukaemia (AMLSG 09-09): A randomised, open-label, multicentre, phase 3 trial. Lancet. Haematol. 2023, 10, e495–e509. [Google Scholar] [CrossRef]
- Schlenk, R.F.; Weber, D.; Krzykalla, J.; Kindler, T.; Wulf, G.; Hertenstein, B.; Salih, H.R.; Südhoff, T.; Krauter, J.; Martens, U.; et al. Randomized phase-III study of low-dose cytarabine and etoposide + /- all-trans retinoic acid in older unfit patients with NPM1-mutated acute myeloid leukemia. Sci. Rep. 2023, 13, 14809. [Google Scholar] [CrossRef] [PubMed]
- Orvain, C.; Bertoli, S.; Peterlin, P.; Desbrosses, Y.; Dumas, P.Y.; Iat, A.; Hospital, M.A.; Carre, M.; Tavernier, E.; Riou, J.; et al. Molecular relapse after first-line intensive therapy in patients with CBF or NPM1-mutated acute myeloid leukemia-a FILO study. Leukemia 2024, 38, 1949–1957. [Google Scholar] [CrossRef] [PubMed]
- Zeidner, J.F.; Yuda, J.; Watts, J.M.; Levis, M.; Erba, H.; Fukushima, K.; Shima, T.; Palmisiano, N.D.; Wang, E.S.; Borate, U.; et al. Phase 1 Results: First-in-Human Phase 1/2 Study of the Menin-MLL Inhibitor Enzomenib (DSP-5336) in Patients with Relapsed or Refractory Acute Leukemia. Blood 2024, 144 (Suppl. S1), 213–216. [Google Scholar] [CrossRef]
- Recher, C.; O’Nions, J.; Aldoss, I.; Pierola, A.A.; Allred, A.; Alonso-Dominguez, J.M.; Barreyro, L.; Bories, P.; Curtis, M.; Daskalakis, N.; et al. Phase 1b Study of Menin-KMT2A Inhibitor Bleximenib in Combination with Intensive Chemotherapy in Newly Diagnosed Acute Myeloid Leukemia with KMT2Ar or NPM1 Alterations. Blood 2024, 144 (Suppl. S1), 215–218. [Google Scholar] [CrossRef]
- Issa, G.C.; Cuglievan, B.; Daver, N.; DiNardo, C.D.; Farhat, A.; Short, N.J.; McCall, D.; Pike, A.; Tan, S.; Kammerer, B.; et al. Phase I/II Study of the All-Oral Combination of Revumenib (SNDX-5613) with Decitabine/Cedazuridine (ASTX727) and Venetoclax (SAVE) in R/R AML. Blood 2024, 144 (Suppl. S1), 216–218. [Google Scholar] [CrossRef]
- Kretschmer, L.; Ruhnke, L.; Schliemann, C.; Fransecky, L.; Steffen, B.; Kaufmann, M.; Burchert, A.; Schmid, C.; Hanoun, M.; Sauer, T.; et al. Trial in Progress: A Randomized-Controlled Phase 2 Study Evaluating Venetoclax Plus Azacitidine Versus Intensive Chemotherapy in Adult Patients with Newly Diagnosed, NPM1-Mutated AML- the SAL/AMLCG Vincent Trial. Blood 2024, 144 (Suppl. S1), 1527.1521–1527.1523. [Google Scholar] [CrossRef]
- Sartor, C.; Candoni, A.; Piciocchi, A.; Marsili, G.; Fazi, P.; Papayannidis, C.; Paolini, S.; Cristiano, G.; Maurillo, L.; Tiribelli, M.; et al. Venetoclax and Azacitidine for Relapse Prevention in NPM1-Mutated Acute Myeloid Leukemia in Molecular Failure: Results from the Ongoing Gimema AML2521 Phase 2 Trial. Blood 2024, 144 (Suppl. S1), 2893–2894. [Google Scholar] [CrossRef]
- Erba, H.P.; Fathi, A.T.; Issa, G.C.; Altman, J.K.; Montesinos, P.; Patnaik, M.M.; Foran, J.M.; De Botton, S.; Baer, M.R.; Schiller, G.J.; et al. Update on a Phase 1/2 First-in-Human Study of the Menin-KMT2A (MLL) Inhibitor Ziftomenib (KO-539) in Patients with Relapsed or Refractory Acute Myeloid Leukemia. Blood ASH Annu. Meet. Abstr. 2022, 140 (Suppl. S1), 153–156. [Google Scholar] [CrossRef]
- Zeidan, A.M.; Wang, E.S.; Issa, G.C.; Erba, H.; Kaplan Altman, J.; Balasubramanian, S.K.; Strickland, S.A.; Roboz, G.J.; Schiller, G.J.; McMahon, C.M.; et al. Ziftomenib Combined with Intensive Induction (7+3) in Newly Diagnosed NPM1-m or KMT2A-r Acute Myeloid Leukemia: Interim Phase 1a Results from KOMET-007. Blood 2024, 144 (Suppl. S1), 214–216. [Google Scholar] [CrossRef]
- Searle, E.; Recher, C.; Abdul-Hay, M.; Abedin, S.; Aldoss, I.; Pierola, A.A.; Alonso-Dominguez, J.M.; Chevallier, P.; Cost, C.; Daskalakis, N.; et al. Bleximenib Dose Optimization and Determination of RP2D from a Phase 1 Study in Relapsed/Refractory Acute Leukemia Patients with KMT2A and NPM1 Alterations. Blood 2024, 144 (Suppl. S1), 212–214. [Google Scholar] [CrossRef]
- Uckelmann, H.J.; Kim, S.M.; Wong, E.M.; Hatton, C.; Giovinazzo, H.; Gadrey, J.Y.; Krivtsov, A.V.; Rücker, F.G.; Döhner, K.; McGeehan, G.M.; et al. Therapeutic targeting of preleukemia cells in a mouse model of NPM1 mutant acute myeloid leukemia. Science 2020, 367, 586–590. [Google Scholar] [CrossRef]
- Hu, Z.B.; Minden, M.D.; McCulloch, E.A. Phosphorylation of BCL-2 after exposure of human leukemic cells to retinoic acid. Blood 1998, 92, 1768–1775. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.B.; Minden, M.D.; McCulloch, E.A. Direct evidence for the participation of bcl-2 in the regulation by retinoic acid of the Ara-C sensitivity of leukemic stem cells. Leukemia 1995, 9, 1667–1673. [Google Scholar] [PubMed]
- Balusu, R.; Fiskus, W.; Rao, R.; Chong, D.G.; Nalluri, S.; Mudunuru, U.; Ma, H.; Chen, L.; Venkannagari, S.; Ha, K.; et al. Targeting levels or oligomerization of nucleophosmin 1 induces differentiation and loss of survival of human AML cells with mutant NPM1. Blood 2011, 118, 3096–3106. [Google Scholar] [CrossRef]
- Martelli, M.P.; Gionfriddo, I.; Mezzasoma, F.; Milano, F.; Pierangeli, S.; Mulas, F.; Pacini, R.; Tabarrini, A.; Pettirossi, V.; Rossi, R.; et al. Arsenic trioxide and all-trans retinoic acid target NPM1 mutant oncoprotein levels and induce apoptosis in NPM1-mutated AML cells. Blood 2015, 125, 3455–3465. [Google Scholar] [CrossRef]
- El Hajj, H.; Dassouki, Z.; Berthier, C.; Raffoux, E.; Ades, L.; Legrand, O.; Hleihel, R.; Sahin, U.; Tawil, N.; Salameh, A.; et al. Retinoic acid and arsenic trioxide trigger degradation of mutated NPM1, resulting in apoptosis of AML cells. Blood 2015, 125, 3447–3454. [Google Scholar] [CrossRef]
- Grant, S. ATRA and ATO team up against NPM1. Blood 2015, 125, 3369–3371. [Google Scholar] [CrossRef]
- Burnett, A.K.; Hills, R.K.; Green, C.; Jenkinson, S.; Koo, K.; Patel, Y.; Guy, C.; Gilkes, A.; Milligan, D.W.; Goldstone, A.H.; et al. The impact on outcome of the addition of all-trans retinoic acid to intensive chemotherapy in younger patients with nonacute promyelocytic acute myeloid leukemia: Overall results and results in genotypic subgroups defined by mutations in NPM1, FLT3, and CEBPA. Blood 2010, 115, 948–956. [Google Scholar] [CrossRef]
- Schlenk, R.F.; Lübbert, M.; Benner, A.; Lamparter, A.; Krauter, J.; Herr, W.; Martin, H.; Salih, H.R.; Kündgen, A.; Horst, H.A.; et al. All-trans retinoic acid as adjunct to intensive treatment in younger adult patients with acute myeloid leukemia: Results of the randomized AMLSG 07-04 study. Ann. Hematol. 2016, 95, 1931–1942. [Google Scholar] [CrossRef]
- Schlenk, R.F.; Fröhling, S.; Hartmann, F.; Fischer, J.T.; Glasmacher, A.; del Valle, F.; Grimminger, W.; Götze, K.; Waterhouse, C.; Schoch, R.; et al. Phase III study of all-trans retinoic acid in previously untreated patients 61 years or older with acute myeloid leukemia. Leukemia 2004, 18, 1798–1803. [Google Scholar] [CrossRef]
- Mouchel, P.L.; Bérard, E.; Tavitian, S.; Gadaud, N.; Vergez, F.; Rieu, J.B.; Luquet, I.; Sarry, A.; Huguet, F.; Largeaud, L.; et al. Vitamin C and D supplementation in acute myeloid leukemia. Blood Adv. 2023, 7, 6886–6897. [Google Scholar] [CrossRef] [PubMed]
- Wei, A.H.; Döhner, H.; Pocock, C.; Montesinos, P.; Afanasyev, B.; Dombret, H.; Ravandi, F.; Sayar, H.; Jang, J.H.; Porkka, K.; et al. Oral Azacitidine Maintenance Therapy for Acute Myeloid Leukemia in First Remission. New Engl. J. Med. 2020, 383, 2526–2537. [Google Scholar] [CrossRef]
- Othman, J.; Potter, N.; Ivey, A.; Tazi, Y.; Papaemmanuil, E.; Jovanovic, J.; Freeman, S.D.; Gilkes, A.; Gale, R.; Rapoz-D’Silva, T.; et al. Molecular, clinical, and therapeutic determinants of outcome in NPM1-mutated AML. Blood 2024, 144, 714–728. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.; Hills, R.; Freeman, S.; Potter, N.; Jovanovic, J.; Ivey, A.; Kanda, A.S.; Runglall, M.; Foot, N.; Valganon, M.; et al. Molecular MRD status and outcome after transplantation in NPM1-mutated AML. Blood 2020, 135, 680–688. [Google Scholar] [CrossRef]
- Lambert, J.; Pautas, C.; Terré, C.; Raffoux, E.; Turlure, P.; Caillot, D.; Legrand, O.; Thomas, X.; Gardin, C.; Gogat-Marchant, K.; et al. Gemtuzumab ozogamicin for de novo acute myeloid leukemia: Final efficacy and safety updates from the open-label, phase III ALFA-0701 trial. Haematologica 2019, 104, 113–119. [Google Scholar] [CrossRef]
- Kapp-Schwoerer, S.; Weber, D.; Corbacioglu, A.; Gaidzik, V.I.; Paschka, P.; Krönke, J.; Theis, F.; Rücker, F.G.; Teleanu, M.V.; Panina, E.; et al. Impact of gemtuzumab ozogamicin on MRD and relapse risk in patients with NPM1-mutated AML: Results from the AMLSG 09-09 trial. Blood 2020, 136, 3041–3050. [Google Scholar] [CrossRef] [PubMed]
- Burnett, A.K.; Hills, R.K.; Milligan, D.; Kjeldsen, L.; Kell, J.; Russell, N.H.; Yin, J.A.; Hunter, A.; Goldstone, A.H.; Wheatley, K. Identification of patients with acute myeloblastic leukemia who benefit from the addition of gemtuzumab ozogamicin: Results of the MRC AML15 trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 369–377. [Google Scholar] [CrossRef]
- Burnett, A.K.; Russell, N.H.; Hills, R.K.; Kell, J.; Freeman, S.; Kjeldsen, L.; Hunter, A.E.; Yin, J.; Craddock, C.F.; Dufva, I.H.; et al. Addition of gemtuzumab ozogamicin to induction chemotherapy improves survival in older patients with acute myeloid leukemia. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2012, 30, 3924–3931. [Google Scholar] [CrossRef]
- Jimenez-Chillon, C.; Dillon, R.; Russell, N. Optimal post-remission consolidation therapy in patients with AML. Acta Haematol. 2023, 147, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Mayer, R.J.; Davis, R.B.; Schiffer, C.A.; Berg, D.T.; Powell, B.L.; Schulman, P.; Omura, G.A.; Moore, J.O.; McIntyre, O.R.; Frei, E. Cancer and Leukemia Group B. Intensive postremission chemotherapy in adults with acute myeloid leukemia. New Engl. J. Med. 1994, 331, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Sartor, C.; Brunetti, L.; Audisio, E.; Cignetti, A.; Zannoni, L.; Cristiano, G.; Nanni, J.; Ciruolo, R.; Zingarelli, F.; Ottaviani, E.; et al. A venetoclax and azacitidine bridge-to-transplant strategy for NPM1-mutated acute myeloid leukaemia in molecular failure. Br. J. Haematol. 2023, 202, 599–607. [Google Scholar] [CrossRef]
- Huang, Z.W.; Zhang, X.N.; Zhang, L.; Liu, L.L.; Zhang, J.W.; Sun, Y.X.; Xu, J.Q.; Liu, Q.; Long, Z.J. STAT5 promotes PD-L1 expression by facilitating histone lactylation to drive immunosuppression in acute myeloid leukemia. Signal Transduct. Target. Ther. 2023, 8, 391. [Google Scholar] [CrossRef] [PubMed]
- Greiner, J.; Mohamed, E.; Fletcher, D.M.; Schuler, P.J.; Schrezenmeier, H.; Götz, M.; Guinn, B.A. Immunotherapeutic Potential of Mutated NPM1 for the Treatment of Acute Myeloid Leukemia. Cancers 2024, 16, 3443. [Google Scholar] [CrossRef]
- Abaza, Y.; Zeidan, A.M. Immune Checkpoint Inhibition in Acute Myeloid Leukemia and Myelodysplastic Syndromes. Cells 2022, 11, 2249. [Google Scholar] [CrossRef]
- Daver, N. Immune checkpoint inhibitors in acute myeloid leukemia. Best. Pr. Res. Clin. Haematol. 2021, 34, 101247. [Google Scholar] [CrossRef]
- Daver, N.; Basu, S.; Garcia-Manero, G.; Cortes, J.E.; Ravandi, F.; Jabbour, E.J.; Hendrickson, S.; Pierce, S.; Ning, J.; Konopleva, M.; et al. Phase IB/II Study of Nivolumab in Combination with Azacytidine (AZA) in Patients (pts) with Relapsed Acute Myeloid Leukemia (AML). Blood 2016, 128, 763. [Google Scholar] [CrossRef]
- Stempel, J.M.; Uy, G.L.; Dinner, S.N.; Gojo, I.; Reed, D.; Roy, R.; Byrd, K.P.; Yerrabothala, S.; Lai, C.E.; Doucette, K.; et al. Efficacy and Safety of Pembrolizumab Added to Azacitidine Plus Venetoclax for Patients with Acute Myeloid Leukemia: Results from an Investigator-Initiated, Multi-Center, CTEP-Sponsored Randomized, Phase II Trial (BLAST AML-2). Blood 2024, 144 (Suppl. S1), 736–738. [Google Scholar] [CrossRef]
- Fehniger, T.A.; Uy, G.L.; Trinkaus, K.; Nelson, A.D.; Demland, J.; Abboud, C.N.; Cashen, A.F.; Stockerl-Goldstein, K.E.; Westervelt, P.; DiPersio, J.F.; et al. A phase 2 study of high-dose lenalidomide as initial therapy for older patients with acute myeloid leukemia. Blood 2011, 117, 1828–1833. [Google Scholar] [CrossRef]
- Hütter-Krönke, M.L.; Fiedler, W.; Kündgen, A.; Krauter, J.; von Lilienfeld-Toal, M.; Döhner, H.; Schlenk, R.F. Continuous high dosing of lenalidomide in relapsed, refractory or older newly diagnosed acute myeloid leukemia patients not suitable for other treatment options-results from a phase I study. Haematologica 2019, 104, e63–e64. [Google Scholar] [CrossRef]
- Löwenberg, B.; Pabst, T.; Maertens, J.; Gradowska, P.; Biemond, B.J.; Spertini, O.; Vellenga, E.; Griskevicius, L.; Tick, L.W.; Jongen-Lavrencic, M.; et al. Addition of lenalidomide to intensive treatment in younger and middle-aged adults with newly diagnosed AML: The HOVON-SAKK-132 trial. Blood Adv. 2021, 5, 1110–1121. [Google Scholar] [CrossRef]
- Guinn, B.A.; Schuler, P.J.; Schrezenmeier, H.; Hofmann, S.; Weiss, J.; Bulach, C.; Götz, M.; Greiner, J. A Combination of the Immunotherapeutic Drug Anti-Programmed Death 1 with Lenalidomide Enhances Specific T Cell Immune Responses against Acute Myeloid Leukemia Cells. Int. J. Mol. Sci. 2023, 24, 9285. [Google Scholar] [CrossRef]
- Bourgeois, W.; Cutler, J.A.; Aubrey, B.J.; Wenge, D.V.; Perner, F.; Martucci, C.; Henrich, J.A.; Klega, K.; Nowak, R.P.; Donovan, K.A.; et al. Mezigdomide is effective alone and in combination with menin inhibition in preclinical models of KMT2A-r and NPM1c AML. Blood 2024, 143, 1513–1527. [Google Scholar] [CrossRef]
- Skayneh, H.; Jishi, B.; Hleihel, R.; Hamie, M.; El Hajj, R.; Deleuze-Masquefa, C.; Bonnet, P.A.; El Sabban, M.; El Hajj, H. EAPB0503, an Imidazoquinoxaline Derivative Modulates SENP3/ARF Mediated SUMOylation, and Induces NPM1c Degradation in NPM1 Mutant AML. Int. J. Mol. Sci. 2022, 23, 3421. [Google Scholar] [CrossRef]
- Makhoul, P.; Hleihel, R.; Itani, S.; Hamie, M.; Pagniagua-Gayraud, S.; Patinote, C.; Richaud, M.; Merhi, R.A.; El-Sabban, M.; Galas, S.; et al. The Novel Imiqualine EAPB02303 Is a Potent Drug for Treating Acute Myeloid Leukemia. Biomolecules 2025, 15, 741. [Google Scholar] [CrossRef]
- Mardiana, S.; Gill, S. CAR T Cells for Acute Myeloid Leukemia: State of the Art and Future Directions. Front. Oncol. 2020, 10, 697. [Google Scholar] [CrossRef]
- Fiorenza, S.; Turtle, C.J. CAR-T Cell Therapy for Acute Myeloid Leukemia: Preclinical Rationale, Current Clinical Progress, and Barriers to Success. BioDrugs 2021, 35, 281–302. [Google Scholar] [CrossRef]
- Xie, G.; Ivica, N.A.; Jia, B.; Li, Y.; Dong, H.; Liang, Y.; Brown, D.; Romee, R.; Chen, J. CAR-T cells targeting a nucleophosmin neoepitope exhibit potent specific activity in mouse models of acute myeloid leukaemia. Nat. Biomed. Eng. 2021, 5, 399–413. [Google Scholar] [CrossRef]
- Dong, H.; Ham, J.D.; Hu, G.; Xie, G.; Vergara, J.; Liang, Y.; Ali, A.; Tarannum, M.; Donner, H.; Baginska, J.; et al. Memory-like NK cells armed with a neoepitope-specific CAR exhibit potent activity against NPM1 mutated acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2022, 119, e2122379119. [Google Scholar] [CrossRef]
- Wang, R.; Xu, P.; Chang, L.L.; Zhang, S.Z.; Zhu, H.H. Targeted therapy in NPM1-mutated AML: Knowns and unknowns. Front. Oncol. 2022, 12, 972606. [Google Scholar] [CrossRef]
- Perriello, V.M.; Gionfriddo, I.; Rossi, R.; Milano, F.; Mezzasoma, F.; Marra, A.; Spinelli, O.; Rambaldi, A.; Annibali, O.; Avvisati, G.; et al. CD123 Is Consistently Expressed on NPM1-Mutated AML Cells. Cancers 2021, 13, 496. [Google Scholar] [CrossRef]
- Perriello, V.M.; Rotiroti, M.C.; Pisani, I.; Galimberti, S.; Alberti, G.; Pianigiani, G.; Ciaurro, V.; Marra, A.; Sabino, M.; Tini, V.; et al. IL-3-zetakine combined with a CD33 costimulatory receptor as a dual CAR approach for safer and selective targeting of AML. Blood Adv. 2023, 7, 2855–2871. [Google Scholar] [CrossRef]
- van der Lee, D.I.; Reijmers, R.M.; Honders, M.W.; Hagedoorn, R.S.; de Jong, R.C.; Kester, M.G.; van der Steen, D.M.; de Ru, A.H.; Kweekel, C.; Bijen, H.M.; et al. Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia. J. Clin. Investig. 2019, 129, 774–785. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).