Comprehensive Profiling of Serum Exosomes by a Multi-Omics Approach Reveals Potential Diagnostic Markers for Brain Metastasis in Lung Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Mouse Experiments
2.3. H&E Staining
2.4. Exosome Isolation
2.5. Nanoparticle Tracking Analysis (NTA)
2.6. Transmission Electron Microscopy (TEM)
2.7. Western Blot Analysis
2.8. RNA Extraction and RT-qPCR
2.9. Small RNA Library Preparation and Sequencing
2.10. In Gel Digestion and Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS)
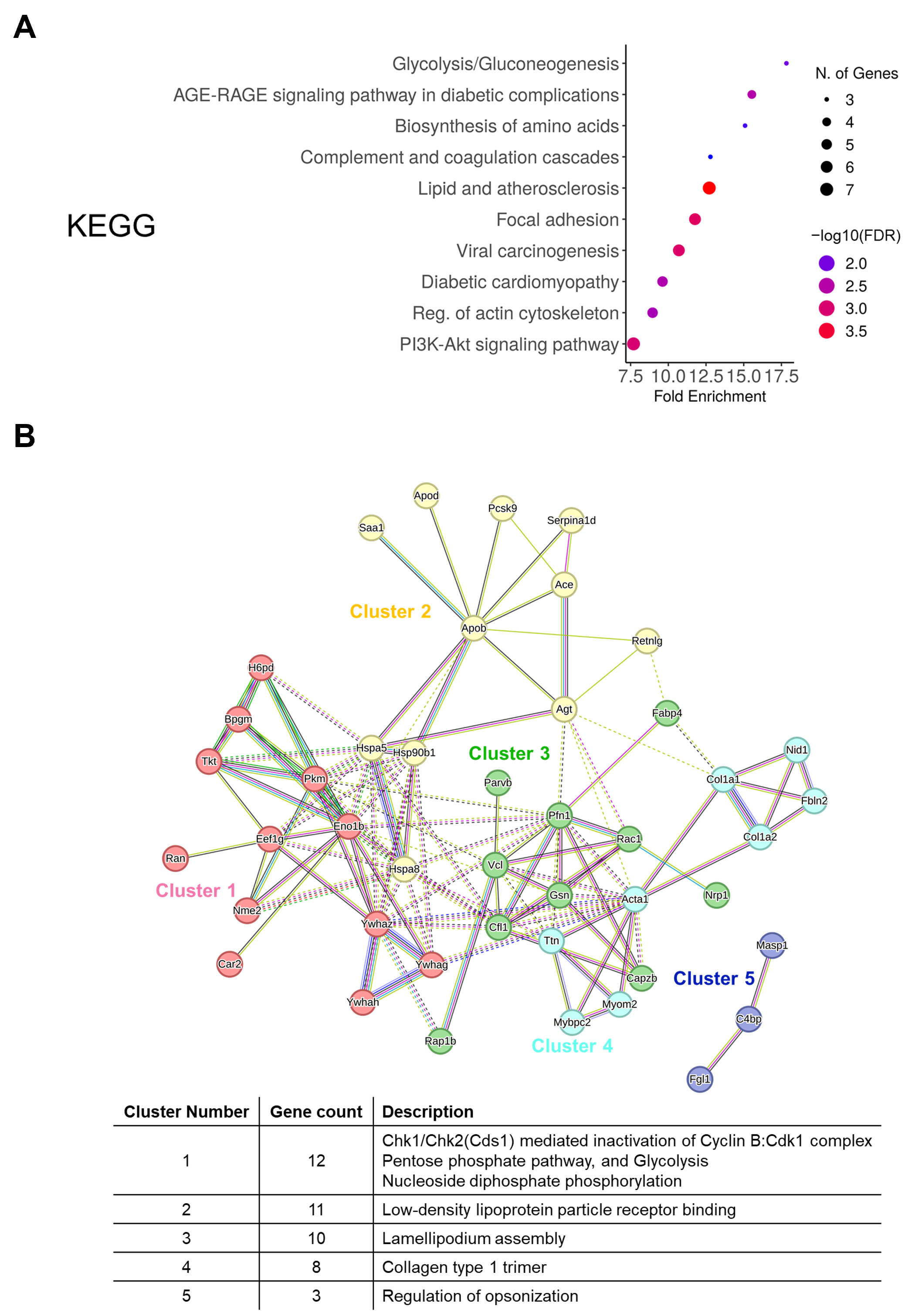
2.11. Gene Ontology, KEGG, and STRING Analysis
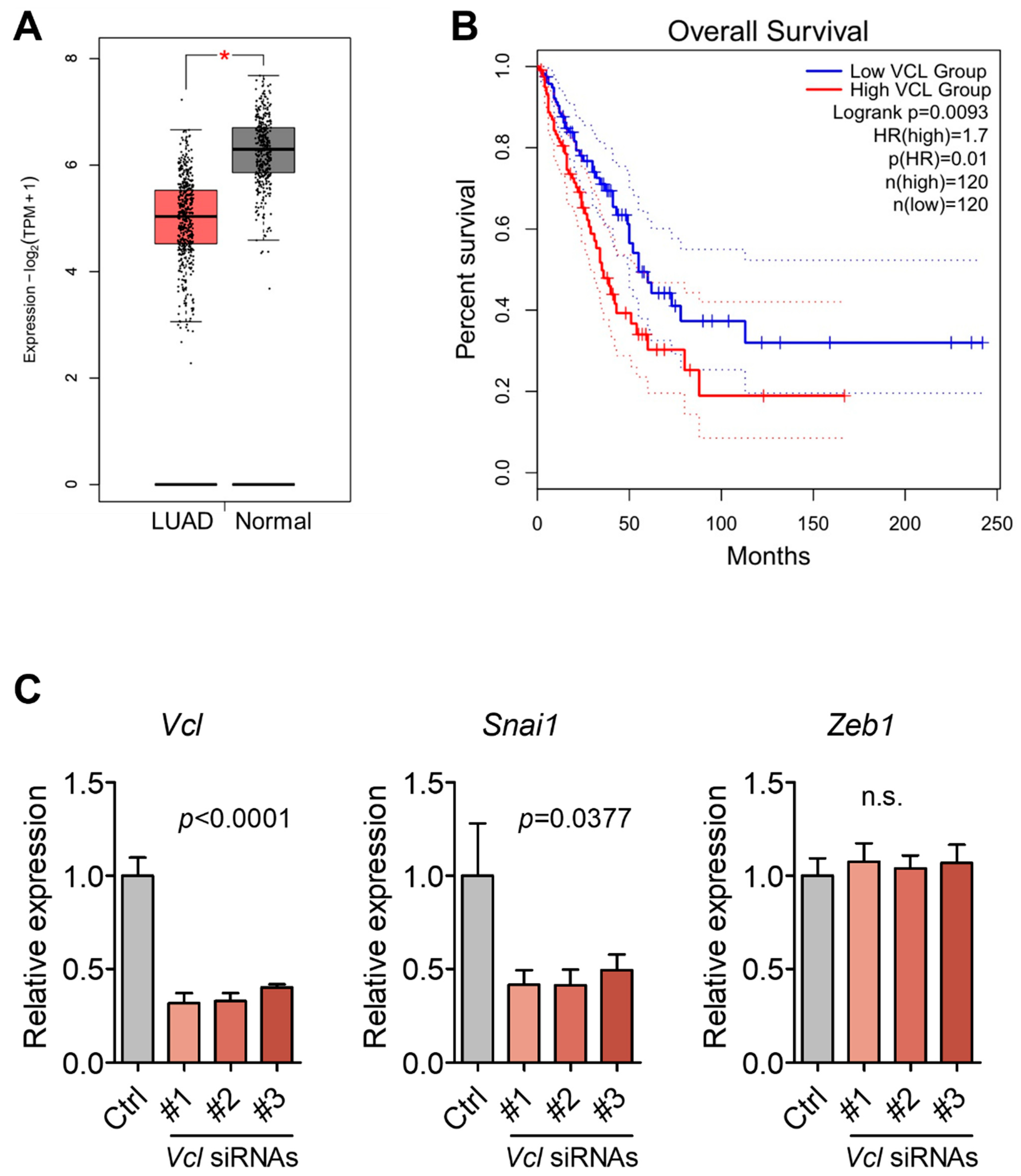
2.12. Gene Expression and Survival Analysis
2.13. Statistical Analysis
3. Results
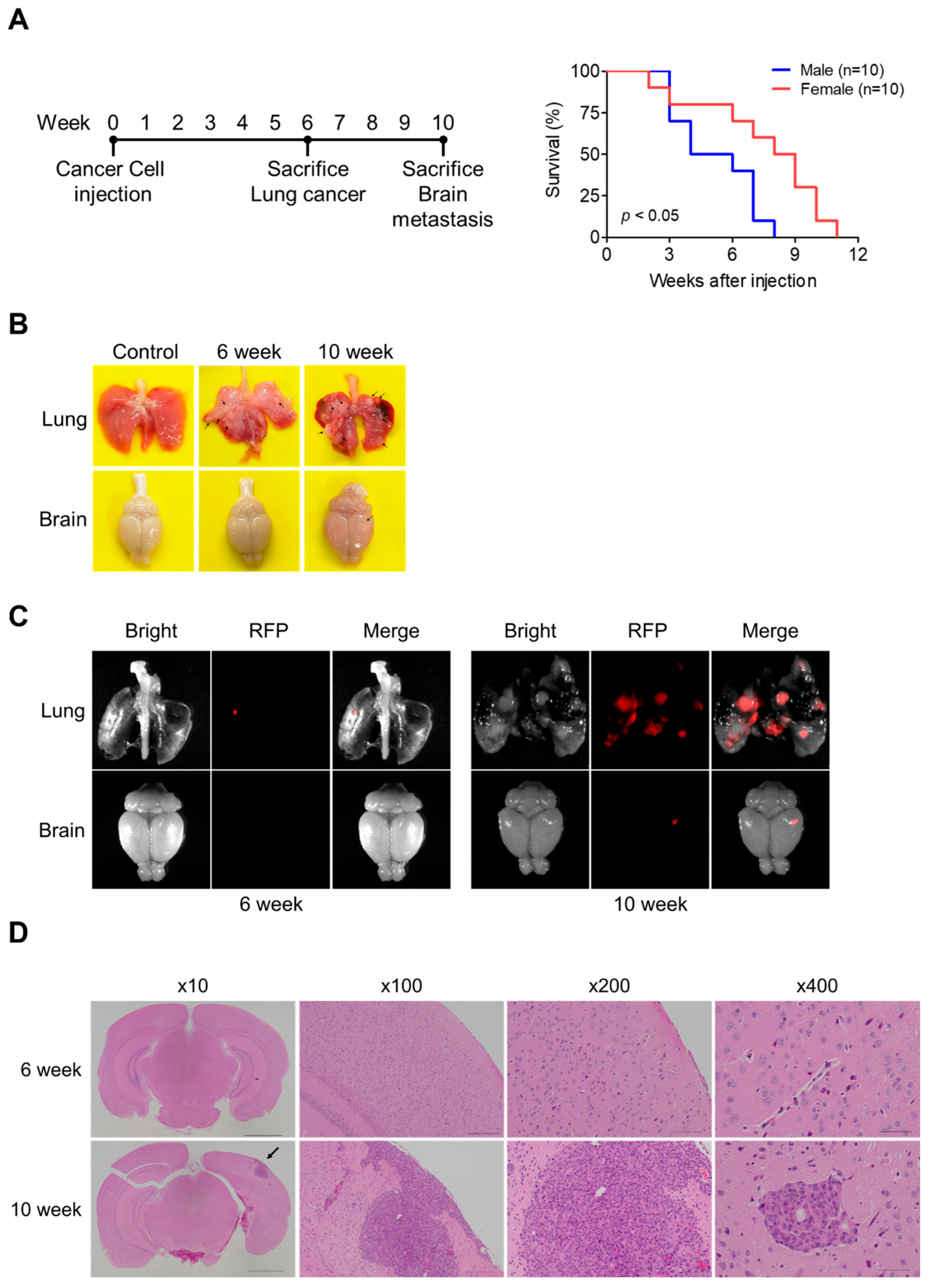
3.1. Development of a Lung Cancer Brain Metastasis Mouse Model
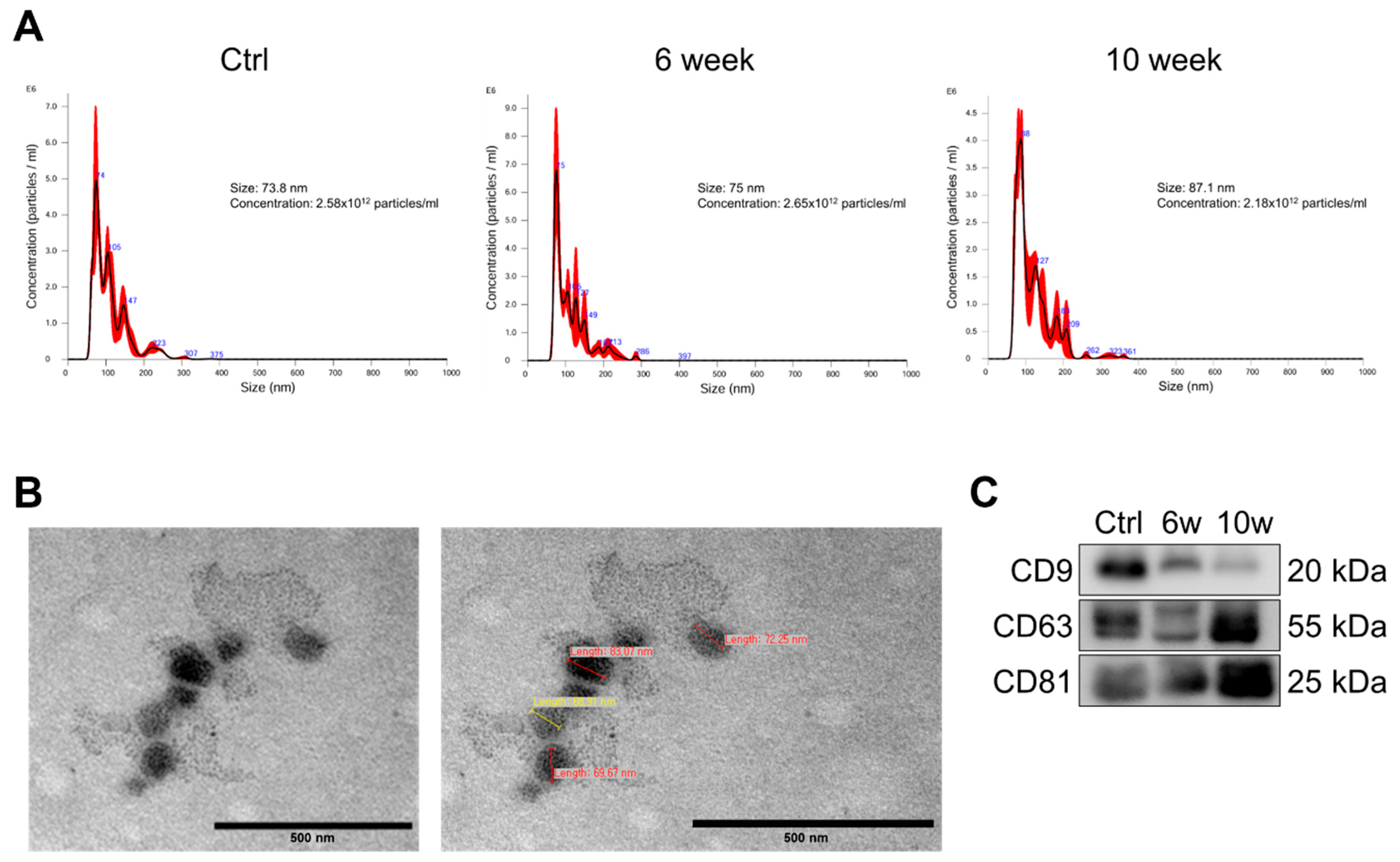
3.2. Characterization and Validation of Isolated Serum Exosomes
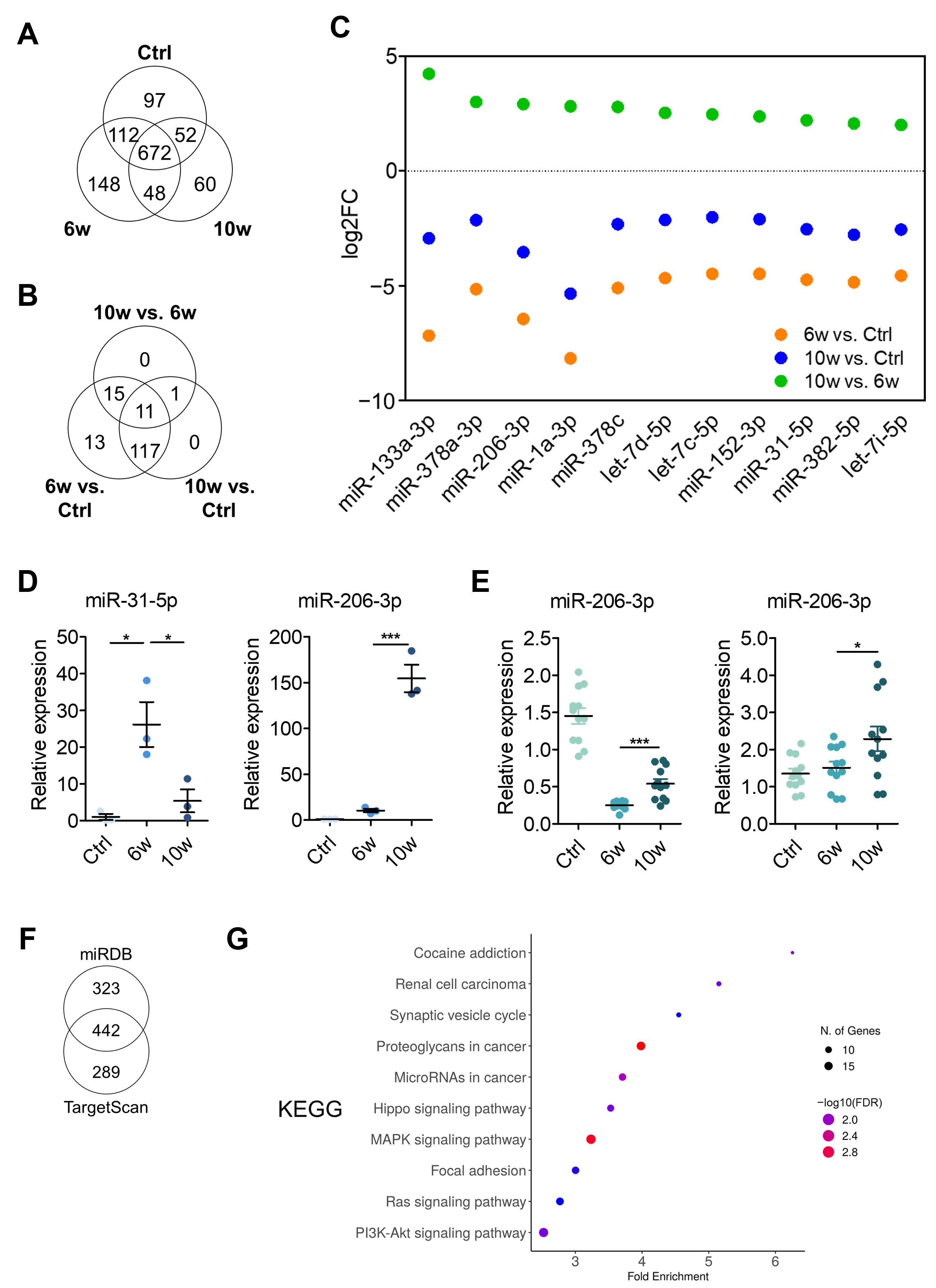
3.3. Comparison of miRNA Profiling and Bioinformatics Analysis of EV-Derived miRNAs Among the Control, 6w, and 10w Groups
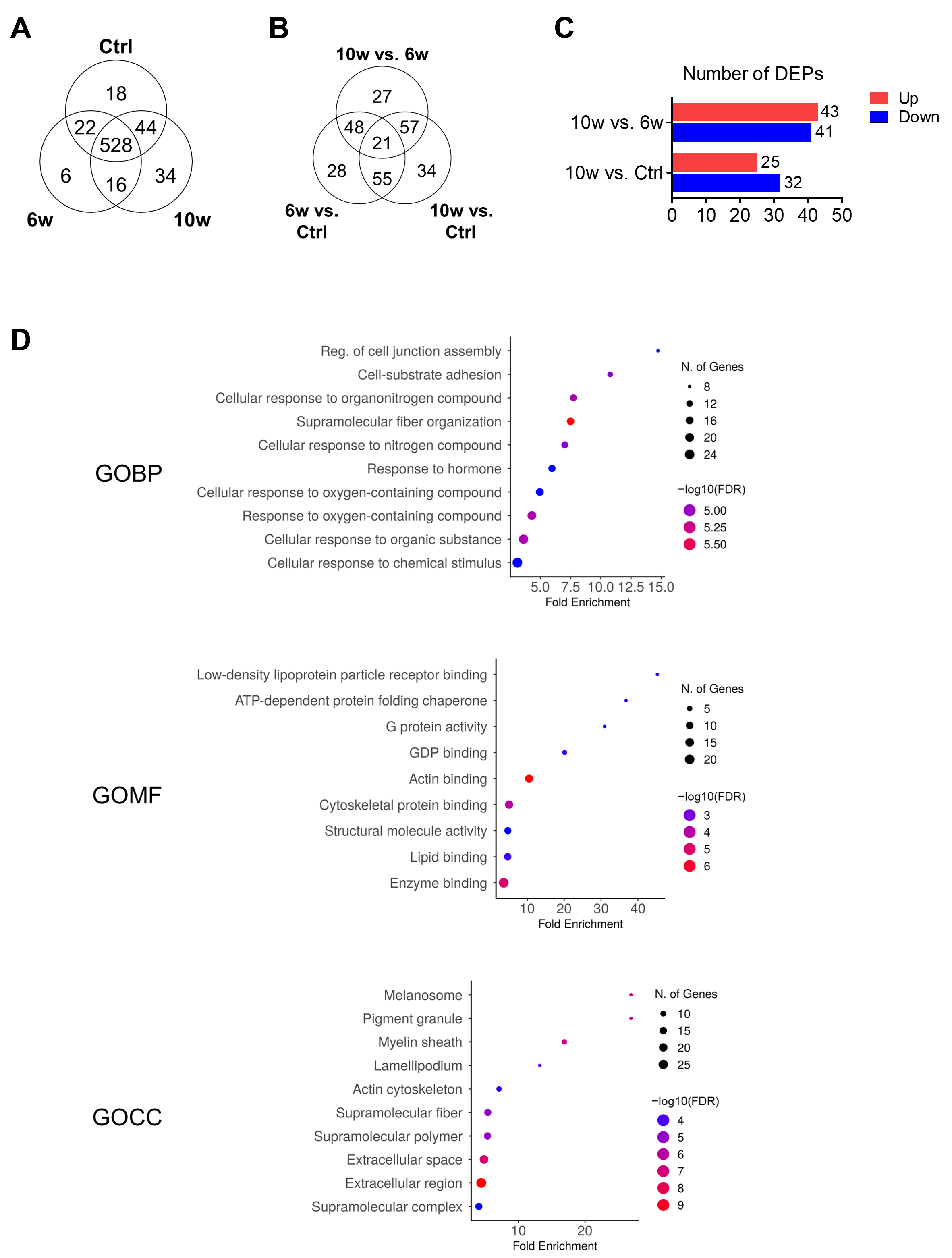
3.4. Comparison of Protein Profiling and Bioinformatics Analysis of EV-Derived Proteins Among the Control, 6w, and 10w Groups
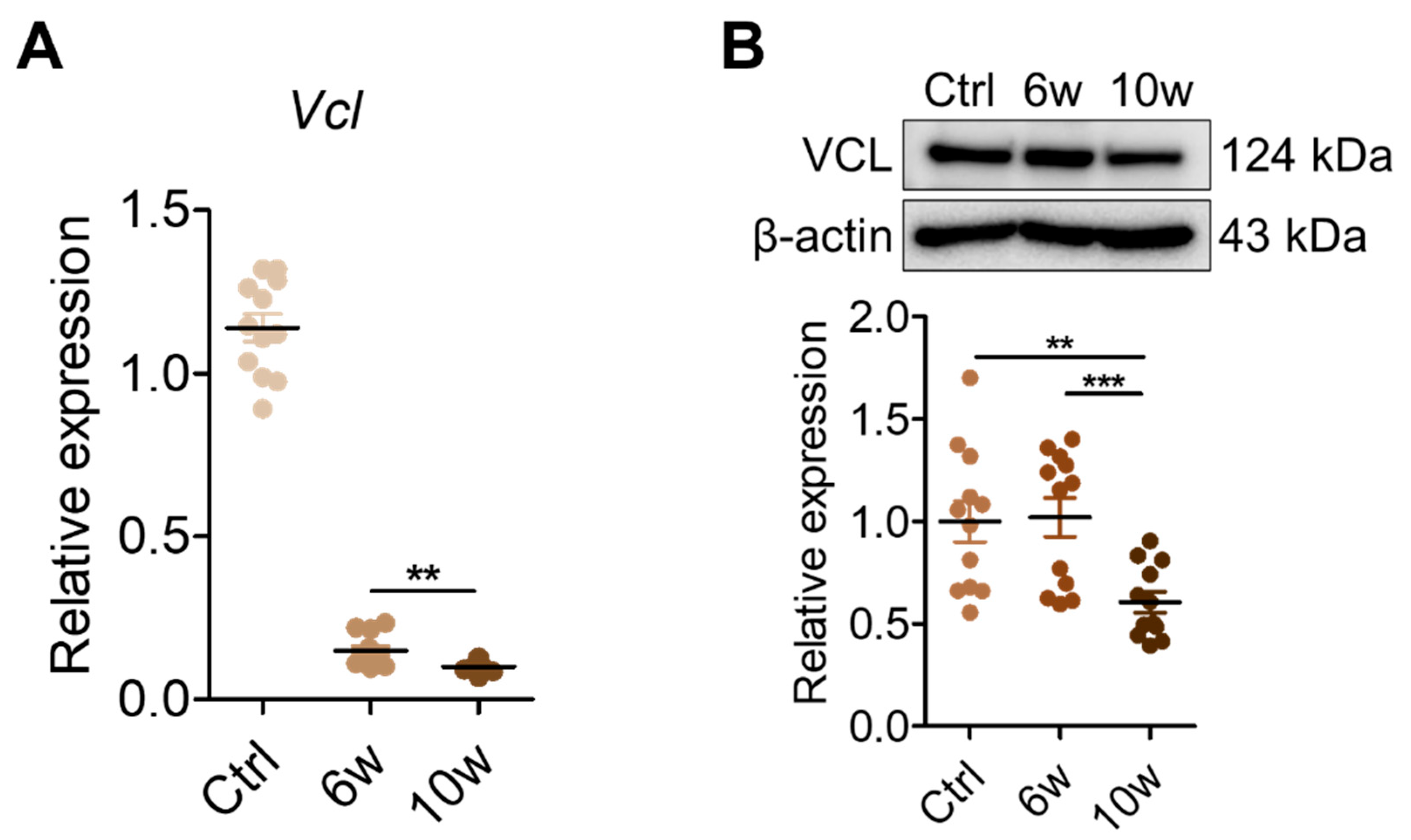
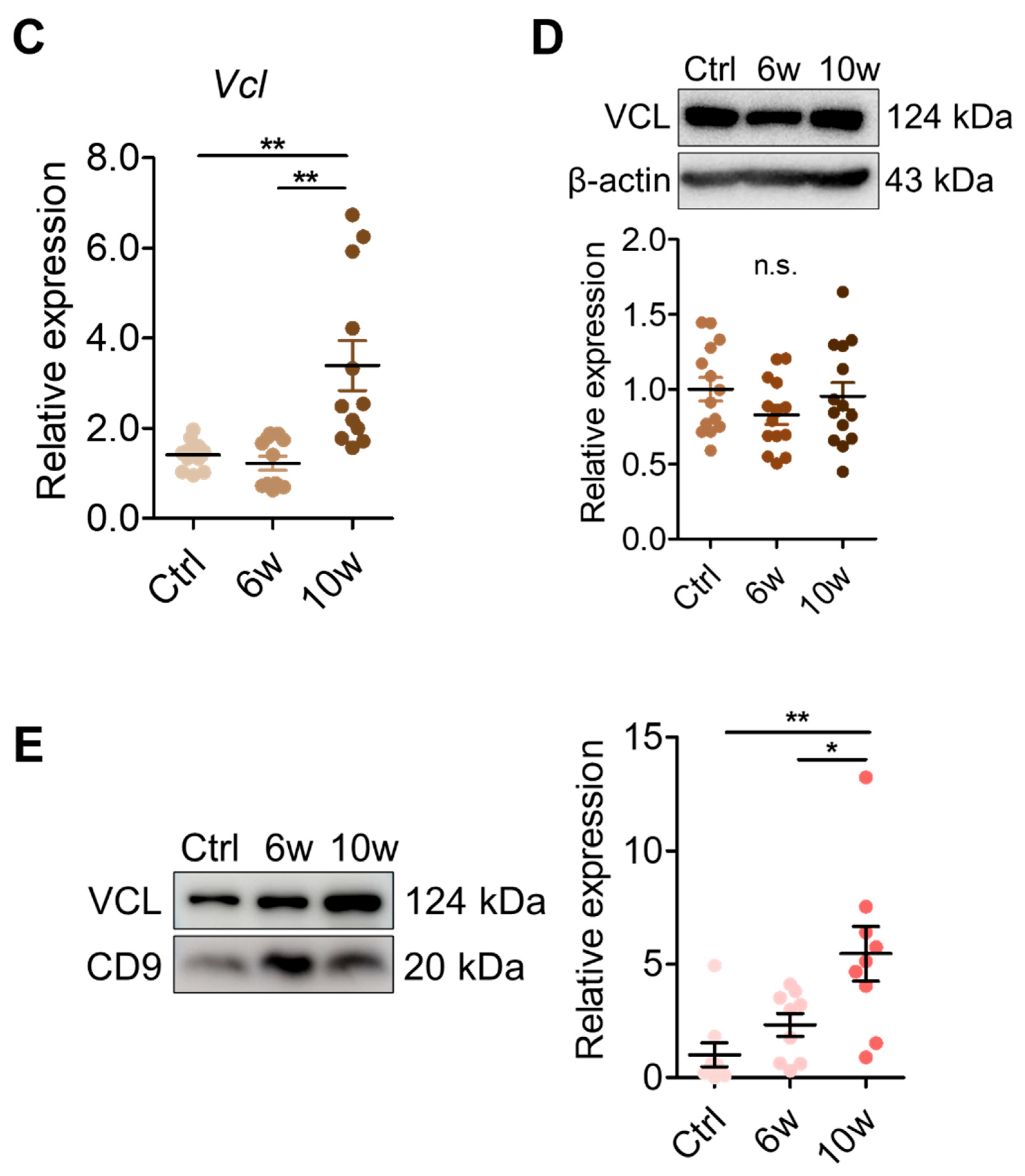
3.5. Validation of VCL Expression in Tissues and Exosomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Achrol, A.S.; Rennert, R.C.; Anders, C.; Soffietti, R.; Ahluwalia, M.S.; Nayak, L.; Peters, S.; Arvold, N.D.; Harsh, G.R.; Steeg, P.S.; et al. Brain metastases. Nat. Rev. Dis. Primers 2019, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Klotz, R.; Yu, M. Insights into brain metastasis: Recent advances in circulating tumor cell research. Cancer Rep. 2022, 5, e1239. [Google Scholar] [CrossRef]
- Valiente, M.; Ahluwalia, M.S.; Boire, A.; Brastianos, P.K.; Goldberg, S.B.; Lee, E.Q.; Le Rhun, E.; Preusser, M.; Winkler, F.; Soffietti, R. The Evolving Landscape of Brain Metastasis. Trends Cancer 2018, 4, 176–196. [Google Scholar] [CrossRef]
- Najjary, S.; de Koning, W.; Kros, J.M.; Mustafa, D.A.M. Unlocking molecular mechanisms and identifying druggable targets in matched-paired brain metastasis of breast and lung cancers. Front. Immunol. 2023, 14, 1305644. [Google Scholar] [CrossRef]
- McKay, M.J. Brain metastases: Increasingly precision medicine—A narrative review. Ann. Transl. Med. 2021, 9, 1629. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhou, L.; Zhao, M.; Cao, K.; Li, Y.; Liu, X.; Hou, Y.; Li, L.; Wang, L.; Chang, L.; et al. Real-world analysis of different intracranial radiation therapies in non-small cell lung cancer patients with 1–4 brain metastases. BMC Cancer 2022, 22, 1010. [Google Scholar] [CrossRef]
- Nishino, M.; Soejima, K.; Mitsudomi, T. Brain metastases in oncogene-driven non-small cell lung cancer. Transl. Lung Cancer Res. 2019, 8 (Suppl. S3), S298–S307. [Google Scholar] [CrossRef]
- Steindl, A.; Yadavalli, S.; Gruber, K.A.; Seiwald, M.; Gatterbauer, B.; Dieckmann, K.; Frischer, J.M.; Klikovits, T.; Zochbauer-Muller, S.; Grisold, A.; et al. Neurological symptom burden impacts survival prognosis in patients with newly diagnosed non-small cell lung cancer brain metastases. Cancer 2020, 126, 4341–4352. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, K.J. Epidemiology and prognosis of brain metastases. Surg. Neurol. Int. 2013, 4 (Suppl. S4), S192–S202. [Google Scholar] [CrossRef]
- Dilsiz, N. Hallmarks of exosomes. Future Sci. OA 2022, 8, FSO764. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Li, I.; Nabet, B.Y. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol. Cancer 2019, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.M.; Xu, Y.M.; Huang, L.F.; Wang, X.Z. Exosomes: Novel biomarkers for clinical diagnosis. ScientificWorldJournal 2015, 2015, 657086. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target. Ther. 2020, 5, 144. [Google Scholar] [CrossRef]
- Wang, X.; Tian, L.; Lu, J.; Ng, I.O. Exosomes and cancer—Diagnostic and prognostic biomarkers and therapeutic vehicle. Oncogenesis 2022, 11, 54. [Google Scholar] [CrossRef]
- Kalluri, R.; McAndrews, K.M. The role of extracellular vesicles in cancer. Cell 2023, 186, 1610–1626. [Google Scholar] [CrossRef]
- Zhong, D.; Wang, Z.; Ye, Z.; Wang, Y.; Cai, X. Cancer-derived exosomes as novel biomarkers in metastatic gastrointestinal cancer. Mol. Cancer 2024, 23, 67. [Google Scholar] [CrossRef]
- Baldasici, O.; Pileczki, V.; Cruceriu, D.; Gavrilas, L.I.; Tudoran, O.; Balacescu, L.; Vlase, L.; Balacescu, O. Breast Cancer-Delivered Exosomal miRNA as Liquid Biopsy Biomarkers for Metastasis Prediction: A Focus on Translational Research with Clinical Applicability. Int. J. Mol. Sci. 2022, 23, 9371. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Li, S.; Qu, Y.; Liu, L.; Zhang, X.; He, Y.; Wang, C.; Guo, Y.; Yuan, L.; Ma, Z.; Bai, H.; et al. Comparative proteomic profiling of plasma exosomes in lung cancer cases of liver and brain metastasis. Cell Biosci. 2023, 13, 180. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kim, E.J.; Lee, S.; Tan, X.; Liu, X.; Park, S.; Kang, K.; Yoon, J.S.; Ko, Y.H.; Kurie, J.M.; et al. MiR-34a and miR-34b/c have distinct effects on the suppression of lung adenocarcinomas. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Lange, T.; Stracke, S.; Rettig, R.; Lendeckel, U.; Kuhn, J.; Schluter, R.; Rippe, V.; Endlich, K.; Endlich, N. Identification of miR-16 as an endogenous reference gene for the normalization of urinary exosomal miRNA expression data from CKD patients. PLoS ONE 2017, 12, e0183435. [Google Scholar] [CrossRef] [PubMed]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 2020, 17, 41–44. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Chen, Q.Y.; Jiao, D.M.; Wang, J.; Hu, H.; Tang, X.; Chen, J.; Mou, H.; Lu, W. miR-206 regulates cisplatin resistance and EMT in human lung adenocarcinoma cells partly by targeting MET. Oncotarget 2016, 7, 24510–24526. [Google Scholar] [CrossRef]
- Davenport, M.L.; Echols, J.B.; Silva, A.D.; Anderson, J.C.; Owens, P.; Yates, C.; Wei, Q.; Harada, S.; Hurst, D.R.; Edmonds, M.D. miR-31 Displays Subtype Specificity in Lung Cancer. Cancer Res. 2021, 81, 1942–1953. [Google Scholar] [CrossRef]
- Zheng, X.; Xu, H.; Gong, L.; Cao, D.; Jin, T.; Wang, Y.; Pi, J.; Yang, Y.; Yi, X.; Liao, D.; et al. Vinculin orchestrates prostate cancer progression by regulating tumor cell invasion, migration, and proliferation. Prostate 2021, 81, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, C.; Lan, L.; Behrens, A.; Tomaschko, M.; Ruiz, J.; Su, Q.; Zhao, G.; Yuan, C.; Xiao, X.; et al. High expression of vinculin predicts poor prognosis and distant metastasis and associates with influencing tumor-associated NK cell infiltration and epithelial-mesenchymal transition in gastric cancer. Aging 2021, 13, 5197–5225. [Google Scholar] [CrossRef] [PubMed]
- Rubashkin, M.G.; Cassereau, L.; Bainer, R.; DuFort, C.C.; Yui, Y.; Ou, G.; Paszek, M.J.; Davidson, M.W.; Chen, Y.Y.; Weaver, V.M. Force engages vinculin and promotes tumor progression by enhancing PI3K activation of phosphatidylinositol (3,4,5)-triphosphate. Cancer Res. 2014, 74, 4597–4611. [Google Scholar] [CrossRef] [PubMed]
- Pope, W.B. Brain metastases: Neuroimaging. Handb. Clin. Neurol. 2018, 149, 89–112. [Google Scholar] [CrossRef]
- Vachha, B.A.; Huang, S.Y.; Massoud, T.F. Editorial: Advanced Neuroimaging of Brain Metastases. Front. Neurol. 2021, 12, 668310. [Google Scholar] [CrossRef]
- Cao, C.F.; Ma, K.L.; Shan, H.; Liu, T.F.; Zhao, S.Q.; Wan, Y.; Jun, Z.; Wang, H.Q. CT Scans and Cancer Risks: A Systematic Review and Dose-response Meta-analysis. BMC Cancer 2022, 22, 1238. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Marchetti, D.; Lang, J.E. Liquid biopsy: From concept to clinical application. Sci. Rep. 2023, 13, 21685. [Google Scholar] [CrossRef]
- Noorwati Sutandyo, R.H.; Wuyung, P.E.; Rahmadi, A.; Mulawarman, A.; Herawati, C.; Ramli, R. Expression of Circulating Mir-206 in Patients with Lung and Head and Neck Cancers and its Association with Cancer Cachexia. J. Clin. Exp. Oncol. 2017, 6, 4. [Google Scholar] [CrossRef]
- Machesky, L.M. Lamellipodia and filopodia in metastasis and invasion. FEBS Lett. 2008, 582, 2102–2111. [Google Scholar] [CrossRef]
- Nawaz, S.; Sanchez, P.; Schmitt, S.; Snaidero, N.; Mitkovski, M.; Velte, C.; Bruckner, B.R.; Alexopoulos, I.; Czopka, T.; Jung, S.Y.; et al. Actin filament turnover drives leading edge growth during myelin sheath formation in the central nervous system. Dev. Cell 2015, 34, 139–151. [Google Scholar] [CrossRef]
- Goldmann, W.H.; Auernheimer, V.; Thievessen, I.; Fabry, B. Vinculin, cell mechanics and tumour cell invasion. Cell Biol. Int. 2013, 37, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fan, J.; Xu, T.; Ahmadinejad, N.; Hess, K.; Lin, S.H.; Zhang, J.; Liu, X.; Liu, L.; Ning, B.; et al. Extracellular vesicle tetraspanin-8 level predicts distant metastasis in non-small cell lung cancer after concurrent chemoradiation. Sci. Adv. 2020, 6, eaaz6162. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, J.; Kang, M.; Ahn, Y.-H.; Cho, M.-S.; Lee, J.H.; Kang, J.L.; Choi, Y.-H. Comprehensive Profiling of Serum Exosomes by a Multi-Omics Approach Reveals Potential Diagnostic Markers for Brain Metastasis in Lung Cancer. Cancers 2025, 17, 1929. https://doi.org/10.3390/cancers17121929
Lim J, Kang M, Ahn Y-H, Cho M-S, Lee JH, Kang JL, Choi Y-H. Comprehensive Profiling of Serum Exosomes by a Multi-Omics Approach Reveals Potential Diagnostic Markers for Brain Metastasis in Lung Cancer. Cancers. 2025; 17(12):1929. https://doi.org/10.3390/cancers17121929
Chicago/Turabian StyleLim, Jiwoo, Mia Kang, Young-Ho Ahn, Min-Sun Cho, Jin Hwa Lee, Jihee Lee Kang, and Youn-Hee Choi. 2025. "Comprehensive Profiling of Serum Exosomes by a Multi-Omics Approach Reveals Potential Diagnostic Markers for Brain Metastasis in Lung Cancer" Cancers 17, no. 12: 1929. https://doi.org/10.3390/cancers17121929
APA StyleLim, J., Kang, M., Ahn, Y.-H., Cho, M.-S., Lee, J. H., Kang, J. L., & Choi, Y.-H. (2025). Comprehensive Profiling of Serum Exosomes by a Multi-Omics Approach Reveals Potential Diagnostic Markers for Brain Metastasis in Lung Cancer. Cancers, 17(12), 1929. https://doi.org/10.3390/cancers17121929






