Molecular Characterization of Atypical Fibroxanthoma and Pleomorphic Dermal Sarcoma
Simple Summary
Abstract
1. Introduction
2. Main Body
2.1. History of Atypical Fibroxanthoma and Pleomorphic Dermal Sarcoma
2.2. DNA Sequencing of Targeted Gene Panels
2.3. Exome Sequencing
2.4. DNA Methylation
2.5. Copy Number Variation
2.6. Bulk RNA Sequencing
2.7. Single-Cell RNA Sequencing
3. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFX | Atypical fibroxanthoma |
| PDS | Pleomorphic dermal sarcoma |
| UPS | Undifferentiated pleomorphic sarcoma |
| cSCC | Cutaneous squamous cell carcinoma |
| scRNA-seq | Single-cell RNA sequencing |
| GOF | Gain of function |
| LOF | Loss of function |
References
- Katz, D.; Palmerini, E.; Pollack, S.M. More than 50 Subtypes of Soft Tissue Sarcoma: Paving the Path for Histology-Driven Treatments. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Sbaraglia, M.; Bellan, E.; Dei Tos, A.P. The 2020 WHO Classification of Soft Tissue Tumours: News and Perspectives. Pathologica 2021, 113, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Gamboa, A.C.; Gronchi, A.; Cardona, K. Soft-Tissue Sarcoma in Adults: An Update on the Current State of Histiotype-Specific Management in an Era of Personalized Medicine. CA Cancer J. Clin. 2020, 70, 200–229. [Google Scholar] [CrossRef] [PubMed]
- Zambo, I.; Veselý, K. WHO classification of tumours of soft tissue and bone 2013: The main changes compared to the 3rd edition. Cesk. Patol. 2014, 50, 64–70. [Google Scholar]
- Ørholt, M.; Abebe, K.; Rasmussen, L.E.; Aaberg, F.L.; Lindskov, L.J.; Schmidt, G.; Wagenblast, A.L.; Petersen, M.M.; Loya, A.C.; Daugaard, S.; et al. Atypical Fibroxanthoma and Pleomorphic Dermal Sarcoma: Local Recurrence and Metastasis in a Nationwide Population-Based Cohort of 1118 Patients. J. Am. Acad. Dermatol. 2023, 89, 1177–1184. [Google Scholar] [CrossRef]
- Beer, T.W.; Drury, P.; Heenan, P.J. Atypical Fibroxanthoma: A Histological and Immunohistochemical Review of 171 Cases. Am. J. Dermatopathol. 2010, 32, 533–540. [Google Scholar] [CrossRef]
- Klein, J.C.; Wang, L.; Strand, D.; Lastufka, C.; Hosler, G.A.; Hon, G.C. Single-Cell and Spatial Transcriptomics Identify COL6A3 as a Prognostic Biomarker in Undifferentiated Pleomorphic Sarcoma. Mol. Cancer 2024, 23, 257. [Google Scholar] [CrossRef]
- Atypical Fibroxanthoma: Causes, Diagnosis and Outcomes—DermNet. Available online: https://dermnetnz.org/topics/atypical-fibroxanthoma (accessed on 23 May 2025).
- Pleomorphic Dermal Sarcoma. Available online: https://dermnetnz.org/topics/pleomorphic-dermal-sarcoma (accessed on 23 May 2025).
- Weidner, N.; Cote, R.J.; Suster, S. Modern Surgical Pathology; Elsevier Health Sciences: Amsterdam, The Netherlands, 2009; ISBN 9781336229228. [Google Scholar]
- Miller, K.; Goodlad, J.R.; Brenn, T. Pleomorphic Dermal Sarcoma. Am. J. Surg. Pathol. 2012, 36, 1317–1326. [Google Scholar] [CrossRef]
- Anderson, M.E.; Rodic, N.; Subtil, A.; Queen, D.; Arcasoy, S.; Niedt, G.W.; Heald, P.W.; Geskin, L.J. Multifocal Pleomorphic Dermal Sarcoma and the Role of Inflammation and Immunosuppression in a Lung Transplant Patient: A Case Report. J. Med. Case Rep. 2019, 13, 169. [Google Scholar] [CrossRef]
- Llombart, B.; Serra-Guillén, C.; Requena, C.; Alsina, M.; Morgado-Carrasco, D.; Machado, I.; Sanmartín, O. Leiomyosarcoma and Pleomorphic Dermal Sarcoma: Guidelines for Diagnosis and Treatment. Actas Dermosifiliogr. 2019, 110, 4–11. [Google Scholar] [CrossRef]
- Helbig, D.; Klein, S. Immune Checkpoint Inhibitors for Unresectable or Metastatic Pleomorphic Dermal Sarcomas. Front. Oncol. 2022, 12, 975342. [Google Scholar] [CrossRef] [PubMed]
- Ak, M.; Kahraman, A.; Arnold, F.M.; Turko, P.; Levesque, M.P.; Zoche, M.; Ramelyte, E.; Dummer, R. Clinicopathological and Genomic Profiles of Atypical Fibroxanthoma and Pleomorphic Dermal Sarcoma Identify Overlapping Signatures with a High Mutational Burden. Genes 2021, 12, 974. [Google Scholar] [CrossRef] [PubMed]
- Griewank, K.G.; Wiesner, T.; Murali, R.; Pischler, C.; Müller, H.; Koelsche, C.; Möller, I.; Franklin, C.; Cosgarea, I.; Sucker, A.; et al. Atypical Fibroxanthoma and Pleomorphic Dermal Sarcoma Harbor Frequent NOTCH1/2 and FAT1 Mutations and Similar DNA Copy Number Alteration Profiles. Mod. Pathol. 2018, 31, 418–428. [Google Scholar] [CrossRef]
- Helbig, D.; Ihle, M.A.; Pütz, K.; Tantcheva-Poor, I.; Mauch, C.; Büttner, R.; Quaas, A. Oncogene and Therapeutic Target Analyses in Atypical Fibroxanthomas and Pleomorphic Dermal Sarcomas. Oncotarget 2016, 7, 21763–21774. [Google Scholar] [CrossRef]
- Lai, K.; Harwood, C.A.; Purdie, K.J.; Proby, C.M.; Leigh, I.M.; Ravi, N.; Mully, T.W.; Brooks, L.; Sandoval, P.M.; Rosenblum, M.D.; et al. Genomic Analysis of Atypical Fibroxanthoma. PLoS ONE 2017, 12, e0188272. [Google Scholar] [CrossRef]
- Klein, S.; Quaas, A.; Noh, K.-W.; Cartolano, M.; Abedpour, N.; Mauch, C.; Quantius, J.; Reinhardt, H.C.; Buettner, R.; Peifer, M.; et al. Integrative Analysis of Pleomorphic Dermal Sarcomas Reveals Fibroblastic Differentiation and Susceptibility to Immunotherapy. Clin. Cancer Res. 2020, 26, 5638–5645. [Google Scholar] [CrossRef]
- Lim, Y.H.; Zaki, T.D.; Levinsohn, J.L.; Galan, A.; Choate, K.A.; Hanlon, A.M. Somatic Mutation Profile of Atypical Fibroxanthoma and Cutaneous Undifferentiated Pleomorphic Sarcoma. Dermatol. Surg. 2021, 47, 290–292. [Google Scholar] [CrossRef]
- Mihic-Probst, D.; Zhao, J.; Saremaslani, P.; Baer, A.; Oehlschlegel, C.; Paredes, B.; Komminoth, P.; Heitz, P.U. CGH Analysis Shows Genetic Similarities and Differences in Atypical Fibroxanthoma and Undifferentiated High Grade Pleomorphic Sarcoma. Anticancer Res. 2004, 24, 19–26. [Google Scholar]
- Koelsche, C.; Stichel, D.; Griewank, K.G.; Schrimpf, D.; Reuss, D.E.; Bewerunge-Hudler, M.; Vokuhl, C.; Dinjens, W.N.M.; Petersen, I.; Mittelbronn, M.; et al. Genome-Wide Methylation Profiling and Copy Number Analysis in Atypical Fibroxanthomas and Pleomorphic Dermal Sarcomas Indicate a Similar Molecular Phenotype. Clin. Sarcoma Res. 2019, 9, 2. [Google Scholar] [CrossRef]
- Doyle, L.A. Sarcoma Classification: An Update Based on the 2013 World Health Organization Classification of Tumors of Soft Tissue and Bone. Cancer 2014, 120, 1763–1774. [Google Scholar] [CrossRef]
- Thum, C.; Hollowood, K.; Birch, J.; Goodlad, J.R.; Brenn, T. Aberrant Melan-A Expression in Atypical Fibroxanthoma and Undifferentiated Pleomorphic Sarcoma of the Skin. J. Cutan. Pathol. 2011, 38, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Lukács, J.; Schliemann, S.; Elsner, P. Undifferentiated Pleomorphic Sarcoma of the Skin—A UV-Induced Occupational Skin Disease? J. Dtsch. Dermatol. Ges. 2017, 15, 338–340. [Google Scholar] [CrossRef] [PubMed]
- Logan, I.T.; Vroobel, K.M.; le Grange, F.; Perrett, C.M. Pleomorphic Dermal Sarcoma: Clinicopathological Features and Outcomes from a 5-Year Tertiary Referral Centre Experience. Cancer Rep. 2022, 5, e1583. [Google Scholar] [CrossRef]
- Brenn, T. Soft Tissue Special Issue: Cutaneous Pleomorphic Spindle Cell Tumors. Head Neck Pathol. 2020, 14, 109–120. [Google Scholar] [CrossRef]
- Dei Tos, A.P.; Maestro, R.; Doglioni, C.; Gasparotto, D.; Boiocchi, M.; Laurino, L.; Fletcher, C.D. Ultraviolet-Induced P53 Mutations in Atypical Fibroxanthoma. Am. J. Pathol. 1994, 145, 11–17. [Google Scholar]
- Sakamoto, A.; Oda, Y.; Itakura, E.; Oshiro, Y.; Nikaido, O.; Iwamoto, Y.; Tsuneyoshi, M. Immunoexpression of Ultraviolet Photoproducts and P53 Mutation Analysis in Atypical Fibroxanthoma and Superficial Malignant Fibrous Histiocytoma. Mod. Pathol. 2001, 14, 581–588. [Google Scholar] [CrossRef]
- Sakamoto, A.; Oda, Y.; Itakura, E.; Oshiro, Y.; Tamiya, S.; Honda, Y.; Ishihara, A.; Iwamoto, Y.; Tsuneyoshi, M. H-, K-, and N-Ras Gene Mutation in Atypical Fibroxanthoma and Malignant Fibrous Histiocytoma. Hum. Pathol. 2001, 32, 1225–1231. [Google Scholar] [CrossRef]
- Griewank, K.G.; Schilling, B.; Murali, R.; Bielefeld, N.; Schwamborn, M.; Sucker, A.; Zimmer, L.; Hillen, U.; Schaller, J.; Brenn, T.; et al. TERT Promoter Mutations Are Frequent in Atypical Fibroxanthomas and Pleomorphic Dermal Sarcomas. Mod. Pathol. 2014, 27, 502–508. [Google Scholar] [CrossRef]
- Shendure, J.; Ji, H. Next-Generation DNA Sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef]
- Bamshad, M.J.; Ng, S.B.; Bigham, A.W.; Tabor, H.K.; Emond, M.J.; Nickerson, D.A.; Shendure, J. Exome Sequencing as a Tool for Mendelian Disease Gene Discovery. Nat. Rev. Genet. 2011, 12, 745–755. [Google Scholar] [CrossRef]
- Nagahashi, M.; Shimada, Y.; Ichikawa, H.; Kameyama, H.; Takabe, K.; Okuda, S.; Wakai, T. Next Generation Sequencing-Based Gene Panel Tests for the Management of Solid Tumors. Cancer Sci. 2019, 110, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, A.; Antonaros, F.; Vitale, L.; Strippoli, P.; Pelleri, M.C.; Caracausi, M. Human Protein-Coding Genes and Gene Feature Statistics in 2019. BMC Res. Notes 2019, 12, 315. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.C.; Chen, W.; Gasperini, M.; Shendure, J. Identifying Novel Enhancer Elements with CRISPR-Based Screens. ACS Chem. Biol. 2018, 13, 326–332. [Google Scholar] [CrossRef]
- Ng, S.B.; Turner, E.H.; Robertson, P.D.; Flygare, S.D.; Bigham, A.W.; Lee, C.; Shaffer, T.; Wong, M.; Bhattacharjee, A.; Eichler, E.E.; et al. Targeted Capture and Massively Parallel Sequencing of 12 Human Exomes. Nature 2009, 461, 272–276. [Google Scholar] [CrossRef]
- Klein, J.C.; Keith, A.; Rice, S.J.; Shepherd, C.; Agarwal, V.; Loughlin, J.; Shendure, J. Functional Testing of Thousands of Osteoarthritis-Associated Variants for Regulatory Activity. Nat. Commun. 2019, 10, 2434. [Google Scholar] [CrossRef]
- Klein, J.C.; Keith, A.; Agarwal, V.; Durham, T.; Shendure, J. Functional Characterization of Enhancer Evolution in the Primate Lineage. Genome Biol. 2018, 19, 99. [Google Scholar] [CrossRef]
- Kohli, R.M.; Zhang, Y. TET Enzymes, TDG and the Dynamics of DNA Demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef]
- Seki, M.; Nishimura, R.; Yoshida, K.; Shimamura, T.; Shiraishi, Y.; Sato, Y.; Kato, M.; Chiba, K.; Tanaka, H.; Hoshino, N.; et al. Integrated Genetic and Epigenetic Analysis Defines Novel Molecular Subgroups in Rhabdomyosarcoma. Nat. Commun. 2015, 6, 7557. [Google Scholar] [CrossRef]
- Koelsche, C.; Schrimpf, D.; Tharun, L.; Roth, E.; Sturm, D.; Jones, D.T.W.; Renker, E.-K.; Sill, M.; Baude, A.; Sahm, F.; et al. Histone 3.3 Hotspot Mutations in Conventional Osteosarcomas: A Comprehensive Clinical and Molecular Characterization of Six H3F3A Mutated Cases. Clin. Sarcoma Res. 2017, 7, 9. [Google Scholar] [CrossRef]
- Silva, F.L.T.; Euzébio, M.F.; Ruas, J.S.; Franco, M.T.; Cassone, A.E.; Junqueira, T.; Lucon, D.R.; Cardinalli, I.A.; Pereira, L.H.; Zenatti, P.P.; et al. Classification of Pediatric Soft and Bone Sarcomas Using DNA Methylation-Based Profiling. BMC Cancer 2024, 24, 1428. [Google Scholar] [CrossRef]
- Shlien, A.; Malkin, D. Copy Number Variations and Cancer. Genome Med. 2009, 1, 62. [Google Scholar] [CrossRef] [PubMed]
- Steele, C.D.; Abbasi, A.; Islam, S.M.A.; Bowes, A.L.; Khandekar, A.; Haase, K.; Hames-Fathi, S.; Ajayi, D.; Verfaillie, A.; Dhami, P.; et al. Signatures of Copy Number Alterations in Human Cancer. Nature 2022, 606, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.-T.; Huang, X.; Huang, S.; Xie, S.-J.; Xiao, Z.-D.; Zhang, H. RNA Sequencing: New Technologies and Applications in Cancer Research. J. Hematol. Oncol. 2020, 13, 166. [Google Scholar] [CrossRef]
- Sannino, G.; Marchetto, A.; Kirchner, T.; Grünewald, T.G.P. Epithelial-to-Mesenchymal and Mesenchymal-to-Epithelial Transition in Mesenchymal Tumors: A Paradox in Sarcomas? Cancer Res. 2017, 77, 4556–4561. [Google Scholar] [CrossRef]
- Kahlert, U.D.; Joseph, J.V.; Kruyt, F.A.E. EMT- and MET-Related Processes in Nonepithelial Tumors: Importance for Disease Progression, Prognosis, and Therapeutic Opportunities. Mol. Oncol. 2017, 11, 860–877. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-Associated Macrophages as Major Players in the Tumor Microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef]
- Lazova, R.; May, R.M.D.; Scott, G. LN-2 (CD74). Cancer 1997, 79, 2115–2124. [Google Scholar] [CrossRef]
- Hollmig, S.T.; Rieger, K.E.; Henderson, M.T.; West, R.B.; Sundram, U.N. Reconsidering the Diagnostic and Prognostic Utility of LN-2 for Undifferentiated Pleomorphic Sarcoma and Atypical Fibroxanthoma. Am. J. Dermatopathol. 2013, 35, 176–179. [Google Scholar] [CrossRef]
- Klein, S.; Persa, O.-D.; Mauch, C.; Noh, K.-W.; Pappesch, R.; Wagener-Ryczek, S.; Buettner, R.; Quaas, A.; Helbig, D. First Report on Two Cases of Pleomorphic Dermal Sarcoma Successfully Treated with Immune Checkpoint Inhibitors. Oncoimmunology 2019, 8, e1665977. [Google Scholar] [CrossRef]
- Cheuque, R.P.R.; Romero, I.V.; Jiménez-Gallo, D.; Cayetano, J.F.M.; Martínez, M.V.; Antón, A.J.; Albarrán, R.M.; Barrios, M.L. Pembrolizumab Treatment for Advanced Pleomorphic Dermal Sarcoma, a Three-Case Series in a Spanish Tertiary-Level Hospital. EJC Skin Cancer 2024, 2, 100205. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, T.; Su, W.; Dou, Z.; Zhao, D.; Jin, X.; Lei, H.; Wang, J.; Xie, X.; Cheng, B.; et al. Mutant P53 in Cancer: From Molecular Mechanism to Therapeutic Modulation. Cell Death Dis. 2022, 13, 974. [Google Scholar] [CrossRef] [PubMed]
- Vinagre, J.; Almeida, A.; Pópulo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT Promoter Mutations in Human Cancers. Nat. Commun. 2013, 4, 2185. [Google Scholar] [CrossRef] [PubMed]
- Boscolo-Rizzo, P.; Tirelli, G.; Polesel, J.; Sia, E.; Phillips, V.; Borsetto, D.; De Rossi, A.; Giunco, S. TERT Promoter Mutations in Head and Neck Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis on Prevalence and Prognostic Significance. Oral Oncol. 2023, 140, 106398. [Google Scholar] [CrossRef] [PubMed]
- Lennox, A.L.; Huang, F.; Behrs, M.K.; González-Sales, M.; Bhise, N.; Wan, Y.; Sun, L.; Berry, T.; Feller, F.; Morcos, P.N. Imetelstat, a Novel, First-in-Class Telomerase Inhibitor: Mechanism of Action, Clinical, and Translational Science. Clin. Transl. Sci. 2024, 17, e70076. [Google Scholar] [CrossRef]
- Shi, Q.; Xue, C.; Zeng, Y.; Yuan, X.; Chu, Q.; Jiang, S.; Wang, J.; Zhang, Y.; Zhu, D.; Li, L. Notch Signaling Pathway in Cancer: From Mechanistic Insights to Targeted Therapies. Signal Transduct. Target. Ther. 2024, 9, 128. [Google Scholar] [CrossRef]
- Chen, Z.; Guo, Y.; Zhao, D.; Zou, Q.; Yu, F.; Zhang, L.; Xu, L. Comprehensive Analysis Revealed That CDKN2A Is a Biomarker for Immune Infiltrates in Multiple Cancers. Front. Cell Dev. Biol. 2021, 9, 808208. [Google Scholar] [CrossRef]
- Pastushenko, I.; Mauri, F.; Song, Y.; de Cock, F.; Meeusen, B.; Swedlund, B.; Impens, F.; Van Haver, D.; Opitz, M.; Thery, M.; et al. Fat1 Deletion Promotes Hybrid EMT State, Tumour Stemness and Metastasis. Nature 2021, 589, 448–455. [Google Scholar] [CrossRef]
- Chen, Z.G.; Saba, N.F.; Teng, Y. The Diverse Functions of FAT1 in Cancer Progression: Good, Bad, or Ugly? J. Exp. Clin. Cancer Res. 2022, 41, 248. [Google Scholar] [CrossRef]
- Cassetta, L.; Pollard, J.W. A Timeline of Tumour-Associated Macrophage Biology. Nat. Rev. Cancer 2023, 23, 238–257. [Google Scholar] [CrossRef]
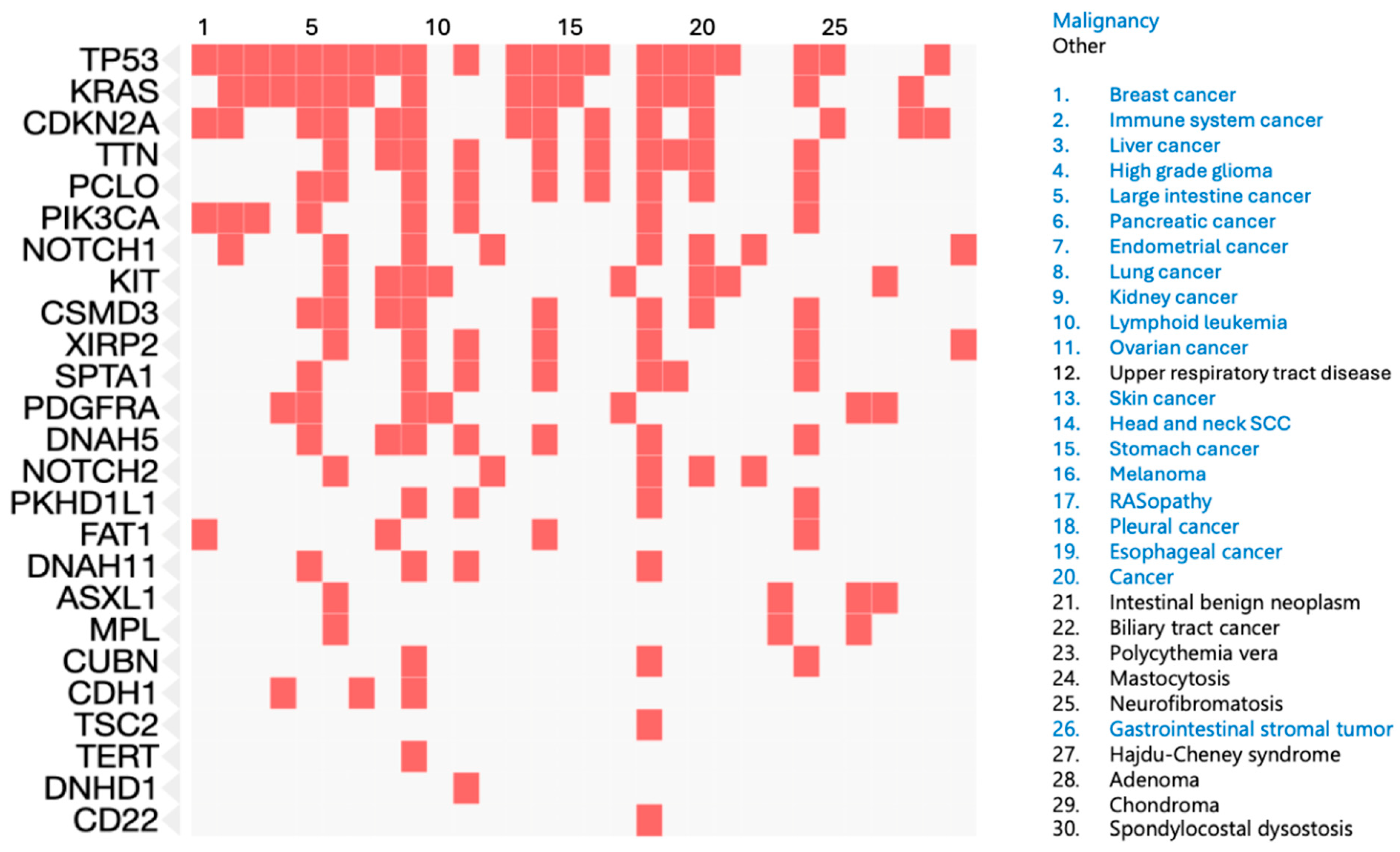
| Study | Tumors Examined | Assay Used | Key Findings |
|---|---|---|---|
| Ak et al. [15] | AFX and PDS | Targeted Gene Panel | Recurrent TP53, TERT promoter, NOTCH1, CDKN2A mutations |
| Griewank et al. [16] | AFX | Targeted Gene Panel and Copy Number Analysis | Recurrent TP53, TERT promoter, NOTCH1, CDKN2A, FAT1 mutations. Recurrent losses of 8p, 9p, 9q, 13, 16, and 18. |
| Helbig et al. [17] | AFX and PDS | Targeted Gene Panel | Recurrent TP53 mutations |
| Lai et al. [18] | AFX | Exome Sequencing and RNA sequencing | Recurrent FAT1, XIRP2, TTN, CUBN, DNAH11, DNAH5, PCLO, SPTA1, and TDRD15. KRAS signaling upregulated and p53 signaling downregulated. |
| Klein et al. [19] | PDS | Exome Sequencing and RNA sequencing | Recurrent TP53, CDKN2A, PDGFRA, DNHD1, and KIT. Expression profiling separates from cSCC, most similar to fibroblasts. |
| Lim et al. [20] | AFX and PDS | Exome Sequencing | Recurrent TP53 mutations |
| Mihic-Probst et al. [21] | AFX | Copy Number Analysis | Recurrent losses of 9p and 13q |
| Koelsche et al. [22] | AFX and PDS | Copy Number Analysis and Methylation Profiling | Recurrent losses in 9p and 13q and gains in 8q. AFX and PDS indistinguishable from each other by methylation profiling. |
| Klein et al. [7] | AFX and PDS | Single-cell RNA sequencing | COL6A3 as a potential prognostic biomarker |
| Gene | Gene Function | Mutation Type | Study Design and Citation | Frequency |
|---|---|---|---|---|
| TP53 | Tumor suppressor | LOF | Exome Sequencing [19,20] + Targeted Gene Panels [15,16,17] | 108/149 |
| TERT (promoter) | Telomere maintenance and cell immortalization | GOF | Targeted Gene Panels [15,16] | 70/80 |
| NOTCH1 | Notch signaling | LOF | Targeted Gene Panels [15,16] | 54/149 |
| CDKN2A | Tumor suppressor | LOF | Exome Sequencing [19] + Targeted Gene Panels [15,16,17] | 53/149 |
| FAT1 | Tumor suppressor | LOF | Exome Sequencing [18] + Targeted Gene Panels [16] | 51/126 |
| NOTCH2 | Notch signaling | LOF | Targeted Gene Panels [15,16] | 36/149 |
| XIRP2 | Actin filament organization | LOF | Exome Sequencing [18,20] | 7/52 |
| CSMD3 | Associated with tumor mutational burden | LOF | Exome Sequencing [18,20] | 6/52 |
| PKHD1L1 | Immune response | LOF | Exome Sequencing [18,20] | 6/52 |
| PIK3CA | Cell growth | GOF | Targeted Gene Panels [15,17] | 4/149 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klein, J.C.; Wilky, B.A.; Ford, H.L. Molecular Characterization of Atypical Fibroxanthoma and Pleomorphic Dermal Sarcoma. Cancers 2025, 17, 1785. https://doi.org/10.3390/cancers17111785
Klein JC, Wilky BA, Ford HL. Molecular Characterization of Atypical Fibroxanthoma and Pleomorphic Dermal Sarcoma. Cancers. 2025; 17(11):1785. https://doi.org/10.3390/cancers17111785
Chicago/Turabian StyleKlein, Jason C., Breelyn A. Wilky, and Heide L. Ford. 2025. "Molecular Characterization of Atypical Fibroxanthoma and Pleomorphic Dermal Sarcoma" Cancers 17, no. 11: 1785. https://doi.org/10.3390/cancers17111785
APA StyleKlein, J. C., Wilky, B. A., & Ford, H. L. (2025). Molecular Characterization of Atypical Fibroxanthoma and Pleomorphic Dermal Sarcoma. Cancers, 17(11), 1785. https://doi.org/10.3390/cancers17111785






