The Role of the Gut Microbiome in Non-Hodgkin Lymphoma (NHL): A Focus on Diffuse Large B-Cell Lymphoma, Follicular Lymphoma, Cutaneous T-Cell Lymphoma, and NK/T-Cell Lymphoma
Simple Summary
Abstract
1. Introduction
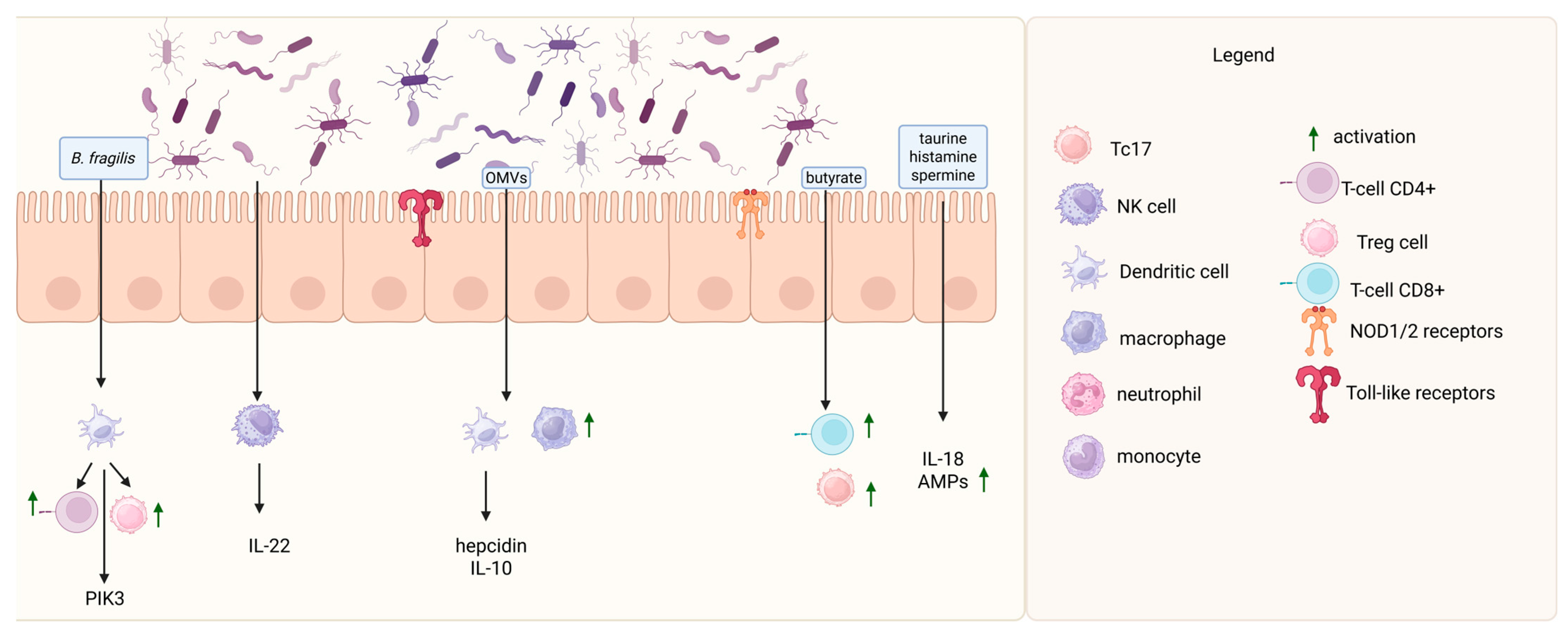
2. Gastrointestinal Microbiota in Health
3. Gut Microbiome Disbalance and Lymphoma Development
4. The Role of the Microbiome in B-Cell Lymphomas
4.1. The Role of the Gut Microbiome in DLBCL
4.2. The Role of the Gut Microbiome in FL
4.3. Evidence for a Causal Relationship Between Gut Microbiota and B-Cell Lymphomas
5. The Role of the Microbiome in Mucosa-Associated Lymphoid Tissue (MALT) Lymphoma
6. The Role of the Microbiome in NK/T-Cell Lymphomas
7. The Association Between the Gut Microbiome and Anticancer Treatment
7.1. The Gut Microbiome Composition and Chemotherapy (CHTH)
7.2. The Gut Microbiome Composition and CAR T-Cell Therapy
7.3. Antibiotics, Anticancer Treatment, and the Microbiome
8. Future Directions
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome Definition Re-Visited: Old Concepts and New Challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science 2009, 326, 1694. [Google Scholar] [CrossRef] [PubMed]
- Tremaroli, V.; Bäckhed, F. Functional Interactions between the Gut Microbiota and Host Metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The Microbiome and Innate Immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Tilg, H.; Zmora, N.; Adolph, T.E.; Elinav, E. The Intestinal Microbiota Fuelling Metabolic Inflammation. Nat. Rev. Immunol. 2020, 20, 40–54. [Google Scholar] [CrossRef]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef]
- Sinha, S.; Lin, G.; Ferenczi, K. The Skin Microbiome and the Gut-Skin Axis. Clin. Dermatol. 2021, 39, 829–839. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Riordan, K.J.; Sandhu, K.; Peterson, V.; Dinan, T.G. The Gut Microbiome in Neurological Disorders. Lancet Neurol. 2020, 19, 179–194. [Google Scholar] [CrossRef]
- Li, C.; Liu, H.; Lin, Y.; Pan, J.; Su, J. The Gut Microbiota and Respiratory Diseases: New Evidence. J. Immunol. Res. 2020. [Google Scholar] [CrossRef]
- Wu, J.; Wang, K.; Wang, X.; Pang, Y.; Jiang, C. The Role of the Gut Microbiome and Its Metabolites in Metabolic Diseases. Protein Cell 2021, 12, 360–373. [Google Scholar] [CrossRef]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe 2018, 24, 133–145.e5. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota across Multiple Body Habitats in Newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 852. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human Genetics Shape the Gut Microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Wang, S.; Charbonnier, L.M.; Noval Rivas, M.; Georgiev, P.; Li, N.; Gerber, G.; Bry, L.; Chatila, T.A. MyD88 Adaptor-Dependent Microbial Sensing by Regulatory T Cells Promotes Mucosal Tolerance and Enforces Commensalism. Immunity 2015, 43, 289–303. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Dahl, W.J.; Rivero Mendoza, D.; Lambert, J.M. Diet, Nutrients and the Microbiome. Prog. Mol. Biol. Transl. Sci. 2020, 171, 237–263. [Google Scholar] [CrossRef]
- Maurice, C.F.; Haiser, H.J.; Turnbaugh, P.J. Xenobiotics Shape the Physiology and Gene Expression of the Active Human Gut Microbiome. Cell 2013, 152, 39–50. [Google Scholar] [CrossRef]
- Stewart, C.J.; Embleton, N.D.; Marrs, E.C.L.; Smith, D.P.; Fofanova, T.; Nelson, A.; Skeath, T.; Perry, J.D.; Petrosino, J.F.; Berrington, J.E.; et al. Longitudinal Development of the Gut Microbiome and Metabolome in Preterm Neonates with Late Onset Sepsis and Healthy Controls. Microbiome 2017, 5, 75. [Google Scholar] [CrossRef]
- Cheng, J.; Ringel-Kulka, T.; Heikamp-De Jong, I.; Ringel, Y.; Carroll, I.; De Vos, W.M.; Salojärvi, J.; Satokari, R. Discordant Temporal Development of Bacterial Phyla and the Emergence of Core in the Fecal Microbiota of Young Children. ISME J. 2016, 10, 1002–1014. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’connor, E.M.; Cusack, S.; Harris, H.M.B.; Coakley, M.; Lakshminarayanan, B.; O’sullivan, O.; et al. Gut Microbiota Composition Correlates with Diet and Health in the Elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- Jeffery, I.B.; Lynch, D.B.; O’Toole, P.W. Composition and Temporal Stability of the Gut Microbiota in Older Persons. ISME J. 2016, 10, 170–182. [Google Scholar] [CrossRef]
- Sagan, L. On the Origin of Mitosing Cells. J. Theor. Biol. 1967, 14, 225–274. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. Towards a Natural System of Organisms: Proposal for the Domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef]
- Baruch, E.N.; Wang, J.; Wargo, J.A. Gut Microbiota and Antitumor Immunity: Potential Mechanisms for Clinical Effect. Cancer Immunol. Res. 2021, 9, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Caruso, R.; Warner, N.; Inohara, N.; Núñez, G. NOD1 and NOD2: Signaling, Host Defense, and Inflammatory Disease. Immunity 2014, 41, 898. [Google Scholar] [CrossRef]
- Li, D.; Wu, M. Pattern Recognition Receptors in Health and Diseases. Signal Transduct. Target. Ther. 2021, 6, 131. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Toll-like Receptors and Their Crosstalk with Other Innate Receptors in Infection and Immunity. Immunity 2011, 34, 637–650. [Google Scholar] [CrossRef]
- Verstak, B.; Nagpal, K.; Bottomley, S.P.; Golenbock, D.T.; Hertzog, P.J.; Mansell, A. MyD88 Adapter-like (Mal)/TIRAP Interaction with TRAF6 Is Critical for TLR2- and TLR4-Mediated NF-KappaB Proinflammatory Responses. J. Biol. Chem. 2009, 284, 24192–24203. [Google Scholar] [CrossRef] [PubMed]
- Platnich, J.M.; Muruve, D.A. NOD-like Receptors and Inflammasomes: A Review of Their Canonical and Non-Canonical Signaling Pathways. Arch. Biochem. Biophys. 2019, 670, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Mikkelsen, H.; Jungersen, G. Intracellular Pathogens: Host Immunity and Microbial Persistence Strategies. J. Immunol. Res. 2019. [Google Scholar] [CrossRef]
- Taniguchi, K.; Karin, M. NF-ΚB, Inflammation, Immunity and Cancer: Coming of Age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Brevi, A.; Cogrossi, L.L.; Grazia, G.; Masciovecchio, D.; Impellizzieri, D.; Lacanfora, L.; Grioni, M.; Bellone, M. Much More Than IL-17A: Cytokines of the IL-17 Family Between Microbiota and Cancer. Front. Immunol. 2020, 11, 565470. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T. The Nuclear Factor NF-KappaB Pathway in Inflammation. Cold. Spring Harb. Perspect. Biol. 2009, 1. [Google Scholar] [CrossRef]
- Kawari, M.; Akhtar, M.; Sager, M.; Basbous, Z.; Baydoun, I.; Kabanja, J.; Darweesh, M.; Mokhtar, N.; Kanfar, S.; Mutahar, E.; et al. Alterations of Gut Microbiome in Untreated Chronic Lymphocytic Leukemia (CLL); Future Therapeutic Potentials. Blood 2019, 134, 5455. [Google Scholar] [CrossRef]
- Elalaoui, K.; Weihe, C.; Oliver, A.; Craver, B.; Lai, H.Y.; Brooks, S.; Kim, D.; Martiny, J.; Whiteson, K.; Fleischman, A. Investigating the Role of the Gut Microbiome in the Inflammatory State of Myeloproliferative Neoplasms. Blood 2018, 132, 3051. [Google Scholar] [CrossRef]
- Calcinotto, A.; Brevi, A.; Chesi, M.; Ferrarese, R.; Garcia Perez, L.; Grioni, M.; Kumar, S.; Garbitt, V.M.; Sharik, M.E.; Henderson, K.J.; et al. Microbiota-Driven Interleukin-17-Producing Cells and Eosinophils Synergize to Accelerate Multiple Myeloma Progression. Nat. Commun. 2018, 9, 4832. [Google Scholar] [CrossRef]
- Uribe-Herranz, M.; Klein-González, N.; Rodríguez-Lobato, L.G.; Juan, M.; de Larrea, C.F. Gut Microbiota Influence in Hematological Malignancies: From Genesis to Cure. Int. J. Mol. Sci. 2021, 22, 1026. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.L.; Maier, I.; Dang, A.T.; Berry, D.; Liu, J.; Ruegger, P.M.; Yang, J.I.; Soto, P.A.; Presley, L.L.; Reliene, R.; et al. Intestinal Bacteria Modify Lymphoma Incidence and Latency by Affecting Systemic Inflammatory State, Oxidative Stress, and Leucocyte Genotoxicity. Cancer Res. 2013, 73, 4222. [Google Scholar] [CrossRef]
- Reiman, A.; Srinivasan, V.; Barone, G.; Last, J.I.; Wootton, L.L.; Davies, E.G.; Verhagen, M.M.; Willemsen, M.A.; Weemaes, C.M.; Byrd, P.J.; et al. Lymphoid Tumours and Breast Cancer in Ataxia Telangiectasia; Substantial Protective Effect of Residual ATM Kinase Activity against Childhood Tumours. Br. J. Cancer 2011, 105, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Chiu, B.C.H.; Hou, N. Epidemiology and Etiology of Non-Hodgkin Lymphoma. Cancer Treat. Res. 2015, 165, 1–25. [Google Scholar] [CrossRef]
- Diefenbach, C.S.; Peters, B.A.; Li, H.; Raphael, B.; Moskovits, T.; Hymes, K.; Schluter, J.; Chen, J.; Bennani, N.N.; Witzig, T.E.; et al. Microbial Dysbiosis Is Associated with Aggressive Histology and Adverse Clinical Outcome in B-Cell Non-Hodgkin Lymphoma. Blood Adv. 2021, 5, 1194–1198. [Google Scholar] [CrossRef]
- Yuan, L.; Wang, W.; Zhang, W.; Zhang, Y.; Wei, C.; Li, J.; Zhou, D. Gut Microbiota in Untreated Diffuse Large B Cell Lymphoma Patients. Front. Microbiol. 2021, 12, 646361. [Google Scholar] [CrossRef] [PubMed]
- Schmiester, M.; Maier, R.; Riedel, R.; Durek, P.; Frentsch, M.; Kolling, S.; Mashreghi, M.F.; Jenq, R.; Zhang, L.; Peterson, C.B.; et al. Flow Cytometry Can Reliably Capture Gut Microbial Composition in Healthy Adults as Well as Dysbiosis Dynamics in Patients with Aggressive B-Cell Non-Hodgkin Lymphoma. Gut Microbes 2022, 14, 2081475. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Mao, D.; Jin, C.; Wang, J.; Lai, Y.; Zhang, Y.; Zhou, M.; Ge, Q.; Zhang, P.; Sun, Y.; et al. The Gut Microbiota Correlate with the Disease Characteristics and Immune Status of Patients with Untreated Diffuse Large B-Cell Lymphoma. Front. Immunol. 2023, 14, 1105293. [Google Scholar] [CrossRef]
- Hooper, M.J.; LeWitt, T.M.; Pang, Y.; Veon, F.L.; Chlipala, G.E.; Feferman, L.; Green, S.J.; Sweeney, D.; Bagnowski, K.T.; Burns, M.B.; et al. Gut Dysbiosis in Cutaneous T-cell Lymphoma Is Characterized by Shifts in Relative Abundances of Specific Bacterial Taxa and Decreased Diversity in More Advanced Disease. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 1552–1563. [Google Scholar] [CrossRef]
- Shi, Z.; Hu, G.; Li, M.W.; Zhang, L.; Li, X.; Li, L.; Wang, X.; Fu, X.; Sun, Z.; Zhang, X.; et al. Gut Microbiota as Non-Invasive Diagnostic and Prognostic Biomarkers for Natural Killer/T-Cell Lymphoma. Gut 2023, 72, 1999–2002. [Google Scholar] [CrossRef]
- Zeze, K.; Hirano, A.; Torisu, T.; Esaki, M.; Shibata, H.; Moriyama, T.; Umeno, J.; Fujioka, S.; Okamoto, Y.; Fuyuno, Y.; et al. Mucosal Dysbiosis in Patients with Gastrointestinal Follicular Lymphoma. Hematol. Oncol. 2020, 38, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Matsuno, Y.; Torisu, T.; Shibata, H.; Hirano, A.; Umeno, J.; Kawasaki, K.; Fujioka, S.; Fuyuno, Y.; Moriyama, T.; et al. Gastric Microbiota in Patients with Helicobacter Pylori-Negative Gastric MALT Lymphoma. Medicine 2021, 100, e27287. [Google Scholar] [CrossRef]
- Xu, Z.F.; Zhao, D.; Wei, C.; Wang, W.; Zhang, Y.; Zhang, W.; Zhou, D. Characteristics and Prognostic Value of Gut Microbiota in Follicular Lymphoma. Oncol. Lett. 2024, 27, 207. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Yu, F.; Zhou, X.; Shi, H.; He, Q.; Song, X. Dissecting Causal Links between Gut Microbiota, Inflammatory Cytokines, and DLBCL: A Mendelian Randomization Study. Blood Adv. 2024, 8, 2268. [Google Scholar] [CrossRef]
- Li, B.; Han, Y.; Fu, Z.; Chai, Y.; Guo, X.; Du, S.; Li, C.; Wang, D. The Causal Relationship between Gut Microbiota and Lymphoma: A Two-Sample Mendelian Randomization Study. Front. Immunol. 2024, 15, 1397485. [Google Scholar] [CrossRef]
- Liang, J.; Liu, G.; Wang, W.; Xue, H. Causal Relationships between Gut Microbiota and Lymphoma: A Bidirectional Mendelian Randomization Study. Front. Cell. Infect. Microbiol. 2024, 14, 1374775. [Google Scholar] [CrossRef]
- Qian, J.; Zheng, W.; Fang, J.; Cheng, S.; Zhang, Y.; Zhuang, X.; Song, C. Causal Relationships of Gut Microbiota, Plasma Metabolites, and Metabolite Ratios with Diffuse Large B-Cell Lymphoma: A Mendelian Randomization Study. Front. Microbiol. 2024, 15, 1356437. [Google Scholar] [CrossRef] [PubMed]
- Violeta Filip, P.; Cuciureanu, D.; Sorina Diaconu, L.; Maria Vladareanu, A.; Silvia Pop, C. MALT Lymphoma: Epidemiology, Clinical Diagnosis and Treatment. J. Med. Life 2018, 11, 187–193. [Google Scholar] [CrossRef]
- Retnakumar, R.J.; Nath, A.N.; Nair, G.B.; Chattopadhyay, S. Gastrointestinal Microbiome in the Context of Helicobacter Pylori Infection in Stomach and Gastroduodenal Diseases. Prog. Mol. Biol. Transl. Sci. 2022, 192, 53–95. [Google Scholar] [CrossRef]
- Mirvish, J.J.; Pomerantz, R.G.; Falo, L.D.; Geskin, L.J. Role of Infectious Agents in Cutaneous T-Cell Lymphoma: Facts and Controversies. Clin. Dermatol. 2013, 31, 423–431. [Google Scholar] [CrossRef]
- Nguyen, V.; Huggins, R.H.; Lertsburapa, T.; Bauer, K.; Rademaker, A.; Gerami, P.; Guitart, J. Cutaneous T-Cell Lymphoma and Staphylococcus Aureus Colonization. J. Am. Acad. Dermatol. 2008, 59, 949–952. [Google Scholar] [CrossRef] [PubMed]
- Fujii, K. Pathogenesis of Cutaneous T Cell Lymphoma: Involvement of Staphylococcus Aureus. J. Dermatol. 2022, 49, 202–209. [Google Scholar] [CrossRef]
- Willerslev-Olsen, A.; Krejsgaard, T.; Lindahl, L.M.; Bonefeld, C.M.; Wasik, M.A.; Koralov, S.B.; Geisler, C.; Kilian, M.; Iversen, L.; Woetmann, A.; et al. Bacterial Toxins Fuel Disease Progression in Cutaneous T-Cell Lymphoma. Toxins 2013, 5, 1402–1421. [Google Scholar] [CrossRef] [PubMed]
- Shohat, M.; Shohat, B.; Mimouni, D.; Pauli, G.; Ellerbrok, H.; David, M.; Hodak, E. Human T-Cell Lymphotropic Virus Type 1 Provirus and Phylogenetic Analysis in Patients with Mycosis Fungoides and Their Family Relatives. Br. J. Dermatol. 2006, 155, 372–378. [Google Scholar] [CrossRef]
- Wood, G.S.; Salvekar, A.; Schaffer, J.; Crooks, C.F.; Henghold, W.; Fivenson, D.P.; Kim, Y.H.; Smoller, B.R. Evidence against a Role for Human T-Cell Lymphotrophic Virus Type I (HTLV-I) in the Pathogenesis of American Cutaneous T-Cell Lymphoma. J. Investig. Dermatol. 1996, 107, 301–307. [Google Scholar] [CrossRef]
- Khan, Z.M.; Sebenik, M.; Zucker-Franklin, D. Localization of Human T-Cell Lymphotropic Virus-1 Tax Proviral Sequences in Skin Biopsies of Patients with Mycosis Fungoides by in Situ Polymerase Chain Reaction. J. Investig. Dermatol. 1996, 106, 667–672. [Google Scholar] [CrossRef]
- Zucker-Franklin, D.; Couta Vas, E.E.; Rush, M.G.; Zouzias, D.C. Detection of Human T-Lymphotropic Virus-like Particles in Cultures of Peripheral Blood Lymphocytes from Patients with Mycosis Fungoides. Proc. Natl. Acad. Sci. USA 1991, 88, 7630–7634. [Google Scholar] [CrossRef]
- Pancake, B.A.; Zucker-Franklin, D.; Coutavas, E.E. The Cutaneous T Cell Lymphoma, Mycosis Fungoides, Is a Human T Cell Lymphotropic Virus-Associated Disease: A Study of 50 Patients. J. Clin. Investig. 1995, 95, 547–554. [Google Scholar] [CrossRef]
- Łyko, M.; Jankowska-Konsur, A. The Skin Microbiome in Cutaneous T-Cell Lymphomas (CTCL)-A Narrative Review. Pathogens 2022, 11, 935. [Google Scholar] [CrossRef]
- Polkowska-Pruszyńska, B.; Gerkowicz, A.; Krasowska, D. The Gut Microbiome Alterations in Allergic and Inflammatory Skin Diseases—An Update. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 455–464. [Google Scholar] [CrossRef]
- Nylund, L.; Nermes, M.; Isolauri, E.; Salminen, S.; De Vos, W.M.; Satokari, R. Severity of Atopic Disease Inversely Correlates with Intestinal Microbiota Diversity and Butyrate-Producing Bacteria. Allergy 2015, 70, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.; Boyle, F.; Pavlakis, N.; Clarke, S.; Guminski, A.; Eade, T.; Lamoury, G.; Carroll, S.; Morgia, M.; Kneebone, A.; et al. Emerging Evidence of the Gut Microbiome in Chemotherapy: A Clinical Review. Front. Oncol. 2021, 11, 706331. [Google Scholar] [CrossRef] [PubMed]
- Aarnoutse, R.; Ziemons, J.; Penders, J.; Rensen, S.S.; De Vos-Geelen, J.; Smidt, M.L. The Clinical Link between Human Intestinal Microbiota and Systemic Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 4145. [Google Scholar] [CrossRef]
- Deng, X.; Li, Z.; Li, G.; Li, B.; Jin, X.; Lyu, G. Comparison of Microbiota in Patients Treated by Surgery or Chemotherapy by 16S RRNA Sequencing Reveals Potential Biomarkers for Colorectal Cancer Therapy. Front. Microbiol. 2018, 9, 1607. [Google Scholar] [CrossRef]
- Zuo, F.; Yin, L.; Yang, X.; Wu, W.; Zhong, J.; Da, M.; Han, S. Gut Microbiome Associated with Chemotherapy-Induced Diarrhea from the CapeOX Regimen as Adjuvant Chemotherapy in Resected Stage III Colorectal Cancer. Gut Pathog. 2019, 11, 18. [Google Scholar] [CrossRef]
- Kong, C.; Gao, R.; Yan, X.; Huang, L.; He, J.; Li, H.; You, J.; Qin, H. Alterations in Intestinal Microbiota of Colorectal Cancer Patients Receiving Radical Surgery Combined with Adjuvant CapeOx Therapy. Sci. China Life Sci. 2019, 62, 1178–1193. [Google Scholar] [CrossRef]
- Han, S.; Yang, X.; Pan, Y.; Xu, J. Effects of Postoperative Adjuvant Chemotherapy and Palliative Chemotherapy on the Gut Microbiome in Colorectal Cancer. Microb. Pathog. 2020, 149, 104343. [Google Scholar] [CrossRef]
- Galloway-Peña, J.R.; Smith, D.P.; Sahasrabhojane, P.; Wadsworth, W.D.; Fellman, B.M.; Ajami, N.J.; Shpall, E.J.; Daver, N.; Guindani, M.; Petrosino, J.F.; et al. Characterization of Oral and Gut Microbiome Temporal Variability in Hospitalized Cancer Patients. Genome Med. 2017, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Galloway-Peña, J.R.; Shi, Y.; Peterson, C.B.; Sahasrabhojane, P.; Gopalakrishnan, V.; Brumlow, C.E.; Daver, N.G.; Alfayez, M.; Boddu, P.C.; Khan, M.A.W.; et al. Gut Microbiome Signatures Are Predictive of Infectious Risk Following Induction Therapy for Acute Myeloid Leukemia. Clin. Infect. Dis. 2020, 71, 63–71. [Google Scholar] [CrossRef]
- Montassier, E.; Batard, E.; Massart, S.; Gastinne, T.; Carton, T.; Caillon, J.; Le Fresne, S.; Caroff, N.; Hardouin, J.B.; Moreau, P.; et al. 16S RRNA Gene Pyrosequencing Reveals Shift in Patient Faecal Microbiota during High-Dose Chemotherapy as Conditioning Regimen for Bone Marrow Transplantation. Microb. Ecol. 2014, 67, 690–699. [Google Scholar] [CrossRef]
- Montassier, E.; Al-Ghalith, G.A.; Ward, T.; Corvec, S.; Gastinne, T.; Potel, G.; Moreau, P.; de la Cochetiere, M.F.; Batard, E.; Knights, D. Pretreatment Gut Microbiome Predicts Chemotherapy-Related Bloodstream Infection. Genome Med. 2016, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Uzan-Yulzari, A.; Morr, M.; Tareef-Nabwani, H.; Ziv, O.; Magid-Neriya, D.; Armoni, R.; Muller, E.; Leibovici, A.; Borenstein, E.; Louzoun, Y.; et al. The Intestinal Microbiome, Weight, and Metabolic Changes in Women Treated by Adjuvant Chemotherapy for Breast and Gynecological Malignancies. BMC Med. 2020, 18, 281. [Google Scholar] [CrossRef]
- Zhao, Z.; Fei, K.; Bai, H.; Wang, Z.; Duan, J.; Wang, J. Metagenome Association Study of the Gut Microbiome Revealed Biomarkers Linked to Chemotherapy Outcomes in Locally Advanced and Advanced Lung Cancer. Thorac. Cancer 2021, 12, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Zhang, X.; Fan, Y.; Chen, L.; Ma, X.; Yu, H.; Li, J.; Guan, X.; Zhao, P.; Yang, J. Changes of Intestinal Microbiota in Ovarian Cancer Patients Treated with Surgery and Chemotherapy. Cancer Manag. Res. 2020, 12, 8125–8135. [Google Scholar] [CrossRef]
- Iida, N.; Mizukoshi, E.; Yamashita, T.; Terashima, T.; Arai, K.; Seishima, J.; Kaneko, S. Overuse of Antianaerobic Drug Is Associated with Poor Postchemotherapy Prognosis of Patients with Hepatocellular Carcinoma. Int. J. Cancer 2019, 145, 2701–2711. [Google Scholar] [CrossRef] [PubMed]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline Gut Microbiota Predicts Clinical Response and Colitis in Metastatic Melanoma Patients Treated with Ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The Commensal Microbiome Is Associated with Anti-PD-1 Efficacy in Metastatic Melanoma Patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Pal, S.K.; Li, S.M.; Wu, X.; Qin, H.; Kortylewski, M.; Hsu, J.; Carmichael, C.; Frankel, P. Stool Bacteriomic Profiling in Patients with Metastatic Renal Cell Carcinoma Receiving Vascular Endothelial Growth Factor-Tyrosine Kinase Inhibitors. Clin. Cancer Res. 2015, 21, 5286–5293. [Google Scholar] [CrossRef]
- Xu, Z.F.; Yuan, L.; Zhang, Y.; Zhang, W.; Wei, C.; Wang, W.; Zhao, D.; Zhou, D.; Li, J. The Gut Microbiome Correlated to Chemotherapy Efficacy in Diffuse Large B-Cell Lymphoma Patients. Hematol. Rep. 2024, 16, 63–75. [Google Scholar] [CrossRef]
- Yoon, S.E.; Kang, W.; Choi, S.; Park, Y.; Chalita, M.; Kim, H.; Lee, J.H.; Hyun, D.W.; Ryu, K.J.; Sung, H.; et al. The Influence of Microbial Dysbiosis on Immunochemotherapy-Related Efficacy and Safety in Diffuse Large B-Cell Lymphoma. Blood 2023, 141, 2224–2238. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, J.; Yakoub-Agha, I. Chimeric Antigen-Receptor T-Cell Therapy for Hematological Malignancies and Solid Tumors: Clinical Data to Date, Current Limitations and Perspectives. Curr. Res. Transl. Med. 2017, 65, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Sermer, D.; Brentjens, R. CAR T-Cell Therapy: Full Speed Ahead. Hematol. Oncol. 2019, 37 (Suppl. S1), 95–100. [Google Scholar] [CrossRef] [PubMed]
- Abid, M.B. Could the Menagerie of the Gut Microbiome Really Cure Cancer? Hope or Hype. J. Immunother. Cancer 2019, 7, 92. [Google Scholar] [CrossRef]
- Abid, M.B.; Shah, N.N.; Maatman, T.C.; Hari, P.N. Gut Microbiome and CAR-T Therapy. Exp. Hematol. Oncol. 2019, 8, 31. [Google Scholar] [CrossRef]
- Smith, M.; Littmann, E.; Slingerland, J.; Clurman, A.; Slingerland, A.E.; Taur, Y.; Peled, J.U.; Park, J.; O’Cearbhaill, R.; Mailankody, S.; et al. Intestinal Microbiome Analyses Identify Biomarkers for Patient Response to CAR T Cell Therapy. Biol. Blood Marrow Transplant. 2019, 25, S177. [Google Scholar] [CrossRef][Green Version]
- Smith, M.; Dai, A.; Ghilardi, G.; Amelsberg, K.V.; Devlin, S.M.; Pajarillo, R.; Slingerland, J.B.; Beghi, S.; Herrera, P.S.; Giardina, P.; et al. Gut Microbiome Correlates of Response and Toxicity Following Anti-CD19 CAR T Cell Therapy. Nat. Med. 2022, 28, 713, Erratum in Nat. Med. 2023, 29, 2954. [Google Scholar] [CrossRef]
- Hu, Y.; Li, J.; Ni, F.; Yang, Z.; Gui, X.; Bao, Z.; Zhao, H.; Wei, G.; Wang, Y.; Zhang, M.; et al. CAR-T Cell Therapy-Related Cytokine Release Syndrome and Therapeutic Response Is Modulated by the Gut Microbiome in Hematologic Malignancies. Nat. Commun. 2022, 13, 5313. [Google Scholar] [CrossRef]
- Stein-Thoeringer, C.K.; Saini, N.Y.; Zamir, E.; Blumenberg, V.; Schubert, M.L.; Mor, U.; Fante, M.A.; Schmidt, S.; Hayase, E.; Hayase, T.; et al. A Non-Antibiotic-Disrupted Gut Microbiome Is Associated with Clinical Responses to CD19-CAR-T Cell Cancer Immunotherapy. Nat. Med. 2023, 29, 906–916. [Google Scholar] [CrossRef]
- Ohmoto, A.; Fuji, S. Infection Profiles of Different Chemotherapy Regimens and the Clinical Feasibility of Antimicrobial Prophylaxis in Patients with DLBCL. Blood Rev. 2021, 46, 100738. [Google Scholar] [CrossRef]
- Yıldız, A.; Öztürk, H.B.A.; Albayrak, M.; Pala, Ç.; Şahin, O.; Maral, S.; Okutan, H. Is Antimicrobial Prophylaxis Necessary for Lymphoma Patients? A Single Centre, Real-Life Experience. J. Oncol. Pharm. Pract. 2019, 25, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; Swaminathan, S.; Almyroudis, N.G.; Angarone, M.; Baluch, A.; Barros, N.; Buss, B.; Cohen, S.; Cooper, B.; Chiang, A.D.; et al. Prevention and Treatment of Cancer-Related Infections, Version 3.2024, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2024, 22, 617–644. [Google Scholar] [CrossRef] [PubMed]
- Kuczma, M.P.; Ding, Z.C.; Li, T.; Habtetsion, T.; Chen, T.; Hao, Z.; Bryan, L.; Singh, N.; Kochenderfer, J.N.; Zhou, G. The Impact of Antibiotic Usage on the Efficacy of Chemoimmunotherapy Is Contingent on the Source of Tumor-Reactive T Cells. Oncotarget 2017, 8, 111931–111942. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Rehman, A.; Rehman, L.; Darbaniyan, F.; Blumenberg, V.; Schubert, M.-L.; Mor, U.; Zamir, E.; Schmidt, S.; Hayase, T.; et al. Antibiotic-Induced Loss of Gut Microbiome Metabolic Output Correlates with Clinical Responses to CAR T-Cell Therapy. Blood 2025, 145, 823–839. [Google Scholar] [CrossRef]
- Pernas, B.; Iacoboni, G.; Los-Arcos, I.; Carpio, C.; Márquez-Algaba, E.; Sanchez-Salinas, M.A.; Albasanz, A.; Esperalba, J.; Viñado, B.; Camps, I.R.; et al. Patients with Aggressive B-Cell Lymphoma Receiving CAR T-Cell Therapy Have a Low Rate of Severe Infections despite Lack of Universal Antibacterial and Antifungal Prophylaxis. Eur. J. Haematol. 2024, 113, 227–234. [Google Scholar] [CrossRef]
| Authors (Year) | Study Population and Study Type | Sample Type | Method | Observed Changes in NHL Patients |
|---|---|---|---|---|
| Yuan et al. (2021) [46] | 25 untreated DLBCL patients vs. 26 healthy controls; cross-sectional study | Fecal samples | 16s rRNA sequencing | ↑ abundance of Proteobacteria, Gammaproteobacteria, Enterobacteriales, Enterobacteriaceae, Escherichia–Shigella, Enterococcus, Veillonella, and Prevotella-2 ↓ abundance of Allisonella, Lachnospira, and Roseburiain |
| Diefenbach et al. (2021) [45] | 51 NHL patients vs. 58 healthy controls; cross-sectional study | Fecal samples | 16s rRNA sequencing | ↑ Bacteroidetes with a simultaneous decrease in Firmicutes ↑ relative abundance of Dorea formicigenerans and Faecalibacterium prausnitzii in responders |
| Schmiester et al. (2022) [47] | 10 DLBCL, 1 primary mediastinal large B-cell lymphoma, and 1 high-grade FL vs. 10 controls; cross-sectional study | Fecal samples | single-cell flow cytometry (FCM) and 16s rRNA sequencing | ↑ abundance of Firmicutes, the family Lachnospiraceae, and the genus Roseburia ↓ α-diversity during CHTH |
| Lin et al. (2023) [48] | 35 untreated DLBCL patients vs. 20 healthy controls; cross-sectional study | Fecal samples | 16s rRNA sequencing | ↓ β-diversity *, ↑ abundance of p_Proteobacteria *, p_Verrucomicrobia *, p_Synergistetes *, g_Escherichia–Shigella *, g_Veillonella *, g_Roseburia *, g_Lachnoclostridium *, g_Alistipes * ↓ abundance of p_Bacteroidetes *, g_Bacteroides *, g_Prevotella_9 *, and g_Megamonas * |
| Hooper et al. (2022) [49] | 27 MF, 5 SS, and 6 with non-MF/SS CTCL vs. 13 healthy controls; cross-sectional study | Fecal samples | 16s rRNA sequencing | ↓ α-diversity |
| Shi et al. (2022) [50] | 30 untreated NKTCL patients vs. 20 healthy controls; cross-sectional study | Fecal samples | shotgun metagenomic sequencing | ↑ Veillonella and Streptococcus in NKTCL group |
| Zeze et al. (2020) [51] | 20 GI-FL patients vs. 20 controls; cross-sectional study | Mucosal biopsy samples from the second portion of the duodenum | 16s rRNA sequencing | ↓ α-diversity * ↓ abundance of Sporomusa, Rothia, Prevotella, and the family Gemellacea * |
| Tanaka et al. (2021) [52] | 33 Helicobacter pylori-negative MALT lymphoma patients vs. 27 controls; cross-sectional study | Mucosal biopsy from the gastric body | 16s rRNA sequencing | ↑ abundance of Burkholderia and Sphingomonas ↓ abundance of Prevotella and Veillonella ↓ α-diversity |
| Xu et al. (2024) [53] | 28 treatment-naïve FL patients vs. 18 sex- and age-matched healthy controls; cross-sectional study | Fecal samples | 16s rRNA sequencing | ↑ abundance of Ruminococcaceae ↑ Ruminococcus is a strong indicator of tumor burden ↓ abundance of Coriobacteriaceae |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Łyko, M.; Maj, J.; Jankowska-Konsur, A. The Role of the Gut Microbiome in Non-Hodgkin Lymphoma (NHL): A Focus on Diffuse Large B-Cell Lymphoma, Follicular Lymphoma, Cutaneous T-Cell Lymphoma, and NK/T-Cell Lymphoma. Cancers 2025, 17, 1709. https://doi.org/10.3390/cancers17101709
Łyko M, Maj J, Jankowska-Konsur A. The Role of the Gut Microbiome in Non-Hodgkin Lymphoma (NHL): A Focus on Diffuse Large B-Cell Lymphoma, Follicular Lymphoma, Cutaneous T-Cell Lymphoma, and NK/T-Cell Lymphoma. Cancers. 2025; 17(10):1709. https://doi.org/10.3390/cancers17101709
Chicago/Turabian StyleŁyko, Magdalena, Joanna Maj, and Alina Jankowska-Konsur. 2025. "The Role of the Gut Microbiome in Non-Hodgkin Lymphoma (NHL): A Focus on Diffuse Large B-Cell Lymphoma, Follicular Lymphoma, Cutaneous T-Cell Lymphoma, and NK/T-Cell Lymphoma" Cancers 17, no. 10: 1709. https://doi.org/10.3390/cancers17101709
APA StyleŁyko, M., Maj, J., & Jankowska-Konsur, A. (2025). The Role of the Gut Microbiome in Non-Hodgkin Lymphoma (NHL): A Focus on Diffuse Large B-Cell Lymphoma, Follicular Lymphoma, Cutaneous T-Cell Lymphoma, and NK/T-Cell Lymphoma. Cancers, 17(10), 1709. https://doi.org/10.3390/cancers17101709






