Amplification of Hippo Signaling Pathway Genes Is Governed and Implicated in the Serous Subtype-Specific Ovarian Carcino-Genesis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Expression Profiles
2.2. Pathway Activation Scoring
2.3. Hierarchical Clustering Analysis
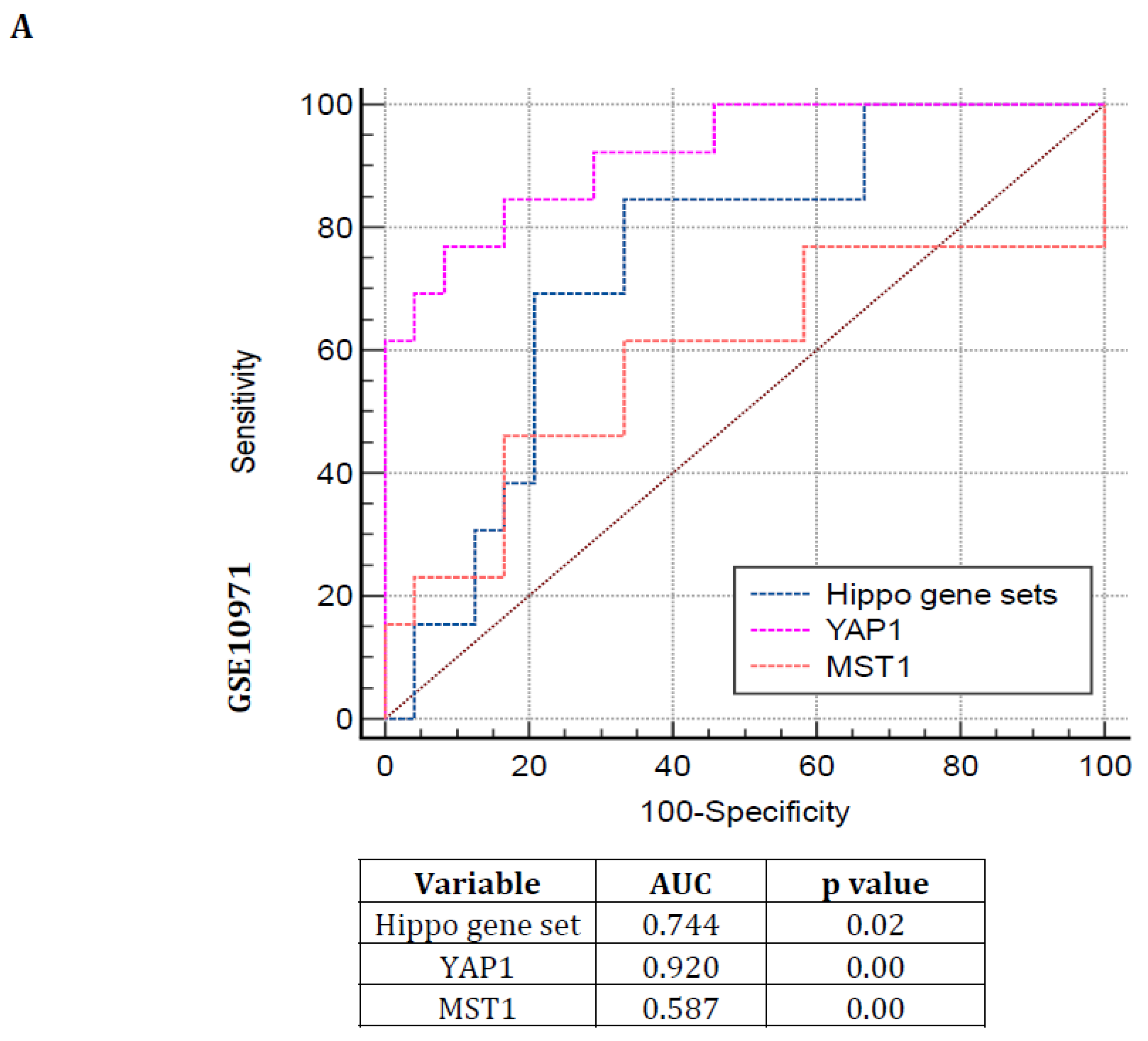
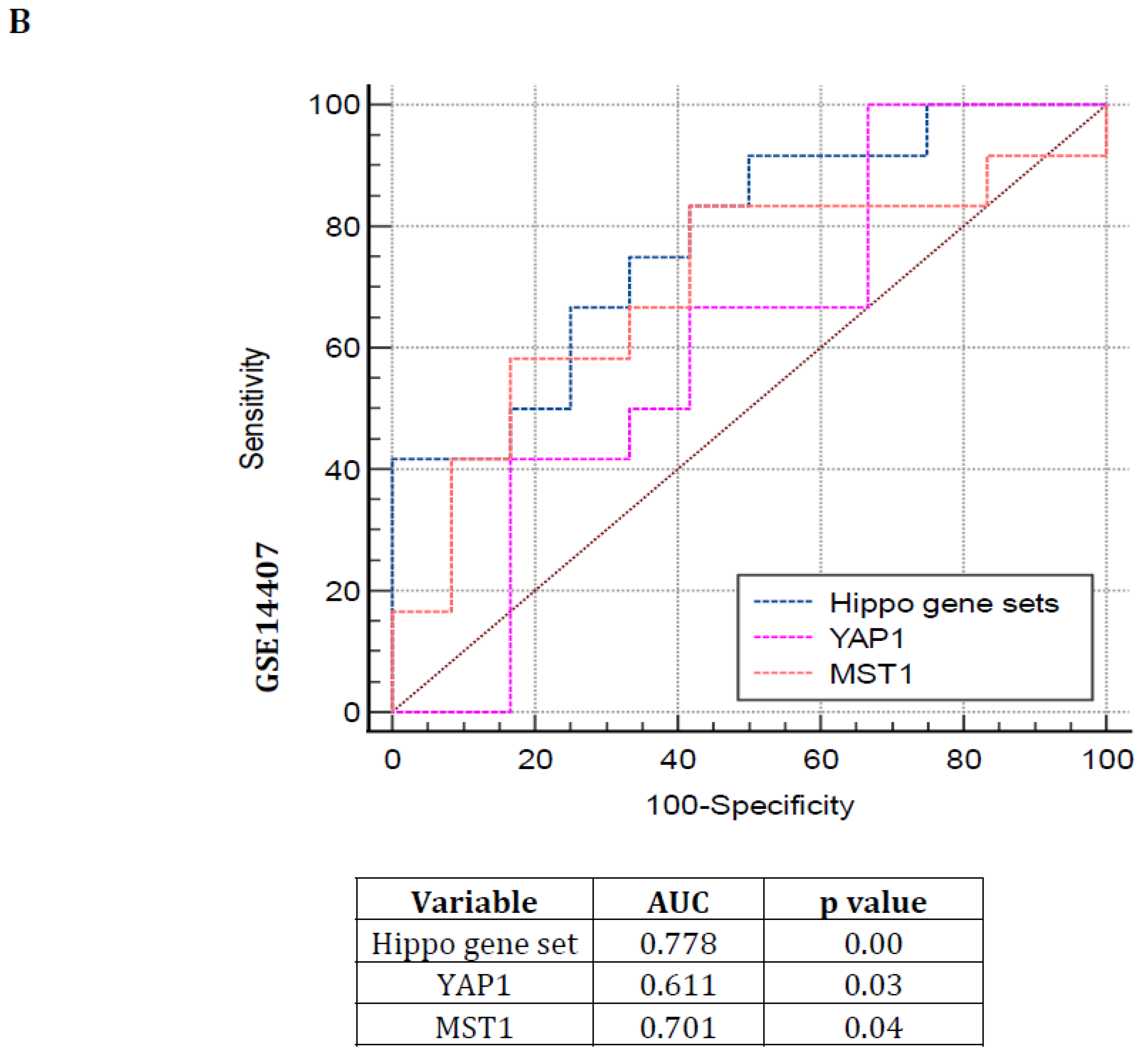
2.4. Receiver Operating Characteristic (ROC) Curve
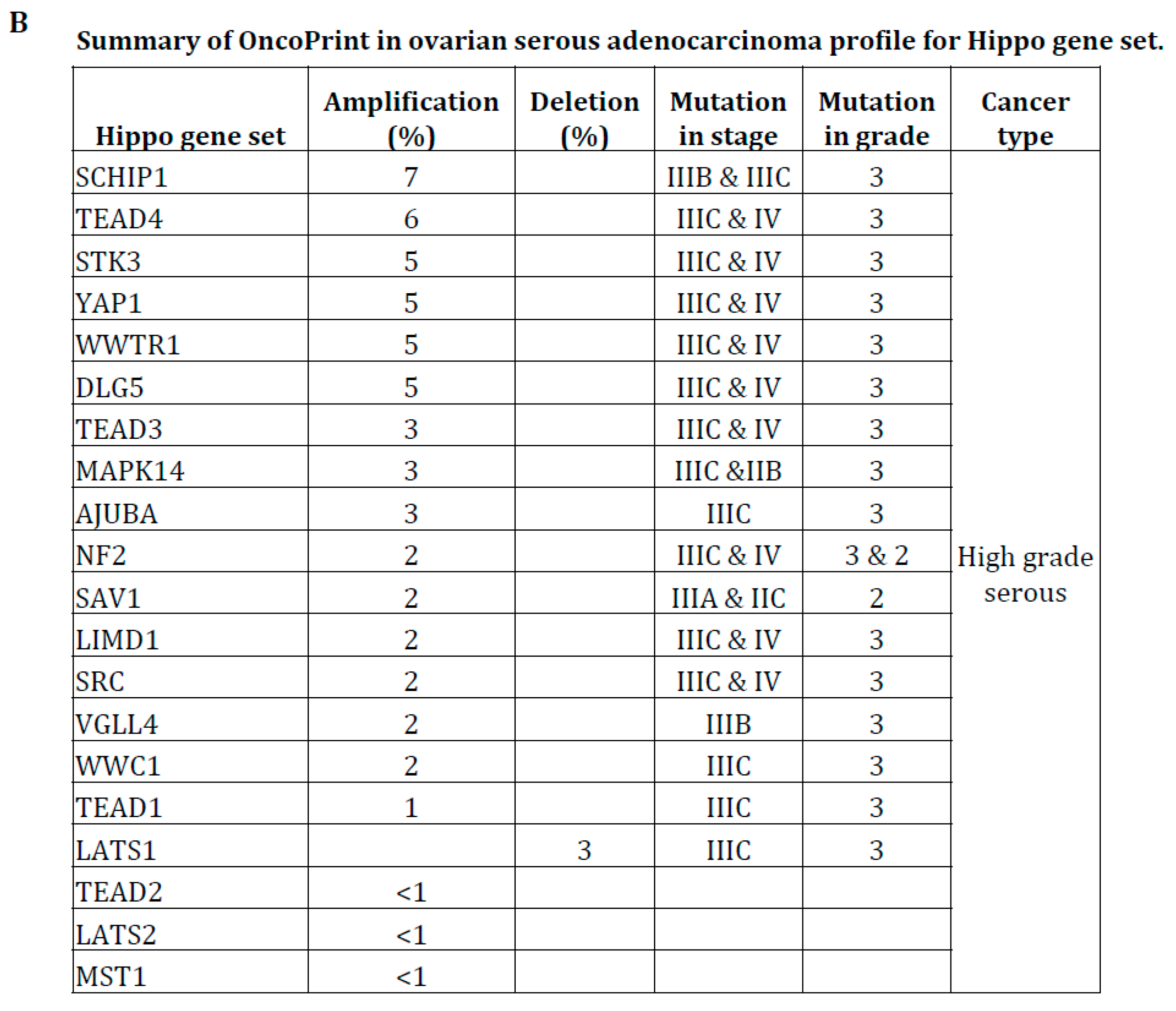
2.5. Mutation Analysis
2.6. Survival Curve
3. Results
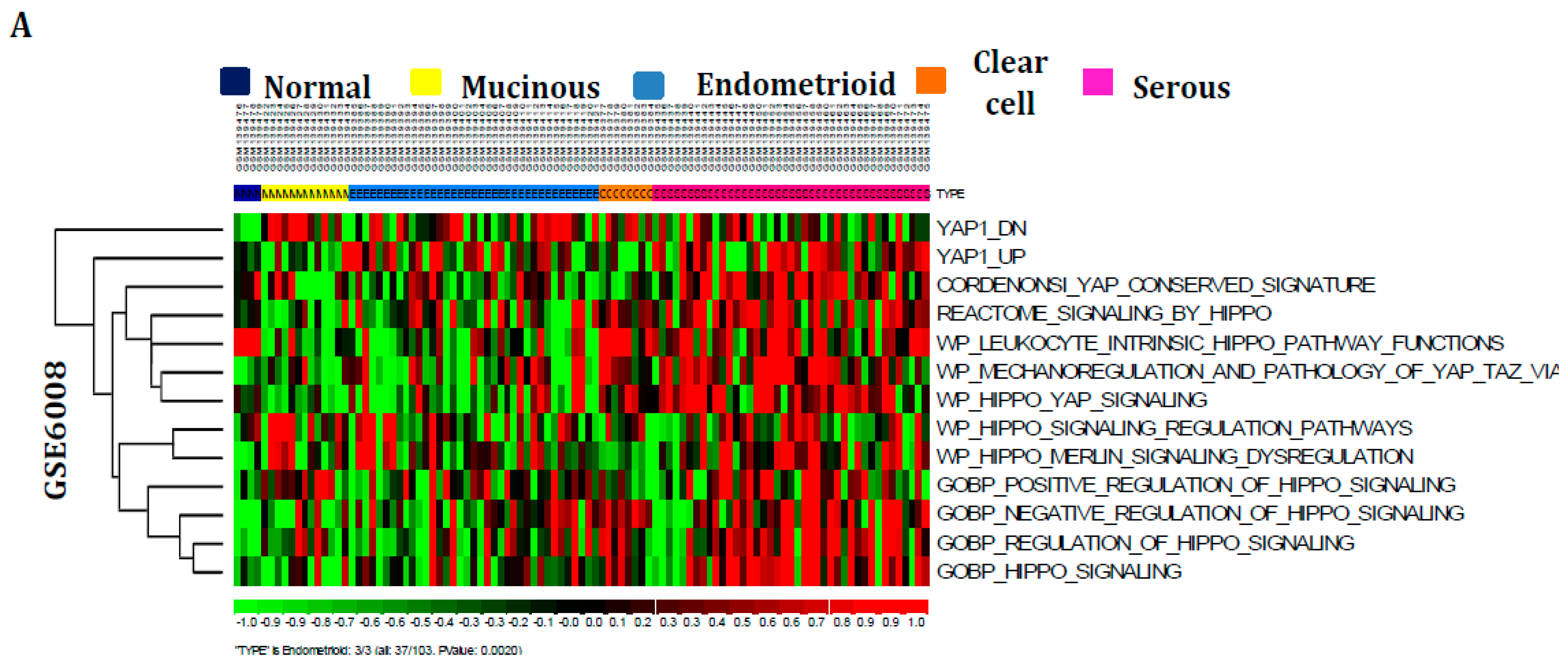
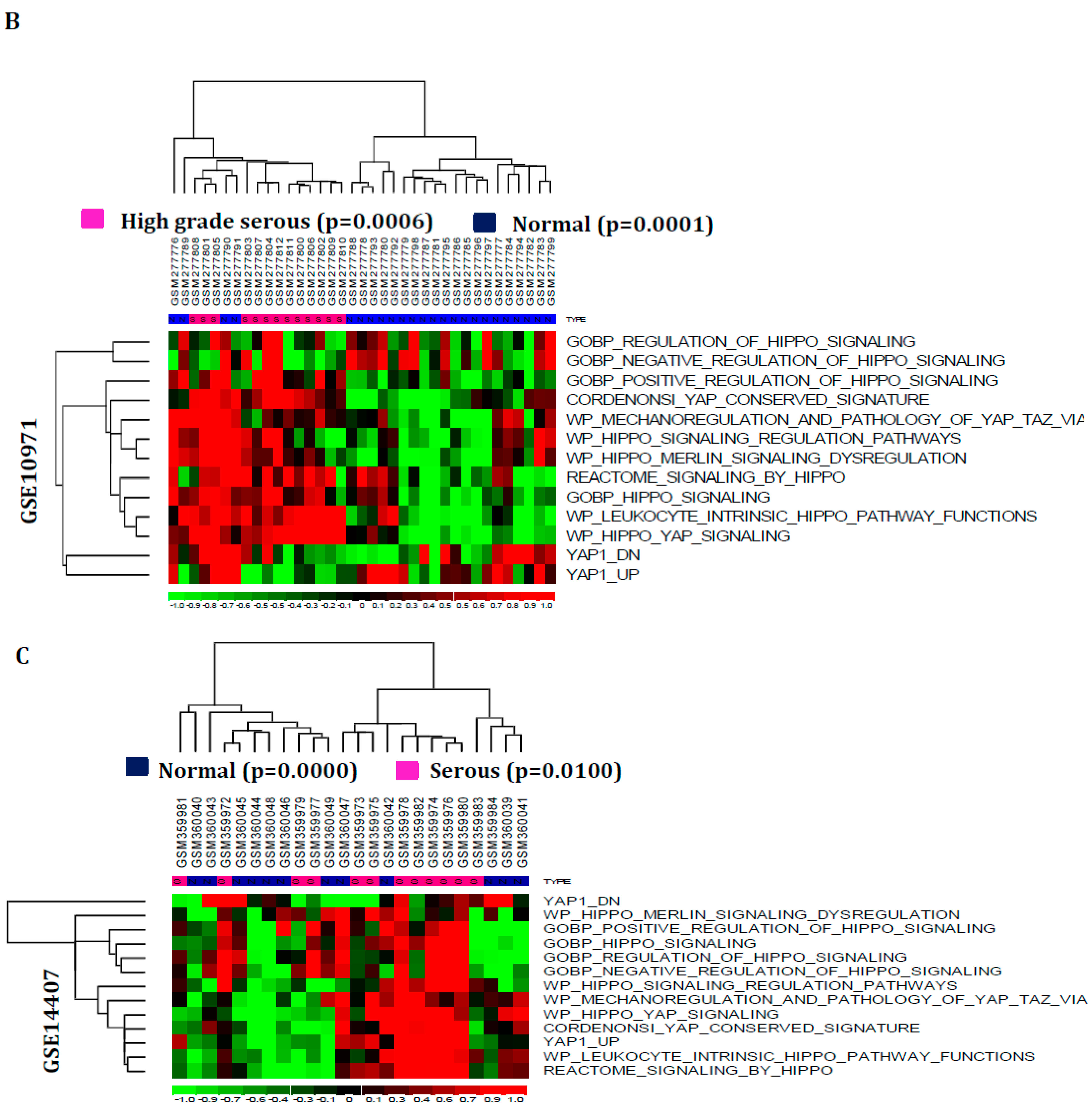
3.1. Hippo Signaling Pathway Is Dysregulated Extensively in Serous Subtypes of Ovarian Cancer
3.2. Hippo Signaling Gene Set Is Also Greatly Enriched in Serous Subtypes
3.3. Hippo Pathway Gene Expressions Are Showing Poor Prognosis with Greater Specificity and Sensitivity in Serous Subtype Ovarian Tumors
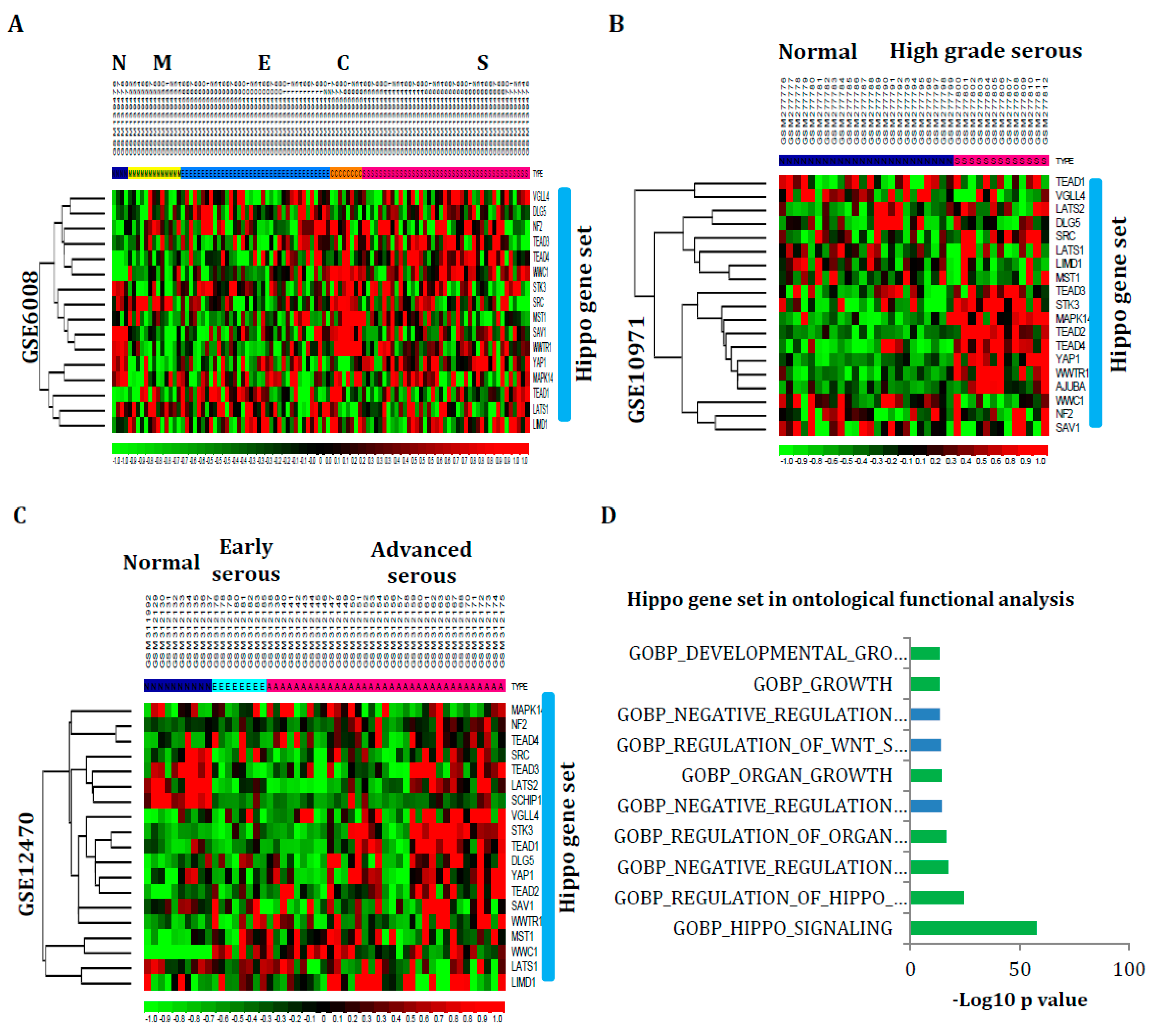
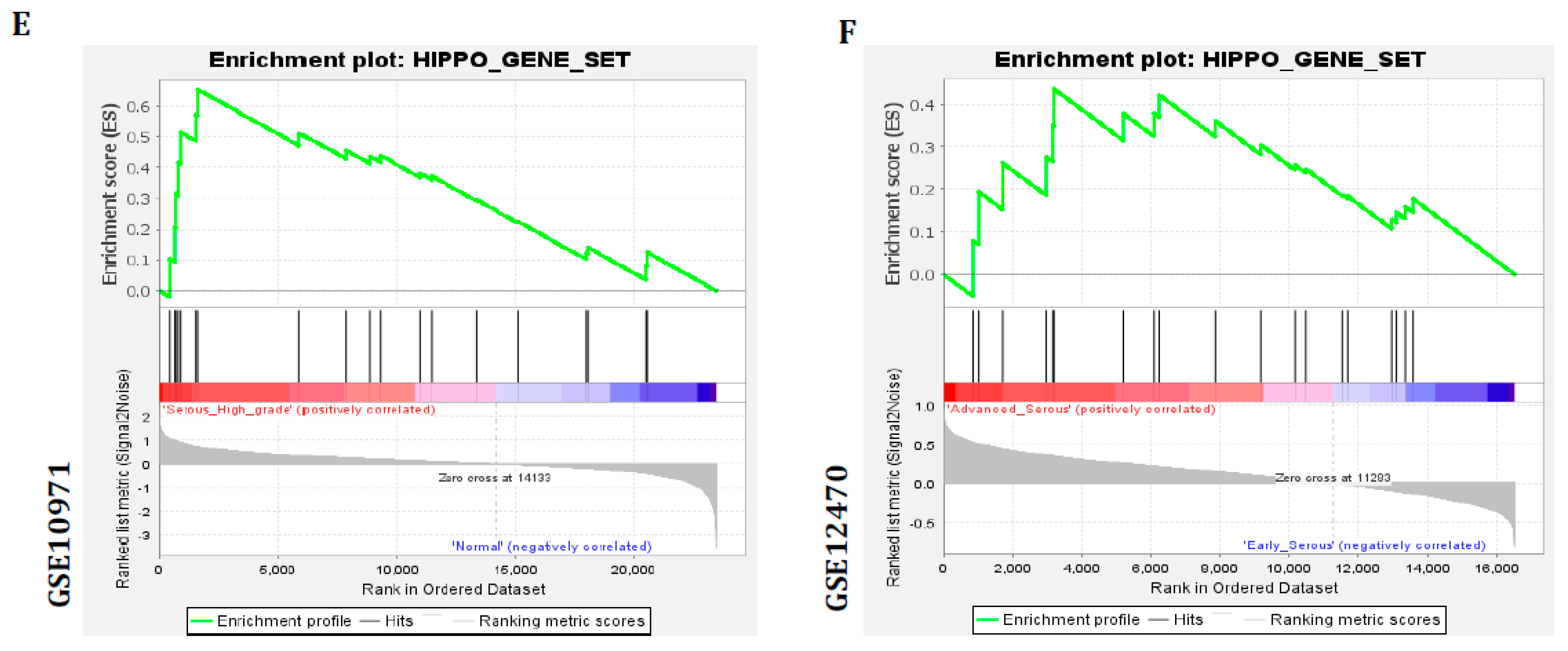
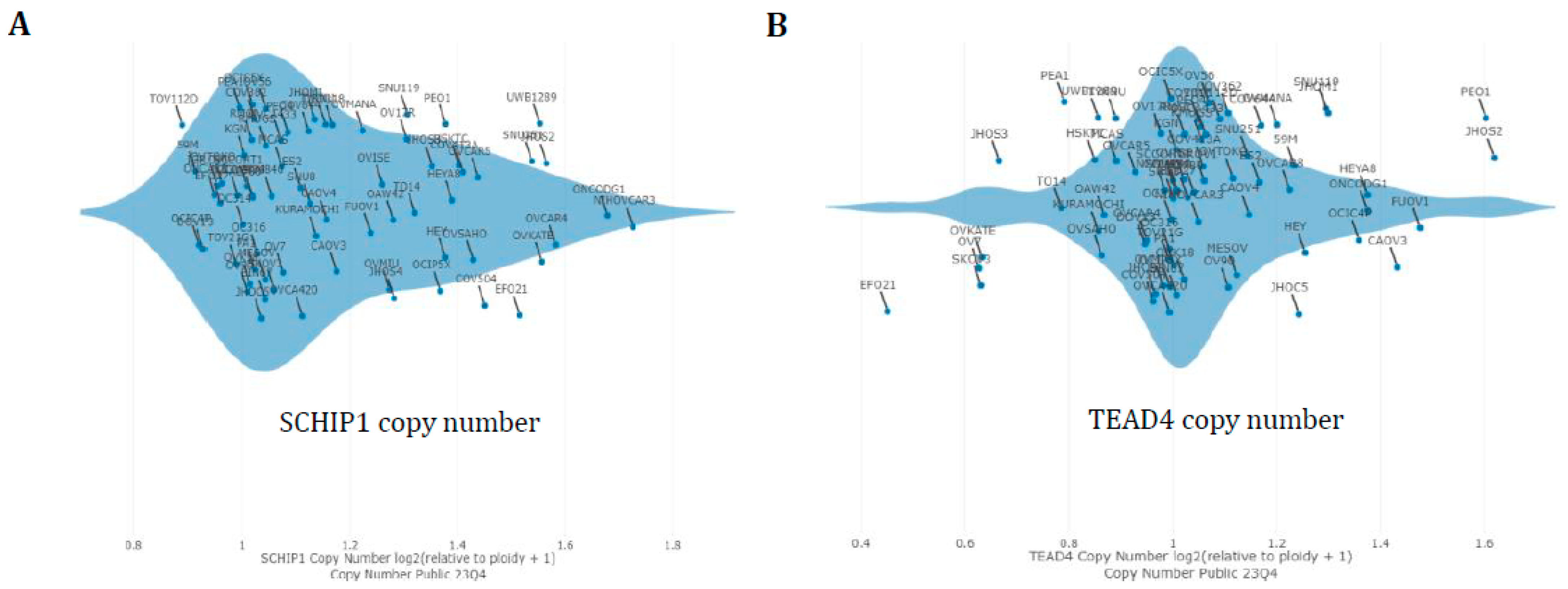
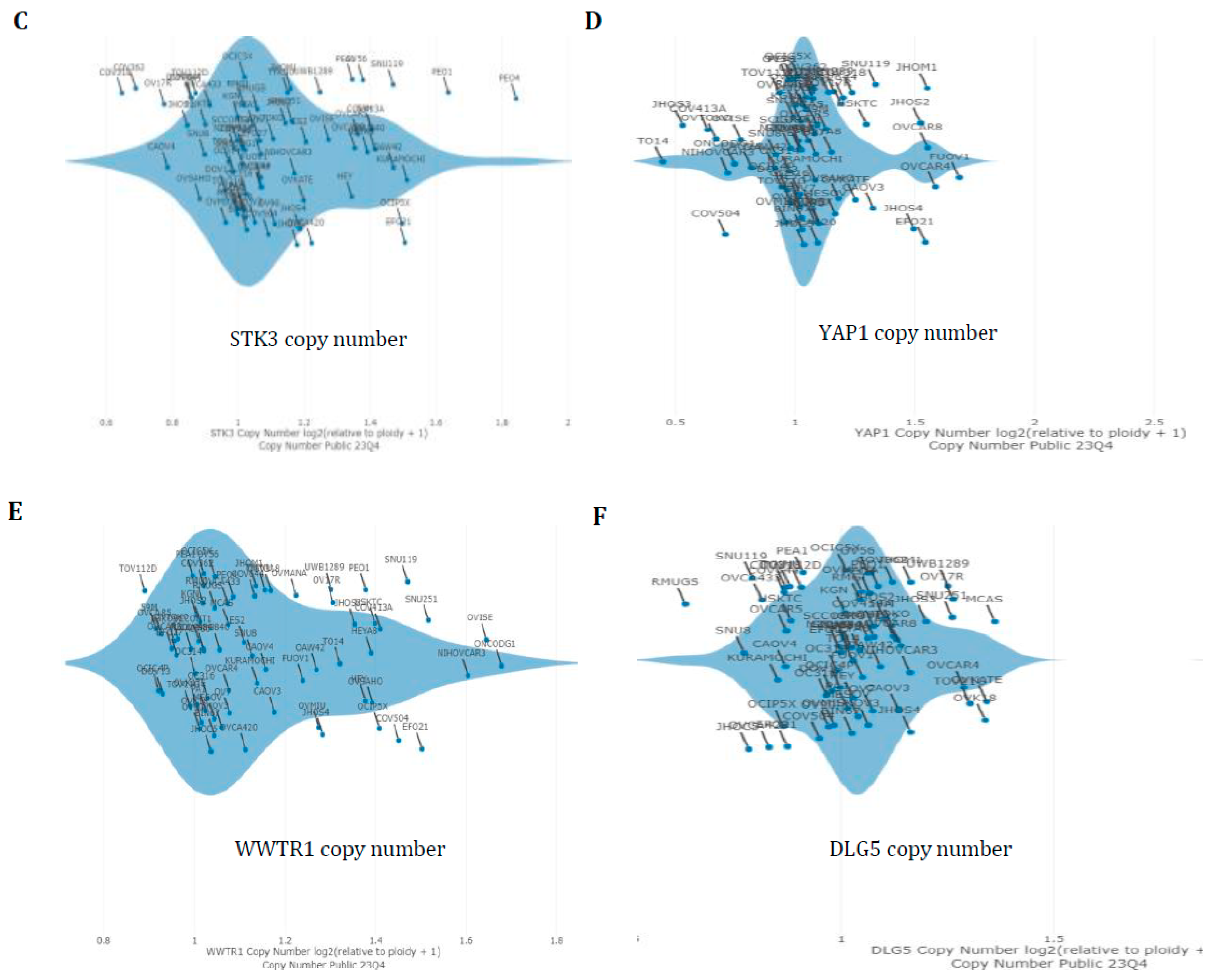
3.4. Hippo Gene Set Is Amplified Significantly in High-Grade Serous Ovarian Cancer
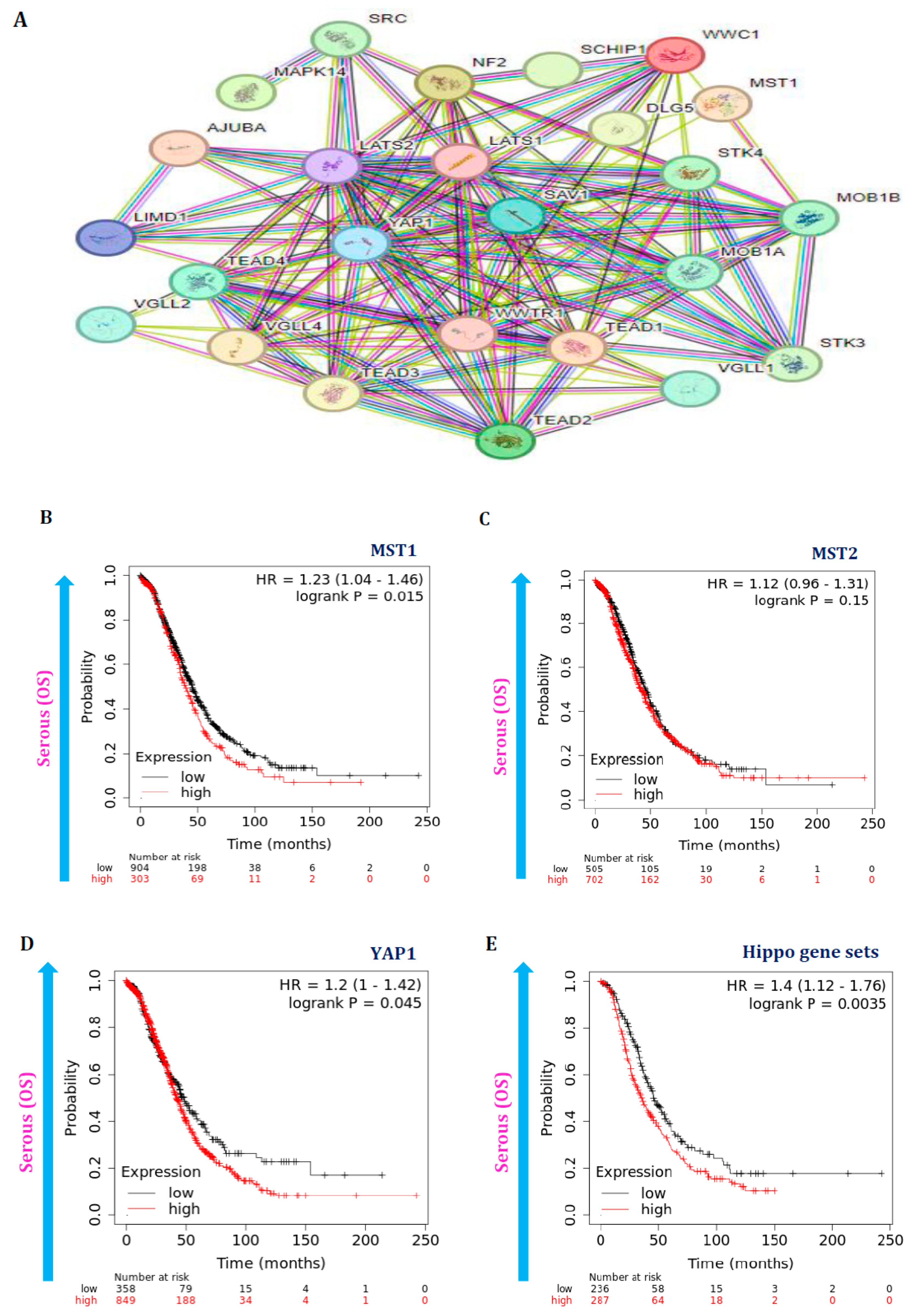
3.5. Hippo Pathway Gene Expressions Are Also Associated with Poor Survival in Serous Ovarian Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clark, K.L.; George, J.W.; Przygrodzka, E.; Plewes, M.R.; Hua, G.; Wang, C.; Davis, J.S. Hippo Signaling in the Ovary: Emerging Roles in Development, Fertility, and Disease. Endocr. Rev. 2022, 43, 1074–1096. [Google Scholar] [CrossRef] [PubMed]
- Vanselow, J.; Christenson, L.K.; Pate, J.L. Editorial: Regulation of Dynamic Changes and Remodeling Events During the Formation, Rescue and Regression of the Corpus Luteum. Front. Endocrinol. 2020, 11, 244. [Google Scholar] [CrossRef] [PubMed]
- Lumish, M.A.; Kohn, E.C.; Tew, W.P. Top Advances of the Year: Ovarian Cancer. Cancer 2024, 130, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; He, C.; Huang, C.; Hua, G.; Chen, X.; Timm, B.K.; Maclin, V.M.; Haggerty, A.A.; Aust, S.K.; Golden, D.M.; et al. Reprogramming of Ovarian Granulosa Cells by YAP1 Leads to Development of High-Grade Cancer with Mesenchymal Lineage and Serous Features. Sci. Bull. 2020, 65, 1281–1296. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fadare, O.; Xiang, L.; Kong, B.; Zheng, W. Ovarian Serous Carcinoma: Recent Concepts on Its Origin and Carcinogenesis. J. Hematol. Oncol. 2012, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Seidman, J.D.; Horkayne-Szakaly, I.; Haiba, M.; Boice, C.R.; Kurman, R.J.; Ronnett, B.M. The Histologic Type and Stage Distribution of Ovarian Carcinomas of Surface Epithelial Origin. Int. J. Gynecol. Pathol. 2004, 23, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Karnezis, A.N.; Cho, K.R.; Gilks, C.B.; Pearce, C.L.; Huntsman, D.G. The Disparate Origins of Ovarian Cancers: Pathogenesis and Prevention Strategies. Nat. Rev. Cancer 2017, 17, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Klinkebiel, D.; Zhang, W.; Akers, S.N.; Odunsi, K.; Karpf, A.R. DNA Methylome Analyses Implicate Fallopian Tube Epithelia as the Origin for High-Grade Serous Ovarian Cancer. Mol. Cancer Res. 2016, 14, 787–794. [Google Scholar] [CrossRef]
- Calses, P.C.; Crawford, J.J.; Lill, J.R.; Dey, A. Hippo Pathway in Cancer: Aberrant Regulation and Therapeutic Opportunities. Trends Cancer 2019, 5, 297–307. [Google Scholar] [CrossRef]
- Taha, Z.; Janse van Rensburg, H.J.; Yang, X. The Hippo Pathway: Immunity and Cancer. Cancers 2018, 10, 94. [Google Scholar] [CrossRef]
- Manno, G.; Filorizzo, C.; Fanale, D.; Brando, C.; Di Lisi, D.; Lunetta, M.; Bazan, V.; Russo, A.; Novo, G. Role of the HIPPO Pathway as Potential Key Player in the Cross Talk between Oncology and Cardiology. Crit. Rev. Oncol. Hematol. 2021, 159, 103246. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Hong, W. Hippo Signaling at the Hallmarks of Cancer and Drug Resistance. Cells 2024, 13, 564. [Google Scholar] [CrossRef] [PubMed]
- Maugeri-Saccà, M.; De Maria, R. The Hippo Pathway in Normal Development and Cancer. Pharmacol. Ther. 2018, 186, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Huang, H.-Y.; Lin, Z.; Ranieri, M.; Li, S.; Sahu, S.; Liu, Y.; Ban, Y.; Guidry, K.; Hu, H.; et al. Genome-Wide CRISPR Screens Identify Multiple Synthetic Lethal Targets That Enhance KRASG12C Inhibitor Efficacy. Cancer Res. 2023, 83, 4095–4111. [Google Scholar] [CrossRef] [PubMed]
- Gujral, T.S.; Kirschner, M.W. Hippo Pathway Mediates Resistance to Cytotoxic Drugs. Proc. Natl. Acad. Sci. USA 2017, 114, E3729–E3738. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Set--Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Wilson, C.L.; Miller, C.J. Simpleaffy: A BioConductor Package for Affymetrix Quality Control and Data Analysis. Bioinformatics 2005, 21, 3683–3685. [Google Scholar] [CrossRef] [PubMed]
- Freedman, J.A.; Tyler, D.S.; Nevins, J.R.; Augustine, C.K. Use of Gene Expression and Pathway Signatures to Characterize the Complexity of Human Melanoma. Am. J. Pathol. 2011, 178, 2513–2522. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Cheadle, C.; Vawter, M.P.; Freed, W.J.; Becker, K.G. Analysis of Microarray Data Using Z Score Transformation. J. Mol. Diagn. 2003, 5, 73–81. [Google Scholar] [CrossRef]
- Balakrishnan, K.; Panneerpandian, P.; Devanandan, H.J.; Sekar, B.T.; Rayala, S.K.; Ganesan, K. Salt-Mediated Transcriptional and Proteasomal Dysregulations Mimic the Molecular Dysregulations of Stomach Cancer. Toxicol. Vitr. 2019, 61, 104588. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, K.; Ganesan, K. Occurrence of Differing Metabolic Dysregulations, a Glucose Driven and Another Fatty Acid Centric in Gastric Cancer Subtypes. Funct. Integr. Genomics 2020, 20, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, K.; Ganesan, K. Identification of Oncogenic Signaling Pathways Associated with the Dimorphic Metabolic Dysregulations in Gastric Cancer Subtypes. Med. Oncol. 2022, 39, 132. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING V10: Protein-Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Tarca, A.L.; Romero, R.; Draghici, S. Analysis of Microarray Experiments of Gene Expression Profiling. Am. J. Obstet. Gynecol. 2006, 195, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wong, W.H. DNA-Chip Analyzer (dChip). In The Analysis of Gene Expression Data: Methods and Software; Parmigiani, G., Garrett, E.S., Irizarry, R.A., Zeger, S.L., Eds.; Springer: New York, NY, USA, 2003; pp. 120–141. ISBN 978-0-387-21679-9. [Google Scholar]
- Balakrishnan, K. The Hepatocellular Carcinoma (HCC) Stage Carcinogenesis Is Associated with Genomic Instability Features. Human. Gene 2023, 38, 201228. [Google Scholar] [CrossRef]
- Murrell, P. R Graphics, 3rd ed.; Chapman and Hall/CRC: New York, NY, USA, 2018; ISBN 978-0-429-42276-8. [Google Scholar]
- Balakrishnan, K. Salt-Driven Chromatin Remodeling Associated with Senescence Dysregulation Plays a Crucial Role in the Carcinogenesis of Gastric Cancer Subtype. Comput. Toxicol. 2023, 25, 100262. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Balakrishnan, K. Hepatocellular Carcinoma Stage: An Almost Loss of Fatty Acid Metabolism and Gain of Glucose Metabolic Pathways Dysregulation. Med. Oncol. 2022, 39, 247. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational Correction of Copy Number Effect Improves Specificity of CRISPR-Cas9 Essentiality Screens in Cancer Cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Lánczky, A.; Menyhárt, O.; Győrffy, B. Validation of miRNA Prognostic Power in Hepatocellular Carcinoma Using Expression Data of Independent Dataset. Sci. Rep. 2018, 8, 9227. [Google Scholar] [CrossRef] [PubMed]
- Menyhárt, O.; Nagy, Á.; Győrffy, B. Determining Consistent Prognostic Biomarkers of Overall Survival and Vascular Invasion in Hepatocellular Carcinoma. R. Soc. Open Sci. 2018, 5, 181006. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, K. Lactate Dehydrogenase Isoform Expressions Differing Impacts on Gastrointestinal Carcinogenesis. Human. Gene 2024, 39, 201243. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. The Logrank Test. BMJ 2004, 328, 1073. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Li, Q.; Sun, Y.; Chen, L.; Xue, F.; Tian, W.; Wang, Y. The Hippo Pathway in Endometrial Cancer: A Potential Therapeutic Target? Front. Oncol. 2023, 13, 1273345. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Pan, D. The Hippo Signaling Pathway in Development and Disease. Dev. Cell 2019, 50, 264–282. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Dong, J. The Hippo Signaling Pathway in Cancer: A Cell Cycle Perspective. Cancers 2021, 13, 6214. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Dong, J. The Hippo Signaling Pathway in Drug Resistance in Cancer. Cancers 2021, 13, 318. [Google Scholar] [CrossRef]
- Meng, Z.; Moroishi, T.; Guan, K.-L. Mechanisms of Hippo Pathway Regulation. Genes. Dev. 2016, 30, 1–17. [Google Scholar] [CrossRef]
- Hua, G.; Lv, X.; He, C.; Remmenga, S.W.; Rodabough, K.J.; Dong, J.; Yang, L.; Lele, S.M.; Yang, P.; Zhou, J.; et al. YAP Induces High-Grade Serous Carcinoma in Fallopian Tube Secretory Epithelial Cells. Oncogene 2016, 35, 2247–2265. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; George, J.; Deb, S.; Degoutin, J.L.; Takano, E.A.; Fox, S.B.; Bowtell, D.D.L.; Harvey, K.F.; AOCS Study group. The Hippo Pathway Transcriptional Co-Activator, YAP, Is an Ovarian Cancer Oncogene. Oncogene 2011, 30, 2810–2822. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Lv, X.; Hua, G.; Lele, S.M.; Remmenga, S.; Dong, J.; Davis, J.S.; Wang, C. YAP Forms Autocrine Loops with the ERBB Pathway to Regulate Ovarian Cancer Initiation and Progression. Oncogene 2015, 34, 6040–6054. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Chang, T.; Wang, Y.; Liu, Y.; Li, W.; Li, M.; Fan, H.-Y. YAP Promotes Ovarian Cancer Cell Tumorigenesis and Is Indicative of a Poor Prognosis for Ovarian Cancer Patients. PLoS ONE 2014, 9, e91770. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Xie, J.; Huang, P.; Yang, Z. Overexpression of TAZ Promotes Cell Proliferation, Migration and Epithelial-Mesenchymal Transition in Ovarian Cancer. Oncol. Lett. 2016, 12, 1821–1825. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Vander Heiden, M.G.; McCormick, F. The Metabolic Landscape of RAS-Driven Cancers from Biology to Therapy. Nat. Cancer 2021, 2, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Therachiyil, L.; Anand, A.; Azmi, A.; Bhat, A.; Korashy, H.M.; Uddin, S. Role of RAS Signaling in Ovarian Cancer. F1000Res 2022, 11, 1253. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-J.; Lee, M.-Y.; Ruan, A.; Chen, C.-K.; Liu, H.-P.; Wang, C.-J.; Chao, W.-R.; Han, C.-P. Multipoint Kras Oncogene Mutations Potentially Indicate Mucinous Carcinoma on the Entire Spectrum of Mucinous Ovarian Neoplasms. Oncotarget 2016, 7, 82097–82103. [Google Scholar] [CrossRef] [PubMed]
- Caduff, R.F.; Svoboda-Newman, S.M.; Ferguson, A.W.; Johnston, C.M.; Frank, T.S. Comparison of Mutations of Ki-RAS and P53 Immunoreactivity in Borderline and Malignant Epithelial Ovarian Tumors. Am. J. Surg. Pathol. 1999, 23, 323–328. [Google Scholar] [CrossRef]
- Tsang, Y.T.; Deavers, M.T.; Sun, C.C.; Kwan, S.-Y.; Kuo, E.; Malpica, A.; Mok, S.C.; Gershenson, D.M.; Wong, K.-K. KRAS (but Not BRAF) Mutations in Ovarian Serous Borderline Tumour Are Associated with Recurrent Low-Grade Serous Carcinoma. J. Pathol. 2013, 231, 449–456. [Google Scholar] [CrossRef]
- Testa, U.; Petrucci, E.; Pasquini, L.; Castelli, G.; Pelosi, E. Ovarian Cancers: Genetic Abnormalities, Tumor Heterogeneity and Progression, Clonal Evolution and Cancer Stem Cells. Medicines 2018, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Emmanuel, C.; Chiew, Y.-E.; George, J.; Etemadmoghadam, D.; Anglesio, M.S.; Sharma, R.; Russell, P.; Kennedy, C.; Fereday, S.; Hung, J.; et al. Genomic Classification of Serous Ovarian Cancer with Adjacent Borderline Differentiates RAS Pathway and TP53-Mutant Tumors and Identifies NRAS as an Oncogenic Driver. Clin. Cancer Res. 2014, 20, 6618–6630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Nandakumar, N.; Shi, Y.; Manzano, M.; Smith, A.; Graham, G.; Gupta, S.; Vietsch, E.E.; Laughlin, S.Z.; Wadhwa, M.; et al. Downstream of Mutant KRAS, the Transcription Regulator YAP Is Essential for Neoplastic Progression to Pancreatic Ductal Adenocarcinoma. Sci. Signal 2014, 7, ra42. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balakrishnan, K.; Chen, Y.; Dong, J. Amplification of Hippo Signaling Pathway Genes Is Governed and Implicated in the Serous Subtype-Specific Ovarian Carcino-Genesis. Cancers 2024, 16, 1781. https://doi.org/10.3390/cancers16091781
Balakrishnan K, Chen Y, Dong J. Amplification of Hippo Signaling Pathway Genes Is Governed and Implicated in the Serous Subtype-Specific Ovarian Carcino-Genesis. Cancers. 2024; 16(9):1781. https://doi.org/10.3390/cancers16091781
Chicago/Turabian StyleBalakrishnan, Karthik, Yuanhong Chen, and Jixin Dong. 2024. "Amplification of Hippo Signaling Pathway Genes Is Governed and Implicated in the Serous Subtype-Specific Ovarian Carcino-Genesis" Cancers 16, no. 9: 1781. https://doi.org/10.3390/cancers16091781
APA StyleBalakrishnan, K., Chen, Y., & Dong, J. (2024). Amplification of Hippo Signaling Pathway Genes Is Governed and Implicated in the Serous Subtype-Specific Ovarian Carcino-Genesis. Cancers, 16(9), 1781. https://doi.org/10.3390/cancers16091781




