Association of miR-146a-5p and miR-21-5p with Prognostic Features in Melanomas
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Sample
2.2. RNA Extraction
2.3. miRNA Expression Analysis
2.4. Statistical Analysis
3. Results
3.1. miR-146a-5p and miR-21-5p Are More Expressed in NM Subtype
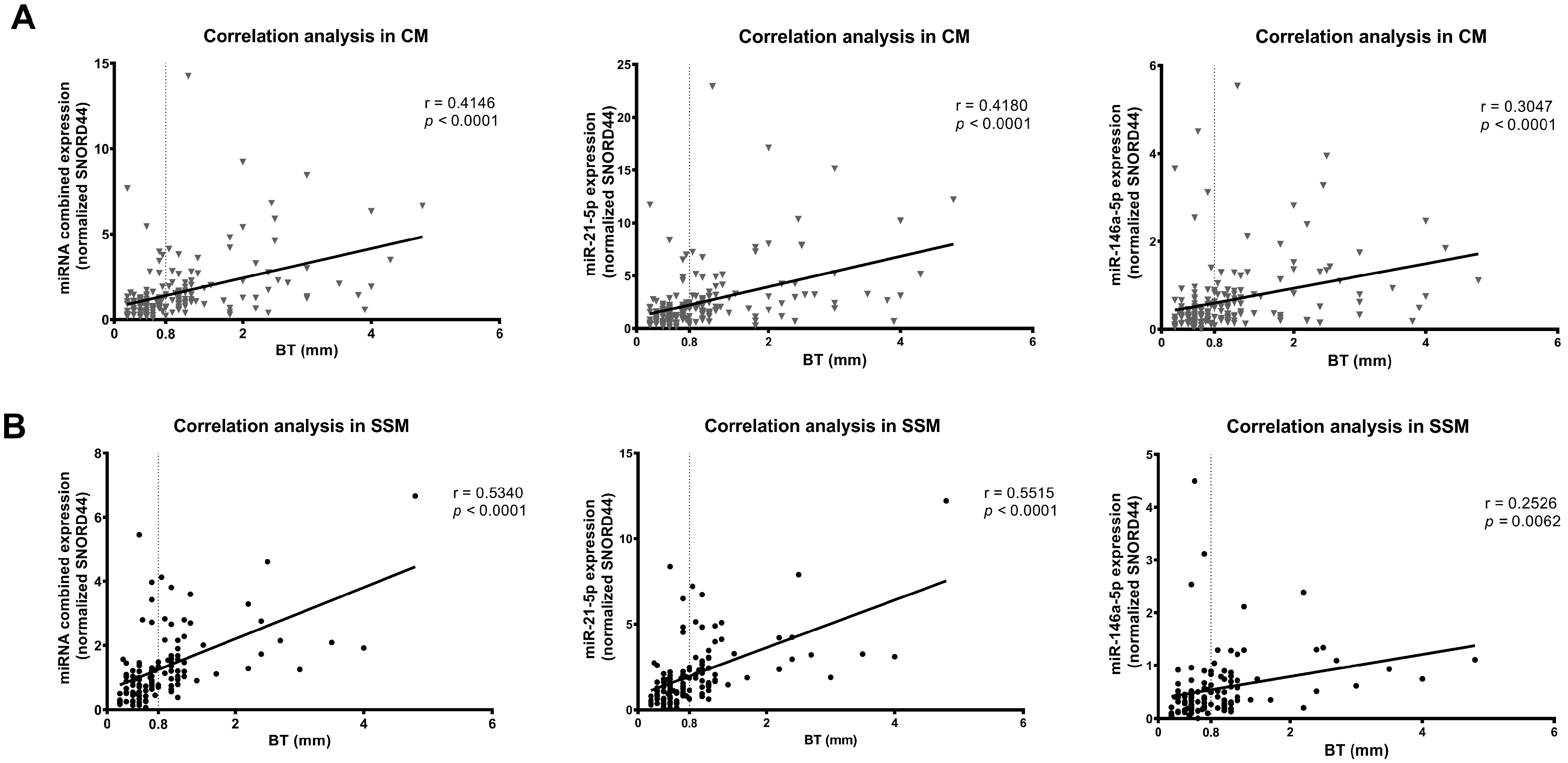
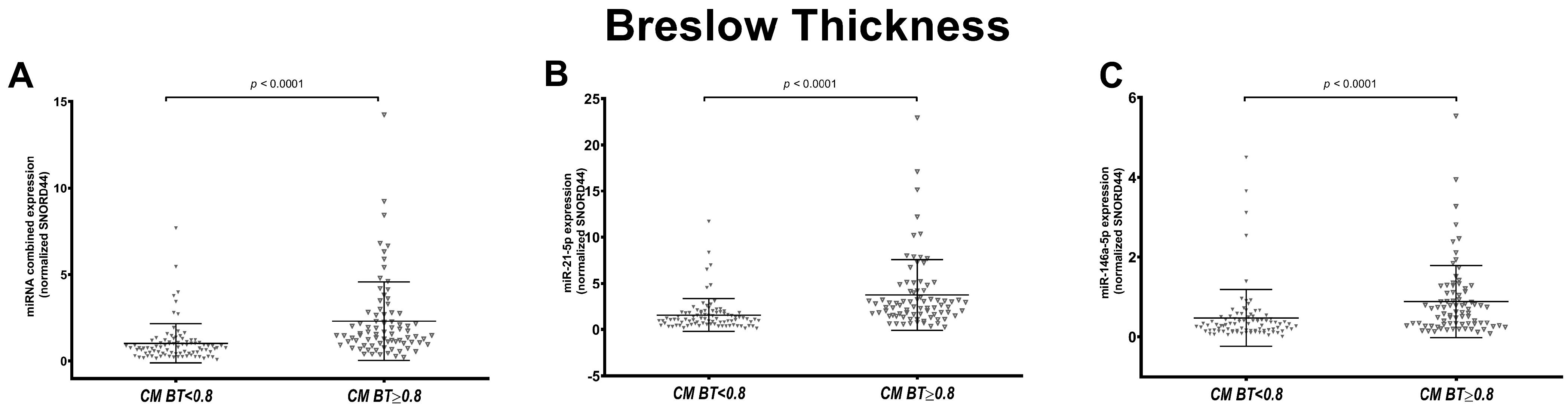
3.2. miR-146a-5p and miR-21-5p Expression Correlates with Breslow Thickness Only in SSM Histological Subtype
3.3. miR-146a-5p and miR-21-5p Expression Is Associated with Ulceration in Melanoma
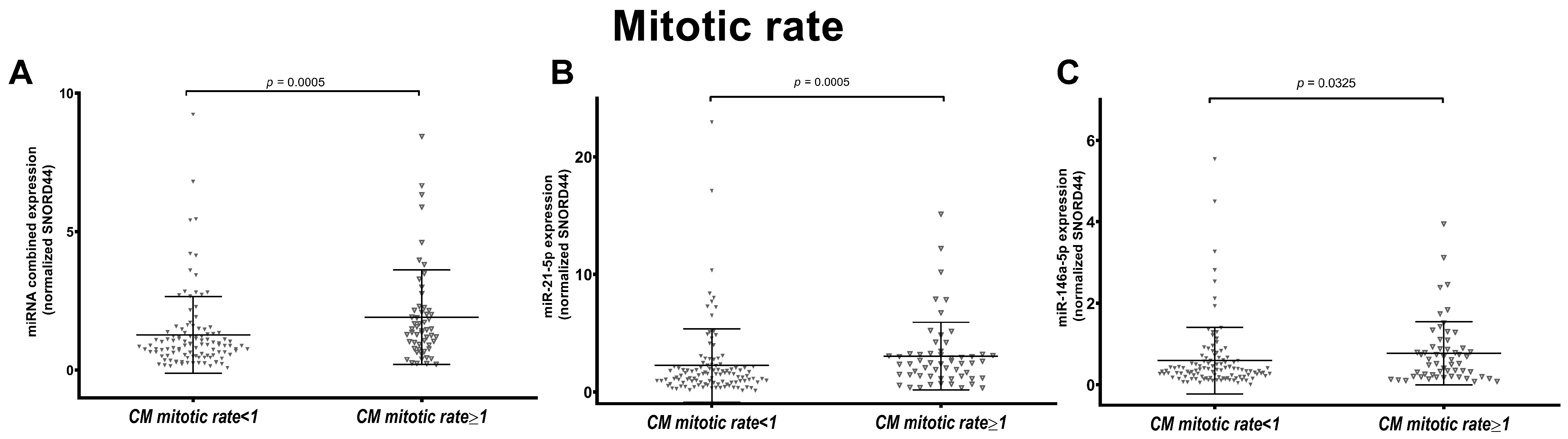
3.4. miR-146a-5p and miR-21-5p Expression Is Associated to Melanoma Mitotic Index in SSM Subtype
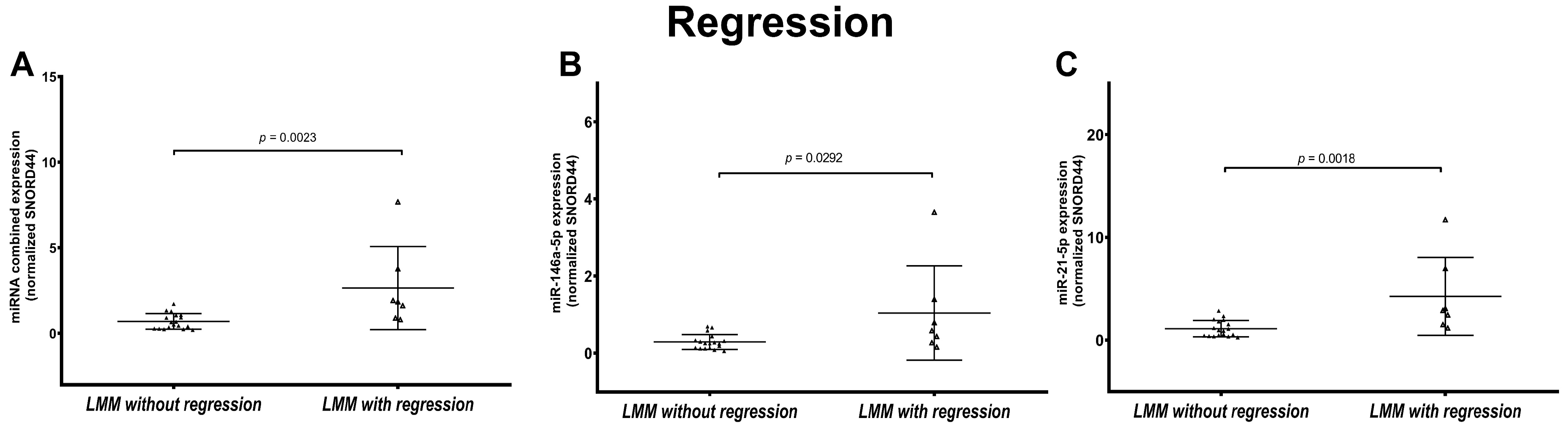
3.5. LMM Present Statistically Significant Different miRNA Expression Based on Regression Status
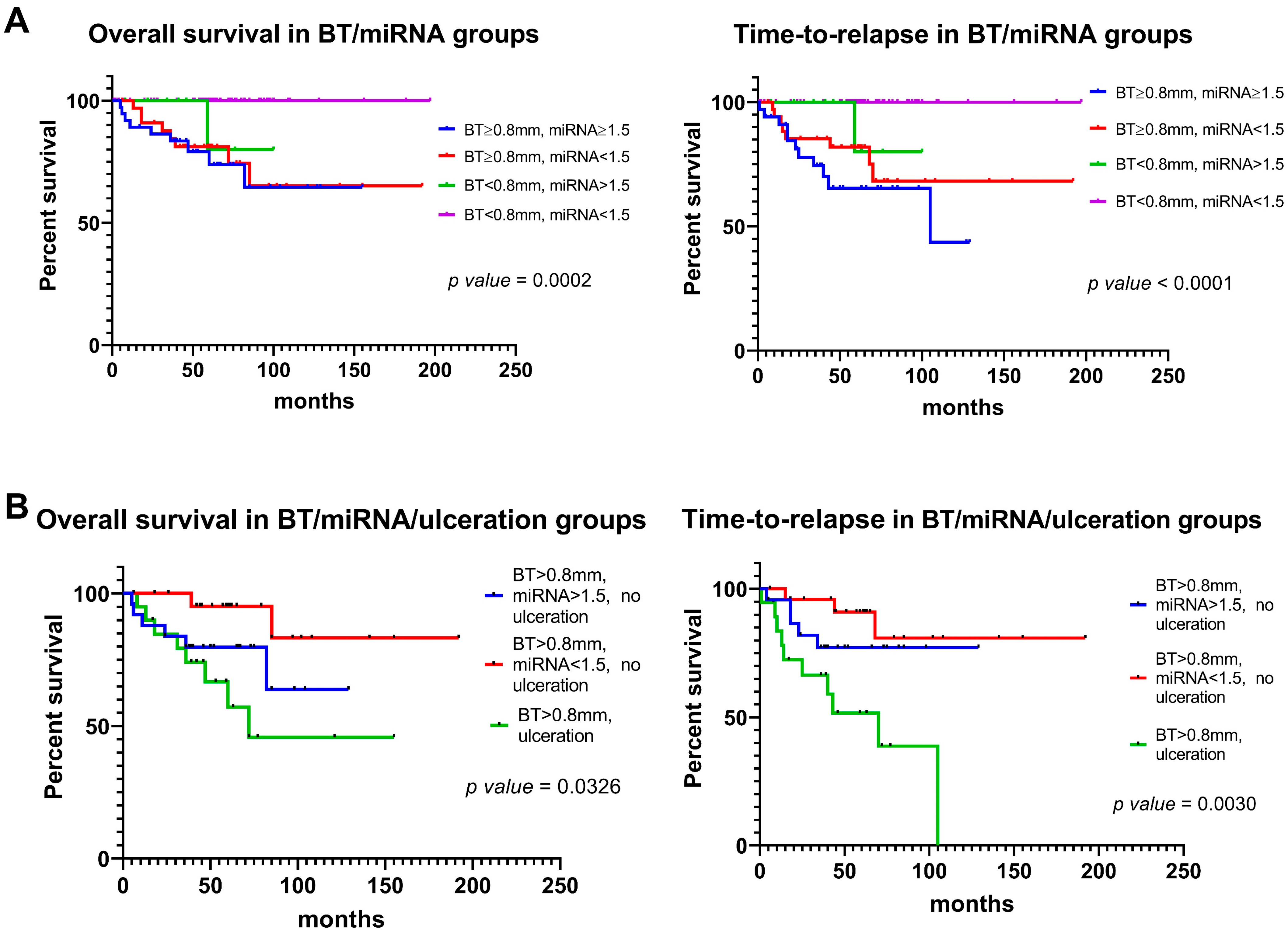
3.6. miR-146a-5p and miR-21-5p Expression and Prognostic Features Can Be Integrated to Improve Outcome Prediction
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gershenwald, J.E.; Scolyer, R.A. Melanoma Staging: American Joint Committee on Cancer (AJCC) 8th Edition and Beyond. Ann. Surg. Oncol. 2018, 25, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Elmore, J.G.; Elder, D.E.; Barnhill, R.L.; Knezevich, S.R.; Longton, G.M.; Titus, L.J.; Weinstock, M.A.; Pepe, M.S.; Nelson, H.D.; Reisch, L.M.; et al. Concordance and Reproducibility of Melanoma Staging According to the 7th vs 8th Edition of the AJCC Cancer Staging Manual. JAMA Netw. Open 2018, 1, e180083. [Google Scholar] [CrossRef]
- Rastrelli, M.; Tropea, S.; Pigozzo, J.; Bezzon, E.; Campana, L.G.; Stramare, R.; Alaibac, M.; Rossi, C.R. Melanoma m1: Diagnosis and therapy. Vivo 2014, 28, 273–285. [Google Scholar]
- Chaudru, V.; Chompret, A.; Bressac-de Paillerets, B.; Spatz, A.; Avril, M.F.; Demenais, F. Influence of genes, nevi, and sun sensitivity on melanoma risk in a family sample unselected by family history and in melanoma-prone families. J. Natl. Cancer Inst. 2004, 96, 785–795. [Google Scholar] [CrossRef]
- Gandini, S.; Sera, F.; Cattaruzza, M.S.; Pasquini, P.; Zanetti, R.; Masini, C.; Boyle, P.; Melchi, C.F. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur. J. Cancer 2005, 41, 2040–2059. [Google Scholar] [CrossRef] [PubMed]
- Dika, E.; Patrizi, A.; Rossi, C.; Turchetti, D.; Miccoli, S.; Ferracin, M.; Veronesi, G.; Scarfi, F.; Lambertini, M. Clinical histopathological features and CDKN2A/CDK4/MITF mutational status of patients with multiple primary melanomas from Bologna: Italy is a fascinating but complex mosaic. Ital. J. Dermatol. Venerol. 2021, 156, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Read, J.; Wadt, K.A.; Hayward, N.K. Melanoma genetics. J. Med. Genet. 2016, 53, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, A.M.; Chan, M.; Harland, M.; Hayward, N.K.; Demenais, F.; Bishop, D.T.; Azizi, E.; Bergman, W.; Bianchi-Scarra, G.; Bruno, W.; et al. Features associated with germline CDKN2A mutations: A GenoMEL study of melanoma-prone families from three continents. J. Med. Genet. 2007, 44, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Moura Brasil Arnaut, J.; Dos Santos Guimaraes, I.; Evangelista Dos Santos, A.C.; de Moraes Lino da Silva, F.; Machado, J.R.; de Melo, A.C. Molecular landscape of Hereditary Melanoma. Crit. Rev. Oncol. Hematol. 2021, 164, 103425. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Pellegrini, C.; Cardelli, L.; Ciciarelli, V.; Di Nardo, L.; Fargnoli, M.C. Familial Melanoma: Diagnostic and Management Implications. Dermatol. Pract. Concept. 2019, 9, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Soura, E.; Eliades, P.J.; Shannon, K.; Stratigos, A.J.; Tsao, H. Hereditary melanoma: Update on syndromes and management: Genetics of familial atypical multiple mole melanoma syndrome. J. Am. Acad. Dermatol. 2016, 74, 395–407, quiz 408–310. [Google Scholar] [CrossRef] [PubMed]
- Potrony, M.; Badenas, C.; Aguilera, P.; Puig-Butille, J.A.; Carrera, C.; Malvehy, J.; Puig, S. Update in genetic susceptibility in melanoma. Ann. Transl. Med. 2015, 3, 210. [Google Scholar] [CrossRef] [PubMed]
- Harland, M.; Cust, A.E.; Badenas, C.; Chang, Y.M.; Holland, E.A.; Aguilera, P.; Aitken, J.F.; Armstrong, B.K.; Barrett, J.H.; Carrera, C.; et al. Prevalence and predictors of germline CDKN2A mutations for melanoma cases from Australia, Spain and the United Kingdom. Hered. Cancer Clin. Pract. 2014, 12, 20. [Google Scholar] [CrossRef]
- Toussi, A.; Mans, N.; Welborn, J.; Kiuru, M. Germline mutations predisposing to melanoma. J. Cutan. Pathol. 2020, 47, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Woods, S.L.; Boyle, G.M.; Aoude, L.G.; MacGregor, S.; Zismann, V.; Gartside, M.; Cust, A.E.; Haq, R.; Harland, M.; et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 2011, 480, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Bertolotto, C.; Lesueur, F.; Giuliano, S.; Strub, T.; de Lichy, M.; Bille, K.; Dessen, P.; d’Hayer, B.; Mohamdi, H.; Remenieras, A.; et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011, 480, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Ciccarese, G.; Dalmasso, B.; Bruno, W.; Queirolo, P.; Pastorino, L.; Andreotti, V.; Spagnolo, F.; Tanda, E.; Ponti, G.; Massone, C.; et al. Clinical, pathological and dermoscopic phenotype of MITF p.E318K carrier cutaneous melanoma patients. J. Transl. Med. 2020, 18, 78. [Google Scholar] [CrossRef] [PubMed]
- Ghiorzo, P.; Pastorino, L.; Queirolo, P.; Bruno, W.; Tibiletti, M.G.; Nasti, S.; Andreotti, V.; Genoa Pancreatic Cancer Study, G.; Paillerets, B.B.; Bianchi Scarra, G. Prevalence of the E318K MITF germline mutation in Italian melanoma patients: Associations with histological subtypes and family cancer history. Pigment. Cell Melanoma Res. 2013, 26, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Aoude, L.G.; Pritchard, A.L.; Robles-Espinoza, C.D.; Wadt, K.; Harland, M.; Choi, J.; Gartside, M.; Quesada, V.; Johansson, P.; Palmer, J.M.; et al. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J. Natl. Cancer Inst. 2015, 107, dju408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.J.; Rush, P.S.; Tsao, H.; Duncan, L.M. BRCA1-associated protein (BAP1)-inactivated melanocytic tumors. J. Cutan. Pathol. 2019, 46, 965–972. [Google Scholar] [CrossRef] [PubMed]
- Harland, M.; Petljak, M.; Robles-Espinoza, C.D.; Ding, Z.; Gruis, N.A.; van Doorn, R.; Pooley, K.A.; Dunning, A.M.; Aoude, L.G.; Wadt, K.A.; et al. Germline TERT promoter mutations are rare in familial melanoma. Fam. Cancer 2016, 15, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Robles-Espinoza, C.D.; Harland, M.; Ramsay, A.J.; Aoude, L.G.; Quesada, V.; Ding, Z.; Pooley, K.A.; Pritchard, A.L.; Tiffen, J.C.; Petljak, M.; et al. POT1 loss-of-function variants predispose to familial melanoma. Nat. Genet. 2014, 46, 478–481. [Google Scholar] [CrossRef] [PubMed]
- Elwood, J.M.; Koh, H.K. Etiology, epidemiology, risk factors, and public health issues of melanoma. Curr. Opin. Oncol. 1994, 6, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Newton-Bishop, J.A.; Chang, Y.M.; Elliott, F.; Chan, M.; Leake, S.; Karpavicius, B.; Haynes, S.; Fitzgibbon, E.; Kukalizch, K.; Randerson-Moor, J.; et al. Relationship between sun exposure and melanoma risk for tumours in different body sites in a large case-control study in a temperate climate. Eur. J. Cancer 2011, 47, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Loras, A.; Gil-Barrachina, M.; Marques-Torrejon, M.A.; Perez-Pastor, G.; Martinez-Cadenas, C. UV-Induced Somatic Mutations Driving Clonal Evolution in Healthy Skin, Nevus, and Cutaneous Melanoma. Life 2022, 12, 1339. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, G.; Argenziano, G. The WHO 2018 Classification of Cutaneous Melanocytic Neoplasms: Suggestions From Routine Practice. Front. Oncol. 2021, 11, 675296. [Google Scholar] [CrossRef] [PubMed]
- Arrington, J.H., 3rd; Reed, R.J.; Ichinose, H.; Krementz, E.T. Plantar lentiginous melanoma: A distinctive variant of human cutaneous malignant melanoma. Am. J. Surg. Pathol. 1977, 1, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Clark, W.H., Jr.; From, L.; Bernardino, E.A.; Mihm, M.C. The histogenesis and biologic behavior of primary human malignant melanomas of the skin. Cancer Res. 1969, 29, 705–727. [Google Scholar]
- McGovern, V.J. The classification of melanoma and its relationship with prognosis. Pathology 1970, 2, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Scolyer, R.A.; Long, G.V.; Thompson, J.F. Evolving concepts in melanoma classification and their relevance to multidisciplinary melanoma patient care. Mol. Oncol. 2011, 5, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Scatena, C.; Murtas, D.; Tomei, S. Cutaneous Melanoma Classification: The Importance of High-Throughput Genomic Technologies. Front. Oncol. 2021, 11, 635488. [Google Scholar] [CrossRef]
- Bobos, M. Histopathologic classification and prognostic factors of melanoma: A 2021 update. Ital. J. Dermatol. Venerol. 2021, 156, 300–321. [Google Scholar] [CrossRef]
- Greenwald, H.S.; Friedman, E.B.; Osman, I. Superficial spreading and nodular melanoma are distinct biological entities: A challenge to the linear progression model. Melanoma Res. 2012, 22, 1–8. [Google Scholar] [CrossRef]
- Dabas, N.; Byrnes, D.M.; Rosa, A.M.; Eller, M.S.; Grichnik, J.M. Diagnostic role of chromosomal instability in melanoma. J. Ski. Cancer 2012, 2012, 914267. [Google Scholar] [CrossRef]
- Timar, J.; Vizkeleti, L.; Doma, V.; Barbai, T.; Raso, E. Genetic progression of malignant melanoma. Cancer Metastasis Rev. 2016, 35, 93–107. [Google Scholar] [CrossRef]
- Kalkhoran, S.; Milne, O.; Zalaudek, I.; Puig, S.; Malvehy, J.; Kelly, J.W.; Marghoob, A.A. Historical, clinical, and dermoscopic characteristics of thin nodular melanoma. Arch. Dermatol. 2010, 146, 311–318. [Google Scholar] [CrossRef]
- Lattanzi, M.; Lee, Y.; Simpson, D.; Moran, U.; Darvishian, F.; Kim, R.H.; Hernando, E.; Polsky, D.; Hanniford, D.; Shapiro, R.; et al. Primary Melanoma Histologic Subtype: Impact on Survival and Response to Therapy. J. Natl. Cancer Inst. 2019, 111, 180–188. [Google Scholar] [CrossRef]
- Xiong, M.; Charifa, A.; Chen, C.S.J. Lentigo Maligna Melanoma. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Sanna, A.; Harbst, K.; Johansson, I.; Christensen, G.; Lauss, M.; Mitra, S.; Rosengren, F.; Hakkinen, J.; Vallon-Christersson, J.; Olsson, H.; et al. Tumor genetic heterogeneity analysis of chronic sun-damaged melanoma. Pigment. Cell Melanoma Res. 2020, 33, 480–489. [Google Scholar] [CrossRef]
- Connolly, K.L.; Nehal, K.S.; Busam, K.J. Lentigo maligna and lentigo maligna melanoma: Contemporary issues in diagnosis and management. Melanoma Manag. 2015, 2, 171–178. [Google Scholar] [CrossRef]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef]
- de Giorgi, V.; Rossari, S.; Gori, A.; Grazzini, M.; Savarese, I.; Crocetti, E.; Cervadoro, E.; Massi, D. The prognostic impact of the anatomical sites in the ‘head and neck melanoma’: Scalp versus face and neck. Melanoma Res. 2012, 22, 402–405. [Google Scholar] [CrossRef]
- Dika, E.; Patrizi, A.; Veronesi, G.; Manuelpillai, N.; Lambertini, M. Malignant cutaneous tumours of the scalp: Always remember to examine the head. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 2208–2215. [Google Scholar] [CrossRef]
- Dika, E.; Veronesi, G.; Misciali, C.; Corti, B.; Dika, I.; Riefolo, M.; Scarfi, F.; Lambertini, M.; Patrizi, A. Malignant Melanoma Cells and Hair Follicles. Am. J. Clin. Pathol. 2019, 152, 109–114. [Google Scholar] [CrossRef]
- Starace, M.; Dika, E.; Fanti, P.A.; Patrizi, A.; Misciali, C.; Alessandrini, A.; Bruni, F.; Piraccini, B.M. Nail apparatus melanoma: Dermoscopic and histopathologic correlations on a series of 23 patients from a single centre. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 164–173. [Google Scholar] [CrossRef]
- Riefolo, M.; Porcellini, E.; Dika, E.; Broseghini, E.; Ferracin, M. Interplay between small and long non-coding RNAs in cutaneous melanoma: A complex jigsaw puzzle with missing pieces. Mol. Oncol. 2019, 13, 74–98. [Google Scholar] [CrossRef]
- Ribero, S.; Lambertini, M.; Ferracin, M.; Dika, E. Non-Coding RNA Investigations in Cutaneous Melanoma: A Step forward in Discovering Novel Biomarkers. J. Investig. Dermatol. 2023, 143, 531–532. [Google Scholar] [CrossRef]
- Durante, G.; Comito, F.; Lambertini, M.; Broseghini, E.; Dika, E.; Ferracin, M. Non-coding RNA dysregulation in skin cancers. Essays Biochem. 2021, 65, 641–655. [Google Scholar] [CrossRef]
- Broseghini, E.; Dika, E.; Londin, E.; Ferracin, M. MicroRNA Isoforms Contribution to Melanoma Pathogenesis. Noncoding RNA 2021, 7, 63. [Google Scholar] [CrossRef]
- Hanniford, D.; Segura, M.F.; Zhong, J.; Philips, E.; Jirau-Serrano, X.; Darvishian, F.; Berman, R.S.; Shapiro, R.L.; Pavlick, A.C.; Brown, B.; et al. Identification of metastasis-suppressive microRNAs in primary melanoma. J. Natl. Cancer Inst. 2015, 107, dju494. [Google Scholar] [CrossRef]
- Howell, P.M., Jr.; Li, X.; Riker, A.I.; Xi, Y. MicroRNA in Melanoma. Ochsner J. 2010, 10, 83–92. [Google Scholar]
- Kozubek, J.; Ma, Z.; Fleming, E.; Duggan, T.; Wu, R.; Shin, D.G.; Dadras, S.S. In-depth characterization of microRNA transcriptome in melanoma. PLoS ONE 2013, 8, e72699. [Google Scholar] [CrossRef]
- Negrini, M.; Calin, G.A.; Croce, C.M. MicroRNA in Human Malignancies; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar] [CrossRef]
- Durante, G.; Broseghini, E.; Comito, F.; Naddeo, M.; Milani, M.; Salamon, I.; Campione, E.; Dika, E.; Ferracin, M. Circulating microRNA biomarkers in melanoma and non-melanoma skin cancer. Expert. Rev. Mol. Diagn. 2022, 22, 305–318. [Google Scholar] [CrossRef]
- Durante, G.; Veronesi, G.; Misciali, C.; Riefolo, M.; Lambertini, M.; Tartari, F.; Ricci, C.; Ferracin, M.; Dika, E. Dysplastic nevi and melanoma: MicroRNAs tell a divergent story. Pathol. Res. Pr. 2022, 235, 153942. [Google Scholar] [CrossRef]
- Negrini, M.; Ferracin, M.; Sabbioni, S.; Croce, C.M. MicroRNAs in human cancer: From research to therapy. J. Cell Sci. 2007, 120, 1833–1840. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- Dika, E.; Broseghini, E.; Porcellini, E.; Lambertini, M.; Riefolo, M.; Durante, G.; Loher, P.; Roncarati, R.; Bassi, C.; Misciali, C.; et al. Unraveling the role of microRNA/isomiR network in multiple primary melanoma pathogenesis. Cell Death Dis. 2021, 12, 473. [Google Scholar] [CrossRef]
- Pegoraro, A.; De Marchi, E.; Ferracin, M.; Orioli, E.; Zanoni, M.; Bassi, C.; Tesei, A.; Capece, M.; Dika, E.; Negrini, M.; et al. P2X7 promotes metastatic spreading and triggers release of miRNA-containing exosomes and microvesicles from melanoma cells. Cell Death Dis. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Chin, A.; Mariscal, J.; Kim, M.; Guerra, G.; Victor, B.; Qian, C.; Broseghini, E.; Posadas, E.; Freeman, M.R.; Sharma, S.; et al. miR-1227 Targets SEC23A to Regulate the Shedding of Large Extracellular Vesicles. Cancers 2021, 13, 5850. [Google Scholar] [CrossRef]
- Cocks, A.; Martinez-Rodriguez, V.; Del Vecchio, F.; Schukking, M.; Broseghini, E.; Giannakopoulos, S.; Fabbri, M. Diverse roles of EV-RNA in cancer progression. Semin. Cancer Biol. 2021, 75, 127–135. [Google Scholar] [CrossRef]
- Ferracin, M.; Pedriali, M.; Veronese, A.; Zagatti, B.; Gafa, R.; Magri, E.; Lunardi, M.; Munerato, G.; Querzoli, G.; Maestri, I.; et al. MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J. Pathol. 2011, 225, 43–53. [Google Scholar] [CrossRef]
- Liu, C.G.; Calin, G.A.; Meloon, B.; Gamliel, N.; Sevignani, C.; Ferracin, M.; Dumitru, C.D.; Shimizu, M.; Zupo, S.; Dono, M.; et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 9740–9744. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Dika, E.; Riefolo, M.; Porcellini, E.; Broseghini, E.; Ribero, S.; Senetta, R.; Osella-Abate, S.; Scarfi, F.; Lambertini, M.; Veronesi, G.; et al. Defining the Prognostic Role of MicroRNAs in Cutaneous Melanoma. J. Investig. Dermatol. 2020, 140, 2260–2267. [Google Scholar] [CrossRef]
- Poniewierska-Baran, A.; Sluczanowska-Glabowska, S.; Malkowska, P.; Sierawska, O.; Zadroga, L.; Pawlik, A.; Niedzwiedzka-Rystwej, P. Role of miRNA in Melanoma Development and Progression. Int. J. Mol. Sci. 2022, 24, 201. [Google Scholar] [CrossRef]
- Varrone, F.; Caputo, E. The miRNAs Role in Melanoma and in Its Resistance to Therapy. Int. J. Mol. Sci. 2020, 21, 878. [Google Scholar] [CrossRef]
- Massi, G.; LeBoit, P.E. Regressing and Regressed Melanoma. In Histological Diagnosis of Nevi and Melanoma; Springer: Berlin/Heidelberg, Germany, 2014; pp. 699–720. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Aksenenko, M.; Palkina, N.; Komina, A.; Tashireva, L.; Ruksha, T. Differences in microRNA expression between melanoma and healthy adjacent skin. BMC Dermatol. 2019, 19, 1. [Google Scholar] [CrossRef]
- Forloni, M.; Dogra, S.K.; Dong, Y.; Conte, D., Jr.; Ou, J.; Zhu, L.J.; Deng, A.; Mahalingam, M.; Green, M.R.; Wajapeyee, N. miR-146a promotes the initiation and progression of melanoma by activating Notch signaling. Elife 2014, 3, e01460. [Google Scholar] [CrossRef]
- Satzger, I.; Mattern, A.; Kuettler, U.; Weinspach, D.; Niebuhr, M.; Kapp, A.; Gutzmer, R. microRNA-21 is upregulated in malignant melanoma and influences apoptosis of melanocytic cells. Exp. Dermatol. 2012, 21, 509–514. [Google Scholar] [CrossRef]
- Babapoor, S.; Wu, R.; Kozubek, J.; Auidi, D.; Grant-Kels, J.M.; Dadras, S.S. Identification of microRNAs associated with invasive and aggressive phenotype in cutaneous melanoma by next-generation sequencing. Lab. Investig. 2017, 97, 636–648. [Google Scholar] [CrossRef][Green Version]
- Grignol, V.; Fairchild, E.T.; Zimmerer, J.M.; Lesinski, G.B.; Walker, M.J.; Magro, C.M.; Kacher, J.E.; Karpa, V.I.; Clark, J.; Nuovo, G.; et al. miR-21 and miR-155 are associated with mitotic activity and lesion depth of borderline melanocytic lesions. Br. J. Cancer 2011, 105, 1023–1029. [Google Scholar] [CrossRef]
- Edwin, F.; Anderson, K.; Ying, C.; Patel, T.B. Intermolecular interactions of Sprouty proteins and their implications in development and disease. Mol. Pharmacol. 2009, 76, 679–691. [Google Scholar] [CrossRef]
- Lu, Z.; Liu, M.; Stribinskis, V.; Klinge, C.M.; Ramos, K.S.; Colburn, N.H.; Li, Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008, 27, 4373–4379. [Google Scholar] [CrossRef]
- Yang, C.H.; Yue, J.; Pfeffer, S.R.; Handorf, C.R.; Pfeffer, L.M. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J. Biol. Chem. 2011, 286, 39172–39178. [Google Scholar] [CrossRef]
- Egger, M.E.; Bhutiani, N.; Farmer, R.W.; Stromberg, A.J.; Martin, R.C., 2nd; Quillo, A.R.; McMasters, K.M.; Scoggins, C.R. Prognostic factors in melanoma patients with tumor-negative sentinel lymph nodes. Surgery 2016, 159, 1412–1421. [Google Scholar] [CrossRef]
- El Sharouni, M.A.; van Diest, P.J.; Witkamp, A.J.; Sigurdsson, V.; van Gils, C.H. Subtyping Cutaneous Melanoma Matters. JNCI Cancer Spectr. 2020, 4, pkaa097. [Google Scholar] [CrossRef]
- Dessinioti, C.; Dimou, N.; Geller, A.C.; Stergiopoulou, A.; Lo, S.; Keim, U.; Gershenwald, J.E.; Haydu, L.E.; Ribero, S.; Quaglino, P.; et al. Distinct Clinicopathological and Prognostic Features of Thin Nodular Primary Melanomas: An International Study from 17 Centers. J. Natl. Cancer Inst. 2019, 111, 1314–1322. [Google Scholar] [CrossRef]
- Boczar, D.; Sisti, A.; Restrepo, D.J.; Huayllani, M.T.; Manrique, O.J.; Lu, X.; Spaulding, A.C.; Bagaria, S.P.; Parker, A.S.; Forte, A.J. National Analysis of Patients With Ulcerated Melanoma in the United States. Anticancer. Res. 2020, 40, 1055–1058. [Google Scholar] [CrossRef]
- Bonnelykke-Behrndtz, M.L.; Steiniche, T. Ulcerated Melanoma: Aspects and Prognostic Impact. In Cutaneous Melanoma: Etiology and Therapy; Ward, W.H., Farma, J.M., Eds.; Codon Publications: Brisbane, Australia, 2017; Chapter 5. [Google Scholar]
- DiVincenzo, M.J.; Schwarz, E.; Ren, C.; Barricklow, Z.; Moufawad, M.; Yu, L.; Fadda, P.; Angell, C.; Sun, S.; Howard, J.H.; et al. Expression Patterns of microRNAs and Associated Target Genes in Ulcerated Primary Cutaneous Melanoma. J. Investig. Dermatol. 2023, 143, 630–638.e3. [Google Scholar] [CrossRef]
- Pinero-Madrona, A.; Ruiz-Merino, G.; Cerezuela Fuentes, P.; Martinez-Barba, E.; Rodriguez-Lopez, J.N.; Cabezas-Herrera, J. Mitotic rate as an important prognostic factor in cutaneous malignant melanoma. Clin. Transl. Oncol. 2019, 21, 1348–1356. [Google Scholar] [CrossRef]
- Aung, P.P.; Nagarajan, P.; Prieto, V.G. Regression in primary cutaneous melanoma: Etiopathogenesis and clinical significance. Lab. Investig. 2017, 97, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.J.; Gimotty, P.A.; Hwang, W.T.; Dawson, P.; Van Belle, P.; Elder, D.E.; Elenitsas, R.; Schuchter, L.; Zhang, P.J.; Guerry, D.; et al. High lymphatic vessel density and lymphatic invasion underlie the adverse prognostic effect of radial growth phase regression in melanoma. Am. J. Surg. Pathol. 2011, 35, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Ribero, S.; Gualano, M.R.; Osella-Abate, S.; Scaioli, G.; Bert, F.; Sanlorenzo, M.; Balagna, E.; Fierro, M.T.; Macripo, G.; Sapino, A.; et al. Association of Histologic Regression in Primary Melanoma With Sentinel Lymph Node Status: A Systematic Review and Meta-analysis. JAMA Dermatol. 2015, 151, 1301–1307. [Google Scholar] [CrossRef]
- Botella-Estrada, R.; Traves, V.; Requena, C.; Guillen-Barona, C.; Nagore, E. Correlation of histologic regression in primary melanoma with sentinel node status. JAMA Dermatol. 2014, 150, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.B.; Edge, S.B.; Greene, F.L.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. (Eds.) Melanoma of the Skin. In AJCC Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2017; pp. 563–585. [Google Scholar]



| SSM | LMM | NM | All Samples | |
|---|---|---|---|---|
| Total (n°) | 116 | 28 | 26 | 170 |
| of which from Dika et al. [39] 1 | 90 | 2 | 25 | 117 |
| Gender (n°) | ||||
| Male | 64 | 13 | 20 | 97 |
| Female | 52 | 15 | 6 | 73 |
| Age | ||||
| <50 years | 24 | 2 | 8 | 34 |
| ≥50 years | 92 | 26 | 18 | 136 |
| Breslow Thickness (n°) | ||||
| <0.8 mm | 65 | 23 | 1 | 89 |
| ≥0.8 mm | 51 | 5 | 25 | 81 |
| Ulceration (n°) | ||||
| Presence | 14 | 0 | 9 | 23 |
| Absence | 102 | 26 | 15 | 143 |
| Not available | 2 | 2 | 4 | |
| Mitosis (n°) | ||||
| <1/mm2 | 81 | 20 | 7 | 108 |
| ≥1/mm2 | 34 | 5 | 17 | 56 |
| Not available | 1 | 3 | 2 | 6 |
| Regression (n°) | ||||
| Presence | 54 | 7 | 4 | 65 |
| Absence | 61 | 18 | 21 | 100 |
| Not available | 1 | 3 | 1 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naddeo, M.; Broseghini, E.; Venturi, F.; Vaccari, S.; Corti, B.; Lambertini, M.; Ricci, C.; Fontana, B.; Durante, G.; Pariali, M.; et al. Association of miR-146a-5p and miR-21-5p with Prognostic Features in Melanomas. Cancers 2024, 16, 1688. https://doi.org/10.3390/cancers16091688
Naddeo M, Broseghini E, Venturi F, Vaccari S, Corti B, Lambertini M, Ricci C, Fontana B, Durante G, Pariali M, et al. Association of miR-146a-5p and miR-21-5p with Prognostic Features in Melanomas. Cancers. 2024; 16(9):1688. https://doi.org/10.3390/cancers16091688
Chicago/Turabian StyleNaddeo, Maria, Elisabetta Broseghini, Federico Venturi, Sabina Vaccari, Barbara Corti, Martina Lambertini, Costantino Ricci, Beatrice Fontana, Giorgio Durante, Milena Pariali, and et al. 2024. "Association of miR-146a-5p and miR-21-5p with Prognostic Features in Melanomas" Cancers 16, no. 9: 1688. https://doi.org/10.3390/cancers16091688
APA StyleNaddeo, M., Broseghini, E., Venturi, F., Vaccari, S., Corti, B., Lambertini, M., Ricci, C., Fontana, B., Durante, G., Pariali, M., Scotti, B., Milani, G., Campione, E., Ferracin, M., & Dika, E. (2024). Association of miR-146a-5p and miR-21-5p with Prognostic Features in Melanomas. Cancers, 16(9), 1688. https://doi.org/10.3390/cancers16091688









