Proteins Involved in Focal Cell Adhesion and Podosome Formation Are Differentially Expressed during Colorectal Tumorigenesis in AOM-Treated Rats
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
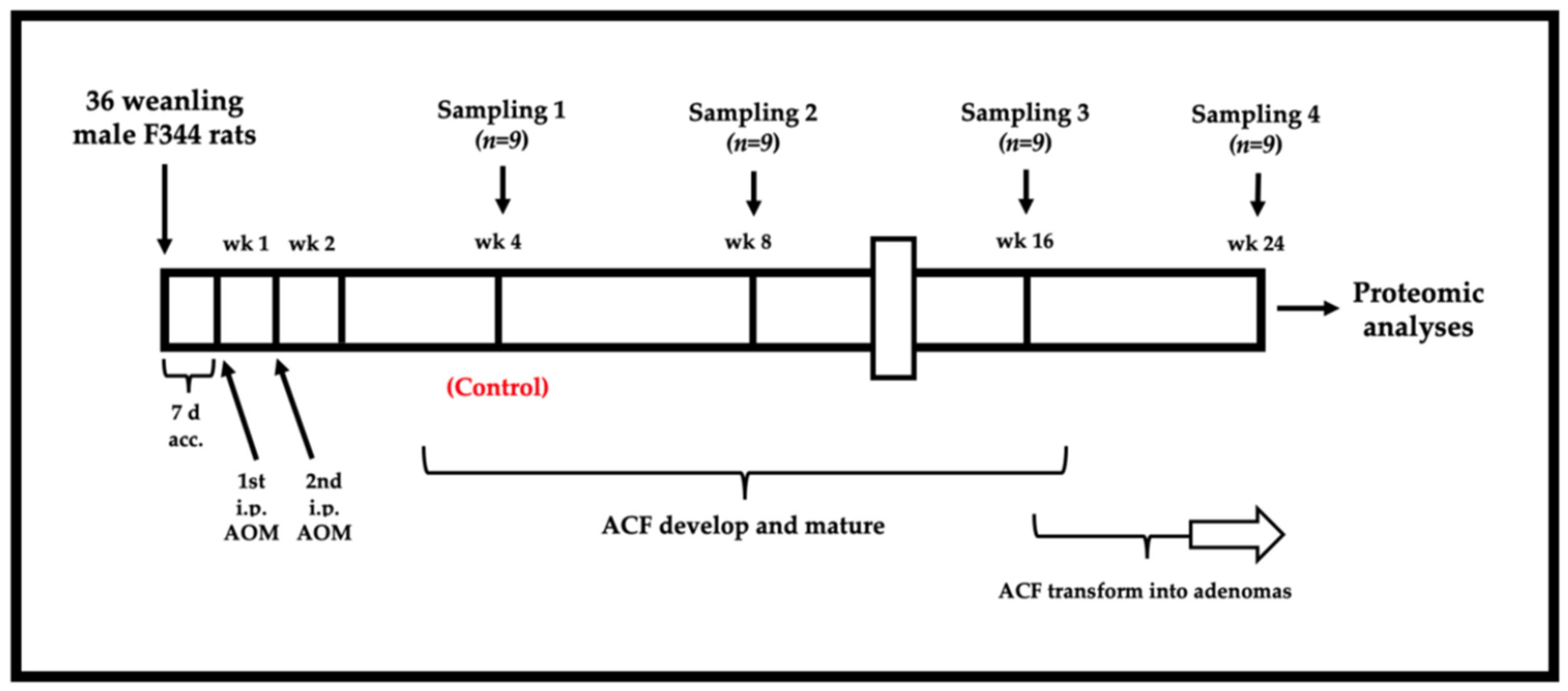
2.2. Animal Preparation and Carcinogen Treatment
2.3. Tissue Collection
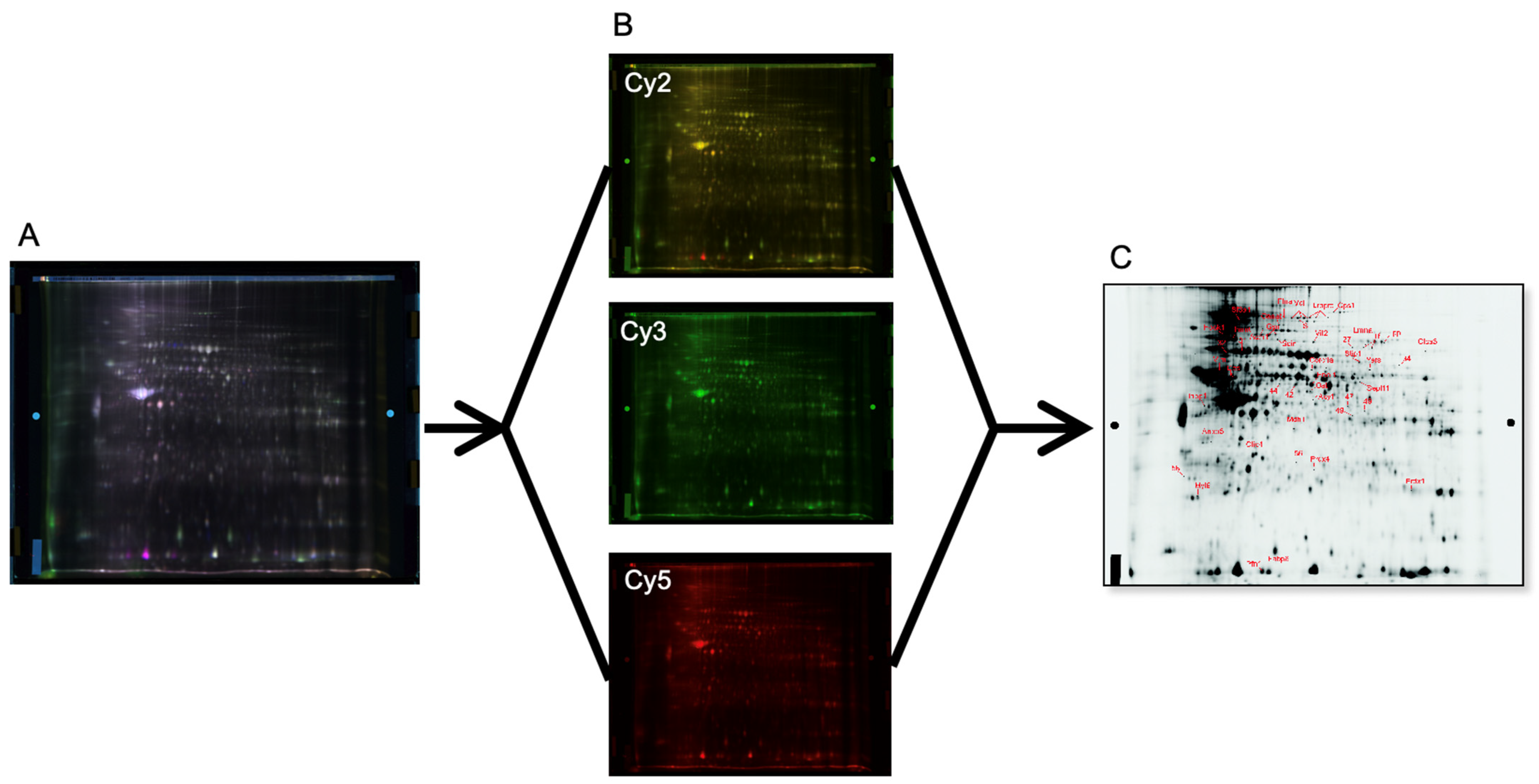
2.4. Protein Extraction and Fluorescence Labeling
2.5. Gel Electrophoresis
2.6. Image Acquisition and Protein Quantification
2.7. In-Gel Digestion and Protein Identification
2.8. Western Blotting
2.9. Network Analysis
2.10. Statistical Analyses
3. Results
3.1. Quantitative Data for ACF and Adenomas
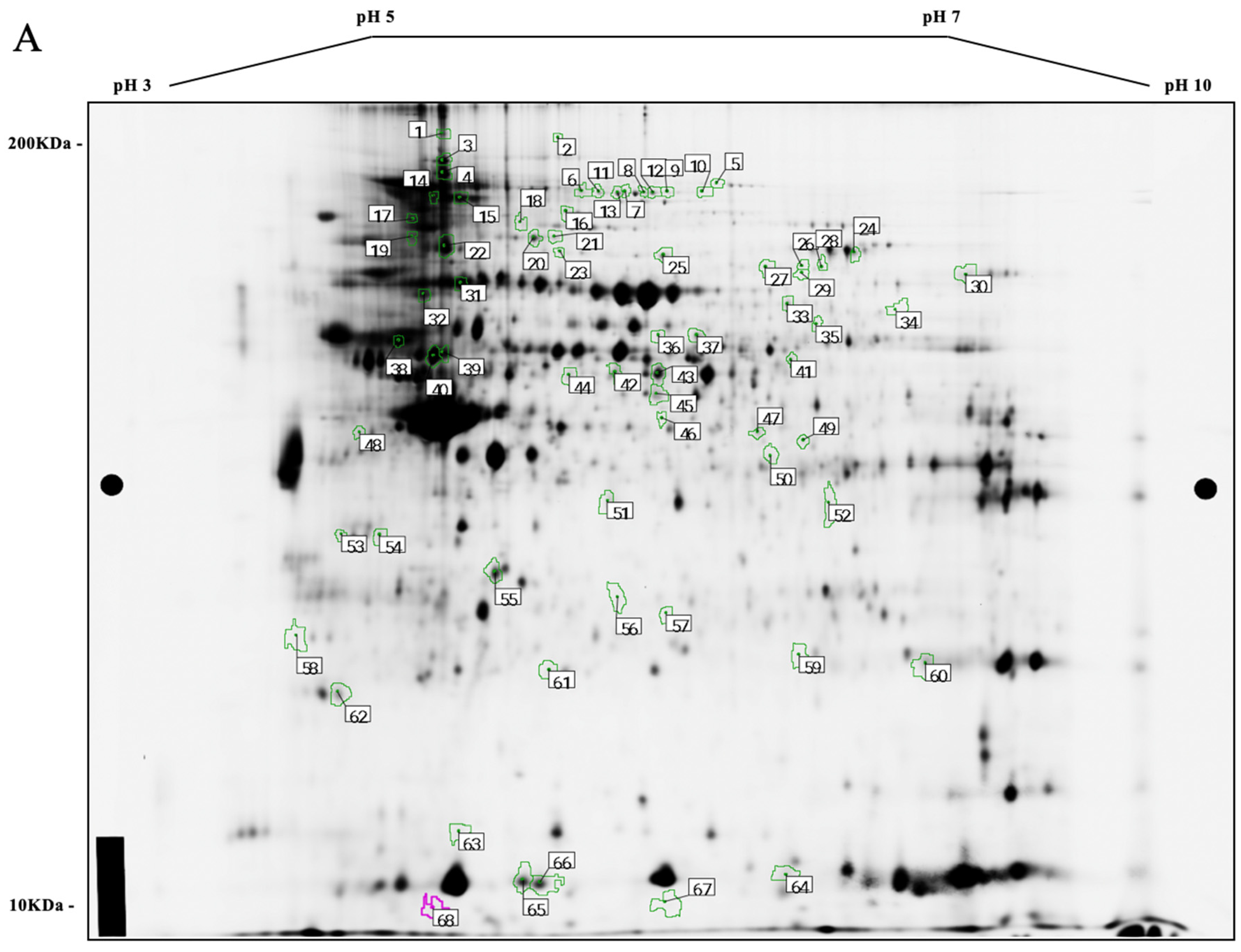
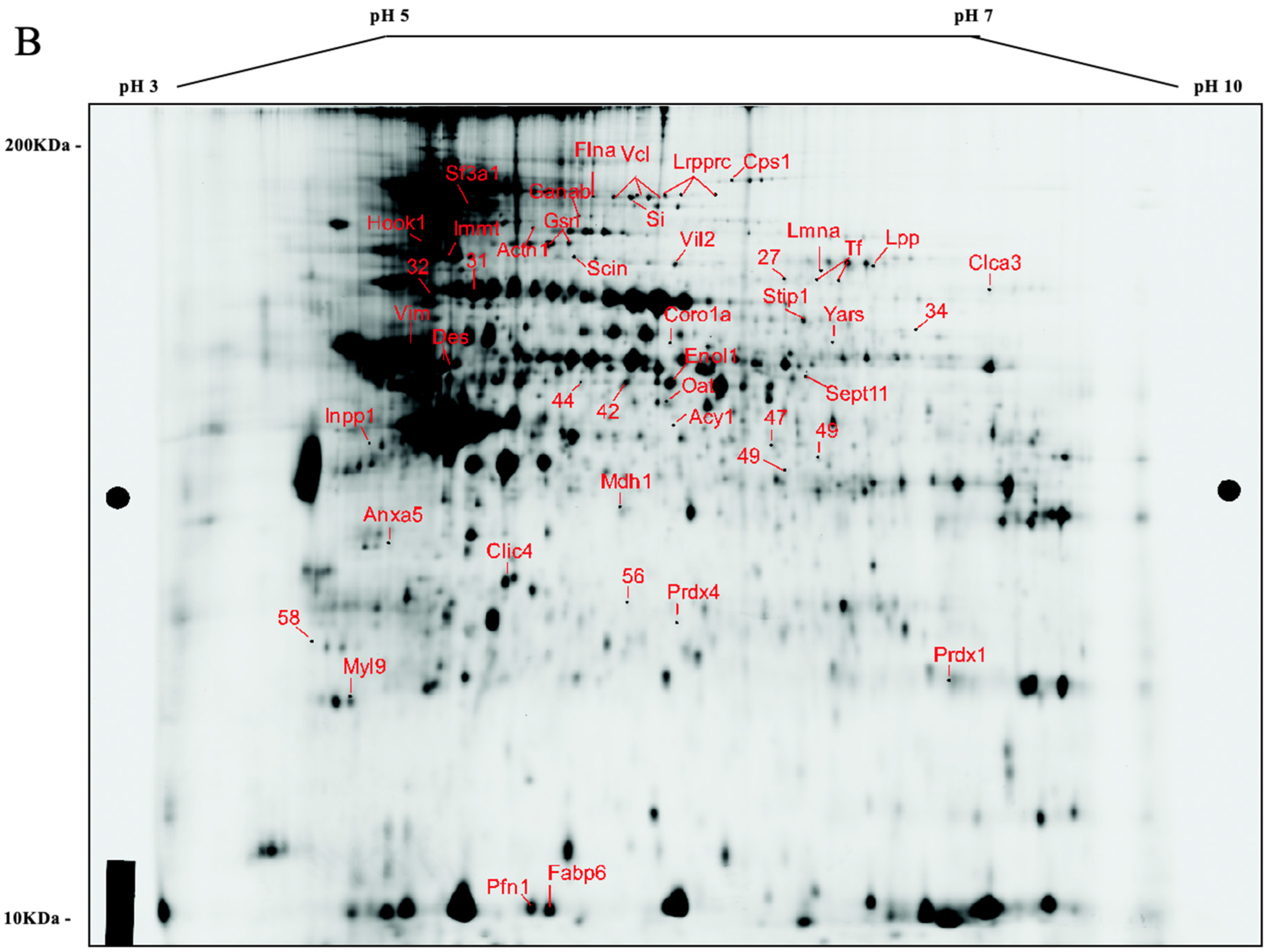
3.2. Comparative Protein Expression Analysis
3.3. Differentially Expressed Proteins
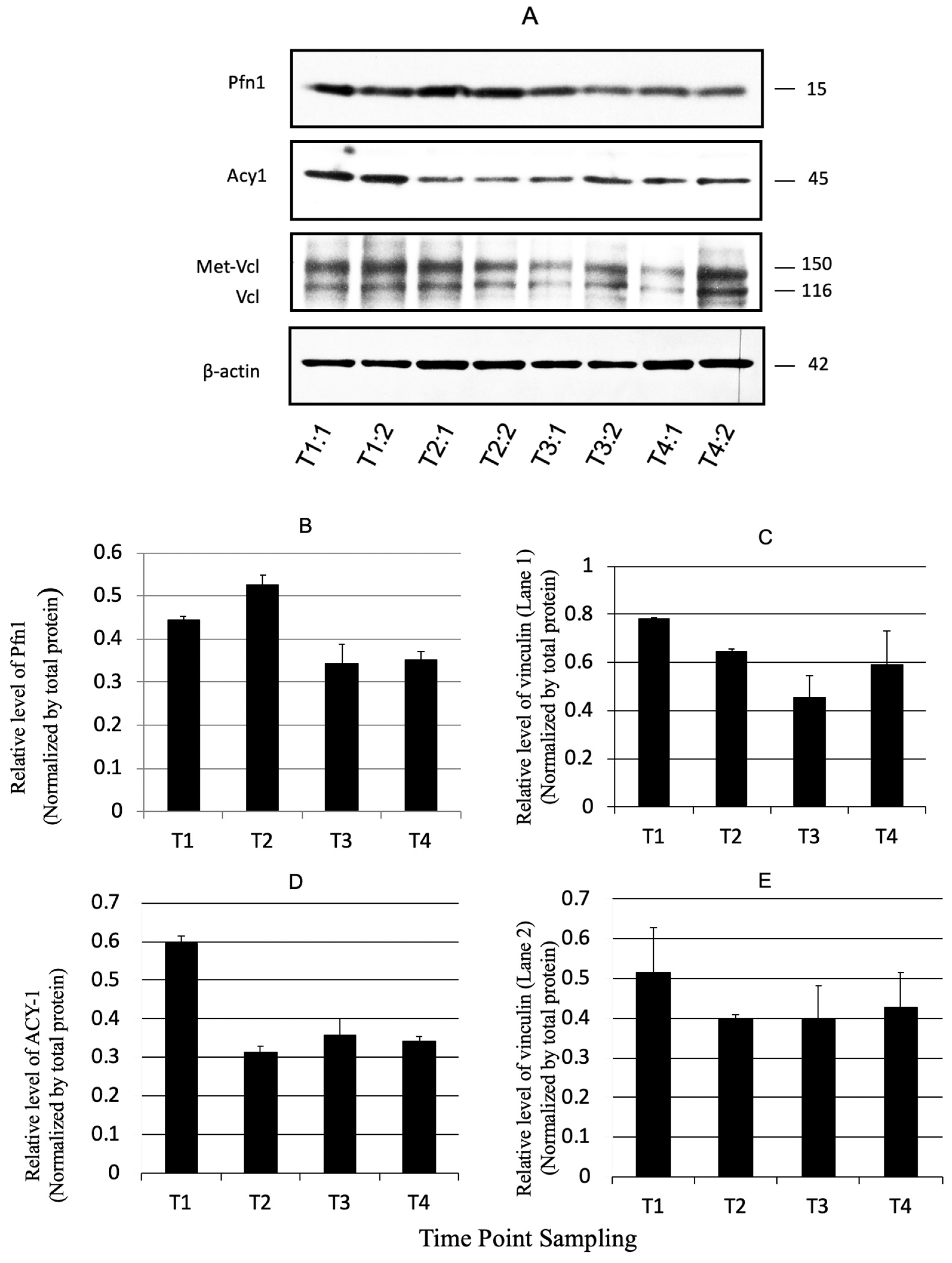
3.4. Validation by Immunoblotting
3.5. Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- de Nooijer, J.; Lechner, L.; Candel, M.; de Vries, H. Short- and long-term effects of tailored information versus general information on determinants and intentions related to early detection of cancer. Prev. Med. 2004, 38, 694–703. [Google Scholar] [CrossRef] [PubMed]
- Bird, R.P. Observation and quantification of aberrant crypts in the murine colon treated with a colon carcinogen: Preliminary findings. Cancer Lett. 1987, 37, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Shivapurkar, N.; Tang, Z.; Ferreira, A.; Nasim, S.; Garett, C.; Alabaster, O. Sequential analysis of K-ras mutations in aberrant crypt foci and colonic tumors induced by azoxymethane in Fischer-344 rats on high-risk diet. Carcinogenesis 1994, 15, 775–778. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Mutoh, M.; Kawamori, T.; Sugimura, T.; Wakabayashi, K. Altered expression of beta-catenin, inducible nitric oxide synthase and cyclooxygenase-2 in azoxymethane-induced rat colon carcinogenesis. Carcinogenesis 2000, 21, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Lai, M.D. Aberrant crypt foci as microscopic precursors of colorectal cancer. World J. Gastroenterol. 2003, 9, 2642–2649. [Google Scholar] [CrossRef]
- Hietanen, E.; Bartsch, H.; Béréziat, J.C.; Camus, A.M.; McClinton, S.; Eremin, O.; Davidson, L.; Boyle, P. Diet and oxidative stress in breast, colon and prostate cancer patients: A case-control study. Eur. J. Clin. Nutr. 1994, 48, 575–586. [Google Scholar] [PubMed]
- Wali, R.K.; Khare, S.; Tretiakova, M.; Cohen, G.; Nguyen, L.; Hart, J.; Wang, J.; Wen, M.; Ramaswamy, A.; Joseph, L.; et al. Ursodeoxycholic acid and F(6)-D(3) inhibit aberrant crypt proliferation in the rat azoxymethane model of colon cancer: Roles of cyclin D1 and E-cadherin. Cancer Epidemiol. Biomarkers Prev. 2002, 11, 1653–1662. [Google Scholar] [PubMed]
- Pretlow, T.P.; Barrow, B.J.; Ashton, W.S.; O’Riordan, M.A.; Pretlow, T.G.; Jurcisek, J.A.; Stellato, T.A. Aberrant crypts: Putative preneoplastic foci in human colonic mucosa. Cancer Res. 1991, 51, 1564–1567. [Google Scholar] [PubMed]
- Konstantakos, A.K.; Siu, I.M.; Pretlow, T.G.; Stellato, T.A.; Pretlow, T.P. Human aberrant crypt foci with carcinoma in situ from a patient with sporadic colon cancer. Gastroenterology 1996, 111, 772–777. [Google Scholar] [CrossRef]
- Siu, I.M.; Pretlow, T.G.; Amini, S.B.; Pretlow, T.P. Identification of dysplasia in human colonic aberrant crypt foci. Am. J. Pathol. 1997, 150, 1805–1813. [Google Scholar] [PubMed]
- Canzian, F.; Ushijima, T.; Serikawa, T.; Wakabayashi, K.; Sugimura, T.; Nagao, M. Instability of microsatellites in rat colon tumors induced by heterocyclic amines. Cancer Res. 1994, 54, 6315–6317. [Google Scholar] [PubMed]
- Rigas, B.; Goldman, I.S.; Levine, L. Altered eicosanoid levels in human colon cancer. J. Lab. Clin. Med. 1993, 122, 518–523. [Google Scholar] [PubMed]
- Kawamori, T.; Uchiya, N.; Sugimura, T.; Wakabayashi, K. Enhancement of colon carcinogenesis by prostaglandin E2 administration. Carcinogenesis 2003, 24, 985–990. [Google Scholar] [CrossRef]
- Bara, J.; Forgue-Lafitte, M.E.; Maurin, N.; Fléjou, J.F.; Zimber, A. Abnormal expression of gastric mucin in human and rat aberrant crypt foci during colon carcinogenesis. Tumour Biol. 2003, 24, 109–115. [Google Scholar] [CrossRef]
- Pierre, F.; Freeman, A.; Taché, S.; Van der Meer, R.; Corpet, D.E. Beef meat and blood sausage promote the formation of azoxymethane-induced mucin-depleted foci and aberrant crypt foci in rat colons. J. Nutr. 2004, 134, 2711–2716. [Google Scholar] [CrossRef]
- Allen, R.G.; Tresini, M. Oxidative stress and gene regulation. Free Radic. Biol. Med. 2000, 28, 463–499. [Google Scholar] [CrossRef]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef]
- Davies, K.J. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life 2000, 50, 279–289. [Google Scholar] [CrossRef]
- Deng, S.S.; Xing, T.Y.; Zhou, H.Y.; Xiong, R.H.; Lu, Y.G.; Wen, B.; Liu, S.Q.; Yang, H.J. Comparative proteome analysis of breast cancer and adjacent normal breast tissues in human. Genom. Proteom. Bioinform. 2006, 4, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.S.; Varambally, S.; Chinnaiyan, A.M. Differential proteomic alterations between localised and metastatic prostate cancer. Br. J. Cancer 2006, 95, 425–430. [Google Scholar] [CrossRef]
- Alexe, G.; Alexe, S.; Liotta, L.A.; Petricoin, E.; Reiss, M.; Hammer, P.L. Ovarian cancer detection by logical analysis of proteomic data. Proteomics 2004, 4, 766–783. [Google Scholar] [CrossRef]
- Mazzanti, R.; Solazzo, M.; Fantappié, O.; Elfering, S.; Pantaleo, P.; Bechi, P.; Cianchi, F.; Ettl, A.; Giulivi, C. Differential expression proteomics of human colon cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, G1329-38. [Google Scholar] [CrossRef]
- Reeves, P.G.; Nielsen, F.H.; Fahey, G.C., Jr. AIN-93 purified diets for laboratory rodents: Final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993, 123, 1939–1951. [Google Scholar] [CrossRef]
- Tannu, N.S.; Hemby, S.E. Two-dimensional fluorescence difference gel electrophoresis for comparative proteomics profiling. Nat Protoc. 2006, 1, 1732–1742. [Google Scholar] [CrossRef]
- Iiizumi, G.; Sadoya, Y.; Hino, S.; Shibuya, N.; Kawabata, H. Proteomic characterization of the site-dependent functional difference in the rat small intestine. Biochim. Biophys. Acta 2007, 1774, 1289–1298. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Duś-Szachniewicz, K.; Ostasiewicz, P.; Ziółkowski, P.; Rakus, D.; Mann, M. Absolute Proteome Analysis of Colorectal Mucosa, Adenoma, and Cancer Reveals Drastic Changes in Fatty Acid Metabolism and Plasma Membrane Transporters. J. Proteome Res. 2015, 14, 4005–4018. [Google Scholar] [CrossRef]
- Herbert, B.; Galvani, M.; Hamdan, M.; Olivieri, E.; MacCarthy, J.; Pedersen, S.; Righetti, P.G. Reduction and alkylation of proteins in preparation of two-dimensional map analysis: Why, when, and how? Electrophoresis 2001, 22, 2046–2057. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, Z.; Niu, Z.; Liang, B.; Niu, J. Epieriocalyxin A Induces Cell Apoptosis Through JNK and ERK1/2 Signaling Pathways in Colon Cancer Cells. Cell Biochem. Biophys. 2015, 73, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, W.; Yan, J.; Zhou, K.; Wan, B.; Shi, P.; Chen, Y.; He, S.; Li, D. Loss of profilin 2 contributes to enhanced epithelial-mesenchymal transition and metastasis of colorectal cancer. Int. J. Oncol. 2018, 53, 1118–1128. [Google Scholar] [CrossRef]
- Miller, Y.E.; Minna, J.D.; Gazdar, A.F. Lack of expression of aminoacylase-1 in small cell lung cancer. Evidence for inactivation of genes encoded by chromosome 3p. J. Clin. Investig. 1989, 83, 2120–2124. [Google Scholar] [CrossRef]
- Gu, S.; Papadopoulou, N.; Nasir, O.; Föller, M.; Alevizopoulos, K.; Lang, F.; Stournaras, C. Activation of membrane androgen receptors in colon cancer inhibits the prosurvival signals Akt/bad in vitro and in vivo and blocks migration via vinculin/actin signaling. Mol. Med. 2011, 17, 48–58. [Google Scholar] [CrossRef]
- Lee, H.T.; Sharek, L.; O’Brien, E.T.; Urbina, F.L.; Gupton, S.L.; Superfine, R.; Burridge, K.; Campbell, S.L. Vinculin and metavinculin exhibit distinct effects on focal adhesion properties, cell migration, and mechanotransduction. PLoS ONE 2019, 14, e0221962. [Google Scholar] [CrossRef]
- Wu, X.; Hasan, M.A.; Chen, J.Y. Pathway and network analysis in proteomics. J. Theor. Biol. 2014, 362, 44–52. [Google Scholar] [CrossRef]
- Bianchi, L.; Bruzzese, F.; Leone, A.; Gagliardi, A.; Puglia, M.; Di Gennaro, E.; Rocco, M.; Gimigliano, A.; Pucci, B.; Armini, A.; et al. Proteomic analysis identifies differentially expressed proteins after HDAC vorinostat and EGFR inhibitor gefitinib treatments in Hep-2 cancer cells. Proteomics 2011, 11, 3725–3742. [Google Scholar] [CrossRef]
- Lin, Q.; Li, J.; Zhu, D.; Niu, Z.; Pan, X.; Xu, P.; Ji, M.; Wei, Y.; Xu, J. Aberrant Scinderin Expression Correlates With Liver Metastasis and Poor Prognosis in Colorectal Cancer. Front. Pharmacol. 2019, 10, 1183. [Google Scholar] [CrossRef]
- Tavabe Ghavami, T.S.; Irani, S.; Mirfakhrai, R.; Shirkoohi, R. Differential expression of Scinderin and Gelsolin in gastric cancer and comparison with clinical and morphological characteristics. EXCLI J. 2020, 19, 750–761. [Google Scholar] [CrossRef]
- Liu, J.J.; Liu, J.Y.; Chen, J.; Wu, Y.X.; Yan, P.; Ji, C.D.; Wang, Y.X.; Xiang, D.F.; Zhang, X.; Zhang, P.; et al. Scinderin promotes the invasion and metastasis of gastric cancer cells and predicts the outcome of patients. Cancer Lett. 2016, 376, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Zunino, R.; Li, Q.; Rosé, S.D.; Romero-Benítez, M.M.; Lejen, T.; Brandan, N.C.; Trifaró, J.M. Expression of scinderin in megakaryoblastic leukemia cells induces differentiation, maturation, and apoptosis with release of plateletlike particles and inhibits proliferation and tumorigenesis. Blood 2001, 98, 2210–2219. [Google Scholar] [CrossRef] [PubMed]
- Luscieti, S.; Galy, B.; Gutierrez, L.; Reinke, M.; Couso, J.; Shvartsman, M.; Di Pascale, A.; Witke, W.; Hentze, M.W.; Pilo Boyl, P.; et al. The actin-binding protein profilin 2 is a novel regulator of iron homeostasis. Blood 2017, 130, 1934–1945. [Google Scholar] [CrossRef]
- Bai, N.; Ma, Y.; Zhao, J.; Li, B. Knockdown of lncRNA HCP5 Suppresses the Progression of Colorectal Cancer by miR-299-3p/PFN1/AKT Axis. Cancer Manag. Res. 2020, 12, 4747–4758. [Google Scholar] [CrossRef]
- Wu, Q.; Yan, T.; Chen, Y.; Chang, J.; Jiang, Y.; Zhu, D.; Wei, Y. Integrated Analysis of Expression and Prognostic Values of Acyl-CoA Dehydrogenase short-chain in Colorectal Cancer. Int. J. Med. Sci. 2021, 18, 3631–3643. [Google Scholar] [CrossRef]
- Yu, D.L.; Li, H.W.; Wang, Y.; Li, C.Q.; You, D.; Jiang, L.; Song, Y.P.; Li, X.H. Acyl-CoA dehydrogenase long chain expression is associated with esophageal squamous cell carcinoma progression and poor prognosis. Oncol. Targets Ther. 2018, 11, 7643–7653. [Google Scholar] [CrossRef]
- Zhan, P.; Wang, Y.; Zhao, S.; Liu, C.; Wang, Y.; Wen, M.; Mao, J.H.; Wei, G.; Zhang, P. FBXW7 negatively regulates ENO1 expression and function in colorectal cancer. Lab. Investig. 2015, 95, 995–1004. [Google Scholar] [CrossRef]
- Hu, T.; Liu, H.; Liang, Z.; Wang, F.; Zhou, C.; Zheng, X.; Zhang, Y.; Song, Y.; Hu, J.; He, X.; et al. Tumor-intrinsic CD47 signal regulates glycolysis and promotes colorectal cancer cell growth and metastasis. Theranostics 2020, 10, 4056–4072. [Google Scholar] [CrossRef]
- Ginguay, A.; Cynober, L.; Curis, E.; Nicolis, I. Ornithine Aminotransferase, an Important Glutamate-Metabolizing Enzyme at the Crossroads of Multiple Metabolic Pathways. Biology 2017, 6, 18. [Google Scholar] [CrossRef]
- Benjamin, D.I.; Louie, S.M.; Mulvihill, M.M.; Kohnz, R.A.; Li, D.S.; Chan, L.G.; Sorrentino, A.; Bandyopadhyay, S.; Cozzo, A.; Ohiri, A.; et al. Inositol phosphate recycling regulates glycolytic and lipid metabolism that drives cancer aggressiveness. ACS Chem. Biol. 2014, 9, 1340–1350. [Google Scholar] [CrossRef]
- Jia, W.; Chen, P.; Cheng, Y. PRDX4 and Its Roles in Various Cancers. Technol. Cancer Res. Treat. 2019, 18, 1533033819864313. [Google Scholar] [CrossRef]
- Zheng, J.; Guo, X.; Nakamura, Y.; Zhou, X.; Yamaguchi, R.; Zhang, J.; Ishigaki, Y.; Uramoto, H.; Yamada, S. Overexpression of PRDX4 Modulates Tumor Microenvironment and Promotes Urethane-Induced Lung Tumorigenesis. Oxid. Med. Cell Longev. 2020, 2020, 8262730. [Google Scholar] [CrossRef]
- Tochhawng, L.; Deng, S.; Pugalenthi, G.; Kumar, A.P.; Lim, K.H.; Tan, T.Z.; Yang, H.; Hooi, S.C.; Goh, Y.C.; Maciver, S.K.; et al. Gelsolin-Cu/ZnSOD interaction alters intracellular reactive oxygen species levels to promote cancer cell invasion. Oncotarget 2016, 7, 52832–52848. [Google Scholar] [CrossRef]
- Kim, J.C.; Ha, Y.J.; Tak, K.H.; Roh, S.A.; Kwon, Y.H.; Kim, C.W.; Yoon, Y.S.; Lee, J.L.; Park, Y.; Kim, S.K.; et al. Opposite functions of GSN and OAS2 on colorectal cancer metastasis, mediating perineural and lymphovascular invasion, respectively. PLoS ONE 2018, 13, e0202856. [Google Scholar] [CrossRef]
- Stanton, S.E.; Gad, E.; Corulli, L.R.; Lu, H.; Disis, M.L. Tumor-associated antigens identified early in mouse mammary tumor development can be effective vaccine targets. Vaccine 2019, 37, 3552–3561. [Google Scholar] [CrossRef]
- Fang, X.; Wu, X.; Xiang, E.; Luo, F.; Li, Q.; Ma, Q.; Yuan, F.; Chen, P. Expression profiling of CPS1 in Correa’s cascade and its association with gastric cancer prognosis. Oncol. Lett. 2021, 21, 441. [Google Scholar] [CrossRef]
- Zhang, W.; Yuan, W.; Song, J.; Wang, S.; Gu, X. LncRna CPS1-IT1 Suppresses Cell Proliferation, Invasion and Metastasis in Colorectal Cancer. Cell Physiol. Biochem. 2017, 44, 567–580. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, X.; Deng, L.; Li, X.; Wang, G.; Li, Y.; Chen, M. High expression of FABP4 and FABP6 in patients with colorectal cancer. World J. Surg. Oncol. 2019, 17, 171. [Google Scholar] [CrossRef]
- Lian, W.; Wang, Z.; Ma, Y.; Tong, Y.; Zhang, X.; Jin, H.; Zhao, S.; Yu, R.; Ju, S.; Zhang, X.; et al. FABP6 Expression Correlates with Immune Infiltration and Immunogenicity in Colorectal Cancer Cells. J. Immunol. Res. 2022, 2022, 3129765. [Google Scholar] [CrossRef]
- Pai, F.C.; Huang, H.W.; Tsai, Y.L.; Tsai, W.C.; Cheng, Y.C.; Chang, H.H.; Chen, Y. Inhibition of FABP6 Reduces Tumor Cell Invasion and Angiogenesis through the Decrease in MMP-2 and VEGF in Human Glioblastoma Cells. Cells 2021, 10, 2782. [Google Scholar] [CrossRef]
- Bertram, J.S. The molecular biology of cancer. Mol. Aspects Med. 2000, 21, 167–223. [Google Scholar] [CrossRef]


| Formula | g/Kg | |
|---|---|---|
| Casein | 200.0 | |
| L-Cysteine | 3.0 | |
| Corn Starch | 397.486 | |
| Maltodextrin | 132.0 | |
| Sucrose | 100.0 | |
| Soybean oil | 70.0 | |
| Cellulose | 50.0 | |
| Mineral Mix, AIN-93G-MX (04046) | 35.0 | |
| Vitamin Mix, AIN-93-VX (94047) | 10.0 | |
| Choline Bitartrate | 2.5 | |
| TBHQ, antioxidant | 0.014 | |
| Macronutrient | % dry weight | % kcal |
| Protein | 17.7 | 18.8 |
| Carbohydrate | 60.1 | 63.9 |
| Fat | 7.2 | 17.2 |
| Gene Name | Protein Identity | Fold Change | One Way ANOVA p-Value | T-Test p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| T2/T1 | T3/T1 | T4/T1 | T2/T1 | T3/T1 | T4/T1 | |||
| Cps1 | Carbamoyl-phosphate synthase, mitochondrial | −1.21 | −1.26 | −1.65 | 0.043 | 0.24 | 0.061 | 0.023 |
| Flna | Filamin, alpha isoform | −1.45 | −1.68 | −1.54 | 0.02 | 0.053 | 0.0015 | 0.0077 |
| Vcl | Vinculin | −2.14 | −3.4 | −3.89 | 0.0072 | 0.034 | 0.0002 | 0.0063 |
| Lrpprc | Leucine-rich PPR motif-containing, mitochondrial | −2.15 | −3.15 | −3.55 | 0.016 | 0.059 | 0.0019 | 0.0012 |
| Si | Si 210 kDa protein | −2.46 | −3.47 | −4.13 | 0.02 | 0.044 | 0.00079 | 0.0088 |
| Sf3a1 | Splicing factor 3a, subunit 1 | −1.44 | −2.14 | −2.19 | 0.03 | 0.071 | 0.00048 | 0.035 |
| Ganab | Putative uncharacterized protein | −1.01 | 1.46 | 1.24 | 0.05 | 0.99 | 0.065 | 0.16 |
| Actn1 | Actinin | −1.53 | −1.47 | −1.91 | 0.041 | 0.17 | 0.21 | 0.049 |
| Hook1 | Hook homolog 1 (predicted) | 1.38 | 1.7 | 1.99 | 0.0029 | 0.041 | 0.0052 | 0.0096 |
| Gsn | Isoform 2 of gelsolin | 1.16 | 1.35 | 1.59 | 0.044 | 0.12 | 0.093 | 0.014 |
| Immt | Inner membrane protein, mitochondrial (predicted) | 1.11 | 1.19 | 1.68 | 0.0096 | 0.014 | 0.071 | 0.018 |
| Scin | Scinderin | −1.13 | −1.38 | −1.48 | 0.028 | 0.44 | 0.04 | 0.016 |
| Lpp | Lipoma-preferred partner homolog | 1.27 | 1.3 | 1.47 | 0.03 | 0.062 | 0.077 | 0.022 |
| Ezr | Ezrin | 1.1 | 1.56 | 1.32 | 0.033 | 0.13 | 0.0064 | 0.16 |
| Lmna | Lamin-A | −1.52 | −1.92 | −1.55 | 0.028 | 0.048 | 0.029 | 0.071 |
| Tf | Isoform 1 of serotransferrin | −1.58 | −1.89 | −1.86 | 0.039 | 0.028 | 0.011 | 0.054 |
| Clca3 | Putative uncharacterized protein | 1.06 | −1.36 | −1.56 | 0.046 | 0.58 | 0.044 | 0.1 |
| Stip1 | Stress-induced-phosphoprotein 1 | 1.15 | 1.5 | 1.25 | 0.0028 | 0.11 | 0.0031 | 0.021 |
| Yars | Tyrosyl-tRNA synthetase, cytoplasmic | 1.23 | 1.73 | 1.4 | 0.025 | 0.14 | 0.017 | 0.061 |
| Coro1a | Coronin-1A | −1.19 | 1.09 | −1.55 | 0.022 | 0.18 | 0.65 | 0.029 |
| Vim | Vimentin | 1.2 | 1.45 | 1.41 | 0.012 | 0.018 | 0.0029 | 0.038 |
| Des | Desmin | 1.37 | 1.61 | 2.76 | 0.025 | 0.07 | 0.016 | 0.04 |
| Sept11 | Isoform 1 of septin-11 | 1.02 | 1.38 | 1.42 | 0.012 | 0.88 | 0.036 | 0.024 |
| Eno1 | Alpha-enolase | −1.55 | −1.85 | −1.89 | 0.043 | 0.056 | 0.012 | 0.024 |
| Oat | Ornithine aminotransferase, mitochondrial | −1.37 | −1.67 | −1.71 | 0.0018 | 0.044 | 0.011 | 0.0098 |
| Acy1 | Aminoacylase-1A | −1.5 | −1.92 | −1.72 | 0.012 | 0.069 | 0.00071 | 0.0052 |
| Inpp1 | Inositol polyphosphate-1-phosphatase | −1.42 | −1.62 | −1.63 | 0.031 | 0.072 | 0.00036 | 0.0088 |
| Acads | Acyl-Coenzyme A dehydrogenase | 1.11 | 1.12 | 1.49 | 0.035 | 0.39 | 0.21 | 0.034 |
| Mdh1 | Malate dehydrogenase, cytoplasmic | −1.33 | −1.41 | −1.48 | 0.02 | 0.049 | 0.025 | 0.023 |
| Clic4 | Chloride intracellular channel protein 4 | 1.48 | 1.38 | 1.58 | 0.034 | 0.062 | 0.056 | 0.031 |
| Prdx4 | Peroxiredoxin-4 | 1.18 | 1.51 | 1.42 | 0.02 | 0.056 | 0.016 | 0.028 |
| Prdx1 | Peroxiredoxin-1 | 1.03 | −1.11 | 1.35 | 0.04 | 0.67 | 0.017 | 0.12 |
| Myl9 | Myosin, light polypeptide 9, regulatory | 1.35 | 1.25 | 1.68 | 0.013 | 0.0075 | 0.064 | 0.016 |
| Pfn1 | Profilin-1 | −1.39 | −1.76 | −2.08 | 0.015 | 0.066 | 0.00067 | 0.02 |
| Fabp6 | Gastrotropin | 8.08 | 5.54 | 4.88 | 0.012 | 0.025 | 0.0046 | 0.0023 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swain, I.X.; Kresak, A.M. Proteins Involved in Focal Cell Adhesion and Podosome Formation Are Differentially Expressed during Colorectal Tumorigenesis in AOM-Treated Rats. Cancers 2024, 16, 1678. https://doi.org/10.3390/cancers16091678
Swain IX, Kresak AM. Proteins Involved in Focal Cell Adhesion and Podosome Formation Are Differentially Expressed during Colorectal Tumorigenesis in AOM-Treated Rats. Cancers. 2024; 16(9):1678. https://doi.org/10.3390/cancers16091678
Chicago/Turabian StyleSwain, Ian X., and Adam M. Kresak. 2024. "Proteins Involved in Focal Cell Adhesion and Podosome Formation Are Differentially Expressed during Colorectal Tumorigenesis in AOM-Treated Rats" Cancers 16, no. 9: 1678. https://doi.org/10.3390/cancers16091678
APA StyleSwain, I. X., & Kresak, A. M. (2024). Proteins Involved in Focal Cell Adhesion and Podosome Formation Are Differentially Expressed during Colorectal Tumorigenesis in AOM-Treated Rats. Cancers, 16(9), 1678. https://doi.org/10.3390/cancers16091678






