Serum Proteomic Signatures in Cervical Cancer: Current Status and Future Directions
Abstract
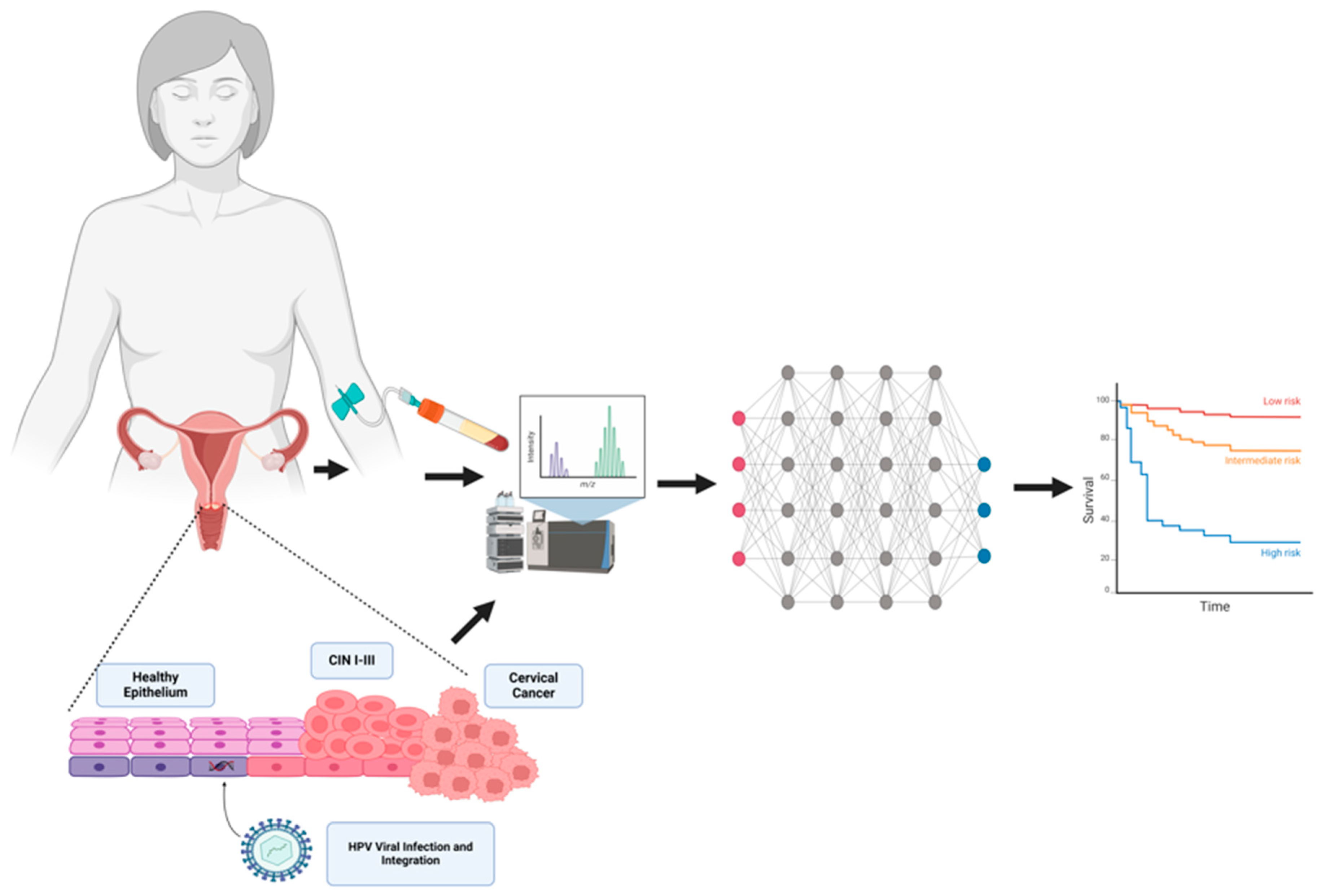
Simple Summary
Abstract
1. Introduction
2. Proteomic Changes in Cervical Cancer Cells
3. Serum Protein Markers in CC
3.1. Proteomic Studies with Diagnostic Potential
3.2. Proteomic Signatures for Prognosis in CC
4. Future Directions in CC Serum Proteomics
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fowler, J.R.; Maani, E.V.; Dunton, C.J.; Jack, B.W. Cervical Cancer. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Safaeian, M.; Solomon, D. Cervical Cancer Prevention—Cervical Screening: Science in Evolution. Obstet. Gynecol. Clin. N. Am. 2007, 34, 739–760. [Google Scholar] [CrossRef]
- Chan, C.K.; Aimagambetova, G.; Ukybassova, T.; Kongrtay, K.; Azizan, A. Human Papillomavirus Infection and Cervical Cancer: Epidemiology, Screening, and Vaccination—Review of Current Perspectives. J. Oncol. 2019, 2019, 3257939. [Google Scholar] [CrossRef]
- Crosbie, E.J.; Einstein, M.H.; Franceschi, S.; Kitchener, H.C. Human Papillomavirus and Cervical Cancer. Lancet 2013, 382, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A. HPV Testing as a Screen for Cervical Cancer. BMJ 2015, 350, h2372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zeng, Q.; Cai, W.; Ruan, W. Trends of Cervical Cancer at Global, Regional, and National Level: Data from the Global Burden of Disease Study 2019. BMC Public Health 2021, 21, 894. [Google Scholar] [CrossRef] [PubMed]
- Go, D.-S.; Kim, Y.-E.; Yoon, S.-J. Subnational Burden of Disease According to the Sociodemographic Index in South Korea. Int. J. Environ. Res. Public Health 2020, 17, 5788. [Google Scholar] [CrossRef] [PubMed]
- Francoeur, A.A.; Liao, C.-I.; Casear, M.A.; Chan, A.; Kapp, D.S.; Cohen, J.G.; Salani, R.; Chan, J.K. The Increasing Incidence of Stage IV Cervical Cancer in the USA: What Factors Are Related? Int. J. Gynecol. Cancer 2022, 32, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Arezzo, F.; Cormio, G.; Loizzi, V.; Cazzato, G.; Cataldo, V.; Lombardi, C.; Ingravallo, G.; Resta, L.; Cicinelli, E. HPV-Negative Cervical Cancer: A Narrative Review. Diagnostics 2021, 11, 952. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Ortiz, A.P.; Pinheiro, P.S.; Bandi, P.; Minihan, A.; Fuchs, H.E.; Martinez Tyson, D.; Tortolero-Luna, G.; Fedewa, S.A.; Jemal, A.M.; et al. Cancer Statistics for the US Hispanic/Latino Population, 2021. CA A Cancer J. Clin. 2021, 71, 466–487. [Google Scholar] [CrossRef]
- Bast, R.C.; Lilja, H.; Urban, N.; Rimm, D.L.; Fritsche, H.; Gray, J.; Veltri, R.; Klee, G.; Allen, A.; Kim, N.; et al. Translational Crossroads for Biomarkers. Clin. Cancer Res. 2005, 11, 6103–6108. [Google Scholar] [CrossRef]
- Güzel, C.; van Sten-van’t Hoff, J.; de Kok, I.M.C.M.; Govorukhina, N.I.; Boychenko, A.; Luider, T.M.; Bischoff, R. Molecular Markers for Cervical Cancer Screening. Expert Rev. Proteom. 2021, 18, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Pouliquen, D.L.; Boissard, A.; Coqueret, O.; Guette, C. Biomarkers of Tumor Invasiveness in Proteomics (Review). Int. J. Oncol. 2020, 57, 409–432. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Rodríguez, F.; Limones-González, J.E.; Mendoza-Almanza, B.; Esparza-Ibarra, E.L.; Gallegos-Flores, P.I.; Ayala-Luján, J.L.; Godina-González, S.; Salinas, E.; Mendoza-Almanza, G. Understanding Cervical Cancer through Proteomics. Cells 2021, 10, 1854. [Google Scholar] [CrossRef] [PubMed]
- Rusling, J.F.; Kumar, C.V.; Gutkind, J.S.; Patel, V. Measurement of Biomarker Proteins for Point-of-Care Early Detection and Monitoring of Cancer. Analyst 2010, 135, 2496–2511. [Google Scholar] [CrossRef] [PubMed]
- Droit, A.; Poirier, G.G.; Hunter, J.M. Experimental and Bioinformatic Approaches for Interrogating Protein-Protein Interactions to Determine Protein Function. J. Mol. Endocrinol. 2005, 34, 263–280. [Google Scholar] [CrossRef] [PubMed]
- Pascovici, D.; Wu, J.X.; McKay, M.J.; Joseph, C.; Noor, Z.; Kamath, K.; Wu, Y.; Ranganathan, S.; Gupta, V.; Mirzaei, M. Clinically Relevant Post-Translational Modification Analyses—Maturing Workflows and Bioinformatics Tools. Int. J. Mol. Sci. 2018, 20, 16. [Google Scholar] [CrossRef] [PubMed]
- Boylan, K.L.M.; Afiuni-Zadeh, S.; Geller, M.A.; Argenta, P.A.; Griffin, T.J.; Skubitz, A.P.N. Evaluation of the Potential of Pap Test Fluid and Cervical Swabs to Serve as Clinical Diagnostic Biospecimens for the Detection of Ovarian Cancer by Mass Spectrometry-Based Proteomics. Clin. Proteom. 2021, 18, 4. [Google Scholar] [CrossRef] [PubMed]
- Al-wajeeh, A.S.; Salhimi, S.M.; Al-Mansoub, M.A.; Khalid, I.A.; Harvey, T.M.; Latiff, A.; Ismail, M.N. Comparative Proteomic Analysis of Different Stages of Breast Cancer Tissues Using Ultra High Performance Liquid Chromatography Tandem Mass Spectrometer. PLoS ONE 2020, 15, e0227404. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.D.; Chen, Z.; Saller, C.; Tarvin, K.; Carvalho, A.L.; Scapulatempo-Neto, C.; Silveira, H.C.; Fregnani, J.H.; Creighton, C.J.; Anderson, M.L.; et al. Integrated Genomic and Molecular Characterization of Cervical Cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Keeratichamroen, S.; Subhasitanont, P.; Chokchaichamnankit, D.; Weeraphan, C.; Saharat, K.; Sritana, N.; Kantathavorn, N.; Wiriyaukaradecha, K.; Sricharunrat, T.; Paricharttanakul, N.M.; et al. Identification of Potential Cervical Cancer Serum Biomarkers in Thai Patients. Oncol. Lett. 2020, 19, 3815–3826. [Google Scholar] [CrossRef]
- Yang, S.; Wang, X.; Cai, J.; Wang, S. Diagnostic Value of Circulating PIGF in Combination with Flt-1 in Early Cervical Cancer. Curr. Med. Sci. 2020, 40, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Ye, M.; Chen, X.; Zhao, H.; Hasim, A.; Guo, X. Discovery and Validation of FBLN1 and ANT3 as Potential Biomarkers for Early Detection of Cervical Cancer. Cancer Cell Int. 2021, 21, 125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dong, D.; Wei, Q.; Ren, L. CXCL10 Serves as a Potential Serum Biomarker Complementing SCC-Ag for Diagnosing Cervical Squamous Cell Carcinoma. BMC Cancer 2022, 22, 1052. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Pang, C.; Li, Z.; Xu, F.; Zhao, L. Serum CXCL8 and CXCR2 as Diagnostic Biomarkers for Noninvasive Screening of Cervical Cancer. Medicine 2023, 102, e34977. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xiong, X.; Wang, Y.; Zhao, J.; Shi, H.; Zhang, H.; Wang, Y.; Wei, Y.; Xue, W.; Zhang, J. Proteomic Screening for Serum Biomarkers for Cervical Cancer and Their Clinical Significance. Med. Sci. Monit. 2019, 25, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Maestri, C.A.; Nisihara, R.; Mendes, H.W.; Jensenius, J.; Thiel, S.; Messias-Reason, I.; de Carvalho, N.S. MASP-1 and MASP-2 Serum Levels Are Associated With Worse Prognostic in Cervical Cancer Progression. Front. Immunol. 2018, 9, 2742. [Google Scholar] [CrossRef] [PubMed]
- Sawada, M.; Oishi, T.; Komatsu, H.; Sato, S.; Chikumi, J.; Nonaka, M.; Kudoh, A.; Osaku, D.; Harada, T. Serum Vascular Endothelial Growth Factor A and Vascular Endothelial Growth Factor Receptor 2 as Prognostic Biomarkers for Uterine Cervical Cancer. Int. J. Clin. Oncol. 2019, 24, 1612–1619. [Google Scholar] [CrossRef] [PubMed]
- Purohit, S.; Zhi, W.; Ferris, D.G.; Alverez, M.; Tran, L.K.H.; Tran, P.M.H.; Dun, B.; Hopkins, D.; Santos, B.D.; Ghamande, S.; et al. Senescence-Associated Secretory Phenotype Determines Survival and Therapeutic Response in Cervical Cancer. Cancers 2020, 12, 2899. [Google Scholar] [CrossRef] [PubMed]
- Harima, Y.; Ariga, T.; Kaneyasu, Y.; Ikushima, H.; Tokumaru, S.; Shimamoto, S.; Takahashi, T.; Ii, N.; Tsujino, K.; Saito, A.I.; et al. Clinical Value of Serum Biomarkers, Squamous Cell Carcinoma Antigen and Apolipoprotein C-II in Follow-up of Patients with Locally Advanced Cervical Squamous Cell Carcinoma Treated with Radiation: A Multicenter Prospective Cohort Study. PLoS ONE 2021, 16, e0259235. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, L.; Chen, K.; Ye, W. Identification and Validation of Serum Tumor-Markers Based Nomogram to Predict the Prognostic Value of Patients with Cervical Adenocarcinoma. Curr. Probl. Cancer 2022, 46, 100899. [Google Scholar] [CrossRef]
- Tan, G.S.; Lim, K.H.; Tan, H.T.; Khoo, M.L.; Tan, S.H.; Toh, H.C.; Ching Ming Chung, M. Novel Proteomic Biomarker Panel for Prediction of Aggressive Metastatic Hepatocellular Carcinoma Relapse in Surgically Resectable Patients. J. Proteome Res. 2014, 13, 4833–4846. [Google Scholar] [CrossRef]
- Ji, X.; Chu, G.; Chen, Y.; Jiao, J.; Lv, T.; Yao, Q. Comprehensive Analysis of Novel Prognosis-Related Proteomic Signature Effectively Improve Risk Stratification and Precision Treatment for Patients with Cervical Cancer. Arch. Gynecol. Obs. 2023, 307, 903–917. [Google Scholar] [CrossRef] [PubMed]
- Ngoi, N.Y.L.; Choong, C.; Lee, J.; Bellot, G.; Wong, A.L.A.; Goh, B.C.; Pervaiz, S. Targeting Mitochondrial Apoptosis to Overcome Treatment Resistance in Cancer. Cancers 2020, 12, 574. [Google Scholar] [CrossRef] [PubMed]
- Betzen, C.; Alhamdani, M.S.S.; Lueong, S.; Schröder, C.; Stang, A.; Hoheisel, J.D. Clinical Proteomics: Promises, Challenges and Limitations of Affinity Arrays. Proteom.—Clin. Appl. 2015, 9, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.L.; Anderson, N.G. The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Mol. Cell. Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef]
- Parker, C.E.; Borchers, C.H. Mass Spectrometry Based Biomarker Discovery, Verification, and Validation—Quality Assurance and Control of Protein Biomarker Assays. Mol. Oncol. 2014, 8, 840–858. [Google Scholar] [CrossRef] [PubMed]
- Abbatiello, S.E.; Schilling, B.; Mani, D.R.; Zimmerman, L.J.; Hall, S.C.; MacLean, B.; Albertolle, M.; Allen, S.; Burgess, M.; Cusack, M.P.; et al. Large-Scale Interlaboratory Study to Develop, Analytically Validate and Apply Highly Multiplexed, Quantitative Peptide Assays to Measure Cancer-Relevant Proteins in Plasma. Mol. Cell. Proteom. 2015, 14, 2357–2374. [Google Scholar] [CrossRef] [PubMed]
- Razavi, M.; Anderson, N.L.; Yip, R.; Pope, M.E.; Pearson, T.W. Multiplexed Longitudinal Measurement of Protein Biomarkers in DBS Using an Automated SISCAPA Workflow. Bioanalysis 2016, 8, 1597–1609. [Google Scholar] [CrossRef] [PubMed]
- Bhawal, R.; Oberg, A.L.; Zhang, S.; Kohli, M. Challenges and Opportunities in Clinical Applications of Blood-Based Proteomics in Cancer. Cancers 2020, 12, 2428. [Google Scholar] [CrossRef]
- Frantzi, M.; Latosinska, A.; Mischak, H. Proteomics in Drug Development: The Dawn of a New Era? Proteom.-Clin. Appl. 2019, 13, 1800087. [Google Scholar] [CrossRef]
- Panis, C.; Pizzatti, L.; Souza, G.F.; Abdelhay, E. Clinical Proteomics in Cancer: Where We Are. Cancer Lett. 2016, 382, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Emilsson, V.; Ilkov, M.; Lamb, J.R.; Finkel, N.; Gudmundsson, E.F.; Pitts, R.; Hoover, H.; Gudmundsdottir, V.; Horman, S.R.; Aspelund, T.; et al. Co-Regulatory Networks of Human Serum Proteins Link Genetics to Disease. Science 2018, 361, 769–773. [Google Scholar] [CrossRef]
- Carlsson, A.; Wingren, C.; Kristensson, M.; Rose, C.; Fernö, M.; Olsson, H.; Jernström, H.; Ek, S.; Gustavsson, E.; Ingvar, C.; et al. Molecular Serum Portraits in Patients with Primary Breast Cancer Predict the Development of Distant Metastases. Proc. Natl. Acad. Sci. USA 2011, 108, 14252–14257. [Google Scholar] [CrossRef]
- Nordström, M.; Wingren, C.; Rose, C.; Bjartell, A.; Becker, C.; Lilja, H.; Borrebaeck, C.A.K. Identification of Plasma Protein Profiles Associated with Risk Groups of Prostate Cancer Patients. Proteom. Clin. Appl. 2014, 8, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Füzéry, A.K.; Levin, J.; Chan, M.M.; Chan, D.W. Translation of Proteomic Biomarkers into FDA Approved Cancer Diagnostics: Issues and Challenges. Clin. Proteom. 2013, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xiao, J.; Wu, S.; Liu, L.; Zeng, X.; Zhao, Q.; Zhang, Z. Clinical Application of Serum-Based Proteomics Technology in Human Tumor Research. Anal. Biochem. 2023, 663, 115031. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wang, W.; Wang, Y.; Liu, C.; Wang, P. The Role of Squamous Cell Carcinoma Antigen (SCC Ag) in Outcome Prediction after Concurrent Chemoradiotherapy and Treatment Decisions for Patients with Cervical Cancer. Radiat. Oncol. 2019, 14, 146. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.M.; Hicklin, D.J. VEGF-Targeted Therapy: Mechanisms of Anti-Tumour Activity. Nat. Rev. Cancer 2008, 8, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Tewari, K.S.; Sill, M.W.; Long, H.J.; Penson, R.T.; Huang, H.; Ramondetta, L.M.; Landrum, L.M.; Oaknin, A.; Reid, T.J.; Leitao, M.M.; et al. Improved Survival with Bevacizumab in Advanced Cervical Cancer. N. Engl. J. Med. 2014, 370, 734–743. [Google Scholar] [CrossRef]
- Degn, S.E.; Jensen, L.; Olszowski, T.; Jensenius, J.C.; Thiel, S. Co-Complexes of MASP-1 and MASP-2 Associated with the Soluble Pattern-Recognition Molecules Drive Lectin Pathway Activation in a Manner Inhibitable by MAp44. J. Immunol. 2013, 191, 1334–1345. [Google Scholar] [CrossRef]
- Bray, F.; Carstensen, B.; Møller, H.; Zappa, M.; Žakelj, M.P.; Lawrence, G.; Hakama, M.; Weiderpass, E. Incidence Trends of Adenocarcinoma of the Cervix in 13 European Countries. Cancer Epidemiol. Biomark. Prev. 2005, 14, 2191–2199. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J Clin 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Iida, M.; Banno, K.; Yanokura, M.; Nakamura, K.; Adachi, M.; Nogami, Y.; Umene, K.; Masuda, K.; Kisu, I.; Iwata, T.; et al. Candidate Biomarkers for Cervical Cancer Treatment: Potential for Clinical Practice (Review). Mol. Clin. Oncol. 2014, 2, 647–655. [Google Scholar] [CrossRef]
- Harima, Y.; Ikeda, K.; Utsunomiya, K.; Komemushi, A.; Kanno, S.; Shiga, T.; Tanigawa, N. Apolipoprotein C-II Is a Potential Serum Biomarker as a Prognostic Factor of Locally Advanced Cervical Cancer After Chemoradiation Therapy. Int. J. Radiat. Oncol.* Biol.* Phys. 2013, 87, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; McCarthy, M.I.; Schwenk, J.M. Genetics Meets Proteomics: Perspectives for Large Population-Based Studies. Nat. Rev. Genet. 2021, 22, 19–37. [Google Scholar] [CrossRef]
- Matzke, L.A.; Watson, P.H. Biobanking for Cancer Biomarker Research: Issues and Solutions. Biomark. Insights 2020, 15, 1177271920965522. [Google Scholar] [CrossRef] [PubMed]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank Resource with Deep Phenotyping and Genomic Data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- German National Cohort (GNC) Consortium. The German National Cohort: Aims, Study Design and Organization. Eur. J. Epidemiol. 2014, 29, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, J.; Collins, R.; Guo, Y.; Peto, R.; Wu, F.; Li, L.; China Kadoorie Biobank (CKB) Collaborative Group. China Kadoorie Biobank of 0.5 Million People: Survey Methods, Baseline Characteristics and Long-Term Follow-Up. Int. J. Epidemiol. 2011, 40, 1652–1666. [Google Scholar] [CrossRef]
- Narita, A.; Ueki, M.; Tamiya, G. Artificial Intelligence Powered Statistical Genetics in Biobanks. J. Hum. Genet. 2021, 66, 61–65. [Google Scholar] [CrossRef]
- Abdulkareem, M.; Aung, N.; Petersen, S.E. Biobanks and Artificial Intelligence. In Artificial Intelligence in Cardiothoracic Imaging; De Cecco, C.N., van Assen, M., Leiner, T., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 81–93. ISBN 978-3-030-92087-6. [Google Scholar]
- Moruz, L.; Käll, L. Peptide Retention Time Prediction. Mass Spectrom. Rev. 2017, 36, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Degroeve, S.; Martens, L. MS2PIP: A Tool for MS/MS Peak Intensity Prediction. Bioinformatics 2013, 29, 3199–3203. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2021, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Shao, W.; Lam, H. Understanding the Improved Sensitivity of Spectral Library Searching over Sequence Database Searching in Proteomics Data Analysis. Proteomics 2011, 11, 1075–1085. [Google Scholar] [CrossRef]
- Bouwmeester, R.; Gabriels, R.; Van Den Bossche, T.; Martens, L.; Degroeve, S. The Age of Data-Driven Proteomics: How Machine Learning Enables Novel Workflows. Proteomics 2020, 20, 1900351. [Google Scholar] [CrossRef] [PubMed]
- Domingos, P. A Few Useful Things to Know about Machine Learning. Commun. ACM 2012, 55, 78–87. [Google Scholar] [CrossRef]
- Bojarski, M.; Del Testa, D.; Dworakowski, D.; Firner, B.; Flepp, B.; Goyal, P.; Jackel, L.D.; Monfort, M.; Muller, U.; Zhang, J.; et al. End to End Learning for Self-Driving Cars. arXiv 2016, arXiv:1604.07316. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Kearney, P.; Boniface, J.J.; Price, N.D.; Hood, L. The Building Blocks of Successful Translation of Proteomics to the Clinic. Curr. Opin. Biotechnol. 2018, 51, 123–129. [Google Scholar] [CrossRef]
- Bengtsson, E.; Malm, P. Screening for Cervical Cancer Using Automated Analysis of PAP-Smears. Comput. Math. Methods Med. 2014, 2014, 842037. [Google Scholar] [CrossRef]
- Hu, L.; Bell, D.; Antani, S.; Xue, Z.; Yu, K.; Horning, M.P.; Gachuhi, N.; Wilson, B.; Jaiswal, M.S.; Befano, B.; et al. An Observational Study of Deep Learning and Automated Evaluation of Cervical Images for Cancer Screening. J. Natl. Cancer Inst. 2019, 111, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Kusy, M.; Obrzut, B.; Kluska, J. Application of Gene Expression Programming and Neural Networks to Predict Adverse Events of Radical Hysterectomy in Cervical Cancer Patients. Med. Biol. Eng. Comput. 2013, 51, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Zhen, X.; Chen, J.; Zhong, Z.; Hrycushko, B.; Zhou, L.; Jiang, S.; Albuquerque, K.; Gu, X. Deep Convolutional Neural Network with Transfer Learning for Rectum Toxicity Prediction in Cervical Cancer Radiotherapy: A Feasibility Study. Phys. Med. Biol. 2017, 62, 8246–8263. [Google Scholar] [CrossRef]
- Kawakami, E.; Tabata, J.; Yanaihara, N.; Ishikawa, T.; Koseki, K.; Iida, Y.; Saito, M.; Komazaki, H.; Shapiro, J.S.; Goto, C.; et al. Application of Artificial Intelligence for Preoperative Diagnostic and Prognostic Prediction in Epithelial Ovarian Cancer Based on Blood Biomarkers. Clin. Cancer Res. 2019, 25, 3006–3015. [Google Scholar] [CrossRef]
- Enshaei, A.; Robson, C.N.; Edmondson, R.J. Artificial Intelligence Systems as Prognostic and Predictive Tools in Ovarian Cancer. Ann. Surg. Oncol. 2015, 22, 3970–3975. [Google Scholar] [CrossRef] [PubMed]
- Mysona, D.; Pyrzak, A.; Purohit, S.; Zhi, W.; Sharma, A.; Tran, L.; Tran, P.; Bai, S.; Rungruang, B.; Ghamande, S.; et al. A Combined Score of Clinical Factors and Serum Proteins Can Predict Time to Recurrence in High Grade Serous Ovarian Cancer. Gynecol. Oncol. 2019, 152, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Mysona, D.P.; Purohit, S.; Richardson, K.P.; Suhner, J.; Brzezinska, B.; Rungruang, B.; Hopkins, D.; Bearden, G.; Higgins, R.; Johnson, M.; et al. Ovarian Recurrence Risk Assessment Using Machine Learning, Clinical Information, and Serum Protein Levels to Predict Survival in High Grade Ovarian Cancer. Sci. Rep. 2023, 13, 20933. [Google Scholar] [CrossRef] [PubMed]
- Mysona, D.P.; Kapp, D.S.; Rohatgi, A.; Lee, D.; Mann, A.K.; Tran, P.; Tran, L.; She, J.-X.; Chan, J.K. Applying Artificial Intelligence to Gynecologic Oncology: A Review. Obstet. Gynecol. Surv. 2021, 76, 292. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Omenn, G.S.; Sun, Z.; Maes, M.; Pernemalm, M.; Palaniappan, K.K.; Letunica, N.; Vandenbrouck, Y.; Brun, V.; Tao, S.; et al. Advances and Utility of the Human Plasma Proteome. J. Proteome Res. 2021, 20, 5241. [Google Scholar] [CrossRef]
| Proteins | Metric | Samples vs. Control | Histological Sample Type | Measure | Outcome | Ref. |
|---|---|---|---|---|---|---|
| ** A1AT, PYCR2, TTR, APOA1, VDBP, MMRN1 | AUC: 0.933 (SN: 84.6%, SP: 87.5%) | 31 vs. 16 | SCC: 22, ACC: 8, HCC: 1 | Diagnostic | HPV + Normal control vs. CC (early stage) | [21] |
| ** P1GF, F1T1 | AUC: 0.8493 (SN: 70.45%, SP: 92.11% | 62 vs. 20 | CIN: 18, SCC: 31, ACC: 13 | Diagnostic | Normal vs. CC | [22] |
| ** ANT3-1, FBLN1-2 | AUC: 0.6885 (SN: 76.92%, SP: 70.00%) | 284 vs. 75 | HSIL/CIN II/III: 88, SCC: 121 | Diagnostic | Normal vs. CC | [23] |
| ** CXCL10, SCC-Ag | AUC: 0.877 | 264 vs. 81 | CIN: 75, SCC: 189 | Diagnostic | Normal vs. CC | [24] |
| ** CXCL8, CXCR2, SCC-Ag | AUC: 0.847 (SN: 85.71%, SP: 70.00%) | 100 vs. 30 | CIN I-III: 30, SCC: 70 | Diagnostic | Normal vs. CC | [25] |
| Proteins | Metric | Samples vs. Control | Histological Sample Type | Measure | Outcome | Ref. |
|---|---|---|---|---|---|---|
| TKT, FGA, APOA1 | AUC = 0.8463, 0.8038, 0.7641 | 67 vs. 50 | SCC: 29, ACC: 10 (pre-surgery) | Prognostic | Pre- vs. Post-operative surgical prognosis | [26] |
| MASP-2, MASP-1, MAp-19 | † SN: 71.3%, SP: 63.2% | 292 vs. 52 | CIN II/III: 214, CC: 78 | Prognostic | Poor survival and Disease Relapse | [27] |
| VEGF-A, VEGFR-2 | HR: 3.42, 6.37 | 107 vs. N/A | SCC: 80, ACC: 23, ASC: 3, SmCC: 1 | Prognostic | Overall Survival | [28] |
| ** CRP, GRO, LEPTIN, MIG, MMP1, SCCA, SAA, sIL2Rα | HR: 1.89, 2.07, 0.61, 1.85, 1.69, 3.55, 1.60, NA | 565 vs. N/A | SCC: 565 | Prognostic | DSS | [29] |
| 1 SCC-Ag, 2 ApoC-II | 1 HR: 1.186, 1.185, 1.260 2 HR: 0.606 | 142 vs. N/A | SCC: 142 | Predictive | (Pre-RT) 1 PFS, OS, DMFS 2 PPFS | [30] |
| CEA, NSE, HCG-ß | 3 HR: 2.281, 2.166, 2.584 4 HR: 2.116, 2.217, 2.478 | 295 vs. N/A | ACC: 295 | Prognostic | 3 OS, 4 PFS | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weaver, C.; Nam, A.; Settle, C.; Overton, M.; Giddens, M.; Richardson, K.P.; Piver, R.; Mysona, D.P.; Rungruang, B.; Ghamande, S.; et al. Serum Proteomic Signatures in Cervical Cancer: Current Status and Future Directions. Cancers 2024, 16, 1629. https://doi.org/10.3390/cancers16091629
Weaver C, Nam A, Settle C, Overton M, Giddens M, Richardson KP, Piver R, Mysona DP, Rungruang B, Ghamande S, et al. Serum Proteomic Signatures in Cervical Cancer: Current Status and Future Directions. Cancers. 2024; 16(9):1629. https://doi.org/10.3390/cancers16091629
Chicago/Turabian StyleWeaver, Chaston, Alisha Nam, Caitlin Settle, Madelyn Overton, Maya Giddens, Katherine P. Richardson, Rachael Piver, David P. Mysona, Bunja Rungruang, Sharad Ghamande, and et al. 2024. "Serum Proteomic Signatures in Cervical Cancer: Current Status and Future Directions" Cancers 16, no. 9: 1629. https://doi.org/10.3390/cancers16091629
APA StyleWeaver, C., Nam, A., Settle, C., Overton, M., Giddens, M., Richardson, K. P., Piver, R., Mysona, D. P., Rungruang, B., Ghamande, S., McIndoe, R., & Purohit, S. (2024). Serum Proteomic Signatures in Cervical Cancer: Current Status and Future Directions. Cancers, 16(9), 1629. https://doi.org/10.3390/cancers16091629





