Targeting Siglec–Sialylated MUC1 Immune Axis in Cancer
Abstract
Simple Summary
Abstract
1. Introduction
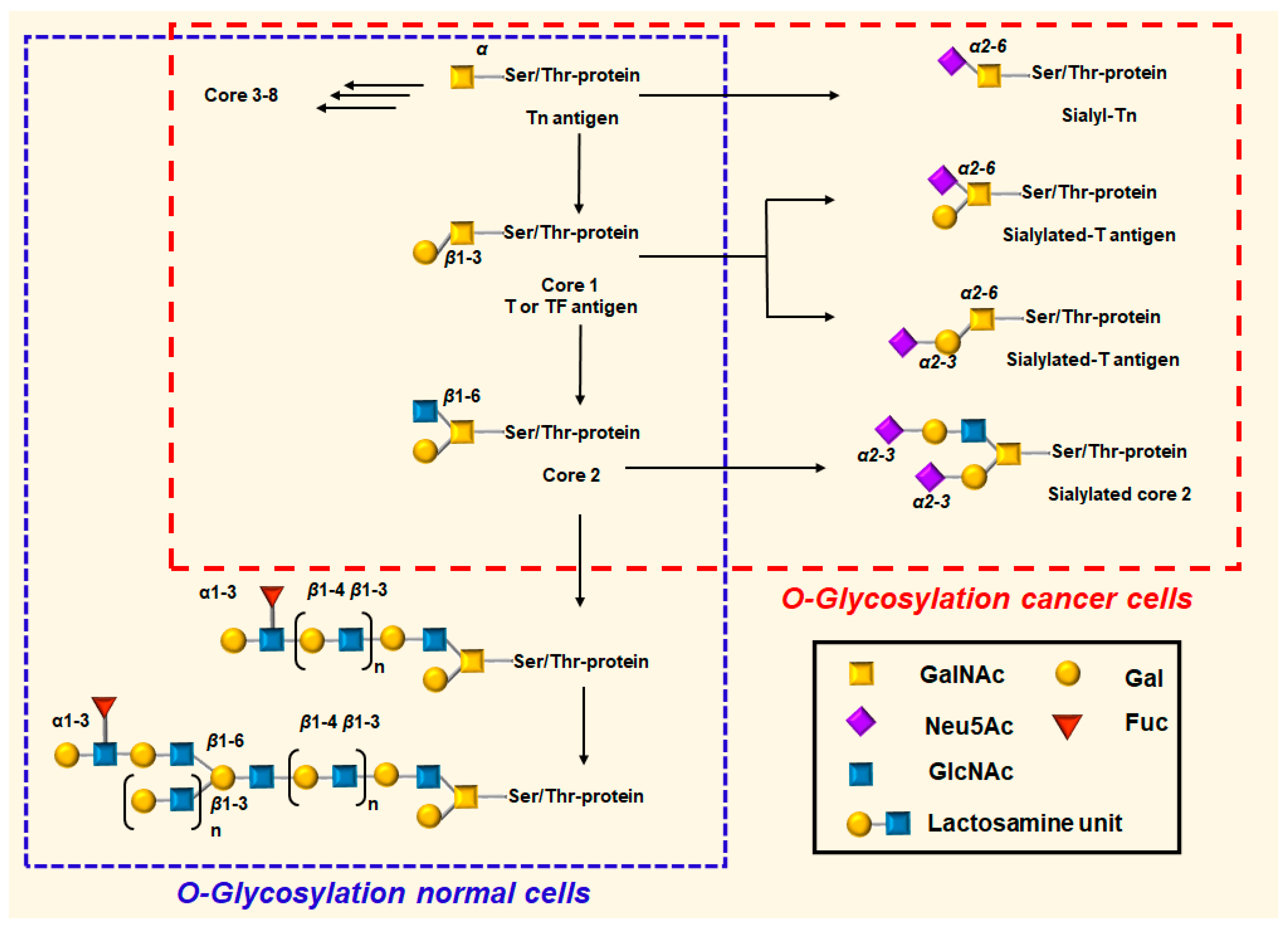
1.1. MUC1 Protein
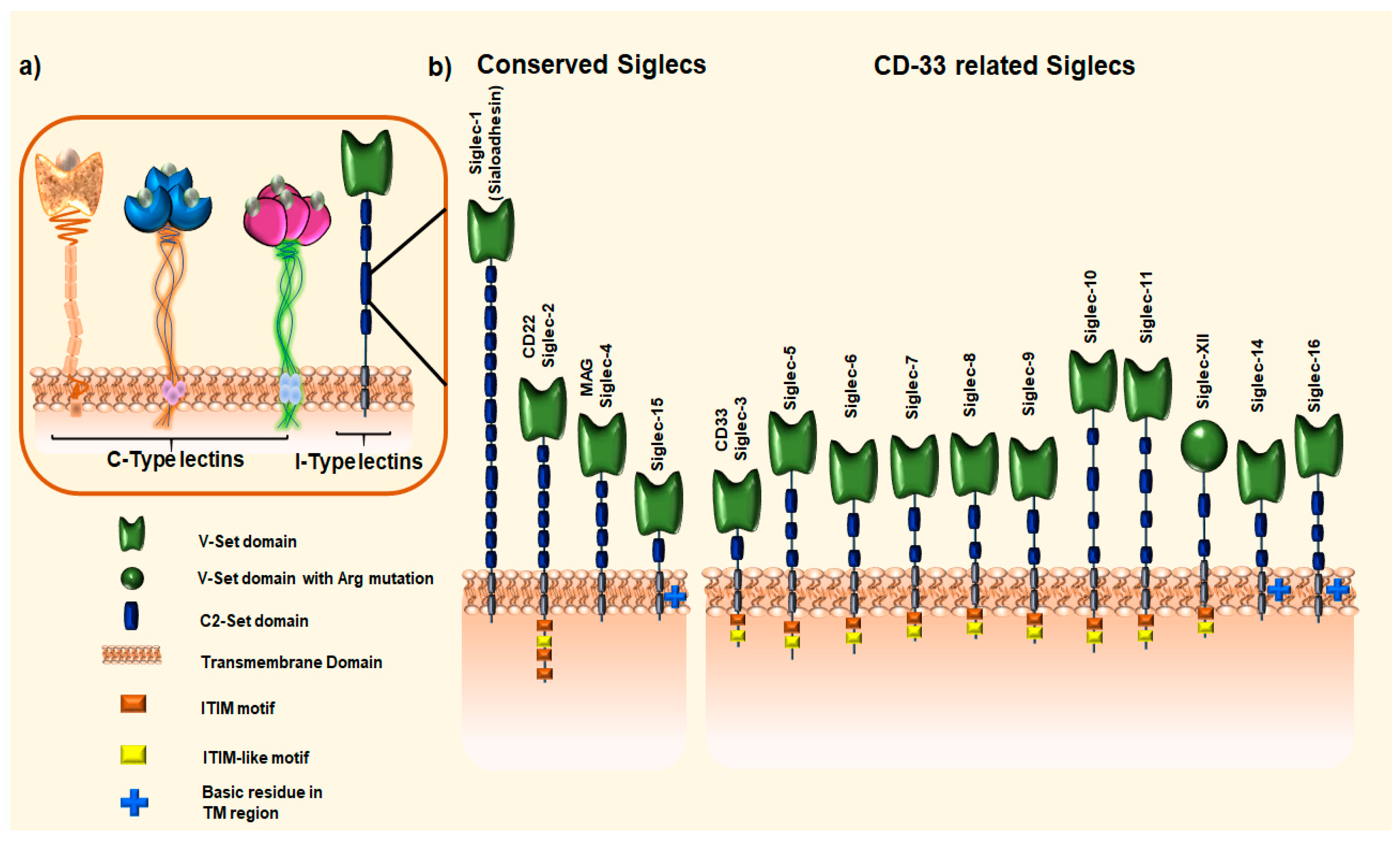
1.2. Sialic Acid-Binding Immunoglobin-like Receptors (Siglecs)
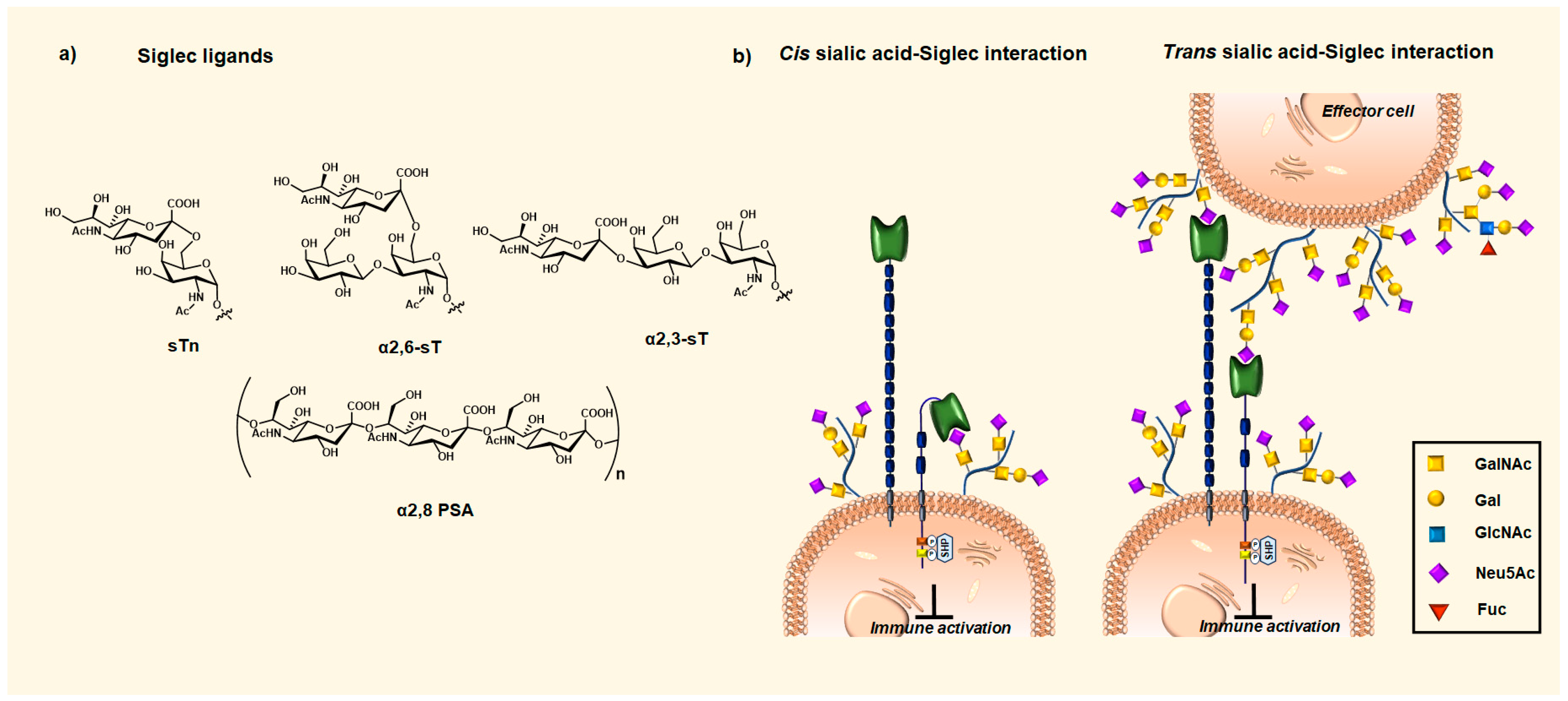
2. Siglecs Immune Evasion via Sialylated MUC1 Glycans Interactions
3. Targeting MUC1–Siglec Axis in Cancer
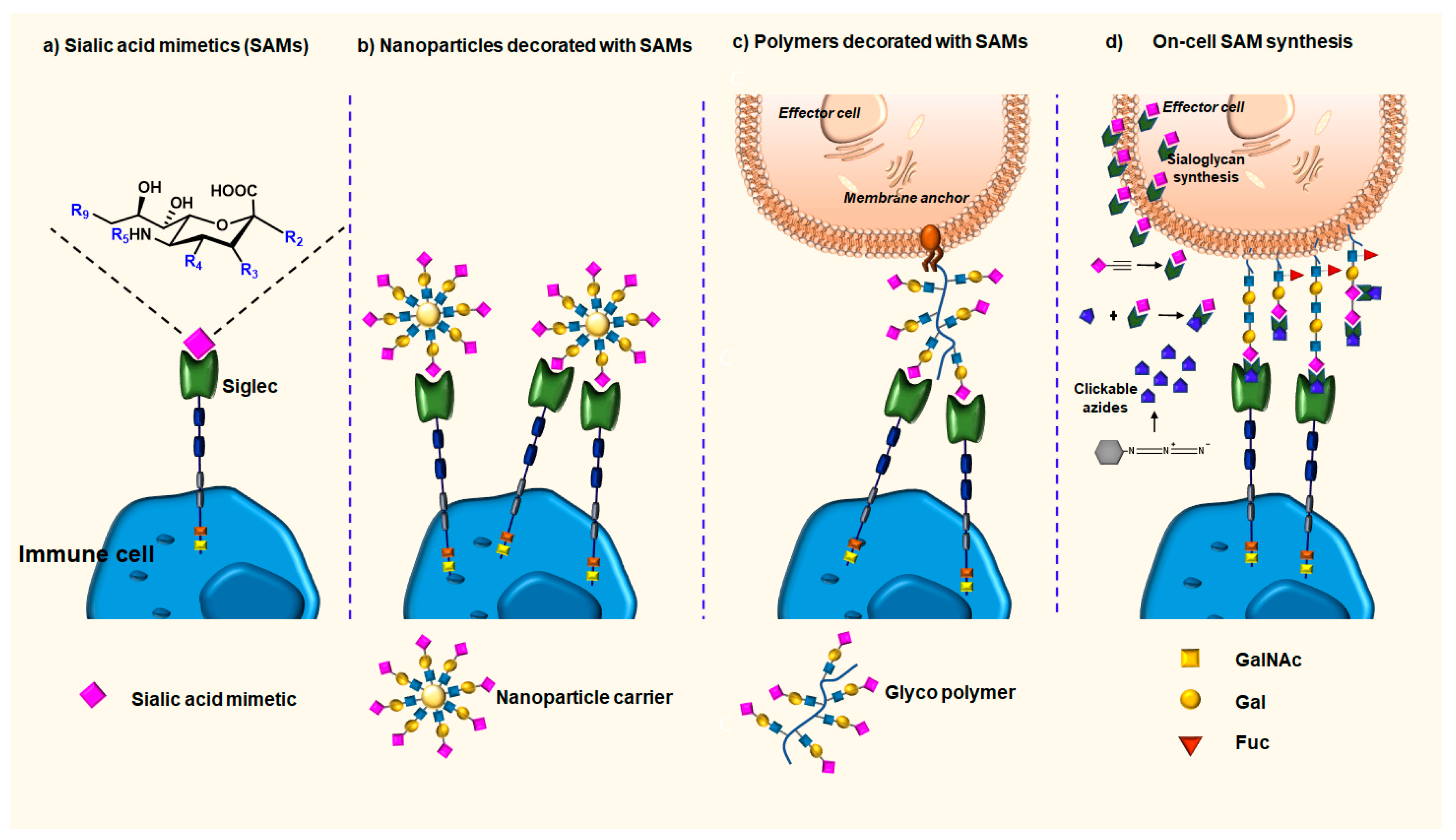
3.1. Sialic Acid Mimetics (SAMs) as High-Affinity Ligands for Siglecs
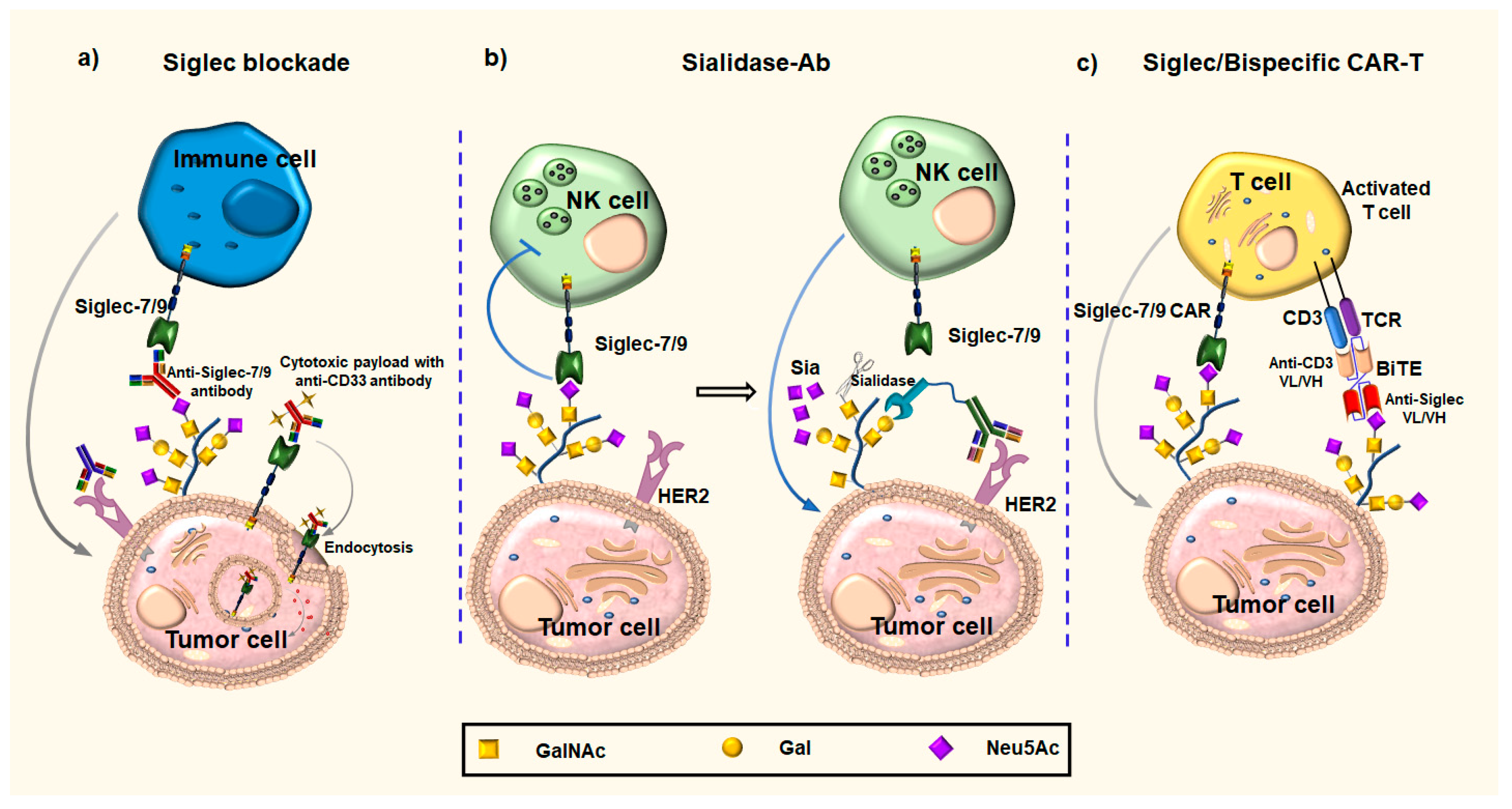
3.2. Anti-Siglecs Antibody-Based Approaches
3.2.1. Anti-Siglecs Antibodies
3.2.2. Anti-Siglecs Antibody–Sialidase Conjugates
3.2.3. Chimeric Antigen Receptors
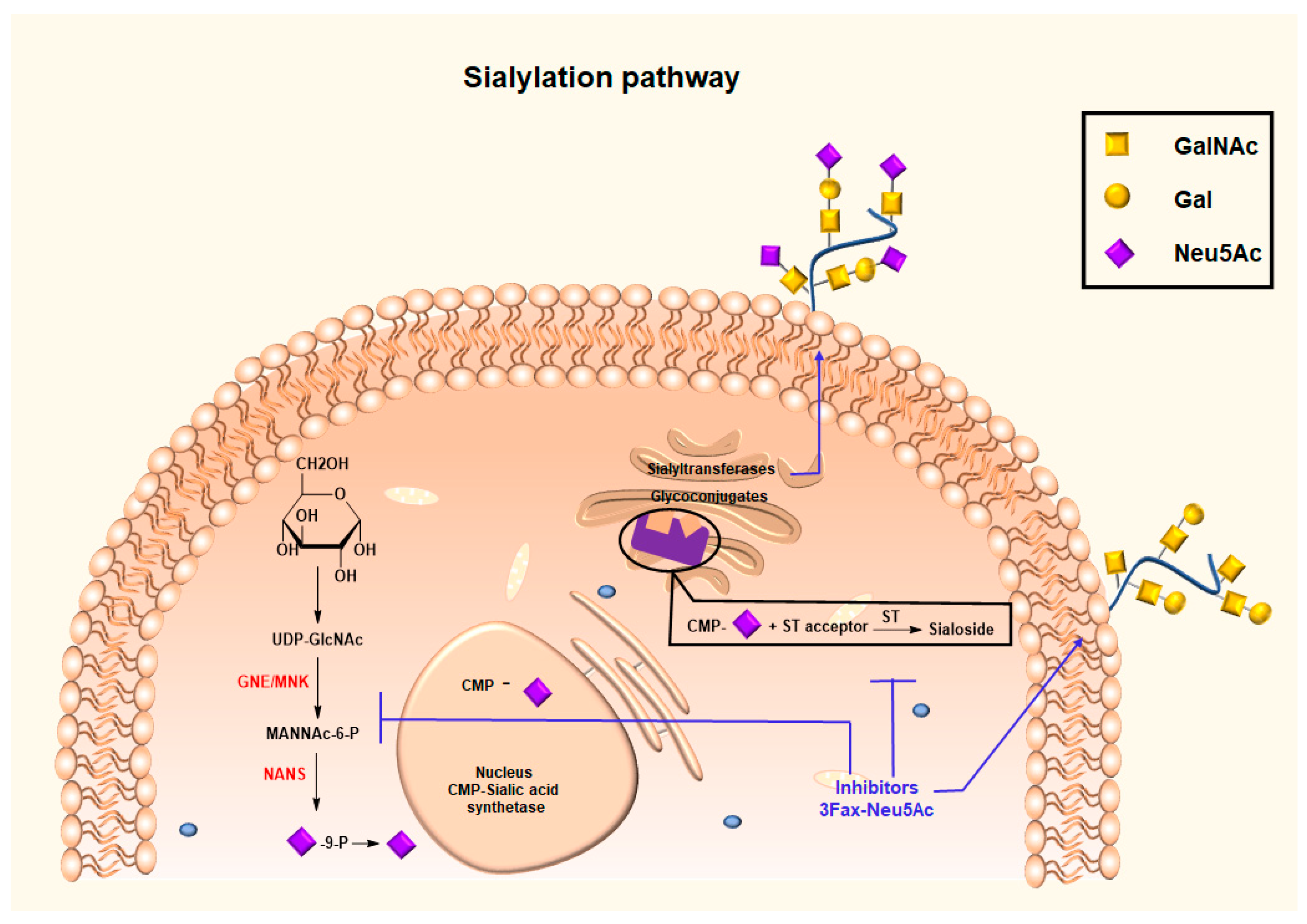
3.3. Glycan Modifying Agents for Altering Synthesis and Expression of the Sialic Acid Glycocalyx
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Munkley, J.; Elliott, D.J. Hallmarks of Glycosylation in Cancer. Oncotarget 2016, 7, 35478–35489. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Reis, C.A. Glycosylation in Cancer: Mechanisms and Clinical Implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Stowell, S.R.; Ju, T.; Cummings, R.D. Protein Glycosylation in Cancer. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 473–510. [Google Scholar] [CrossRef] [PubMed]
- Chugh, S.; Gnanapragassam, V.S.; Jain, M.; Rachagani, S.; Ponnusamy, M.P.; Batra, S.K. Pathobiological Implications of Mucin Glycans in Cancer: Sweet Poison and Novel Targets. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2015, 1856, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Ju, T.; Aryal, R.P.; Kudelka, M.R.; Wang, Y.; Cummings, R.D. The Cosmc Connection to the Tn Antigen in Cancer. Cancer Biomark. 2014, 14, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Hugonnet, M.; Singh, P.; Haas, Q.; von Gunten, S. The Distinct Roles of Sialyltransferases in Cancer Biology and Onco-Immunology. Front. Immunol. 2021, 12, 799861. [Google Scholar] [CrossRef]
- Bastian, K.; Scott, E.; Elliott, D.J.; Munkley, J. FUT8 Alpha-(1,6)-Fucosyltransferase in Cancer. Int. J. Mol. Sci. 2021, 22, 455. [Google Scholar] [CrossRef] [PubMed]
- Boligan, K.F.; Mesa, C.; Fernandez, L.E.; von Gunten, S. Cancer Intelligence Acquired (CIA): Tumor Glycosylation and Sialylation Codes Dismantling Antitumor Defense. Cell. Mol. Life Sci. 2015, 72, 1231–1248. [Google Scholar] [CrossRef] [PubMed]
- Beatson, R.; Maurstad, G.; Picco, G.; Arulappu, A.; Coleman, J.; Wandell, H.H.; Clausen, H.; Mandel, U.; Taylor-Papadimitriou, J.; Sletmoen, M.; et al. The Breast Cancer-Associated Glycoforms of MUC1, MUC1-Tn and Sialyl-Tn, Are Expressed in COSMC Wild-Type Cells and Bind the C-Type Lectin MGL. PLoS ONE 2015, 10, e0125994. [Google Scholar] [CrossRef]
- Beckwith, D.M.; Cudic, M. Tumor-Associated O-Glycans of MUC1: Carriers of the Glyco-Code and Targets for Cancer Vaccine Design. Semin. Immunol. 2020, 47, 101389. [Google Scholar] [CrossRef]
- Engering, A.; Geijtenbeek, T.B.H.; van Kooyk, Y. Immune Escape through C-Type Lectins on Dendritic Cells. Trends Immunol. 2002, 23, 480–485. [Google Scholar] [CrossRef]
- Senapati, S.; Das, S.; Batra, S.K. Mucin-Interacting Proteins: From Function to Therapeutics. Trends Biochem. Sci. 2010, 35, 236–245. [Google Scholar] [CrossRef]
- Nath, S.; Mukherjee, P. MUC1: A Multifaceted Oncoprotein with a Key Role in Cancer Progression. Trends Mol. Med. 2014, 20, 332–342. [Google Scholar] [CrossRef] [PubMed]
- van Putten, J.P.M.; Strijbis, K. Transmembrane Mucins: Signaling Receptors at the Intersection of Inflammation and Cancer. J. Innate Immun. 2017, 9, 281–299. [Google Scholar] [CrossRef]
- Hattrup, C.L.; Gendler, S.J. Structure and Function of the Cell Surface (Tethered) Mucins. Annu. Rev. Physiol. 2008, 70, 431–457. [Google Scholar] [CrossRef]
- Kufe, D.W. Mucins in Cancer: Function, Prognosis and Therapy. Nat. Rev. Cancer 2009, 9, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Rachagani, S.; Torres, M.P.; Moniaux, N.; Batra, S.K. Current Status of Mucins in the Diagnosis and Therapy of Cancer. BioFactors 2009, 35, 509–527. [Google Scholar] [CrossRef] [PubMed]
- Moniaux, N.; Escande, F.; Porchet, N.; Aubert, J.P.; Batra, S.K. Structural Organization and Classification of the Human Mucin Genes. Front. Biosci. Landmark 2001, 6, 1192–1206. [Google Scholar] [CrossRef]
- Johansson, M.E.V.; Sjövall, H.; Hansson, G.C. The Gastrointestinal Mucus System in Health and Disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 352–361. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, Z.; Zhang, S.; Zhu, P.; Ko, J.K.-S.; Yung, K.K.-L. MUC1: Structure, Function, and Clinic Application in Epithelial Cancers. Int. J. Mol. Sci. 2021, 22, 6567. [Google Scholar] [CrossRef]
- Thompson, E.J.; Shanmugam, K.; Hattrup, C.L.; Kotlarczyk, K.L.; Gutierrez, A.; Bradley, J.M.; Mukherjee, P.; Gendler, S.J. Tyrosines in the MUC1 Cytoplasmic Tail Modulate Transcription via the Extracellular Signal-Regulated Kinase 1/2 and Nuclear Factor-ΚB Pathways. Mol. Cancer Res. 2006, 4, 489–497. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bose, M.; Grover, P.; Sanders, A.J.; Zhou, R.; Ahmad, M.; Shwartz, S.; Lala, P.; Nath, S.; Yazdanifar, M.; Brouwer, C.; et al. Overexpression of MUC1 Induces Non-Canonical TGF-β Signaling in Pancreatic Ductal Adenocarcinoma. Front. Cell Dev. Biol. 2022, 10, 821875. [Google Scholar] [CrossRef]
- Hosseinzadeh, A.; Merikhian, P.; Naseri, N.; Eisavand, M.R.; Farahmand, L. MUC1 Is a Potential Target to Overcome Trastuzumab Resistance in Breast Cancer Therapy. Cancer Cell Int. 2022, 22, 110. [Google Scholar] [CrossRef]
- Huang, L.; Chen, D.; Liu, D.; Yin, L.; Kharbanda, S.; Kufe, D. MUC1 Oncoprotein Blocks Glycogen Synthase Kinase 3β–Mediated Phosphorylation and Degradation of β-Catenin. Cancer Res. 2005, 65, 10413–10422. [Google Scholar] [CrossRef]
- Ahmad, R.; Raina, D.; Joshi, M.D.; Kawano, T.; Ren, J.; Kharbanda, S.; Kufe, D. MUC1-C Oncoprotein Functions as a Direct Activator of the Nuclear Factor-ΚB P65 Transcription Factor. Cancer Res. 2009, 69, 7013–7021. [Google Scholar] [CrossRef]
- Joshi, S.; Kumar, S.; Choudhury, A.; Ponnusamy, M.P.; Batra, S.K. Altered Mucins (MUC) Trafficking in Benign and Malignant Conditions. Oncotarget 2014, 5, 7272–7284. [Google Scholar] [CrossRef]
- Cascio, S.; Farkas, A.M.; Hughey, R.P.; Finn, O.J. Altered Glycosylation of MUC1 Influences Its Association with CIN85: The Role of This Novel Complex in Cancer Cell Invasion and Migration. Oncotarget 2013, 4, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Cascio, S.; Zhang, L.; Finn, O.J. MUC1 Protein Expression in Tumor Cells Regulates Transcription of Proinflammatory Cytokines by Forming a Complex with Nuclear Factor-ΚB P65 and Binding to Cytokine Promoters. J. Biol. Chem. 2011, 286, 42248–42256. [Google Scholar] [CrossRef] [PubMed]
- Borsig, L.; Wong, R.; Feramisco, J.; Nadeau, D.R.; Varki, N.M.; Varki, A. Heparin and Cancer Revisited: Mechanistic Connections Involving Platelets, P-Selectin, Carcinoma Mucins, and Tumor Metastasis. Proc. Natl. Acad. Sci. USA 2001, 98, 3352–3357. [Google Scholar] [CrossRef]
- Zhao, Q.; Guo, X.; Nash, G.B.; Stone, P.C.; Hilkens, J.; Rhodes, J.M.; Yu, L.-G. Circulating Galectin-3 Promotes Metastasis by Modifying MUC1 Localization on Cancer Cell Surface. Cancer Res. 2009, 69, 6799–6806. [Google Scholar] [CrossRef]
- Zhao, Q.; Barclay, M.; Hilkens, J.; Guo, X.; Barrow, H.; Rhodes, J.M.; Yu, L.-G. Interaction between Circulating Galectin-3 and Cancer-Associated MUC1 Enhances Tumour Cell Homotypic Aggregation and Prevents Anoikis. Mol. Cancer 2010, 9, 154. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Piyush, T.; Chen, C.; Hollingsworth, M.A.; Hilkens, J.; Rhodes, J.M.; Yu, L.-G. MUC1 Extracellular Domain Confers Resistance of Epithelial Cancer Cells to Anoikis. Cell Death Dis. 2014, 5, e1438. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, R.; Gautam, S.K.; Cannon, A.; Thompson, C.; Hall, B.R.; Aithal, A.; Banerjee, K.; Jain, M.; Solheim, J.C.; Kumar, S.; et al. Cancer-Associated Mucins: Role in Immune Modulation and Metastasis. Cancer Metastasis Rev. 2019, 38, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Ju, T. Aberrant Glycosylation as Immune Therapeutic Targets for Solid Tumors. Cancers 2023, 15, 3536. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Papadimitriou, J.; Burchell, J.M.; Graham, R.; Beatson, R. Latest Developments in MUC1 Immunotherapy. Biochem. Soc. Trans. 2018, 46, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Ni, W.; Tai, G. Expression of MUC1 in Different Tumours and Its Clinical Significance (Review). Mol. Clin. Oncol. 2022, 17, 161. [Google Scholar] [CrossRef] [PubMed]
- Striefler, J.K.; Riess, H.; Lohneis, P.; Bischoff, S.; Kurreck, A.; Modest, D.P.; Bahra, M.; Oettle, H.; Sinn, M.; Bläker, H.; et al. Mucin-1 Protein Is a Prognostic Marker for Pancreatic Ductal Adenocarcinoma: Results From the CONKO-001 Study. Front. Oncol. 2021, 11, 670396. [Google Scholar] [CrossRef] [PubMed]
- Siroy, A.; Abdul-Karim, F.W.; Miedler, J.; Fong, N.; Fu, P.; Gilmore, H.; Baar, J. MUC1 Is Expressed at High Frequency in Early-Stage Basal-like Triple-Negative Breast Cancer. Hum. Pathol. 2013, 44, 2159–2166. [Google Scholar] [CrossRef]
- Jing, X.; Liang, H.; Hao, C.; Yang, X.; Cui, X. Overexpression of MUC1 Predicts Poor Prognosis in Patients with Breast Cancer. Oncol. Rep. 2018, 41, 801–810. [Google Scholar] [CrossRef]
- Levitin, F.; Stern, O.; Weiss, M.; Gil-Henn, C.; Ziv, R.; Prokocimer, Z.; Smorodinsky, N.I.; Rubinstein, D.B.; Wreschner, D.H. The MUC1 SEA Module Is a Self-Cleaving Domain. J. Biol. Chem. 2005, 280, 33374–33386. [Google Scholar] [CrossRef]
- Constantinou, P.E.; Danysh, B.P.; Dharmaraj, N.; Carson, D.D. Transmembrane Mucins as Novel Therapeutic Targets. Expert Rev. Endocrinol. Metab. 2011, 6, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Burchell, J.M.; Mungul, A.; Taylor-Papadimitriou, J. O-Linked Glycosylation in the Mammary Gland: Changes That Occur During Malignancy. J. Mammary Gland Biol. Neoplasia 2001, 6, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Häuselmann, I.; Borsig, L. Altered Tumor-Cell Glycosylation Promotes Metastasis. Front. Oncol. 2014, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Cazet, A.; Julien, S.; Bobowski, M.; Burchell, J.; Delannoy, P. Tumour-Associated Carbohydrate Antigens in Breast Cancer. Breast Cancer Res. 2010, 12, 204. [Google Scholar] [CrossRef] [PubMed]
- Kobata, A.; Amano, J. Altered Glycosylation of Proteins Produced by Malignant Cells, and Application for the Diagnosis and Immunotherapy of Tumours. Immunol. Cell Biol. 2005, 83, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Warren, J.D.; Danishefsky, S.J. Synthetic Carbohydrate-Based Anticancer Vaccines: The Memorial Sloan-Kettering Experience. Expert Rev. Vaccines 2009, 8, 1399–1413. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, Q. Recent Development in Carbohydrate-Based Cancer Vaccines. Curr. Opin. Chem. Biol. 2009, 13, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Borsig, L. Selectins in Cancer Immunity. Glycobiology 2018, 28, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Marcelo, F.; Garcia-Martin, F.; Matsushita, T.; Sardinha, J.; Coelho, H.; Oude-Vrielink, A.; Koller, C.; André, S.; Cabrita, E.J.; Gabius, H.; et al. Delineating Binding Modes of Gal/GalNAc and Structural Elements of the Molecular Recognition of Tumor-Associated Mucin Glycopeptides by the Human Macrophage Galactose-Type Lectin. Chem. A Eur. J. 2014, 20, 16147–16155. [Google Scholar] [CrossRef]
- Monti, P.; Leone, B.E.; Zerbi, A.; Balzano, G.; Cainarca, S.; Sordi, V.; Pontillo, M.; Mercalli, A.; Di Carlo, V.; Allavena, P.; et al. Tumor-Derived MUC1 Mucins Interact with Differentiating Monocytes and Induce IL-10highIL-12low Regulatory Dendritic Cell. J. Immunol. 2004, 172, 7341–7349. [Google Scholar] [CrossRef]
- Macauley, M.S.; Crocker, P.R.; Paulson, J.C. Siglec-Mediated Regulation of Immune Cell Function in Disease. Nat. Rev. Immunol. 2014, 14, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Angata, T.; von Gunten, S.; Schnaar, R.L.; Varki, A. I-Type Lectins. In Essentials of Glycobiology [Internet], 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2022. [Google Scholar]
- Beatson, R.; Tajadura-Ortega, V.; Achkova, D.; Picco, G.; Tsourouktsoglou, T.-D.; Klausing, S.; Hillier, M.; Maher, J.; Noll, T.; Crocker, P.R.; et al. The Mucin MUC1 Modulates the Tumor Immunological Microenvironment through Engagement of the Lectin Siglec-9. Nat. Immunol. 2016, 17, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Beatson, R.; Graham, R.; Grundland Freile, F.; Cozzetto, D.; Kannambath, S.; Pfeifer, E.; Woodman, N.; Owen, J.; Nuamah, R.; Mandel, U.; et al. Cancer-Associated Hypersialylated MUC1 Drives the Differentiation of Human Monocytes into Macrophages with a Pathogenic Phenotype. Commun. Biol. 2020, 3, 644. [Google Scholar] [CrossRef] [PubMed]
- Crocker, P.R.; Paulson, J.C.; Varki, A. Siglecs and Their Roles in the Immune System. Nat. Rev. Immunol. 2007, 7, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Adams, O.J.; Stanczak, M.A.; von Gunten, S.; Läubli, H. Targeting Sialic Acid–Siglec Interactions to Reverse Immune Suppression in Cancer. Glycobiology 2017, 28, 640–647. [Google Scholar] [CrossRef] [PubMed]
- Bochner, B.S.; Zimmermann, N. Role of Siglecs and Related Glycan-Binding Proteins in Immune Responses and Immunoregulation. J. Allergy Clin. Immunol. 2015, 135, 598–608. [Google Scholar] [CrossRef]
- van de Wall, S.; Santegoets, K.C.M.; van Houtum, E.J.H.; Büll, C.; Adema, G.J. Sialoglycans and Siglecs Can Shape the Tumor Immune Microenvironment. Trends Immunol. 2020, 41, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, F.; Landig, C.S.; Siddiqui, S.; Secundino, I.; Olson, J.; Varki, N.; Nizet, V.; Varki, A. Paired Siglec Receptors Generate Opposite Inflammatory Responses to a Human-specific Pathogen. EMBO J. 2017, 36, 751–760. [Google Scholar] [CrossRef] [PubMed]
- May, A.P.; Robinson, R.C.; Vinson, M.; Crocker, P.R.; Jones, E.Y. Crystal Structure of the N-Terminal Domain of Sialoadhesin in Complex with 3′ Sialyllactose at 1.85 Å Resolution. Mol. Cell 1998, 1, 719–728. [Google Scholar] [CrossRef]
- Alphey, M.S.; Attrill, H.; Crocker, P.R.; van Aalten, D.M.F. High Resolution Crystal Structures of Siglec-7. J. Biol. Chem. 2003, 278, 3372–3377. [Google Scholar] [CrossRef]
- Pröpster, J.M.; Yang, F.; Rabbani, S.; Ernst, B.; Allain, F.H.-T.; Schubert, M. Structural Basis for Sulfation-Dependent Self-Glycan Recognition by the Human Immune-Inhibitory Receptor Siglec-8. Proc. Natl. Acad. Sci. USA 2016, 113, E4170–E4179. [Google Scholar] [CrossRef] [PubMed]
- Pronker, M.F.; Lemstra, S.; Snijder, J.; Heck, A.J.R.; Thies-Weesie, D.M.E.; Pasterkamp, R.J.; Janssen, B.J.C. Structural Basis of Myelin-Associated Glycoprotein Adhesion and Signalling. Nat. Commun. 2016, 7, 13584. [Google Scholar] [CrossRef]
- Ereño-Orbea, J.; Sicard, T.; Cui, H.; Mazhab-Jafari, M.T.; Benlekbir, S.; Guarné, A.; Rubinstein, J.L.; Julien, J.-P. Molecular Basis of Human CD22 Function and Therapeutic Targeting. Nat. Commun. 2017, 8, 764. [Google Scholar] [CrossRef] [PubMed]
- Attrill, H.; Takazawa, H.; Witt, S.; Kelm, S.; Isecke, R.; Brossmer, R.; Ando, T.; Ishida, H.; Kiso, M.; Crocker, P.R.; et al. The Structure of Siglec-7 in Complex with Sialosides: Leads for Rational Structure-Based Inhibitor Design. Biochem. J. 2006, 397, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Zhuravleva, M.A.; Trandem, K.; Sun, P.D. Structural Implications of Siglec-5-Mediated Sialoglycan Recognition. J. Mol. Biol. 2008, 375, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Lenza, M.P.; Atxabal, U.; Oyenarte, I.; Jiménez-Barbero, J.; Ereño-Orbea, J. Current Status on Therapeutic Molecules Targeting Siglec Receptors. Cells 2020, 9, 2691. [Google Scholar] [CrossRef]
- Swanson, B.J.; McDermott, K.M.; Singh, P.K.; Eggers, J.P.; Crocker, P.R.; Hollingsworth, M.A. MUC1 Is a Counter-Receptor for Myelin-Associated Glycoprotein (Siglec-4a) and Their Interaction Contributes to Adhesion in Pancreatic Cancer Perineural Invasion. Cancer Res. 2007, 67, 10222–10229. [Google Scholar] [CrossRef] [PubMed]
- Perez-Oliva, A.B.; Martinez-Esparza, M.; Vicente-Fernandez, J.J.; Corral-San Miguel, R.; Garcia-Penarrubia, P.; Hernandez-Caselles, T. Epitope Mapping, Expression and Post-Translational Modifications of Two Isoforms of CD33 (CD33M and CD33m) on Lymphoid and Myeloid Human Cells. Glycobiology 2011, 21, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Munday, J.; Floyd, H.; Crocker, P.R. Sialic Acid Binding Receptors (Siglecs) Expressed by Macrophages. J. Leukoc. Biol. 1999, 66, 705–711. [Google Scholar] [CrossRef]
- Gonzalez-Gil, A.; Schnaar, R.L. Siglec Ligands. Cells 2021, 10, 1260. [Google Scholar] [CrossRef]
- Blixt, O.; Collins, B.E.; van den Nieuwenhof, I.M.; Crocker, P.R.; Paulson, J.C. Sialoside Specificity of the Siglec Family Assessed Using Novel Multivalent Probes. J. Biol. Chem. 2003, 278, 31007–31019. [Google Scholar] [CrossRef] [PubMed]
- van Houtum, E.J.H.; Büll, C.; Cornelissen, L.A.M.; Adema, G.J. Siglec Signaling in the Tumor Microenvironment. Front. Immunol. 2021, 12, 790317. [Google Scholar] [CrossRef] [PubMed]
- McQuillan, A.M.; Byrd-Leotis, L.; Heimburg-Molinaro, J.; Cummings, R.D. Natural and Synthetic Sialylated Glycan Microarrays and Their Applications. Front. Mol. Biosci. 2019, 6, 88. [Google Scholar] [CrossRef] [PubMed]
- Büll, C.; Nason, R.; Sun, L.; Van Coillie, J.; Madriz Sørensen, D.; Moons, S.J.; Yang, Z.; Arbitman, S.; Fernandes, S.M.; Furukawa, S.; et al. Probing the Binding Specificities of Human Siglecs by Cell-Based Glycan Arrays. Proc. Natl. Acad. Sci. USA 2021, 118, e2026102118. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.A.H.; Bertozzi, C.R. The Clinical Impact of Glycobiology: Targeting Selectins, Siglecs and Mammalian Glycans. Nat. Rev. Drug Discov. 2021, 20, 217–243. [Google Scholar] [CrossRef]
- Nason, R.; Büll, C.; Konstantinidi, A.; Sun, L.; Ye, Z.; Halim, A.; Du, W.; Sørensen, D.M.; Durbesson, F.; Furukawa, S.; et al. Display of the Human Mucinome with Defined O-Glycans by Gene Engineered Cells. Nat. Commun. 2021, 12, 4070. [Google Scholar] [CrossRef]
- Yamaji, T.; Teranishi, T.; Alphey, M.S.; Crocker, P.R.; Hashimoto, Y. A Small Region of the Natural Killer Cell Receptor, Siglec-7, Is Responsible for Its Preferred Binding to A2,8-Disialyl and Branched A2,6-Sialyl Residues. J. Biol. Chem. 2002, 277, 6324–6332. [Google Scholar] [CrossRef] [PubMed]
- Cheever, M.A.; Allison, J.P.; Ferris, A.S.; Finn, O.J.; Hastings, B.M.; Hecht, T.T.; Mellman, I.; Prindiville, S.A.; Viner, J.L.; Weiner, L.M.; et al. The Prioritization of Cancer Antigens: A National Cancer Institute Pilot Project for the Acceleration of Translational Research. Clin. Cancer Res. 2009, 15, 5323–5337. [Google Scholar] [CrossRef]
- Díaz-Zaragoza, M.; Hernández-Ávila, R.; Hernández-Ávila, R.; Arenas-Aranda, D.; Ostoa-Saloma, P. Natural and Adaptive IgM Antibodies in the Recognition of Tumor-Associated Antigens of Breast Cancer (Review). Oncol. Rep. 2015, 34, 1106–1114. [Google Scholar] [CrossRef]
- Hamanaka, Y.; Suehiro, Y.; Fukui, M.; Shikichi, K.; Imai, K.; Hinoda, Y. Circulating Anti-MUC1 IgG Antibodies as a Favorable Prognostic Factor for Pancreatic Cancer. Int. J. Cancer 2003, 103, 97–100. [Google Scholar] [CrossRef]
- Wandall, H.H.; Blixt, O.; Tarp, M.A.; Pedersen, J.W.; Bennett, E.P.; Mandel, U.; Ragupathi, G.; Livingston, P.O.; Hollingsworth, M.A.; Taylor-Papadimitriou, J.; et al. Cancer Biomarkers Defined by Autoantibody Signatures to Aberrant O-Glycopeptide Epitopes. Cancer Res. 2010, 70, 1306–1313. [Google Scholar] [CrossRef]
- Fletcher, R.; Wang, Y.-J.; Schoen, R.E.; Finn, O.J.; Yu, J.; Zhang, L. Colorectal Cancer Prevention: Immune Modulation Taking the Stage. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2018, 1869, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Fremd, C.; Stefanovic, S.; Beckhove, P.; Pritsch, M.; Lim, H.; Wallwiener, M.; Heil, J.; Golatta, M.; Rom, J.; Sohn, C.; et al. Mucin 1-Specific B Cell Immune Responses and Their Impact on Overall Survival in Breast Cancer Patients. Oncoimmunology 2016, 5, e1057387. [Google Scholar] [CrossRef] [PubMed]
- Bose, M.; Mukherjee, P. Potential of Anti-MUC1 Antibodies as a Targeted Therapy for Gastrointestinal Cancers. Vaccines 2020, 8, 659. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.; Lakshminarayanan, V.; Supekar, N.T.; Bradley, J.M.; Cohen, P.A.; Wolfert, M.A.; Gendler, S.J.; Boons, G.-J. Linear Synthesis and Immunological Properties of a Fully Synthetic Vaccine Candidate Containing a Sialylated MUC1 Glycopeptide. Chem. Commun. 2015, 51, 10214–10217. [Google Scholar] [CrossRef] [PubMed]
- Rangappa, S.; Artigas, G.; Miyoshi, R.; Yokoi, Y.; Hayakawa, S.; Garcia-Martin, F.; Hinou, H.; Nishimura, S.-I. Effects of the Multiple O-Glycosylation States on Antibody Recognition of the Immunodominant Motif in MUC1 Extracellular Tandem Repeats. Medchemcomm 2016, 7, 1102–1122. [Google Scholar] [CrossRef]
- Wakui, H.; Tanaka, Y.; Ose, T.; Matsumoto, I.; Kato, K.; Min, Y.; Tachibana, T.; Sato, M.; Naruchi, K.; Martin, F.G.; et al. A Straightforward Approach to Antibodies Recognising Cancer Specific Glycopeptidic Neoepitopes. Chem. Sci. 2020, 11, 4999–5006. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, W.; DeDosso, S.; Cresta, S.; Weidmann, J.; Tessari, A.; Salzberg, M.; Dietrich, B.; Baumeister, H.; Goletz, S.; Gianni, L.; et al. A Phase I Study of PankoMab-GEX, a Humanised Glyco-Optimised Monoclonal Antibody to a Novel Tumour-Specific MUC1 Glycopeptide Epitope in Patients with Advanced Carcinomas. Eur. J. Cancer 2016, 63, 55–63. [Google Scholar] [CrossRef] [PubMed]
- de Visser, K.E.; Joyce, J.A. The Evolving Tumor Microenvironment: From Cancer Initiation to Metastatic Outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef]
- RodrÍguez, E.; Schetters, S.T.T.; van Kooyk, Y. The Tumour Glyco-Code as a Novel Immune Checkpoint for Immunotherapy. Nat. Rev. Immunol. 2018, 18, 204–211. [Google Scholar] [CrossRef]
- Nath, D.; Hartnell; Happerfield, A.; Miles, D.W.; Burchell, J.; Taylor-Papadimitriou, J.; Crocker, P.R. Macrophage-Tumour Cell Interactions: Identification of MUC1 on Breast Cancer Cells as a Potential Counter-Receptor for the Macrophage-Restricted Receptor, Sialoadhesin. Immunology 1999, 98, 213–219. [Google Scholar] [CrossRef]
- Duan, S.; Paulson, J.C. Siglecs as Immune Cell Checkpoints in Disease. Annu. Rev. Immunol. 2020, 38, 365–395. [Google Scholar] [CrossRef] [PubMed]
- Stanczak, M.A.; Läubli, H. Siglec Receptors as New Immune Checkpoints in Cancer. Mol. Asp. Med. 2023, 90, 101112. [Google Scholar] [CrossRef] [PubMed]
- Hudak, J.E.; Canham, S.M.; Bertozzi, C.R. Glycocalyx Engineering Reveals a Siglec-Based Mechanism for NK Cell Immunoevasion. Nat. Chem. Biol. 2014, 10, 69–75. [Google Scholar] [CrossRef]
- Jandus, C.; Boligan, K.F.; Chijioke, O.; Liu, H.; Dahlhaus, M.; Démoulins, T.; Schneider, C.; Wehrli, M.; Hunger, R.E.; Baerlocher, G.M.; et al. Interactions between Siglec-7/9 Receptors and Ligands Influence NK Cell–Dependent Tumor Immunosurveillance. J. Clin. Investig. 2014, 124, 1810–1820. [Google Scholar] [CrossRef]
- Christo, S.N.; Diener, K.R.; Hayball, J.D. The Functional Contribution of Calcium Ion Flux Heterogeneity in T Cells. Immunol. Cell Biol. 2015, 93, 694–704. [Google Scholar] [CrossRef]
- Läubli, H.; Pearce, O.M.T.; Schwarz, F.; Siddiqui, S.S.; Deng, L.; Stanczak, M.A.; Deng, L.; Verhagen, A.; Secrest, P.; Lusk, C.; et al. Engagement of Myelomonocytic Siglecs by Tumor-Associated Ligands Modulates the Innate Immune Response to Cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 14211–14216. [Google Scholar] [CrossRef] [PubMed]
- Lustig, M.; Chan, C.; Jansen, J.H.M.; Bräutigam, M.; Kölling, M.A.; Gehlert, C.L.; Baumann, N.; Mester, S.; Foss, S.; Andersen, J.T.; et al. Disruption of the Sialic Acid/Siglec-9 Axis Improves Antibody-Mediated Neutrophil Cytotoxicity towards Tumor Cells. Front. Immunol. 2023, 14, 1178817. [Google Scholar] [CrossRef]
- Rodriguez, E.; Boelaars, K.; Brown, K.; Eveline Li, R.J.; Kruijssen, L.; Bruijns, S.C.M.; van Ee, T.; Schetters, S.T.T.; Crommentuijn, M.H.W.; van der Horst, J.C.; et al. Sialic Acids in Pancreatic Cancer Cells Drive Tumour-Associated Macrophage Differentiation via the Siglec Receptors Siglec-7 and Siglec-9. Nat. Commun. 2021, 12, 1270. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Liu, L.N.; Flies, D.B.; Nie, X.; Toki, M.; Zhang, J.; Song, C.; Zarr, M.; Zhou, X.; et al. Siglec-15 as an Immune Suppressor and Potential Target for Normalization Cancer Immunotherapy. Nat. Med. 2019, 25, 656–666. [Google Scholar] [CrossRef]
- Wculek, S.K.; Cueto, F.J.; Mujal, A.M.; Melero, I.; Krummel, M.F.; Sancho, D. Dendritic Cells in Cancer Immunology and Immunotherapy. Nat. Rev. Immunol. 2020, 20, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Guo, Z.; Liu, Y.; Li, X.; Zhang, Q.; Xu, X.; Gu, Y.; Zhang, Y.; Zhao, D.; Cao, X. The Lectin Siglec-G Inhibits Dendritic Cell Cross-Presentation by Impairing MHC Class I–Peptide Complex Formation. Nat. Immunol. 2016, 17, 1167–1175. [Google Scholar] [CrossRef]
- Perdicchio, M.; Ilarregui, J.M.; Verstege, M.I.; Cornelissen, L.A.M.; Schetters, S.T.T.; Engels, S.; Ambrosini, M.; Kalay, H.; Veninga, H.; den Haan, J.M.M.; et al. Sialic Acid-Modified Antigens Impose Tolerance via Inhibition of T-Cell Proliferation and de Novo Induction of Regulatory T Cells. Proc. Natl. Acad. Sci. USA 2016, 113, 3329–3334. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Manni, M.; Bärenwaldt, A.; Wieboldt, R.; Kirchhammer, N.; Ivanek, R.; Stanczak, M.; Zippelius, A.; König, D.; Rodrigues Manutano, N.; et al. Siglec Receptors Modulate Dendritic Cell Activation and Antigen Presentation to T Cells in Cancer. Front. Cell Dev. Biol. 2022, 10, 828916. [Google Scholar] [CrossRef]
- Büll, C.; Collado-Camps, E.; Kers-Rebel, E.D.; Heise, T.; Søndergaard, J.N.; den Brok, M.H.; Schulte, B.M.; Boltje, T.J.; Adema, G.J. Metabolic Sialic Acid Blockade Lowers the Activation Threshold of MoDCs for TLR Stimulation. Immunol. Cell Biol. 2017, 95, 408–415. [Google Scholar] [CrossRef]
- Rughetti, A.; Pellicciotta, I.; Biffoni, M.; Bäckström, M.; Link, T.; Bennet, E.P.; Clausen, H.; Noll, T.; Hansson, G.C.; Burchell, J.M.; et al. Recombinant Tumor-Associated MUC1 Glycoprotein Impairs the Differentiation and Function of Dendritic Cells. J. Immunol. 2005, 174, 7764–7772. [Google Scholar] [CrossRef]
- Ohta, M.; Ishida, A.; Toda, M.; Akita, K.; Inoue, M.; Yamashita, K.; Watanabe, M.; Murata, T.; Usui, T.; Nakada, H. Immunomodulation of Monocyte-Derived Dendritic Cells through Ligation of Tumor-Produced Mucins to Siglec-9. Biochem. Biophys. Res. Commun. 2010, 402, 663–669. [Google Scholar] [CrossRef]
- Balneger, N.; Cornelissen, L.A.M.; Wassink, M.; Moons, S.J.; Boltje, T.J.; Bar-Ephraim, Y.E.; Das, K.K.; Søndergaard, J.N.; Büll, C.; Adema, G.J. Sialic Acid Blockade in Dendritic Cells Enhances CD8+ T Cell Responses by Facilitating High-Avidity Interactions. Cell. Mol. Life Sci. 2022, 79, 98. [Google Scholar] [CrossRef] [PubMed]
- Kelm, S.; Schauer, R.; Manuguerra, J.-C.; Gross, H.-J.; Crocker, P.R. Modifications of Cell Surface Sialic Acids Modulate Cell Adhesion Mediated by Sialoadhesin and CD22. Glycoconj. J. 1994, 11, 576–585. [Google Scholar] [CrossRef]
- Sjoberg, E.R.; Powell, L.D.; Klein, A.; Varki, A. Natural Ligands of the B Cell Adhesion Molecule CD22 Beta Can Be Masked by 9-O-Acetylation of Sialic Acids. J. Cell Biol. 1994, 126, 549–562. [Google Scholar] [CrossRef]
- Büll, C.; Heise, T.; Adema, G.J.; Boltje, T.J. Sialic Acid Mimetics to Target the Sialic Acid–Siglec Axis. Trends Biochem. Sci. 2016, 41, 519–531. [Google Scholar] [CrossRef]
- Büll, C.; Stoel, M.A.; den Brok, M.H.; Adema, G.J. Sialic Acids Sweeten a Tumor’s Life. Cancer Res. 2014, 74, 3199–3204. [Google Scholar] [CrossRef] [PubMed]
- Angata, T.; Nycholat, C.M.; Macauley, M.S. Therapeutic Targeting of Siglecs Using Antibody- and Glycan-Based Approaches. Trends Pharmacol. Sci. 2015, 36, 645–660. [Google Scholar] [CrossRef]
- Ghosh, S. Nanotechnology and Sialic Acid Biology. In Sialic Acids and Sialoglycoconjugates in the Biology of Life, Health and Disease; Elsevier: Amsterdam, The Netherlands, 2020; pp. 297–325. [Google Scholar]
- Chen, W.C.; Completo, G.C.; Sigal, D.S.; Crocker, P.R.; Saven, A.; Paulson, J.C. In Vivo Targeting of B-Cell Lymphoma with Glycan Ligands of CD22. Blood 2010, 115, 4778–4786. [Google Scholar] [CrossRef] [PubMed]
- Macauley, M.S.; Pfrengle, F.; Rademacher, C.; Nycholat, C.M.; Gale, A.J.; von Drygalski, A.; Paulson, J.C. Antigenic Liposomes Displaying CD22 Ligands Induce Antigen-Specific B Cell Apoptosis. J. Clin. Investig. 2013, 123, 3074–3083. [Google Scholar] [CrossRef] [PubMed]
- Nycholat, C.M.; Duan, S.; Knuplez, E.; Worth, C.; Elich, M.; Yao, A.; O’Sullivan, J.; McBride, R.; Wei, Y.; Fernandes, S.M.; et al. A Sulfonamide Sialoside Analogue for Targeting Siglec-8 and -F on Immune Cells. J. Am. Chem. Soc. 2019, 141, 14032–14037. [Google Scholar] [CrossRef] [PubMed]
- Paszek, M.J.; DuFort, C.C.; Rossier, O.; Bainer, R.; Mouw, J.K.; Godula, K.; Hudak, J.E.; Lakins, J.N.; Wijekoon, A.C.; Cassereau, L.; et al. The Cancer Glycocalyx Mechanically Primes Integrin-Mediated Growth and Survival. Nature 2014, 511, 319–325. [Google Scholar] [CrossRef]
- Büll, C.; Heise, T.; Beurskens, D.M.H.; Riemersma, M.; Ashikov, A.; Rutjes, F.P.J.T.; van Kuppevelt, T.H.; Lefeber, D.J.; den Brok, M.H.; Adema, G.J.; et al. Sialic Acid Glycoengineering Using an Unnatural Sialic Acid for the Detection of Sialoglycan Biosynthesis Defects and On-Cell Synthesis of Siglec Ligands. ACS Chem. Biol. 2015, 10, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Rillahan, C.D.; Schwartz, E.; Rademacher, C.; McBride, R.; Rangarajan, J.; Fokin, V.V.; Paulson, J.C. On-Chip Synthesis and Screening of a Sialoside Library Yields a High Affinity Ligand for Siglec-7. ACS Chem. Biol. 2013, 8, 1417–1422. [Google Scholar] [CrossRef]
- Prescher, J.A.; Bertozzi, C.R. Chemistry in Living Systems. Nat. Chem. Biol. 2005, 1, 13–21. [Google Scholar] [CrossRef]
- Nischan, N.; Kohler, J.J. Advances in Cell Surface Glycoengineering Reveal Biological Function. Glycobiology 2016, 26, 789–796. [Google Scholar] [CrossRef]
- Runcie, K.; Budman, D.R.; John, V.; Seetharamu, N. Bi-Specific and Tri-Specific Antibodies- the next Big Thing in Solid Tumor Therapeutics. Mol. Med. 2018, 24, 50. [Google Scholar] [CrossRef] [PubMed]
- Feldman, E.J.; Brandwein, J.; Stone, R.; Kalaycio, M.; Moore, J.; O’Connor, J.; Wedel, N.; Roboz, G.J.; Miller, C.; Chopra, R.; et al. Phase III Randomized Multicenter Study of a Humanized Anti-CD33 Monoclonal Antibody, Lintuzumab, in Combination With Chemotherapy, Versus Chemotherapy Alone in Patients With Refractory or First-Relapsed Acute Myeloid Leukemia. J. Clin. Oncol. 2005, 23, 4110–4116. [Google Scholar] [CrossRef]
- Kantarjian, H.M.; DeAngelo, D.J.; Stelljes, M.; Martinelli, G.; Liedtke, M.; Stock, W.; Gökbuget, N.; O’Brien, S.; Wang, K.; Wang, T.; et al. Inotuzumab Ozogamicin versus Standard Therapy for Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2016, 375, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Baron, J.; Wang, E.S. Gemtuzumab Ozogamicin for the Treatment of Acute Myeloid Leukemia. Expert Rev. Clin. Pharmacol. 2018, 11, 549–559. [Google Scholar] [CrossRef]
- Drago, J.Z.; Modi, S.; Chandarlapaty, S. Unlocking the Potential of Antibody–Drug Conjugates for Cancer Therapy. Nat. Rev. Clin. Oncol. 2021, 18, 327–344. [Google Scholar] [CrossRef]
- Khongorzul, P.; Ling, C.J.; Khan, F.U.; Ihsan, A.U.; Zhang, J. Antibody–Drug Conjugates: A Comprehensive Review. Mol. Cancer Res. 2020, 18, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Lai, S.; Yu, H.; Ma, L. Suppression of MUC1-Overexpressing Tumors by a Novel MUC1/CD3 Bispecific Antibody. Antibodies 2023, 12, 47. [Google Scholar] [CrossRef] [PubMed]
- Franke, A.-K.; Wessolowski, C.; Thaden, V.; Müller, I.; Cornils, K. Glyco-Binding Domain Chimeric Antigen Receptors as a New Option for Cancer Immunotherapy. Gene Ther. 2023, 30, 603–611. [Google Scholar] [CrossRef]
- Ibarlucea-Benitez, I.; Weitzenfeld, P.; Smith, P.; Ravetch, J.V. Siglecs-7/9 Function as Inhibitory Immune Checkpoints in Vivo and Can Be Targeted to Enhance Therapeutic Antitumor Immunity. Proc. Natl. Acad. Sci. USA 2021, 118, e2107424118. [Google Scholar] [CrossRef]
- Wang, J.H.S.; Jiang, N.; Jain, A.; Lim, J. Development of Effective Siglec-9 Antibodies Against Cancer. Curr. Oncol. Rep. 2023, 25, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Läubli, H.; Varki, A. Sialic Acid–Binding Immunoglobulin-like Lectins (Siglecs) Detect Self-Associated Molecular Patterns to Regulate Immune Responses. Cell. Mol. Life Sci. 2020, 77, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Haas, Q.; Boligan, K.F.; Jandus, C.; Schneider, C.; Simillion, C.; Stanczak, M.A.; Haubitz, M.; Seyed Jafari, S.M.; Zippelius, A.; Baerlocher, G.M.; et al. Siglec-9 Regulates an Effector Memory CD8+ T-Cell Subset That Congregates in the Melanoma Tumor Microenvironment. Cancer Immunol. Res. 2019, 7, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Stanczak, M.A.; Siddiqui, S.S.; Trefny, M.P.; Thommen, D.S.; Boligan, K.F.; von Gunten, S.; Tzankov, A.; Tietze, L.; Lardinois, D.; Heinzelmann-Schwarz, V.; et al. Self-Associated Molecular Patterns Mediate Cancer Immune Evasion by Engaging Siglecs on T Cells. J. Clin. Investig. 2018, 128, 4912–4923. [Google Scholar] [CrossRef] [PubMed]
- Bordoloi, D.; Kulkarni, A.J.; Adeniji, O.S.; Pampena, M.B.; Bhojnagarwala, P.S.; Zhao, S.; Ionescu, C.; Perales-Puchalt, A.; Parzych, E.M.; Zhu, X.; et al. Siglec-7 Glyco-Immune Binding MAbs or NK Cell Engager Biologics Induce Potent Antitumor Immunity against Ovarian Cancers. Sci. Adv. 2023, 9, eadh4379. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Ho, M.; Adeniji, O.S.; Giron, L.; Bordoloi, D.; Kulkarni, A.J.; Puchalt, A.P.; Abdel-Mohsen, M.; Muthumani, K. Development of Siglec-9 Blocking Antibody to Enhance Anti-Tumor Immunity. Front. Oncol. 2021, 11, 778989. [Google Scholar] [CrossRef] [PubMed]
- Anthony, T.; Omid, H.; Jeffrey, S.W.; Patricia, L.; Kathryn, S.; Kevin, N.H. Single Agent Anti-Tumor Activity in PD-1 Refractory NSCLC: Phase 1 Data from the First-in-Human Trial of NC318, a Siglec-15-Targeted Antibody. In Proceedings of the The Society for Immunotherapy of Cancer (SITC) Annual Meeting, National Harbor, MD, USA, 6–10 November 2019. [Google Scholar]
- Läubli, H.; Nalle, S.C.; Maslyar, D. Targeting the Siglec–Sialic Acid Immune Axis in Cancer: Current and Future Approaches. Cancer Immunol. Res. 2022, 10, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Woods, E.C.; Vukojicic, P.; Bertozzi, C.R. Precision Glycocalyx Editing as a Strategy for Cancer Immunotherapy. Proc. Natl. Acad. Sci. USA 2016, 113, 10304–10309. [Google Scholar] [CrossRef]
- Gray, M.A.; Stanczak, M.A.; Mantuano, N.R.; Xiao, H.; Pijnenborg, J.F.A.; Malaker, S.A.; Miller, C.L.; Weidenbacher, P.A.; Tanzo, J.T.; Ahn, G.; et al. Targeted Glycan Degradation Potentiates the Anticancer Immune Response in Vivo. Nat. Chem. Biol. 2020, 16, 1376–1384. [Google Scholar] [CrossRef]
- Rabinovich, G.A.; Toscano, M.A. Turning “sweet” on Immunity: Galectin–Glycan Interactions in Immune Tolerance and Inflammation. Nat. Rev. Immunol. 2009, 9, 338–352. [Google Scholar] [CrossRef]
- Kapetanakis, N.-I.; Busson, P. Galectins as Pivotal Components in Oncogenesis and Immune Exclusion in Human Malignancies. Front. Immunol. 2023, 14, 1145268. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Niu, Q.; Deng, B.; Liu, S.; Wu, T.; Gao, Z.; Liu, Z.; Zhang, Y.; Qu, X.; Zhang, Y.; et al. CD22 CAR T-Cell Therapy in Refractory or Relapsed B Acute Lymphoblastic Leukemia. Leukemia 2019, 33, 2854–2866. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tao, Z.; Xu, Y.; Liu, J.; An, N.; Wang, Y.; Xing, H.; Tian, Z.; Tang, K.; Liao, X.; et al. CD33-Specific Chimeric Antigen Receptor T Cells with Different Co-Stimulators Showed Potent Anti-Leukemia Efficacy and Different Phenotype. Hum. Gene Ther. 2018, 29, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Aigner, M.; Feulner, J.; Schaffer, S.; Kischel, R.; Kufer, P.; Schneider, K.; Henn, A.; Rattel, B.; Friedrich, M.; Baeuerle, P.A.; et al. T Lymphocytes Can Be Effectively Recruited for Ex Vivo and in Vivo Lysis of AML Blasts by a Novel CD33/CD3-Bispecific BiTE Antibody Construct. Leukemia 2013, 27, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Wu, Z.; Jia, H.; Tong, C.; Guo, Y.; Ti, D.; Han, X.; Liu, Y.; Zhang, W.; Wang, C.; et al. Bispecific CAR-T Cells Targeting Both CD19 and CD22 for Therapy of Adults with Relapsed or Refractory B Cell Acute Lymphoblastic Leukemia. J. Hematol. Oncol. 2020, 13, 30. [Google Scholar] [CrossRef]
- Meril, S.; Harush, O.; Reboh, Y.; Matikhina, T.; Barliya, T.; Cohen, C.J. Targeting Glycosylated Antigens on Cancer Cells Using Siglec-7/9-based CAR T-cells. Mol. Carcinog. 2020, 59, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Klichinsky, M.; Ruella, M.; Shestova, O.; Lu, X.M.; Best, A.; Zeeman, M.; Schmierer, M.; Gabrusiewicz, K.; Anderson, N.R.; Petty, N.E.; et al. Human Chimeric Antigen Receptor Macrophages for Cancer Immunotherapy. Nat. Biotechnol. 2020, 38, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Jiang, J.; Wu, C. CAR-NK for Tumor Immunotherapy: Clinical Transformation and Future Prospects. Cancer Lett. 2020, 472, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Yang, L.; Li, Z.; Nalin, A.P.; Dai, H.; Xu, T.; Yin, J.; You, F.; Zhu, M.; Shen, W.; et al. First-in-Man Clinical Trial of CAR NK-92 Cells: Safety Test of CD33-CAR NK-92 Cells in Patients with Relapsed and Refractory Acute Myeloid Leukemia. Am. J. Cancer Res. 2018, 8, 1083–1089. [Google Scholar] [PubMed]
- Zhou, R.; Yazdanifar, M.; Das Roy, L.; Whilding, L.M.; Gavrill, A.; Maher, J.; Mukherjee, P. CAR T Cells Targeting the Tumor MUC1 Glycoprotein Reduce Triple-Negative Breast Cancer Growth. Front. Immunol. 2019, 10, 441662. [Google Scholar] [CrossRef]
- Mei, Z.; Zhang, K.; Lam, A.K.; Huang, J.; Qiu, F.; Qiao, B.; Zhang, Y. MUC1 as a Target for CAR-T Therapy in Head and Neck Squamous Cell Carinoma. Cancer Med. 2020, 9, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Posey, A.D.; Schwab, R.D.; Boesteanu, A.C.; Steentoft, C.; Mandel, U.; Engels, B.; Stone, J.D.; Madsen, T.D.; Schreiber, K.; Haines, K.M.; et al. Engineered CAR T Cells Targeting the Cancer-Associated Tn-Glycoform of the Membrane Mucin MUC1 Control Adenocarcinoma. Immunity 2016, 44, 1444–1454. [Google Scholar] [CrossRef]
- Rillahan, C.D.; Antonopoulos, A.; Lefort, C.T.; Sonon, R.; Azadi, P.; Ley, K.; Dell, A.; Haslam, S.M.; Paulson, J.C. Global Metabolic Inhibitors of Sialyl- and Fucosyltransferases Remodel the Glycome. Nat. Chem. Biol. 2012, 8, 661–668. [Google Scholar] [CrossRef]
- Heise, T.; Pijnenborg, J.F.A.; Büll, C.; van Hilten, N.; Kers-Rebel, E.D.; Balneger, N.; Elferink, H.; Adema, G.J.; Boltje, T.J. Potent Metabolic Sialylation Inhibitors Based on C-5-Modified Fluorinated Sialic Acids. J. Med. Chem. 2019, 62, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Büll, C.; Boltje, T.J.; Balneger, N.; Weischer, S.M.; Wassink, M.; van Gemst, J.J.; Bloemendal, V.R.; Boon, L.; van der Vlag, J.; Heise, T.; et al. Sialic Acid Blockade Suppresses Tumor Growth by Enhancing T-Cell–Mediated Tumor Immunity. Cancer Res. 2018, 78, 3574–3588. [Google Scholar] [CrossRef]
- Büll, C.; Boltje, T.J.; van Dinther, E.A.W.; Peters, T.; de Graaf, A.M.A.; Leusen, J.H.W.; Kreutz, M.; Figdor, C.G.; den Brok, M.H.; Adema, G.J. Targeted Delivery of a Sialic Acid-Blocking Glycomimetic to Cancer Cells Inhibits Metastatic Spread. ACS Nano 2015, 9, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Macauley, M.S.; Arlian, B.M.; Rillahan, C.D.; Pang, P.-C.; Bortell, N.; Marcondes, M.C.G.; Haslam, S.M.; Dell, A.; Paulson, J.C. Systemic Blockade of Sialylation in Mice with a Global Inhibitor of Sialyltransferases. J. Biol. Chem. 2014, 289, 35149–35158. [Google Scholar] [CrossRef] [PubMed]
- Hidari, K.I.P.J.; Oyama, K.; Ito, G.; Nakayama, M.; Inai, M.; Goto, S.; Kanai, Y.; Watanabe, K.; Yoshida, K.; Furuta, T.; et al. Identification and Characterization of Flavonoids as Sialyltransferase Inhibitors. Biochem. Biophys. Res. Commun. 2009, 382, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.-W.; Yu, C.-Y.; Lin, T.-W.; Wang, P.-H.; Tsai, Y.-C. Soyasaponin I Decreases the Expression of A2,3-Linked Sialic Acid on the Cell Surface and Suppresses the Metastatic Potential of B16F10 Melanoma Cells. Biochem. Biophys. Res. Commun. 2006, 341, 614–619. [Google Scholar] [CrossRef]
- Hsu, C.-C.; Lin, T.-W.; Chang, W.-W.; Wu, C.-Y.; Lo, W.-H.; Wang, P.-H.; Tsai, Y.-C. Soyasaponin-I-Modified Invasive Behavior of Cancer by Changing Cell Surface Sialic Acids. Gynecol. Oncol. 2005, 96, 415–422. [Google Scholar] [CrossRef]
- Chang, K.-H.; Lee, L.; Chen, J.; Li, W.-S. Lithocholic Acid Analogues, New and Potent α-2,3-Sialyltransferase Inhibitors. Chem. Commun. 2006, 6, 629–631. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-Y.; Tang, Y.-A.; Huang, S.-M.; Juan, H.-F.; Wu, L.-W.; Sun, Y.-C.; Wang, S.-C.; Wu, K.-W.; Balraj, G.; Chang, T.-T.; et al. A Novel Sialyltransferase Inhibitor Suppresses FAK/Paxillin Signaling and Cancer Angiogenesis and Metastasis Pathways. Cancer Res. 2011, 71, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Niu, Y.; Xiong, D.-C.; Cao, X.; Ye, X.-S. Highly Substituted Cyclopentane–CMP Conjugates as Potent Sialyltransferase Inhibitors. J. Med. Chem. 2015, 58, 7972–7990. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayyalasomayajula, R.; Cudic, M. Targeting Siglec–Sialylated MUC1 Immune Axis in Cancer. Cancers 2024, 16, 1334. https://doi.org/10.3390/cancers16071334
Ayyalasomayajula R, Cudic M. Targeting Siglec–Sialylated MUC1 Immune Axis in Cancer. Cancers. 2024; 16(7):1334. https://doi.org/10.3390/cancers16071334
Chicago/Turabian StyleAyyalasomayajula, Ramya, and Mare Cudic. 2024. "Targeting Siglec–Sialylated MUC1 Immune Axis in Cancer" Cancers 16, no. 7: 1334. https://doi.org/10.3390/cancers16071334
APA StyleAyyalasomayajula, R., & Cudic, M. (2024). Targeting Siglec–Sialylated MUC1 Immune Axis in Cancer. Cancers, 16(7), 1334. https://doi.org/10.3390/cancers16071334





