IL1B Polymorphism (rs1143634) and IL-1β Plasma Concentration as Predictors of Nutritional Disorders and Prognostic Factors in Multiple Myeloma Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Group
2.2. Blood Collection
2.3. Genotyping
2.4. Assessment of IL-1β Plasma Concentration
2.5. Bioelectrical Impendence Analysis
2.6. Statistical Analysis
3. Results
3.1. Characteristics of the Study Group
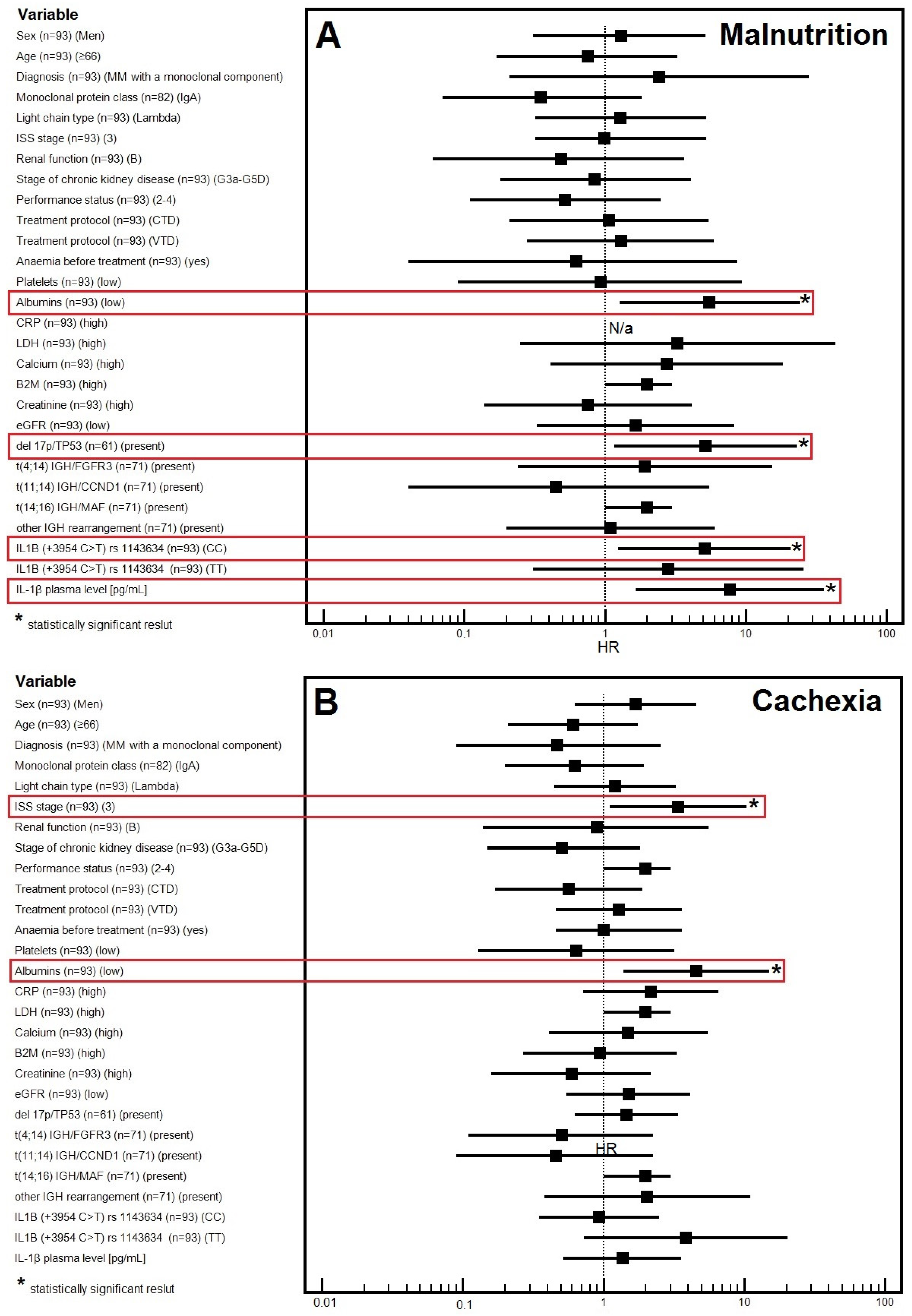
3.2. Factors Affecting the Risk of Malnutrition
3.2.1. Univariable Analysis
3.2.2. Multivariable Analysis
3.3. Factors Affecting the Risk of Cachexia
3.3.1. Univariable Analysis
3.3.2. Multivariable Analysis
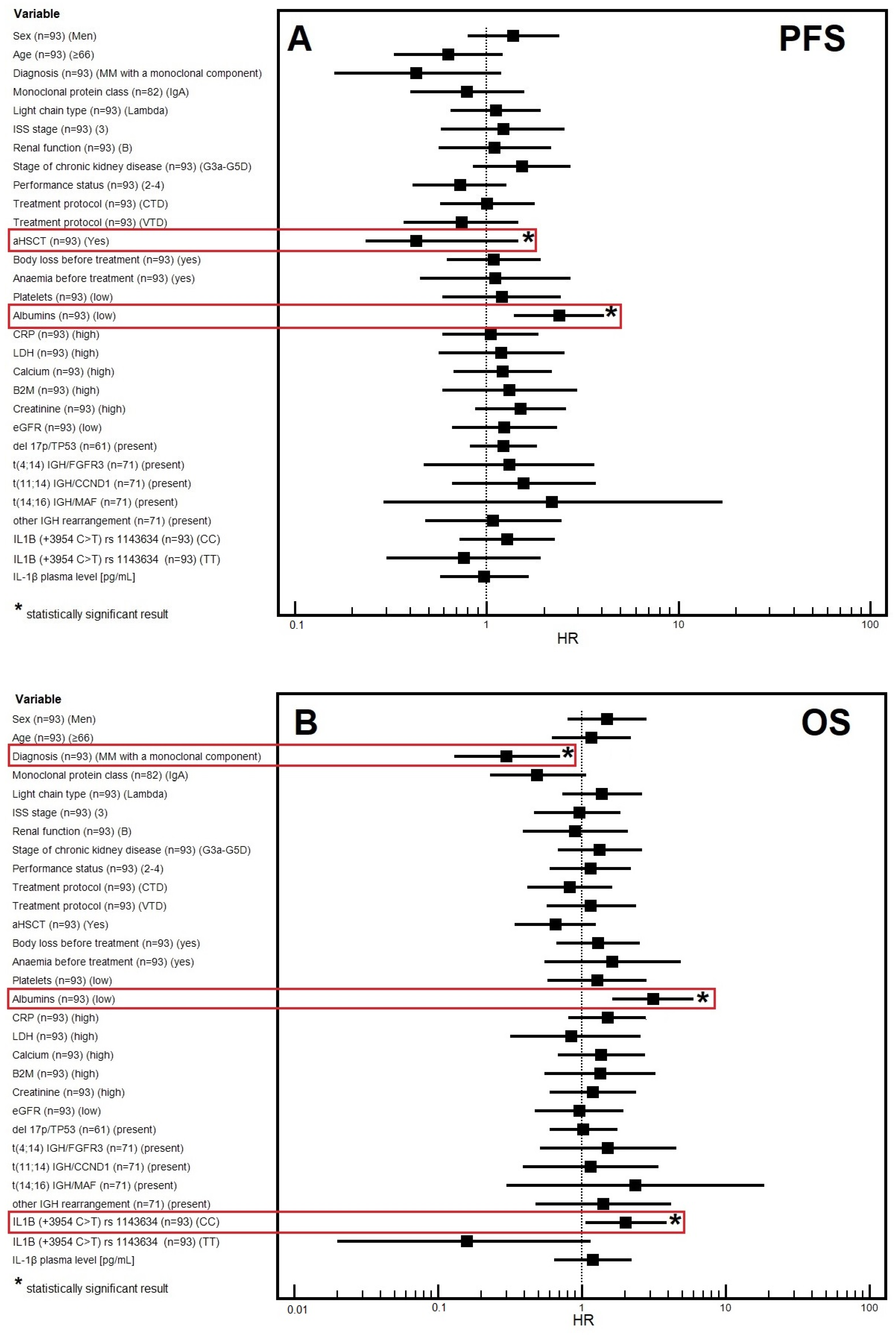
3.4. Progression-Free Survival
3.4.1. Univariable Analysis
3.4.2. Multivariable Analysis
3.5. Overall Survival
3.5.1. Univariable Analysis
3.5.2. Multivariable Analysis
3.6. The Association between Gender and Demographic, Clinical, and Molecular Factors
3.7. The Association between IL1B Genotypes’ Distribution and Demographic, Clinical, and Molecular Factors
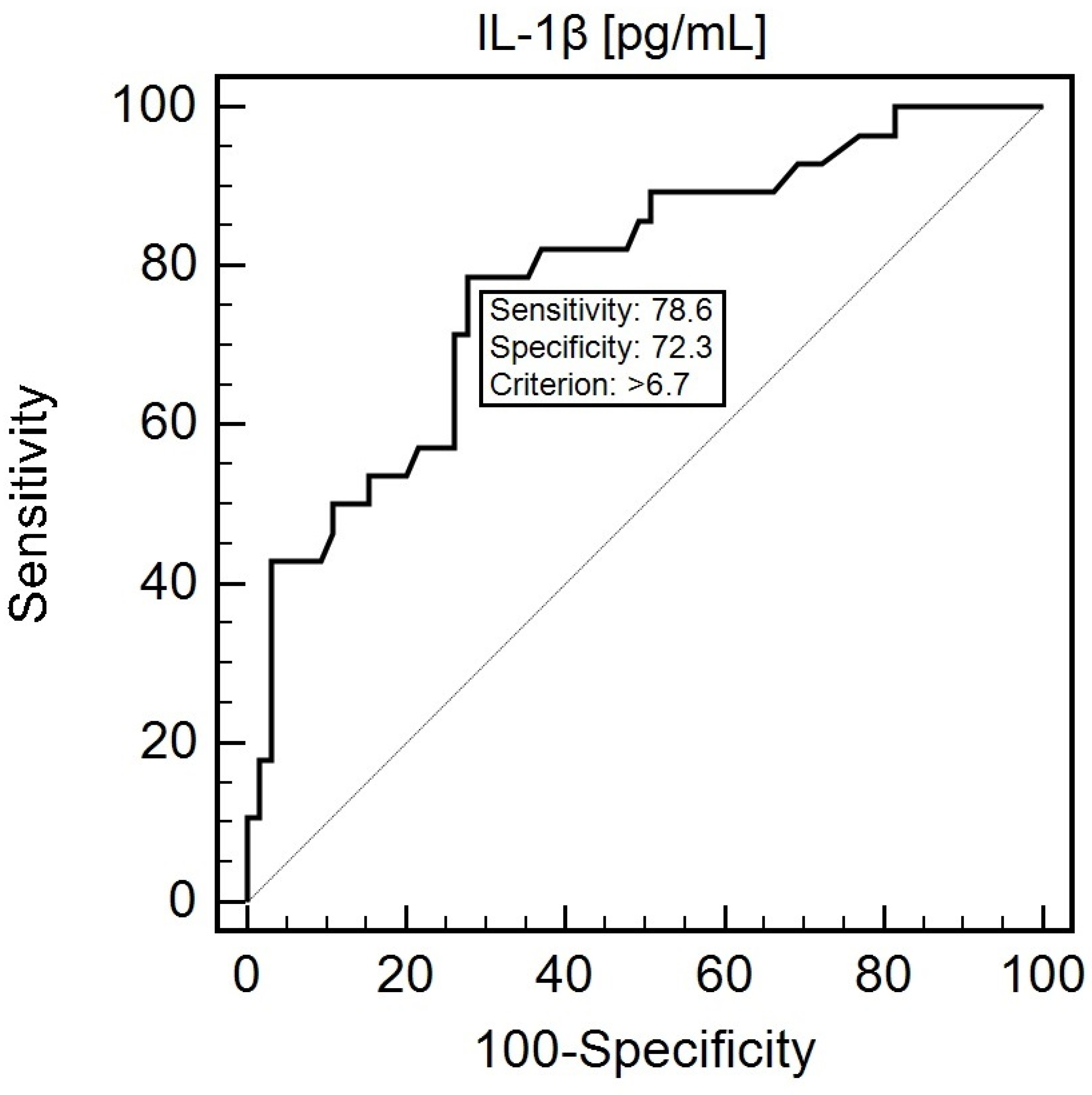
3.8. Diagnostic Usefulness of the Assessment of the IL1B SNP (rs1143634) and IL-1β Concentration in Predicting Nutritional Disorders
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Das, S.; Juliana, N.; Yazit, N.A.A.; Azmani, S.; Abu, I.F. Multiple Myeloma: Challenges Encountered and Future Options for Better Treatment. Int. J. Mol. Sci. 2022, 23, 1649. [Google Scholar] [CrossRef]
- van de Donk, N.W.C.J.; Pawlyn, C.; Yong, K.L. Multiple myeloma. Lancet 2021, 397, 410–427. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V. Multiple myeloma: Every year a new standard? Hematol. Oncol. 2019, 37 (Suppl. S1), 62–65. [Google Scholar] [CrossRef] [PubMed]
- Diamond, E.; Lahoud, O.B.; Landau, H. Managing multiple myeloma in elderly patients. Leuk. Lymphoma 2018, 59, 1300–1311. [Google Scholar] [CrossRef] [PubMed]
- Padala, S.A.; Barsouk, A.; Barsouk, A.; Rawla, P.; Vakiti, A.; Kolhe, R.; Kota, V.; Ajebo, G.H. Epidemiology, Staging, and Management of Multiple Myeloma. Med. Sci. 2021, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Zagouri, F.; Kastritis, E.; Zomas, A.; Terpos, E.; Katodritou, E.; Symeonidis, A.; Delimpasi, S.; Pouli, A.; Vassilakopoulos, T.P.; Michalis, E.; et al. Hypercalcemia remains an adverse prognostic factor for newly diagnosed multiple myeloma patients in the era of novel antimyeloma therapies. Eur. J. Haematol. 2017, 99, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Heider, M.; Nickel, K.; Högner, M.; Bassermann, F. Multiple Myeloma: Molecular Pathogenesis and Disease Evolution. Oncol. Res. Treat. 2021, 44, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, J.Y.; Lim, S.H.; Cho, J.; Kim, S.J.; Jang, J.H.; Kim, W.S.; Jung, C.W.; Kim, K. Patient-Generated Subjective Global Assessment as a prognosis tool in patients with multiple myeloma. Nutrition 2017, 36, 67–71. [Google Scholar] [CrossRef]
- Galán-Olleros, M.; Marco, J.; Oteo, D.; Cristóbal-Bilbao, R.; Manrique, E.; García-Maroto, R.; Marco, F.; Cebrián-Parra, J.L. Orthopedic Surgical Treatment and Perioperative Complications in Multiple Myeloma Bone Disease: Analysis of a Series (2009–2018). Ann. Surg. Oncol. 2021, 28, 1158–1166. [Google Scholar] [CrossRef]
- Garzón Herazo, J.R.; Muñoz Velandia, O.M.; Solano, J.C.; Molina Pimienta, L.; Figueroa Lemus, W.J. The nutrition risk index is associated with bacteremia within 30 days after autologous stem cell transplantation in patients with multiple myeloma. Transpl. Infect. Dis. 2020, 22, e13302. [Google Scholar] [CrossRef]
- Bossi, P.; Delrio, P.; Mascheroni, A.; Zanetti, M. The Spectrum of Malnutrition/Cachexia/Sarcopenia in Oncology According to Different Cancer Types and Settings: A Narrative Review. Nutrients 2021, 13, 1980. [Google Scholar] [CrossRef]
- Kuypers, J.; Simmance, N.; Quach, H.; Hastie, K.; Kiss, N. Nutrition support use and clinical outcomes in patients with multiple myeloma undergoing autologous stem cell transplant. Support. Care Cancer 2022, 30, 9341–9350. [Google Scholar] [CrossRef]
- Meza-Valderrama, D.; Marco, E.; Dávalos-Yerovi, V.; Muns, M.D.; Tejero-Sánchez, M.; Duarte, E.; Sánchez-Rodríguez, D. Sarcopenia, Malnutrition, and Cachexia: Adapting Definitions and Terminology of Nutritional Disorders in Older People with Cancer. Nutrients 2021, 13, 761. [Google Scholar] [CrossRef]
- Muscaritoli, M.; Imbimbo, G.; Jager-Wittenaar, H.; Cederholm, T.; Rothenberg, E.; di Girolamo, F.G.; Amabile, M.I.; Sealy, M.; Schneider, S.; Barazzoni, R.; et al. Disease-related malnutrition with inflammation and cachexia. Clin. Nutr. 2023, 42, 1475–1479. [Google Scholar] [CrossRef]
- Aoyagi, T.; Terracina, K.P.; Raza, A.; Matsubara, H.; Takabe, K. Cancer cachexia, mechanism and treatment. World J. Gastrointest. Oncol. 2015, 7, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Tisdale, M.J. Mechanisms of cancer cachexia. Physiol. Rev. 2009, 89, 381–410. [Google Scholar] [CrossRef] [PubMed]
- Fearon, K.; Strasser, F.; Anker, S.D.; Bosaeus, I.; Bruera, E.; Fainsinger, R.L.; Jatoi, A.; Loprinzi, C.; MacDonald, N.; Mantovani, G.; et al. Definition and classification of cancer cachexia: An international consensus. Lancet Oncol. 2011, 12, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Lobo, D.N.; Gianotti, L.; Adiamah, A.; Barazzoni, R.; Deutz, N.E.P.; Dhatariya, K.; Greenhaff, P.L.; Hiesmayr, M.; Hjort, D.; Klek, S.; et al. Perioperative nutrition: Recommendations from the ESPEN expert group. Clin. Nutr. 2020, 39, 3211–3227. [Google Scholar] [CrossRef] [PubMed]
- Beirer, A. Malnutrition and cancer, diagnosis and treatment. Memo-Mag. Eur. Med. Oncol. 2021, 14, 168–173. [Google Scholar] [CrossRef]
- Sadeghi, M.; Keshavarz-Fathi, M.; Baracos, V.; Arends, J.; Mahmoudi, M.; Rezaei, N. Cancer cachexia: Diagnosis, assessment, and treatment. Crit. Rev. Oncol. Hematol. 2018, 127, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Teimuri Nobari, S.; Rasmi, Y.; Khadem Ansari, M.H. Serum Levels of Interleukin-1β and Disease Progression in Multiple Myeloma Patients: A Case and Control Study. Asian Pac. J. Cancer Prev. 2022, 23, 2937–2942. [Google Scholar] [CrossRef] [PubMed]
- Lust, J.A.; Donovan, K.A. The role of interleukin-1 beta in the pathogenesis of multiple myeloma. Hematol. Oncol. Clin. N. Am. 1999, 13, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Malla, J.; Zahra, A.; Venugopal, S.; Selvamani, T.Y.; Shoukrie, S.I.; Selvaraj, R.; Dhanoa, R.K.; Hamouda, R.K.; Mostafa, J. What Role Do Inflammatory Cytokines Play in Cancer Cachexia? Cureus 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Loumaye, A.; Thissen, J.P. Biomarkers of cancer cachexia. Clin. Biochem. 2017, 50, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Laird, B.J.; McMillan, D.; Skipworth, R.J.E.; Fallon, M.T.; Paval, D.R.; McNeish, I.; Gallagher, I.J. The Emerging Role of Interleukin 1β (IL-1β) in Cancer Cachexia. Inflammation 2021, 44, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Abtahi, S.; Farazmand, A.; Mahmoudi, M.; Ashraf-Ganjouei, A.; Javinani, A.; Nazari, B.; Kavosi, H.; Amirzargar, A.A.; Jamshidi, A.R.; Gharibdoost, F. IL-1A rs1800587, IL-1B rs1143634 and IL-1R1 rs2234650 polymorphisms in Iranian patients with systemic sclerosis. Int. J. Immunogenet. 2015, 42, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.; Dinis-Oliveira, R.J.; Nogueira, A.; Gonçalves, F.; Silva, P.; Vieira, C.; Silvestre, R.; Carvalho, F.; Medeiros, R. Interleukin-1β genotype and circulating levels in cancer patients: Metastatic status and pain perception. Clin. Biochem. 2014, 47, 1209–1213. [Google Scholar] [CrossRef] [PubMed]
- Abed, A.S.; Mokdad-Gargouri, R.; Raoof, W.M. Association between interleuleukin-1β polymorphism (rs16944) and biomarkers levels in Iraqi patients with prostate cancer. Mol. Biol. Rep. 2023, 50, 1157–1165. [Google Scholar] [CrossRef]
- Jafrin, S.; Aziz, M.A.; Islam, M.S. Role of IL-1β rs1143634 (+3954C>T) polymorphism in cancer risk: An updated meta-analysis and trial sequential analysis. J. Int. Med. Res. 2021, 49, 3000605211060144. [Google Scholar] [CrossRef]
- Wang, J.; Shi, Y.; Wang, G.; Dong, S.; Yang, D.; Zuo, X. The association between interleukin-1 polymorphisms and their protein expression in Chinese Han patients with breast cancer. Mol. Genet. Genom. Med. 2019, 7, e804. [Google Scholar] [CrossRef]
- Zhang, D.; Zheng, H.; Zhou, Y.; Tang, X.; Yu, B.; Li, J. Association of IL-1beta gene polymorphism with cachexia from locally advanced gastric cancer. BMC Cancer 2007, 7, 45. [Google Scholar] [CrossRef]
- Szudy-szczyrek, A.; Mlak, R.; Mazurek, M.; Krajka, T.; Chocholska, S.; Małecka-massalska, T.; Hus, M. The TT Genotype of the KIAA1524 rs2278911 Polymorphism Is Associated with Poor Prognosis in Multiple Myeloma. Cells 2023, 12, 1029. [Google Scholar] [CrossRef]
- Joshi, R.; Bajracharya, S.; Gurung, S.; Shrestha, D. Burden of Anemia: A profile of a tertiary care hospital. J. Nepalgunj Med. Coll. 2019, 16, 54–57. [Google Scholar] [CrossRef]
- Faiman, B.M.; Mangan, P.; Spong, J.; Tariman, J.D. Renal complications in multiple myeloma and related disorders: Survivorship care plan of the International Myeloma Foundation Nurse Leadership Board. Clin. J. Oncol. Nurs. 2011, 15, 66–76. [Google Scholar] [CrossRef]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, e538–e548. [Google Scholar] [CrossRef] [PubMed]
- Mallard, J.; Gagez, A.-L.; Baudinet, C.; Herbinet, A.; Maury, J.; Bernard, P.L.; Cartron, G. C-Reactive Protein Level: A Key Predictive Marker of Cachexia in Lymphoma and Myeloma Patients. J. Hematol. 2019, 8, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Shyh-Chang, N. Metabolic Changes During Cancer Cachexia Pathogenesis. Adv. Exp. Med. Biol. 2017, 1026, 233–249. [Google Scholar] [CrossRef]
- Wu, Q.; Sun, S.; Li, Z.; Yang, Q.; Li, B.; Zhu, S.; Wang, L.; Wu, J.; Yuan, J.; Wang, C.; et al. Breast cancer-released exosomes trigger cancer-associated cachexia to promote tumor progression. Adipocyte 2019, 8, 31–45. [Google Scholar] [CrossRef]
- Daas, S.I.; Rizeq, B.R.; Nasrallah, G.K. Adipose tissue dysfunction in cancer cachexia. J. Cell. Physiol. 2018, 234, 13–22. [Google Scholar] [CrossRef]
- Gherardi, R.K.; Bélec, L.; Fromont, G.; Divine, M.; Malapert, D.; Gaulard, P.; Degos, J.D. Elevated levels of interleukin-1 beta (IL-1 beta) and IL-6 in serum and increased production of IL-1 beta mRNA in lymph nodes of patients with polyneuropathy, organomegaly, endocrinopathy, M protein, and skin changes (POEMS) syndrome. Blood 1994, 83, 2587–2593. [Google Scholar] [CrossRef]
- Ozkurt, Z.N.; Yağci, M.; Sucak, G.T.; Kirazli, S.; Haznedar, R. Thrombopoietic cytokines and platelet count in multiple myeloma. Platelets 2010, 21, 33–36. [Google Scholar] [CrossRef]
- Bębnowska, D.; Hrynkiewicz, R.; Grywalska, E.; Pasiarski, M.; Sosnowska-Pasiarska, B.; Smarz-Widelska, I.; Góźdź, S.; Roliński, J.; Niedźwiedzka-Rystwej, P. Immunological Prognostic Factors in Multiple Myeloma. Int. J. Mol. Sci. 2021, 22, 3587. [Google Scholar] [CrossRef] [PubMed]
- Kline, M.; Donovan, K.; Wellik, L.; Lust, C.; Jin, W.; Moon-Tasson, L.; Xiong, Y.; Witzig, T.E.; Kumar, S.; Rajkumar, S.V.; et al. Cytokine and chemokine profiles in multiple myeloma; significance of stromal interaction and correlation of IL-8 production with disease progression. Leuk. Res. 2007, 31, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin, Y. Tumor Necrosis Factor Alpha-308G/A Polymorphism and the Risk of Multiple Myeloma: A Meta-Analysis of Pooled Data from Twelve Case-Control Studies. Turkish J. Haematol. Off. J. Turkish Soc. Haematol. 2019, 36, 72–80. [Google Scholar] [CrossRef]
- Chakraborty, B.; Vishnoi, G.; Gowda, S.H.; Goswami, B. Interleukin-6 gene-174 G/C promoter polymorphism and its association with clinical profile of patients with multiple myeloma. Asia. Pac. J. Clin. Oncol. 2017, 13, e402–e407. [Google Scholar] [CrossRef] [PubMed]
- Vangsted, A.J.; Klausen, T.W.; Abildgaard, N.; Andersen, N.F.; Gimsing, P.; Gregersen, H.; Nexø, B.A.; Vogel, U. Single nucleotide polymorphisms in the promoter region of the IL1B gene influence outcome in multiple myeloma patients treated with high-dose chemotherapy independently of relapse treatment with thalidomide and bortezomib. Ann. Hematol. 2011, 90, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Banu, C.; Moise, A.; Arion, C.V.; Coriu, D.; Tănase, A.; Constantinescu, I. Cytokine gene polymorphisms support diagnostic monitoring of Romanian multiple myeloma patients. J. Med. Life 2011, 4, 264–268. [Google Scholar]
- Vangsted, A.J.; Nielsen, K.R.; Klausen, T.W.; Haukaas, E.; Tjønneland, A.; Vogel, U. A functional polymorphism in the promoter region of the IL1B gene is associated with risk of multiple myeloma. Br. J. Haematol. 2012, 158, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Jatoi, A.; Nguyen, P.L.; Foster, N.; Sun, D.; Stella, P.J.; Campbell, M.; Tschetter, L.K.; Dakhil, S.R.; Mailliard, J.A.; Nikcevich, D.A. Interleukin-1 genetic polymorphisms and their relationship to the cancer anorexia/weight loss syndrome in metastatic gastric and gastroesophageal junction adenocarcinoma. J. Support. Oncol. 2007, 5, 41–46. [Google Scholar]
- Broekhuizen, R.; Grimble, R.F.; Howell, W.M.; Shale, D.J.; Creutzberg, E.C.; Wouters, E.F.; Schols, A.M. Pulmonary cachexia, systemic inflammatory profile, and the interleukin 1beta-511 single nucleotide polymorphism. Am. J. Clin. Nutr. 2005, 82, 1059–1064. [Google Scholar] [CrossRef]
- Shi, Q.; Wang, X.S.; Li, G.; Shah, N.D.; Orlowski, R.Z.; Williams, L.A.; Mendoza, T.R.; Cleeland, C.S. Racial/ethnic disparities in inflammatory gene single-nucleotide polymorphisms as predictors of a high risk for symptom burden in patients with multiple myeloma 1 year after diagnosis. Cancer 2015, 121, 1138–1146. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E. Understanding cachexia as a cancer metabolism syndrome. Oncogenesis 2016, 5, e200. [Google Scholar] [CrossRef] [PubMed]
- Vangsted, A.J.; Klausen, T.W.; Ruminski, W.; Gimsing, P.; Andersen, N.F.; Gang, A.O.; Abildgaard, N.; Knudsen, L.M.; Nielsen, J.L.; Gregersen, H.; et al. The polymorphism IL-1beta T-31C is associated with a longer overall survival in patients with multiple myeloma undergoing auto-SCT. Bone Marrow Transplant. 2009, 43, 539–545. [Google Scholar] [CrossRef] [PubMed]

| Factor | Study Group (n = 93) | |
|---|---|---|
| Gender | Male | 44 (47.3%) |
| Female | 49 (52.7%) | |
| Age (years) | Mean ± standard deviation, median (range) | 64.3 ± 9.83 66 (37–87) |
| ≥65 | 49 (52.7%) | |
| <65 | 44 (47.3%) | |
| Myeloma type | IgG | 55 (59.1%) |
| IgA | 27 (29%) | |
| Light chains | 11 (11.8%) | |
| Light chain type | Kappa | 57 (61.3%) |
| Lambda | 36 (38.7%) | |
| ISS stage | 1 | 29 (31.2%) |
| 2 | 29 (31.2%) | |
| 3 | 35 (37.6%) | |
| Renal function | A | 80 (86%) |
| B | 13 (14%) | |
| Performance status | 0 | 7 (7.5%) |
| 1 | 37 (39.8%) | |
| 2 | 37 (39.8%) | |
| 3 | 10 (10.8%) | |
| 4 | 2 (2.2%) | |
| BMI (kg/m2) | Mean ± standard deviation, median (range) | 27.06 ± 5.98 26.59 (14.53–56.82) |
| Body weight loss | Yes | 46 (49.5%) |
| No | 47 (50.5%) | |
| 5% | 14 (30.4%) | |
| 10% | 32 (69.6%) | |
| Body weight loss (kg) | Mean ± standard deviation, median (range) | 5.6 ± 2.4 5 (0–17) |
| Anemia grade before treatment (WHO) | Absent or I° | 51 (54.8%) |
| II° | 27 (29%) | |
| III° | 14 (15.1%) | |
| IV° | 1 (1.1%) | |
| Treatment protocol | CTD | 27 (29%) |
| V(C)D | 26 (28%) | |
| VTD | 40 (43%) | |
| aHSCT | No | 55 (59.1%) |
| Yes | 38 (40.9%) | |
| del 17p/TP53 No data: n = 32 | Absent | 45 (73.8%) |
| Present | 16 (26.2%) | |
| t(4;14) IGH/FGFR3 No data: n = 22 | Absent | 61 (85.9%) |
| Present | 10 (14.1%) | |
| t(11;14) IGH/CCND1 No data: n = 22 | Absent | 63 (88.7%) |
| Present | 8 (11.3%) | |
| t(14;16) IGH/MAF No data: n = 31 | Absent | 61 (98.4%) |
| Present | 1 (1.6%) | |
| Other IGH rearrangement No data: n = 22 | Absent | 60 (84.5%) |
| Present | 11 (15.5%) | |
| Variable | Malnutrition | Cancer Cachexia | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariable | Multivariable | Univariable | Multivariable | |||||
| No (n = 29) | Yes (n = 64) | OR [95% CI] p | OR [95% CI] p | No (n = 65) | Yes (n = 28) | OR [95% CI] p | OR [95% CI] p | |
| Gender | ||||||||
| Men | 11 (25%) | 33 (75%) | 1.74 [0.71–4.27] | 1.70 [0.63–4.58] | 28 (63.6%) | 16 (36.4%) | 1.76 [0.72–4.31] | 1.31 [0.31–5.21] |
| Women | 18 (36.7%) | 31 (63.3%) | 0.2248 | 0.2891 | 37 (75.5%) | 12 (24.5%) | 0.2149 | 0.7170 |
| Age | ||||||||
| ≥65 | 14 (28.6%) | 35 (71.4%) | 1.29 [0.54–3.11] | 0.61 [0.21–1.76] | 31 (63.3%) | 18 (36.7%) | 1.97 [0.79–4.92] | 0.76 [0.17–3.28] |
| <65 | 15 (34.1%) | 29 (65.9%) | 0.5666 | 0.3586 | 34 (77.3%) | 10 (22.7%) | 0.1445 | 0.7101 |
| Diagnosis | ||||||||
| MM with a monoclonal component | 27 (32.9%) | 55 (67.1%) | 0.45 [0.09–2.24] | 0.47 [0.09–2.57] | 57 (69.55) | 25 (30.5%) | 1.17 [0.29–4.78] | 2.43 [0.21–28.07] |
| Light chain disease | 2 (18.2%) | 9 (81.8%) | 0.3316 | 0.3856 | 8 (72.7%) | 3 (27.3%) | 0.8273 | 0.4757 |
| Monoclonal protein class | ||||||||
| IgA | 8 (29.6%) | 19 (70.4%) | 0.80 [0.29–2.16] | 0.63 [0.2–1.94] | 17 (63%) | 10 (37%) | 0.64 [0.24–1.70] | 0.35 [0.07–1.82] |
| IgG | 19 (34.5%) | 36 (65.5%) | 0.6565 | 0.4192 | 40 (72.7%) | 15 (27.3%) | 0.3684 | 0.2138 |
| N/a: n = 11 | ||||||||
| Light chain type | ||||||||
| Lambda | 10 (27.8%) | 26 (72.2%) | 1.30 [0.52–3.24] | 1.21 [0.45–3.28] | 23 (63.9%) | 13 (36.1%) | 1.58 [0.64–3.89] | 1.29 [0.32–5.28] |
| Kappa | 19 (33.3%) | 38 (66.7%) | 0.5736 | 0.7047 | 42 (73.7%) | 15 (26.3%) | 0.3175 | 0.7206 |
| ISS stage | ||||||||
| 3 | 5 (14.3%) | 30 (85.7%) | 4.23 [1.43–12.49] | 3.39 [1.11–10.41] | 20 (57.1%) | 15 (42.9%) | 2.60 [1.04–6.45] | 0.99 [0.22–4.45] |
| 1, 2 | 24 (41.4%) | 34 (58.6%) | 0.0089 * | 0.0327 * | 45 (77.6%) | 13 (22.4%) | 0.0400 * | 0.9880 |
| Renal function | ||||||||
| B | 2 (15.4%) | 11 (54.6%) | 2.80 [0.58–13.55] | 0.90 [0.14–5.57] | 7 (53.8%) | 6 (46.2%) | 2.56 [0.68–7.47] | 0.49 [0.06–3.66] |
| A | 27 (33.7%) | 53 (66.2%) | 0.2002 | 0.9067 | 58 (72.5%) | 22 (27.5%) | 0.1815 | 0.4857 |
| Stage of chronic kidney disease | ||||||||
| G3a,G3b, G4, G5D | 8 (24.2%) | 25 (75.8%) | 1.68 [0.64–4.38] | 0.51 [0.15–1.82] | 20 (60.6%) | 13 (39.4%) | 1.95 [0.78–4.85] | 0.85 [0.18–4.12] |
| G1,G2 | 21 (35%) | 39 (65%) | 0.2864 | 0.3017 | 45 (75%) | 15 (25%) | 0.1506 | 0.8456 |
| Performance status | ||||||||
| 2–4 | 0 (0%) | 49 (100%) | 188.42 [10.87–3266.72] | -[-] | 30 (61.2%) | 19 (38.8%) | 2.46 [0.97–6.24] | 0.52 [0.11–2.48] |
| 0, 1 | 29 (65.9%) | 15 (34.1%) | 0.0003 * | 0.9939 | 35 (79.5%) | 9 (20.5%) | 0.0578 | 0.4152 |
| Treatment protocol (1) | ||||||||
| CTD | 5 (18.5%) | 22 (81.5%) | 2.51 [0.84–7.50] | 0.57 [0.17–1.89] | 18 (66.7%) | 9 (33.3%) | 1.24 [0.47–3.23] | 1.07 [0.21–5.46] |
| V(C)D, VTD | 24 (36.4%) | 42 (63.6%) | 0.0982 | 0.3625 | 47 (71.2%) | 19 (28.8%) | 0.6647 | 0.9381 |
| Treatment protocol (2) | ||||||||
| VTD | 10 (35.7%) | 18 (64.3%) | 0.74 [0.29–1.90] | 1.29 [0.46–3.64] | 20 (71.4%) | 8 (28.6%) | 0.90 [0.34–2.38] | 1.30 [0.28–5.98] |
| CTD, V(C)D | 19 (29.2%) | 46 (70.8%) | 0.5365 | 0.6288 | 45 (69.2%) | 20 (30.8%) | 0.8322 | 0.7354 |
| Anemia before treatment (WHO) | ||||||||
| Yes | 21 (28%) | 54 (72%) | 2.06 [0.71–5.92] | 1.01 [0.28–3.61] | 49 (65.3%) | 26 (34.7%) | 4.24 [0.90–19.90] | 0.63 [0.04–8.71] |
| No | 8 (44.4%) | 10 (55.6%) | 0.1813 | 0.9923 | 16 (88.9%) | 2 (11.1%) | 0.0666 | 0.7317 |
| Platelets | ||||||||
| Low | 3 (25%) | 9 (75%) | 1.42 [0.35–5.68] | 0.64 [0.13–3.18] | 7 (58.3%) | 5 (41.7%) | 1.80 [0.52–6.26] | 0.93 [0.09–9.46] |
| Normal | 26 (32.1%) | 55 (67.9%) | 0.6216 | 0.5885 | 58 (71.6%) | 23 (28.4%) | 0.3543 | 0.9547 |
| Albumins | ||||||||
| Low | 4 (11.8%) | 30 (88.2%) | 5.14 [1.72–17.66] | 4.56 [1.39–15.03] | 15 (44.1%) | 19 (55.9%) | 7.04 [2.64–18.76] | 5.54 [1.27–24.1] |
| Normal | 25 (42.4%) | 34 (57.6%) | 0.0040 * | 0.0125 * | 50 (84.7%) | 9 (15.3%) | 0.0001 * | 0.0225 * |
| CRP | ||||||||
| High | 6 (17.6%) | 28 (82.4%) | 2.98 [1.07–8.31] | 2.17 [0.72–6.59] | 6 (17.6%) | 28 (82.4%) | N/a | N/a |
| Normal | 23 (39%) | 36 (61%) | 0.0368 * | 0.1705 | 59 (100%) | 0 (0%) | ||
| LDH | ||||||||
| High | 0 (0%) | 10 (100%) | 11.37 [0.64–200.93] | -[-] | 4 (40%) | 6 (60%) | 4.16 [1.07–16.14] | 3.28 [0.25–43.74] |
| Normal | 29 (34.9%) | 54 (65.1%) | 0.0972 | 0.9937 | 61 (73.5%) | 22 (26.5%) | 0.0394 * | 0.3678 |
| Calcium | ||||||||
| High | 4 (19%) | 17 (81%) | 2.26 [0.68–7.44] | 1.5 [0.41–5.51] | 11 (52.4%) | 10 (47.6%) | 2.72 [0.99–7.48] | 2.75 [0.41–18.33] |
| Normal | 25 (34.7%) | 47 (65.3%) | 0.1800 | 0.5400 | 54 (75%) | 18 (25%) | 0.0513 | 0.2944 |
| B2M | ||||||||
| High | 23 (29.5%) | 55 (70.5%) | 1.59 [0.51–4.99] | 0.95 [0.27–3.32] | 52 (66.7%) | 26 (33.3%) | 3.25 [0.68–15.49] | -[-] |
| Normal | 6 (40%) | 9 (60%) | 0.4234 | 0.9332 | 13 (86.7%) | 2 (13.3%) | 0.1390 | 0.9946 |
| Creatinine | ||||||||
| High | 7 (21.9%) | 25 (78.1%) | 2.01 [0.75–5.41] | 0.6 [0.16–2.17] | 18 (56.2%) | 14 (43.7%) | 2.61 [1.04–6.54] | 0.76 [0.14–4.16] |
| Normal | 22 (36.1%) | 39 (63.9%) | 0.1645 | 0.4355 | 47 (77%) | 14 (23%) | 0.0406 * | 0.7545 |
| eGFR | ||||||||
| Low | 15 (24.6%) | 46 (75.4%) | 2.38 [0.96–5.92] | 1.51 [0.55–4.13] | 41 (67.2%) | 20 (32.8%) | 1.46 [0.56–3.83] | 1.65 [0.33–8.28] |
| Normal | 14 (43.7%) | 18 (56.2%) | 0.0611 | 0.4221 | 24 (75%) | 8 (25%) | 0.4380 | 0.5425 |
| del 17p/TP53 | ||||||||
| Present | 2 (12.5%) | 14 (87.5%) | 4.25 [0.86–21.04] | 1.47 [0.63–3.41] | 8 (50%) | 8 (50%) | 4.62 [1.33–16.02] | 5.2 [1.17–23.21] |
| Absent | 17 (37.8%) | 28 (62.2%) | 0.0762 | 0.3678 | 37 (82.2%) | 89 (17.8%) | 0.0157 * | 0.0307 * |
| No data: n = 32 | ||||||||
| t(4;14) IGH/FGFR3 | ||||||||
| Present | 4 (40%) | 6 (60%) | 0.63 [0.16–2.50] | 0.51 [0.11–2.27] | 7 (70%) | 3 (30%) | 1.11 [0.26–4.79] | 1.92 [0.24–15.49] |
| Absent | 18 (29.5%) | 43 (70.5%) | 0.5085 | 0.3736 | 44 (72.1%) | 17 (27.9%) | 0.8896 | 0.5393 |
| No data: n = 22 | ||||||||
| t(11;14) IGH/CCND1 | ||||||||
| Present | 4 (50%) | 4 (50%) | 0.40 [0.09–1.77] | 0.46 [0.09–2.27] | 6 (75%) | 2 (25%) | 0.83 [0.15–4.52] | 0.45 [0.04–5.54] |
| Absent | 18 (28.6%) | 45 (71.4%) | 0.2280 | 0.3404 | 45 (71.4%) | 18 (28.6%) | 0.8326 | 0.5366 |
| No data: n = 22 | ||||||||
| t(14;16) IGH/MAF | ||||||||
| Present | 1 (100%) | 0 (0%) | 0.14 [0.005–3.70] | -[-] | 1 (100%) | 0 (0%) | 0.82 [0.03–20.99] | -[-] |
| Absent | 21 (30%) | 49 (70%) | 0.2424 | 0.9920 | 50 (71.4%) | 20 (28.6%) | 0.9052 | 0.9950 |
| No data: n = 22 | ||||||||
| Other IGH rearrangement | ||||||||
| Present | 2 (18.2%) | 9 (81.8%) | 2.25 [0.44–11.41] | 2.05 [0.38–11.03] | 7 (63.6%) | 4 (36.4%) | 1.57 [0.40–6.09] | 1.1 [0.2–6.01] |
| Absent | 20 (33.3%) | 40 (66.7%) | 0.3276 | 0.4008 | 44 (73.3%) | 16 (26.7%) | 0.5133 | 0.9094 |
| No data: n = 22 | ||||||||
| IL1B genotype (rs1143634) | ||||||||
| CC | 15 (29.4%) | 36 (70.6%) | 1.20 [0.50–2.89] | 0.93 [0.35–2.48] | 27 (52.9%) | 24 (47.1%) | 8.44 [2.63–27.15] | 5.11 [1.25–20.92] |
| TT or TC | 14 (33.3%) | 28 (66.7%) | 0.6847 | 0.8935 | 38 (90.5%) | 4 (9.5%) | 0.0003 * | 0.0233 * |
| IL1B genotype (rs1143634) | ||||||||
| TT | 2 (15.4%) | 11 (84.6%) | 2.80 [0.58–13.55] | 3.87 [0.73–20.43] | 11 (84.6%) | 2 (15.4%) | 0.38 [0.08–1.83] | 2.81 [0.31–25.77] |
| TC or CC | 27 (33.7%) | 53 (66.2%) | 0.2002 | 0.1110 | 54 (67.5%) | 26 (32.5%) | 0.2263 | 0.3607 |
| IL-1β plasma level [pg/mL] | ||||||||
| Low | 17 (36.2%) | 30 (63.8%) | 0.62 [0.256–1.51] | 1.37 [0.52–3.59] | 42 (89.4%) | 5 (10.6%) | 8.40 [2.82–25.05] | 7.76 [1.66–36.31] |
| High | 12 (26.1%) | 34 (73.9%) | 0.2956 | 0.5229 | 23 (50%) | 23 (50%) | 0.0001 * | 0.0092 * |
| Variable | Progression Free Survival | Overall Survival | ||||
|---|---|---|---|---|---|---|
| Univariable | Multivariable | Univariable | Multivariable | |||
| mPFS (Months) 18 | HR (95% CI) p | HR (95% CI) p | mOS (Months) 25 | HR (95% CI) p | HR (95% CI) p | |
| Gender | ||||||
| Men | 24 | 1.7 (1.01–2.87) | 1.38 (0.8–2.39) | 34 | 1.50 (0.81–2.77) | 1.5 (0.8–2.81) |
| Women | 25 | 0.0402 * | 0.2485 | 47 | 0.1911 | 0.2125 |
| Age | ||||||
| ≥65 | 17 | 1.49 (0.89–2.50) | 0.63 (0.33–1.21) | 38 | 1.30 (0.71–2.41) | 1.17 (0.62–2.2) |
| <65 | 30 | 0.1287 | 0.1687 | 45 | 0.3971 | 0.6286 |
| Diagnosis | ||||||
| MM with a monoclonal component | 25 | 0.64 (0.2–1.48) | 0.43 (0.16–1.19) | 47 | 0.33 (0.12–0.93) | 0.30 (0.13–0.72) |
| Light chain disease | 15 | 0.2082 | 0.1056 | 16 | 0.0011 * | 0.0076 * |
| Monoclonal protein class | ||||||
| IgA | 24 | 1.10 (0.59–2.02) | 0.79 (0.40–1.58) | 47 | 0.62 (0.29–1.33) | 0.49 (0.23–1.07) |
| IgG | 25 | 0.7522 | 0.5162 | 45 | 0.1780 | 0.0766 |
| N/a: n = 11 | ||||||
| Light chain type | ||||||
| Lambda | 17 | 1.10 (0.64–1.90) | 1.12 (0.65–1.92) | 33 | 1.57 (0.82–3) | 1.38 (0.73–2.62) |
| Kappa | 26 | 0.7174 | 0.6845 | 48 | 0.1446 | 0.3214 |
| ISS stage | ||||||
| 3 | 17 | 1.39 (0.8–2.40) | 1.22 (0.58–2.56) | 33 | 1.3 (0.68–2.46) | 0.96 (0.8–1.86) |
| 1, 2 | 26 | 0.2114 | 0.6018 | 45 | 0.4061 | 0.9156 |
| Renal function | ||||||
| B | 15 | 1.75 (0.79–3.89) | 1.1 (0.56–2.17) | 24 | 1.82 (0.74–4.48) | 0.9 (0.39–2.09) |
| A | 25 | 0.0854 | 0.7773 | 45 | 0.1018 | 0.8109 |
| Stage of chronic kidney disease | ||||||
| G3a, G3b,G4,G5D | 13 | 2.42 (1.36–4.31) | 1.53 (0.85–2.74) | 25 | 2.08 (1.09–3.98) | 1.33 (0.68–2.62) |
| G1, G2 | 34 | 0.0004 * | 0.1568 | 47 | 0.0153 * | 0.4091 |
| Performance status | ||||||
| 2–4 | 25 | 1.25 (0.75–2.11) | 0.73 (0.41–1.27) | 33 | 1.7 (0.92–3.15) | 1.15 (0.6–2.21) |
| 0, 1 | 24 | 0.3872 | 0.2653 | 47 | 0.0849 | 0.6632 |
| Treatment protocol (1) | ||||||
| CTD | 24 | 1.68 (0.67–2.04) | 1.01 (0.57–1.79) | 33 | 0.96 (0.51–1.8) | 0.83 (0.42–1.63) |
| V(C)D, VTD | 25 | 0.5624 | 0.9609 | 43 | 0.8874 | 0.5900 |
| Treatment protocol (2) | ||||||
| VTD | 36 | 0.51 (0.29–0.90) | 0.74 (0.37–1.47) | 38 | 1.03 (0.51–2.07) | 1.16 (0.57–2.38) |
| CTD, V(C)D | 17 | 0.0387 * | 0.3923 | 47 | 0.9232 | 0.6752 |
| aHSCT | ||||||
| Yes | 42 | 0.37 (0.22–0.62) | 0.43 (0.24–0.79) | 45 | 0.60 (0.32–1.1) | 0.66 (0.34–1.26) |
| No | 15 | 0.0005 * | 0.0065 * | 38 | 0.1094 | 0.2111 |
| Body weight loss before treatment | ||||||
| Yes | 18 | 1.46 (0.87–2.45) | 1.09 (0.62–1.91) | 33 | 1.36 (0.73–2.52) | 1.3 (0.67–2.53) |
| No | 34 | 0.1471 | 0.7602 | 46 | 0.3181 | 0.4368 |
| Anemia before treatment (WHO) | ||||||
| Yes | 18 | 2.21 (1.2–4.07) | 1.11 (0.45–2.75) | 38 | 1.84 (0.92–3.65) | 1.64 (0.55–4.88) |
| No | - | 0.0385 * | 0.8137 | 48 | 0.1263 | 0.3719 |
| Platelets | ||||||
| Low | 12 | 1.82 (0.78–4.24) | 1.2 (0.59–2.43) | 24 | 1.60 (0.65–3.94) | 1.28 (0.58–2.83) |
| Normal | 25 | 0.0753 | 0.6086 | 43 | 0.2253 | 0.5421 |
| Albumins | ||||||
| Low | 12 | 2.69 (1.49–4.84) | 2.4 (1.39–4.14) | 28 | 2.68 (1.33–5.37) | 3.14 (1.64–6.02) |
| Normal | 37 | 0.0001 * | 0.0017 * | 48 | 0.0007 * | 0.0006* |
| CRP | ||||||
| High | 24 | 1.48 (0.85–2.58) | 1.05 (0.59–1.86) | 30 | 1.79 (0.92–3.49) | 1.51 (0.81–2.84) |
| Normal | 26 | 0.1268 | 0.8611 | 46 | 0.0560 | 0.1973 |
| LDH | ||||||
| High | 12 | 1.71 (0.68–4.29) | 1.19 (0.56–2.54) | 22 | 1.55 (0.51–4.74) | 0.85 (0.32–2.56) |
| Normal | 25 | 0.1468 | 0.6489 | 45 | 0.3468 | 0.7491 |
| Calcium | ||||||
| High | 25 | 1.24 (0.66–2.31) | 1.21 (0.67–2.19) | 25 | 1.62 (0.46–3.45) | 1.37 (0.68–2.77) |
| Normal | 25 | 0.4689 | 0.5284 | 46 | 0.1541 | 0.3748 |
| B2M | ||||||
| High | 24 | 1.85 (0.97–3.52) | 1.32 (0.59–2.96) | 38 | 1.76 (0.85–3.64) | 1.34 (0.55–3.27) |
| Normal | - | 0.1093 | 0.4951 | 52 | 0.1826 | 0.5242 |
| Creatinine | ||||||
| High | 14 | 2.21 (1.25–3.92) | 1.5 (0.87–2.59) | 28 | 2.04 (1.06–3.95) | 1.2 (0.6–2.4) |
| Normal | 34 | 0.0017 * | 0.1499 | 47 | 0.0182 * | 0.6110 |
| eGFR | ||||||
| Low | 24 | 1.76 (1.02–3.05) | 1.24 (0.66–2.33) | 41 | 1.29 (0.67–2.49) | 0.96 (0.47–1.96) |
| Normal | - | 0.0634 | 0.5088 | 45 | 0.4557 | 0.9055 |
| del 17p/TP53 | ||||||
| Present | 15 | 1.69 (0.78–3.68) | 1.23 (0.82–1.84) | 30 | 1.97 (0.68–5.71) | 1.03 (0.6–1.78) |
| Absent | 25 | 0.1191 | 0.3247 | 52 | 0.1106 | 0.9128 |
| No data: n = 32 | ||||||
| t(4;14) IGH/FGFR3 | ||||||
| Present | 42 | 0.87 (0.36–2.12) | 1.32 (0.47–3.65) | - | 1.26 (0.40–4.02) | 1.52 (0.51–4.55) |
| Absent | 24 | 0.7707 | 0.5994 | 46 | 0.6608 | 0.4514 |
| No data: n = 22 | ||||||
| t(11;14) IGH/CCND1 | ||||||
| Present | 9 | 1.61 (0.57–4.56) | 1.56 (0.66–3.7) | 46 | 1.08 (0.36–3.22) | 1.16 (0.39–3.43) |
| Absent | 25 | 0.2657 | 0.3166 | 45 | 0.8852 | 0.7950 |
| No data: n = 22 | ||||||
| t(14;16) IGH/MAF | ||||||
| Present | 9 | 4.68 (0.07–315.0.4) | 2.2 (0.29–17.08) | 28 | 3.3 (0.1–113.16) | 2.35 (0.3–18.53) |
| Absent | 25 | 0.0872 | 0.4534 | 46 | 0.2082 | 0.4189 |
| No data: n = 22 | ||||||
| Other IGH rearrangement | ||||||
| Present | 17 | 1.13 (0.48–2.64) | 1.08 (0.48–2.46) | - | 1.15 (0.38–3.51) | 1.42 (0.48–4.19) |
| Absent | 25 | 0.7620 | 0.8487 | 45 | 0.7936 | 0.5287 |
| No data: n = 22 | ||||||
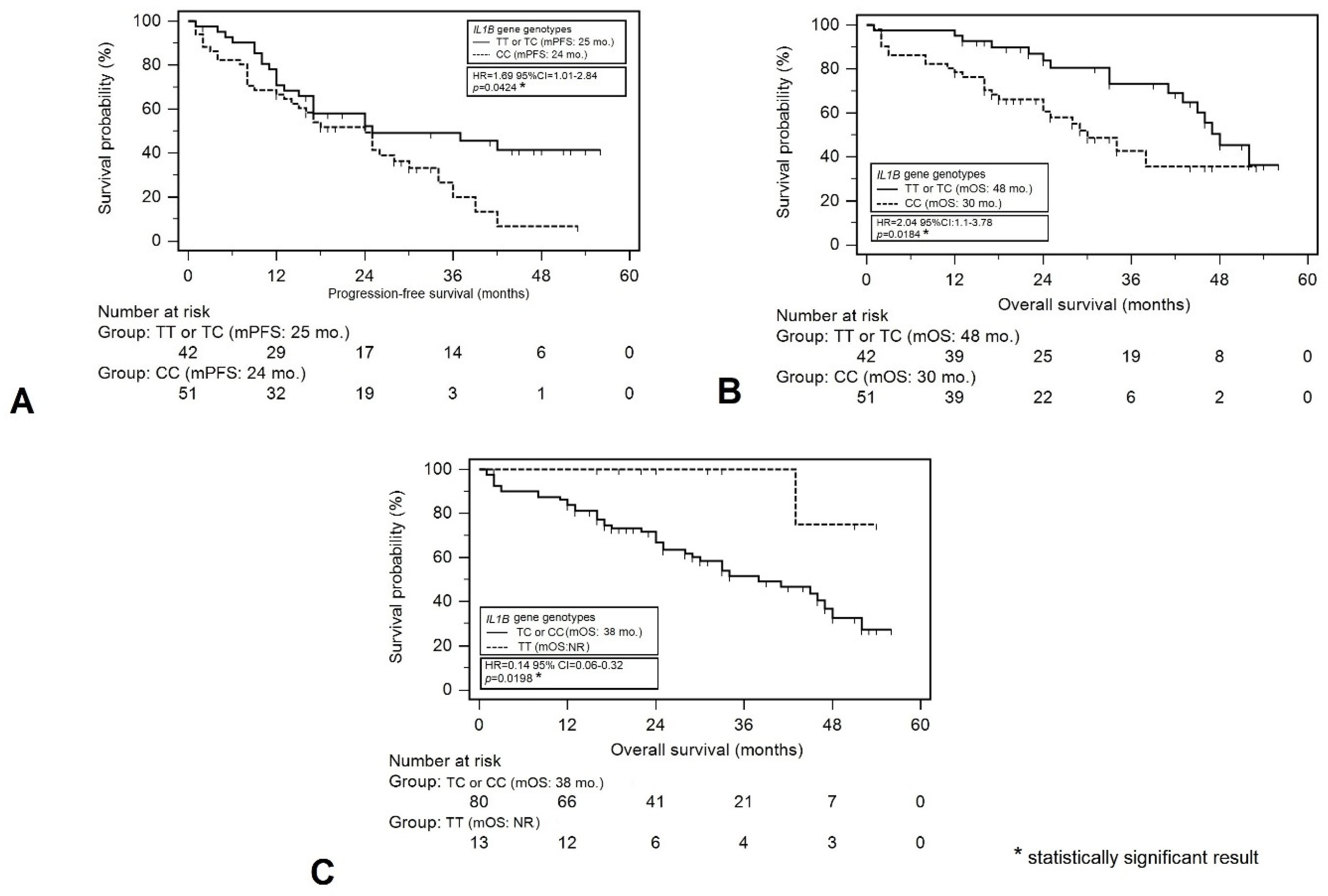
| IL1B genotype (rs1143634) | ||||||
| CC | 24 | 1.69 (1.01–2.84) | 1.28 (0.72–2.27) | 30 | 2.04 (1.1–3.78) | 2.03 [1.06–3.88] |
| TT or TC | 25 | 0.0424 * | 0.4064 | 48 | 0.0184 * | 0.0337 * |
| IL1B genotype (rs1143634) | ||||||
| TT | - | 0.60 (0.28–1.26) | 0.76 (0.3–1.92) | - | 0.14 (0.06–0.32) | 0.16 (0.02–1.15) |
| TC or CC | 24 | 0.2549 | 0.5695 | 38 | 0.0198 * | 0.0700 |
| IL-1β plasma level [pg/mL] | ||||||
| Low | 25 | 0.89 (0.53–1.5) | 0.97 (0.57–1.66) | 46 | 0.70 (0.38–1.31) | 1.20 (0.64–2.23) |
| High | 24 | 0.6655 | 0.9164 | 34 | 0.2507 | 0.5724 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazurek, M.; Szudy-Szczyrek, A.; Homa-Mlak, I.; Hus, M.; Małecka-Massalska, T.; Mlak, R. IL1B Polymorphism (rs1143634) and IL-1β Plasma Concentration as Predictors of Nutritional Disorders and Prognostic Factors in Multiple Myeloma Patients. Cancers 2024, 16, 1263. https://doi.org/10.3390/cancers16071263
Mazurek M, Szudy-Szczyrek A, Homa-Mlak I, Hus M, Małecka-Massalska T, Mlak R. IL1B Polymorphism (rs1143634) and IL-1β Plasma Concentration as Predictors of Nutritional Disorders and Prognostic Factors in Multiple Myeloma Patients. Cancers. 2024; 16(7):1263. https://doi.org/10.3390/cancers16071263
Chicago/Turabian StyleMazurek, Marcin, Aneta Szudy-Szczyrek, Iwona Homa-Mlak, Marek Hus, Teresa Małecka-Massalska, and Radosław Mlak. 2024. "IL1B Polymorphism (rs1143634) and IL-1β Plasma Concentration as Predictors of Nutritional Disorders and Prognostic Factors in Multiple Myeloma Patients" Cancers 16, no. 7: 1263. https://doi.org/10.3390/cancers16071263
APA StyleMazurek, M., Szudy-Szczyrek, A., Homa-Mlak, I., Hus, M., Małecka-Massalska, T., & Mlak, R. (2024). IL1B Polymorphism (rs1143634) and IL-1β Plasma Concentration as Predictors of Nutritional Disorders and Prognostic Factors in Multiple Myeloma Patients. Cancers, 16(7), 1263. https://doi.org/10.3390/cancers16071263







