Role of Non-Coding RNAs in Diagnosis, Prediction and Prognosis of Multiple Myeloma
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Multiple Myeloma
1.2. Diagnostic and Prognostic Approaches for MM
1.3. miRNA
2. The Role of miRNA Expression in Diagnosis and Prognosis of MM
2.1. The Role of miRNA as a Potential Diagnostic Factor for MM
2.1.1. Studies Showing the Role of Increased miRNA Expression
2.1.2. Studies Showing the Role of Decreased miRNA Expression
2.1.3. Studies Showing the Role of Both Increased and Decreased miRNA Expression
2.1.4. Other Non-Coding RNA
2.2. The Role of miRNAs as a Potential Predictive and Prognostic Factor for MM
2.2.1. Studies Showing the Role of the Increased miRNA Expression
2.2.2. Studies Showing the Role of the Decreased miRNA Expression
2.2.3. Other Non-Coding RNA
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Soliman, A.M.; Lin, T.S.; Mahakkanukrauh, P.; Das, S. Role of microRNAs in Diagnosis, Prognosis and Management of Multiple Myeloma. Int. J. Mol. Sci. 2020, 21, 7539. [Google Scholar] [CrossRef] [PubMed]
- Teras, L.R.; DeSantis, C.E.; Cerhan, J.R.; Morton, L.M.; Jemal, A.; Flowers, C.R. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J. Clin. 2016, 66, 443–459. [Google Scholar] [CrossRef]
- Padala, S.A.; Barsouk, A.; Barsouk, A.; Rawla, P.; Vakiti, A.; Kolhe, R.; Kota, V.; Ajebo, G.H. Epidemiology, Staging, and Management of Multiple Myeloma. Med. Sci. 2021, 9, 3. [Google Scholar] [CrossRef]
- van de Donk, N.W.C.J.; Pawlyn, C.; Yong, K.L. Multiple myeloma. Lancet 2021, 397, 410–427. [Google Scholar] [CrossRef]
- Castaneda, O.; Baz, R. Multiple Myeloma Genomics—A Concise Review. Acta Med. Acad. 2019, 48, 57–67. [Google Scholar] [CrossRef]
- Kazandjian, D. Multiple myeloma epidemiology and survival: A unique malignancy. Semin. Oncol. 2016, 43, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Landgren, O. Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma: Biological insights and early treatment strategies. Hematol. Am. Soc. Hematol. Educ. Program 2013, 2013, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V.; Gupta, V.; Fonseca, R.; Dispenzieri, A.; Gonsalves, W.I.; Larson, D.; Ketterling, R.P.; Lust, J.A.; Kyle, R.A.; Kumar, S.K. Impact of primary molecular cytogenetic abnormalities and risk of progression in smoldering multiple myeloma. Leukemia 2013, 27, 1738–1744. [Google Scholar] [CrossRef]
- Firth, J. Haematology: Multiple myeloma. Clin. Med. 2019, 19, 58–60. [Google Scholar]
- Hillengass, J.; Fechtner, K.; Weber, M.A.; Bäuerle, T.; Ayyaz, S.; Heiss, C.; Hielscher, T.; Moehler, T.M.; Egerer, G.; Neben, K.; et al. Prognostic significance of focal lesions in whole-body magnetic resonance imaging in patients with asymptomatic multiple myeloma. J. Clin. Oncol. 2010, 28, 1606–1610. [Google Scholar] [CrossRef]
- Rajkumar, S.V.; Kumar, S. Multiple Myeloma: Diagnosis and Treatment. Mayo Clin. Proc. 2016, 91, 101–119. [Google Scholar] [CrossRef]
- Rajkumar, S.V. Multiple myeloma: 2022 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2022, 97, 1086–1107. [Google Scholar] [CrossRef]
- Liang, J.; Yang, Y.; He, P.; Mandizadza, O.O.; Zhang, W.; Lin, S.; Ji, C. Treatment of multiple myeloma based on autologous stem cell transplant: An overview of systematic reviews. Medicine 2023, 102, e35456. [Google Scholar] [CrossRef]
- Xu, L.; Wu, S. New diagnostic strategy for multiple myeloma: A review. Medicine 2023, 102, e36660. [Google Scholar] [CrossRef]
- Hanamura, I. Multiple myeloma with high-risk cytogenetics and its treatment approach. Int. J. Hematol. 2022, 115, 762–777. [Google Scholar] [CrossRef] [PubMed]
- Soliman, A.M.; Das, S.; Abd Ghafar, N.; Teoh, S.L. Role of microRNA in proliferation Phase of wound healing. Front. Genet. 2018, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct Target. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef]
- Rizk, N.I.; Midan, H.M.; Helal, G.K.; Abulsoud, A.I.; Elshaer, S.S.; El-Husseiny, A.A.; Fathi, D.; Abdelmaksoud, N.M.; Abdel Mageed, S.S.; Elballal, M.S.; et al. The emerging role of miRNAs in Merkel cell carcinoma pathogenesis: Signaling pathway crosstalk. Pathol. Res. Pract. 2023, 249, 154771. [Google Scholar] [CrossRef]
- Mazziotta, C.; Cervellera, C.F.; Lanzillotti, C.; Touzé, A.; Gaboriaud, P.; Tognon, M.; Martini, F.; Rotondo, J.C. MicroRNA dysregulations in Merkel cell carcinoma: Molecular mechanisms and clinical applications. J. Med. Virol. 2023, 95, e28375. [Google Scholar] [CrossRef] [PubMed]
- Pourhanifeh, M.H.; Mahjoubin-Tehran, M.; Shafiee, A.; Hajighadimi, S.; Moradizarmehri, S.; Mirzaei, H.; Asemi, Z. MicroRNAs and exosomes: Small molecules with big actions in multiple myeloma pathogenesis. IUBMB Life 2020, 72, 314–333. [Google Scholar] [CrossRef]
- Li, G.; Yin, J.; Wu, Z.; Li, S.; He, A.; Sun, Z. Expression level of miRNA in the peripheral blood of patients with multiple myeloma and its clinical significance. Am. J. Transl. Res. 2021, 13, 5343–5349. [Google Scholar]
- Szudy-Szczyrek, A.; Ahern, S.; Krawczyk, J.; Szczyrek, M.; Hus, M. MiRNA as a Potential Target for Multiple Myeloma Therapy-Current Knowledge and Perspectives. J. Pers. Med. 2022, 12, 1428. [Google Scholar] [CrossRef]
- Al-Masri, A.; Price-Troska, T.; Chesi, M.; Chung, T.H.; Kim, S.; Carpten, J.; Bergsagel, P.L.; Fonseca, R. MicroRNA Expression Analysis in Multiple Myeloma. Blood 2005, 106, 1554. [Google Scholar] [CrossRef]
- Chen, D.; Yang, X.; Liu, M.; Zhang, Z.; Xing, E. Roles of miRNA dysregulation in the pathogenesis of multiple myeloma. Cancer Gene Ther. 2021, 28, 1256–1268. [Google Scholar] [CrossRef]
- Kubiczkova, L.; Kryukov, F.; Slaby, O.; Dementyeva, E.; Jarkovsky, J.; Nekvindova, J.; Radova, L.; Greslikova, H.; Kuglik, P.; Vetesnikova, E.; et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica 2014, 99, 511–518. [Google Scholar] [CrossRef]
- Jiang, Y.; Luan, Y.; Chang, H.; Chen, G. The diagnostic and prognostic value of plasma microRNA-125b-5p in patients with multiple myeloma. Oncol. Lett. 2018, 16, 4001–4007. [Google Scholar] [CrossRef] [PubMed]
- Handa, H.; Murakami, Y.; Ishihara, R.; Kimura-Masuda, K.; Masuda, Y. The Role and Function of microRNA in the Pathogenesis of Multiple Myeloma. Cancers 2019, 11, 1738. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tian, D.; Chen, H.; Cai, Y.; Chen, S.; Duan, S. MicroRNA-490-3p and -490-5p in carcinogenesis: Separate or the same goal? Oncol. Lett. 2021, 22, 678. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Jing, X.; Li, Z.; Wu, X.; Gao, Z.; Ma, R. The diagnostic and prognostic values of circulating miRNA-1246 in multiple myeloma. Hematology 2022, 27, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Chen, T.; Wang, L.; Zhang, Z.; Xing, E. Circulating miR-448 acts as a potential diagnostic biomarker for multiple myeloma. Hematology 2022, 27, 847–852. [Google Scholar] [CrossRef]
- Fan, C.B.; Yan, X.H.; Tian, M.; Zhang, S.; Liu, J.L.; Sheng, Y.X.; Dong, L.; Zhang, W.L. Long non-coding RNA NEAT1 regulates Hodgkin’s lymphoma cell proliferation and invasion via miR-448 mediated regulation of DCLK1. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 6219–6227. [Google Scholar] [PubMed]
- Shen, X.; Ye, Y.; Qi, J.; Shi, W.; Wu, X.; Ni, H.; Cong, H.; Ju, S. Identification of a novel microRNA, miR-4449, as a potential blood based marker in multiple myeloma. Clin. Chem. Lab. Med. 2017, 55, 748–754. [Google Scholar] [CrossRef]
- Li, J.; Zhang, M.; Wang, C. Circulating miRNAs as diagnostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. J. Clin. Lab. Anal. 2020, 34, e23233. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Kumar, R.; Seth, T.; Garg, B.; Sati, H.C.; Sharma, A. Clinical significance of circulatory microRNA-203 in serum as novel potential diagnostic marker for multiple myeloma. J. Cancer Res. Clin. Oncol. 2019, 145, 1601–1611. [Google Scholar] [CrossRef] [PubMed]
- Wight, T.N.; Kang, I.; Evanko, S.P.; Harten, I.A.; Chang, M.Y.; Pearce, O.M.T.; Allen, C.E.; Frevert, C.W. Versican-A Critical Extracellular Matrix Regulator of Immunity and Inflammation. Front. Immunol. 2020, 11, 512. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xu, Y.; Deng, S.; Li, Z.; Zou, D.; Yi, S.; Sui, W.; Hao, M.; Qiu, L. MicroRNA-15a/16-1 cluster located at chromosome 13q14 is down-regulated but displays different expression pattern and prognostic significance in multiple myeloma. Oncotarget 2015, 6, 38270–38282. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Chen, H.; Zhang, X.; Pan, Y.; Jing, R.; Shen, L.; Wang, X.; Ju, S.; Jin, C.; Cong, H. Serum miR-30d as a novel biomarker for multiple myeloma and its antitumor role in U266 cells through the targeting of the MTDH/PI3K/Akt signaling pathway. Int. J. Oncol. 2018, 53, 2131–2144. [Google Scholar] [CrossRef]
- Papanota, A.M.; Karousi, P.; Kontos, C.K.; Artemaki, P.I.; Liacos, C.; Papadimitriou, M.A.; Bagratuni, T.; Eleutherakis-Papaiakovou, E.; Malandrakis, P.; Ntanasis-Stathopoulos, I.; et al. A Cancer-Related microRNA Signature Shows Biomarker Utility in Multiple Myeloma. Int. J. Mol. Sci. 2021, 22, 13144. [Google Scholar] [CrossRef]
- Li, J.; Wang, C.; Meng, Q.; Hu, Z.; Hu, M.; Zhang, M. MicroRNAs in urine as diagnostic biomarkers for multiple myeloma. Int. J. Lab. Hematol. 2021, 43, 227–234. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, L.; Xiang, B.; Zhu, H.; Wu, Y.; Chen, M.; Guan, P.; Zou, X.; Valencia, C.A.; Dong, B.; et al. Potential role of exosome-associated microRNA panels and in vivo environment to predict drug resistance for patients with multiple myeloma. Oncotarget 2016, 7, 30876–30891. [Google Scholar] [CrossRef]
- Besse, L.; Sedlarikova, L.; Kryukov, F.; Nekvindova, J.; Radova, L.; Slaby, O.; Kuglik, P.; Almasi, M.; Penka, M.; Krejci, M.; et al. Circulating Serum MicroRNA-130a as a Novel Putative Marker of Extramedullary Myeloma. PLoS ONE 2015, 10, e0137294. [Google Scholar] [CrossRef]
- Guo, B.; Xiao, C.; Liu, Y.; Zhang, N.; Bai, H.; Yang, T.; Xiang, Y.; Nan, Y.; Li, Q.; Zhang, W.; et al. miR-744-5p Inhibits Multiple Myeloma Proliferation, Epithelial Mesenchymal Transformation and Glycolysis by Targeting SOX12/Wnt/β-Catenin Signaling. Onco Targets Ther. 2021, 14, 1161–1172. [Google Scholar] [CrossRef]
- Federico, C.; Sacco, A.; Belotti, A.; Ribolla, R.; Cancelli, V.; Giacomini, A.; Ronca, R.; Chiarini, M.; Imberti, L.; Marini, M.; et al. Circulating microRNAs and Their Role in Multiple Myeloma. Noncoding RNA 2019, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Zang, M.; Zhao, L.; Deng, S.; Xu, Y.; Qi, F.; An, G.; Qin, Y.; Sui, W.; Li, F.; et al. Serum high expression of miR-214 and miR-135b as novel predictor for myeloma bone disease development and prognosis. Oncotarget 2016, 7, 19589–19600. [Google Scholar] [CrossRef]
- Tavakoli Pirzaman, A.; Ebrahimi, P.; Hasanpour, A.H.; Shakeri, M.; Babajani, B.; Pourali Ganji, Z.; Babaei, H.; Rahmati, A.; Hosseinzadeh, R.; Doostmohamadian, S.; et al. miRNAs and Multiple Myeloma: Focus on the Pathogenesis, Prognosis, and Drug Resistance. Technol. Cancer Res. Treat. 2023, 22, 15330338231202391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Li, Y.C.; Geng, C.Y.; Zhou, H.X.; Gao, W.; Chen, W.M. Serum exosomal microRNAs as novel biomarkers for multiple myeloma. Hematol. Oncol. 2019, 37, 409–417. [Google Scholar] [CrossRef]
- Yang, C.; Liang, Y.; Shu, J.; Wang, S.; Hong, Y.; Chen, K.; Sun, M. Long non-coding RNAs in multiple myeloma (Review). Int. J. Oncol. 2023, 62, 69. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Cicero, N.; Tonacci, A.; Musolino, C.; Gangemi, S. Circular RNA as a Novel Biomarker for Diagnosis and Prognosis and Potential Therapeutic Targets in Multiple Myeloma. Cancers 2022, 14, 1700. [Google Scholar] [CrossRef]
- Zhu, C.; Guo, A.; Sun, B.; Zhou, Z. Comprehensive elaboration of circular RNA in multiple myeloma. Front. Pharmacol. 2022, 13, 971070. [Google Scholar] [CrossRef]
- Xie, B.; Li, L.; Zhang, Z.; Zhao, L.; Cheng, J.; Zhou, C.; Cheng, J.; Yan, J.; Chen, J.; Yi, J.; et al. MicroRNA-1246 by Targeting AXIN2 and GSK-3β Overcomes Drug Resistance and Induces Apoptosis in Chemo-resistant Leukemia Cells. J. Cancer 2021, 12, 4196–4208. [Google Scholar] [CrossRef]
- Torii, C.; Maishi, N.; Kawamoto, T.; Morimoto, M.; Akiyama, K.; Yoshioka, Y.; Minami, T.; Tsumita, T.; Alam, M.T.; Ochiya, T.; et al. miRNA-1246 in extracellular vesicles secreted from metastatic tumor induces drug resistance in tumor endothelial cells. Sci. Rep. 2021, 11, 13502. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Li, X.; Wang, W.; He, W.; Wang, J.; Wang, Y. Expression of Peripheral Blood miRNA-720 and miRNA-1246 Can Be Used as a Predictor for Outcome in Multiple Myeloma Patients. Clin. Lymphoma Myeloma Leuk. 2017, 17, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Szudy-Szczyrek, A.; Mlak, R.; Mielnik, M.; Mazurek, M.; Chocholska, S.; Podgajna, M.; Szczyrek, M.; Homa-Mlak, I.; Małecka-Massalska, T.; Hus, M. Circulating Serum MiRNA-8074 as a Novel Prognostic Biomarker for Multiple Myeloma. Cells 2022, 11, 752. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martín, S.; Soucek, L. MYC inhibitors in multiple myeloma. Cancer Drug Resist. 2021, 4, 842–865. [Google Scholar] [CrossRef] [PubMed]
- Roseth Aass, K.; Nedal, T.M.V.; Anshushaug Bouma, S.; Tryggestad, S.S.; Haukås, E.; Slørdahl, T.S.; Waage, A.; Standal, T.; Mjelle, R. Comprehensive small RNA-sequencing of primary myeloma cells identifies miR-105-5p as a predictor of patient survival. Br. J. Cancer 2023, 128, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Miliotis, C.; Slack, F.J. miR-105-5p regulates PD-L1 expression and tumor immunogenicity in gastric cancer. Cancer Lett. 2021, 518, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, M.A.; Soureas, K.; Papanota, A.M.; Tsiakanikas, P.; Adamopoulos, P.G.; Ntanasis-Stathopoulos, I.; Malandrakis, P.; Gavriatopoulou, M.; Sideris, D.C.; Kastritis, E.; et al. miRNA-seq identification and clinical validation of CD138+ and circulating miR-25 in treatment response of multiple myeloma. J. Transl. Med. 2023, 21, 245. [Google Scholar] [CrossRef] [PubMed]
- Mehlich, D.; Garbicz, F.; Włodarski, P.K. The emerging roles of the polycistronic miR-106b∼25 cluster in cancer—A comprehensive review. Biomed. Pharmacother. 2018, 107, 1183–1195. [Google Scholar] [CrossRef]
- Du, L.; Liu, W.; Aldana-Masangkay, G.; Pozhnitkov, A.; Pichiorri, F.; Chen, Y.; Rosen, S.T. SUMOylation inhibition enhances dexamethasone sensitivity in multiple myeloma. J. Exp. Clin. Cancer Res. 2022, 41, 8. [Google Scholar] [CrossRef]
- Manier, S.; Liu, C.J.; Avet-Loiseau, H.; Park, J.; Shi, J.; Campigotto, F.; Salem, K.Z.; Huynh, D.; Glavey, S.V.; Rivotto, B.; et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood 2017, 129, 2429–2436. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Díaz, T.; Tovar, N.; Pedrosa, F.; Tejero, R.; Cibeira, M.T.; Magnano, L.; Rosiñol, L.; Monzó, M.; Bladé, J.; et al. A serum microRNA signature associated with complete remission and progression after autologous stem-cell transplantation in patients with multiple myeloma. Oncotarget 2015, 6, 1874–1883. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Shen, L.; Yang, H.; Gong, H.; Du, X. Circular RNA circPSAP functions as an efficient miR-331-3p sponge to regulate proliferation, apoptosis and bortezomib sensitivity of human multiple myeloma cells by upregulating HDAC4. J. Pharmacol. Sci. 2022, 149, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Buranjiang, G.; Kuerban, R.; Abuduwanke, A.; Li, X.; Kuerban, G. MicroRNA-331-3p inhibits proliferation and metastasis of ovarian cancer by targeting RCC2. Arch. Med. Sci. 2019, 15, 1520–1529. [Google Scholar] [CrossRef]
- Carrasco-Leon, A.; Ezponda, T.; Meydan, C.; Valcárcel, L.V.; Ordoñez, R.; Kulis, M.; Garate, L.; Miranda, E.; Segura, V.; Guruceaga, E.; et al. Characterization of complete lncRNAs transcriptome reveals the functional and clinical impact of lncRNAs in multiple myeloma. Leukemia 2021, 35, 1438–1450. [Google Scholar] [CrossRef]
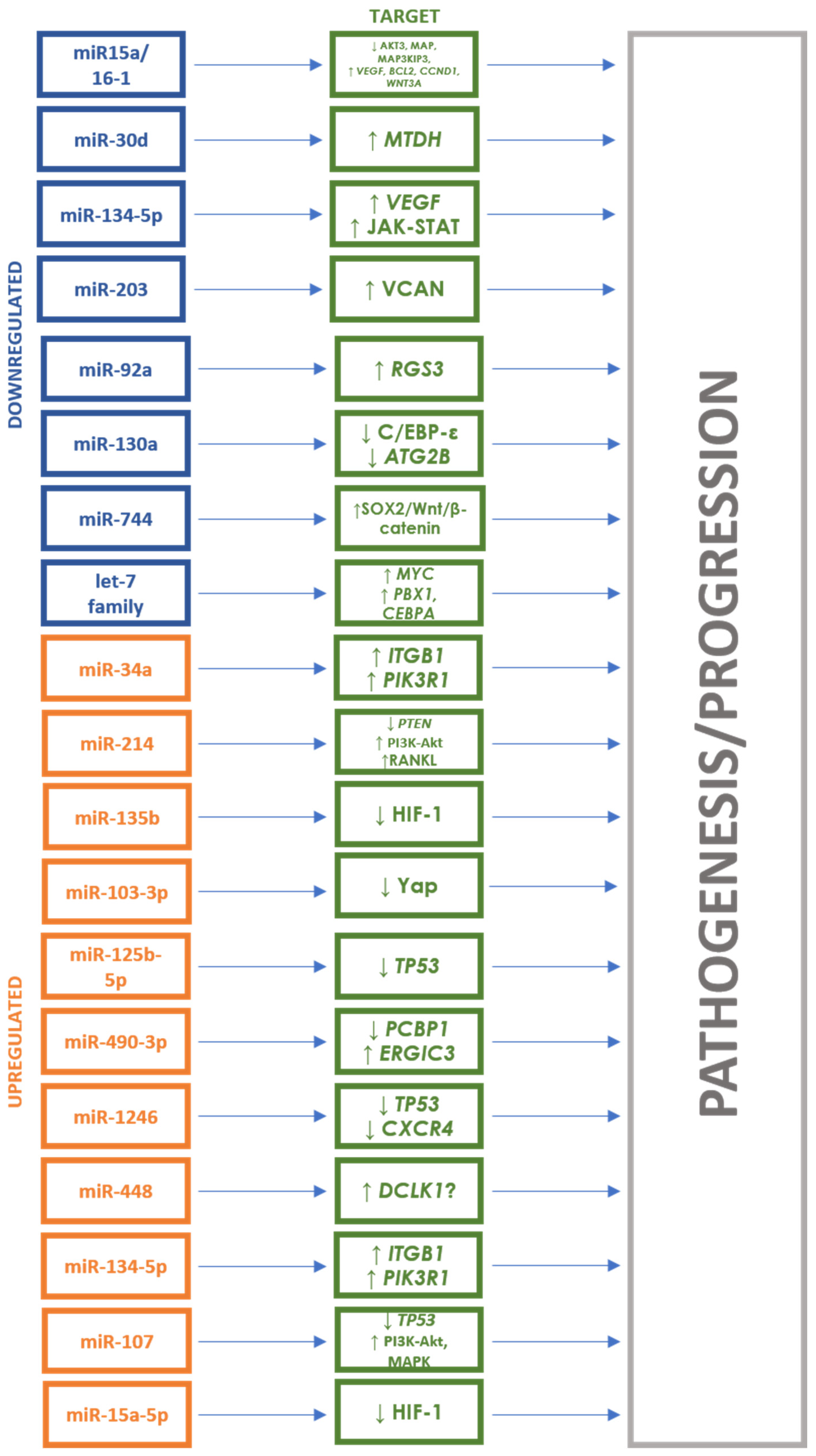
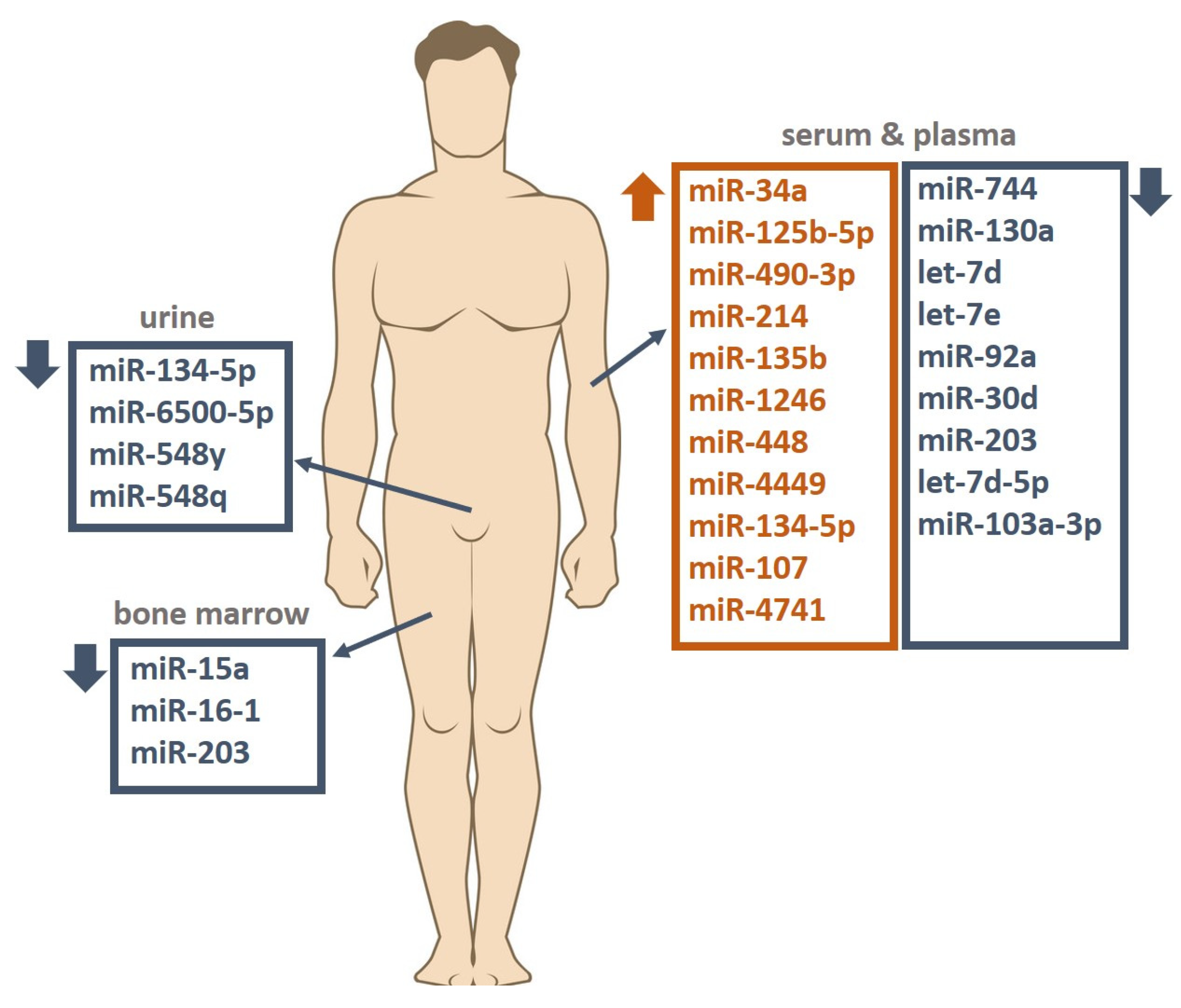
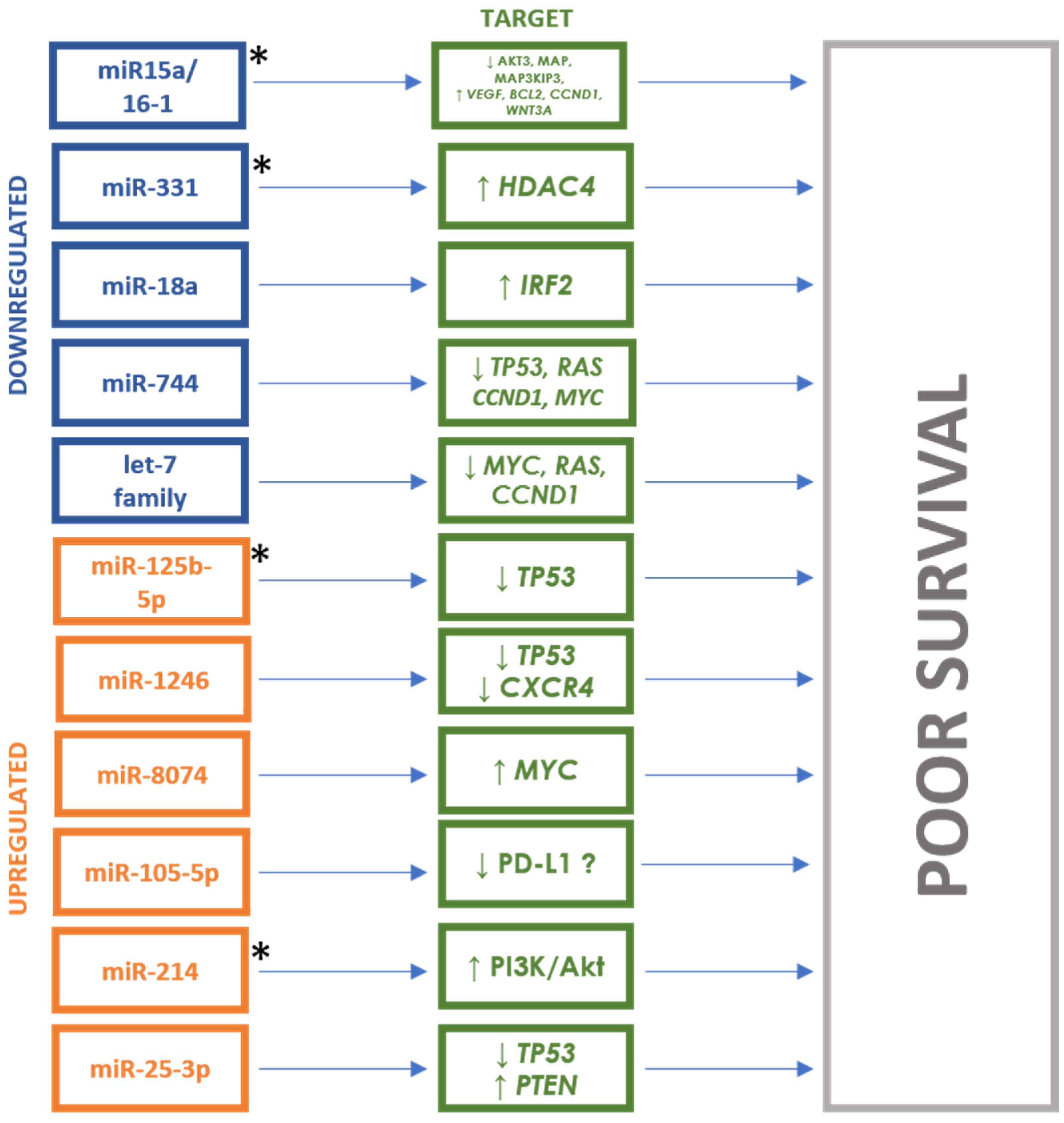
| Authors | Study Group | Race/ Nationality | Study Material | Studied miRNA | miRNA Change | miRNA Targets |
|---|---|---|---|---|---|---|
| Kubiczkova et al. (2014) [26] | Newly diagnosed MM (n = 103), patients in relapse (n = 18), MGUS (n = 57), healthy donors (n = 30) | Czech | Serum | miR-744 | Downregulated | TUBB4, APC2, JUNB |
| miR-130a | IGF1, CCND2, TGFβ | |||||
| let-7d | APC2, TGFβRI, CDC25A, TP53 | |||||
| let-7e | MAPK6, IGF1, MYCN, CDK6, APC2, TP53 | |||||
| miR-34a | Upregulated | MYCN, E2F3, BCL2, CDK6 | ||||
| Jiang et al. (2018) [27] | MM (n = 35), healthy donors (n = 20) | China | Plasma | miR-125b-5p | Upregulated | TP53, IRF4 |
| miR-490-3p | ERGIC3, PCBP1 | |||||
| Zhao et al. (2022) [30] | MM (n = 90), healthy donors (n = 30) | China | Serum | miR-1246 | Upregulated | TP53, CXCR4 |
| Chen et al. (2022) [31] | Newly diagnosed MM (n = 30), patients in relapse (n = 30), patients in remission (n = 30), healthy donors (n = 40) | China | Serum | miR-448 | Upregulated | - |
| Shen et al. (2017) [33] | Newly diagnosed MM (n = 71), healthy donors (n = 46) | China | Serum | miR-4449 | Upregulated | - |
| Gupta et al. (2019) [35] | Newly diagnosed MM (n = 30), healthy donors (n = 30) | India | Bone marrow, serum | miR-203 | Downregulated | VCAN |
| Li et al. (2020) [34] | MM (n = 23), MGUS (n = 16), healthy donors (n = 18) | China | Serum | miR-134-5p | Upregulated | ITGB1, PIK3R1 |
| Li et al. (2015) [37] | Newly diagnosed MM (n = 90), patients in remission (n = 16), patients in relapse (n = 11), healthy donors (n = 19) | China | Bone marrow | miR-15a | Downregulated | BCL2, MCL1, ETS1, JUN, TP53 |
| miR-16-1 | BCL2, MCL1, CCND1, WNT3A, VEGF | |||||
| Zhu et al. (2018) [38] | Newly diagnosed MM (n = 81), healthy donors (n = 78) | China | Serum | miR-30d | Downregulated | MTDH |
| Li et al. (2020) [40] | Newly diagnosed MM (n = 12), patients in relapse (n = 6), patients in remission (n = 9), healthy donors (n = 12) | China | Serum, urine | miR-134-5p | VEGF, JAK-STAT, CML pathway | |
| miR-6500-5p | Downregulated | Lysosomal pathway | ||||
| miR-548q | MAPK, RAS, PI3K-Akt, Hippo | |||||
| miR-548y | MAPK, RAS, PI3K-Akt, Hippo | |||||
| Hao et al. (2016) [45] | Newly diagnosed MM (n = 108), healthy donors (n = 44) | China | Serum | miR-214 | Upregulated | FBXW7, PTEN, AKT, GSK3 |
| miR-135b | GSK3, FIH1 | |||||
| miR-92a | Downregulated | RGS3 | ||||
| Zhang et al. (2019) [47] | MM (n = 20), sMM (n = 20), healthy donors (n = 16) | China | Serum | let-7d-5p | Downregulated | MYC |
| miR-103a-3p | YAP, Hippo | |||||
| miR-4741 | Upregulated | - |
| Authors | Study Group | Race/ Nationality | Study Material | Assessed Study Endpoints (Associated Change) | Studied miRNA | miRNA Change | miRNA Targets |
|---|---|---|---|---|---|---|---|
| Kubiczkova et al. (2014) [26] | Newly diagnosed MM (n = 103), patients in relapse (n = 18), MGUS (n = 57), healthy donors (n = 30) | Czech | Serum | 1-year survival rate (↓), TTP (↓) | miR-744 | Downregulated | TUBB4, APC2, JUNB |
| let-7e | MAPK6, IGF1, MYCN, CDK6, APC2, TP53 | ||||||
| Jiang et al. (2018) [27] | MM (n = 35), healthy donors (n = 20) | China | Plasma | EFS (↓) | miR-125b-5p | Upregulated | TP53, IRF4 |
| Zhao et al. (2022) [31] | MM (n = 90), healthy donors (n = 30) | China | Serum | PFS (↓), OS (↓) | miR-1246 | Upregulated | TP53, CXCR4 |
| Li et al. (2015) [37] | Newly diagnosed MM (n = 90), patients in remission (n = 16), patients in relapse (n = 11), healthy donors (n = 19) | China | Bone marrow | PFS (↓), OS (↓) | miR-15a | Downregulated | BCL2, MCL1, ETS1, JUN, TP53 |
| miR-16-1 | BCL2, MCL1, CCND1, WNT3A, VEGF | ||||||
| Hao et al. (2016) [45] | Newly diagnosed MM (n = 108), healthy donors (n = 44) | China | Serum | PFS (↓), OS (↓) | miR-214 | Upregulated | FBXW7, PTEN, AKT, GSK3 |
| Ren et al. (2017) [53] | Newly diagnosed MM (n = 60), healthy donors (n = 16) | China | Serum | PFS (↓) | miR-720 | Upregulated | TP53 |
| miR-1246 | |||||||
| Szudy-Szczyrek et al. (2022) [54] | MM (n = 105) | Poland | Serum | PFS (↓), OS (↓) | miR-8074 | Upregulated | TP53, MYC, MAPK1 KIAA |
| Roseth Aass et al. (2023) [56] | MM (n = 86) | Norway | Bone marrow | OS (↓) | miR-105-5p | Upregulated | PD-L1, F522 |
| Papadimitriou et al. (2023) [58] | MM (n = 69) | Greece | Serum | OS (↓), PFS (ns) | miR-25-3p | Upregulated | TP53, PTEN, PI3K/Akt, MYC |
| Manier et al. (2017) [61] | Newly diagnosed MM (n = 156), healthy donors (n = 5) | France | Serum | PFS (↓), OS (↓) | miR-18a | Downregulated | IRF2 |
| let-7b | MYC, RAS, CCND1 | ||||||
| Navarro et al. (2015) [62] | Newly diagnosed MM and after aHSCT (n = 33), MGUS (n = 8), healthy donors (n = 8) | Spain | Serum | PFS (↓) | miR-19b | Downregulated | PTEN, IL6R |
| miR-331 | HDAC4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubaj, M.; Bigosiński, K.; Dembowska, A.; Mlak, R.; Szudy-Szczyrek, A.; Małecka-Massalska, T.; Homa-Mlak, I. Role of Non-Coding RNAs in Diagnosis, Prediction and Prognosis of Multiple Myeloma. Cancers 2024, 16, 1033. https://doi.org/10.3390/cancers16051033
Dubaj M, Bigosiński K, Dembowska A, Mlak R, Szudy-Szczyrek A, Małecka-Massalska T, Homa-Mlak I. Role of Non-Coding RNAs in Diagnosis, Prediction and Prognosis of Multiple Myeloma. Cancers. 2024; 16(5):1033. https://doi.org/10.3390/cancers16051033
Chicago/Turabian StyleDubaj, Maciej, Karol Bigosiński, Aleksandra Dembowska, Radosław Mlak, Aneta Szudy-Szczyrek, Teresa Małecka-Massalska, and Iwona Homa-Mlak. 2024. "Role of Non-Coding RNAs in Diagnosis, Prediction and Prognosis of Multiple Myeloma" Cancers 16, no. 5: 1033. https://doi.org/10.3390/cancers16051033
APA StyleDubaj, M., Bigosiński, K., Dembowska, A., Mlak, R., Szudy-Szczyrek, A., Małecka-Massalska, T., & Homa-Mlak, I. (2024). Role of Non-Coding RNAs in Diagnosis, Prediction and Prognosis of Multiple Myeloma. Cancers, 16(5), 1033. https://doi.org/10.3390/cancers16051033







