Tissue-Based Diagnostic Biomarkers of Aggressive Variant Prostate Cancer: A Narrative Review
Abstract
Simple Summary
Abstract
1. Introduction
2. AVPC Definition
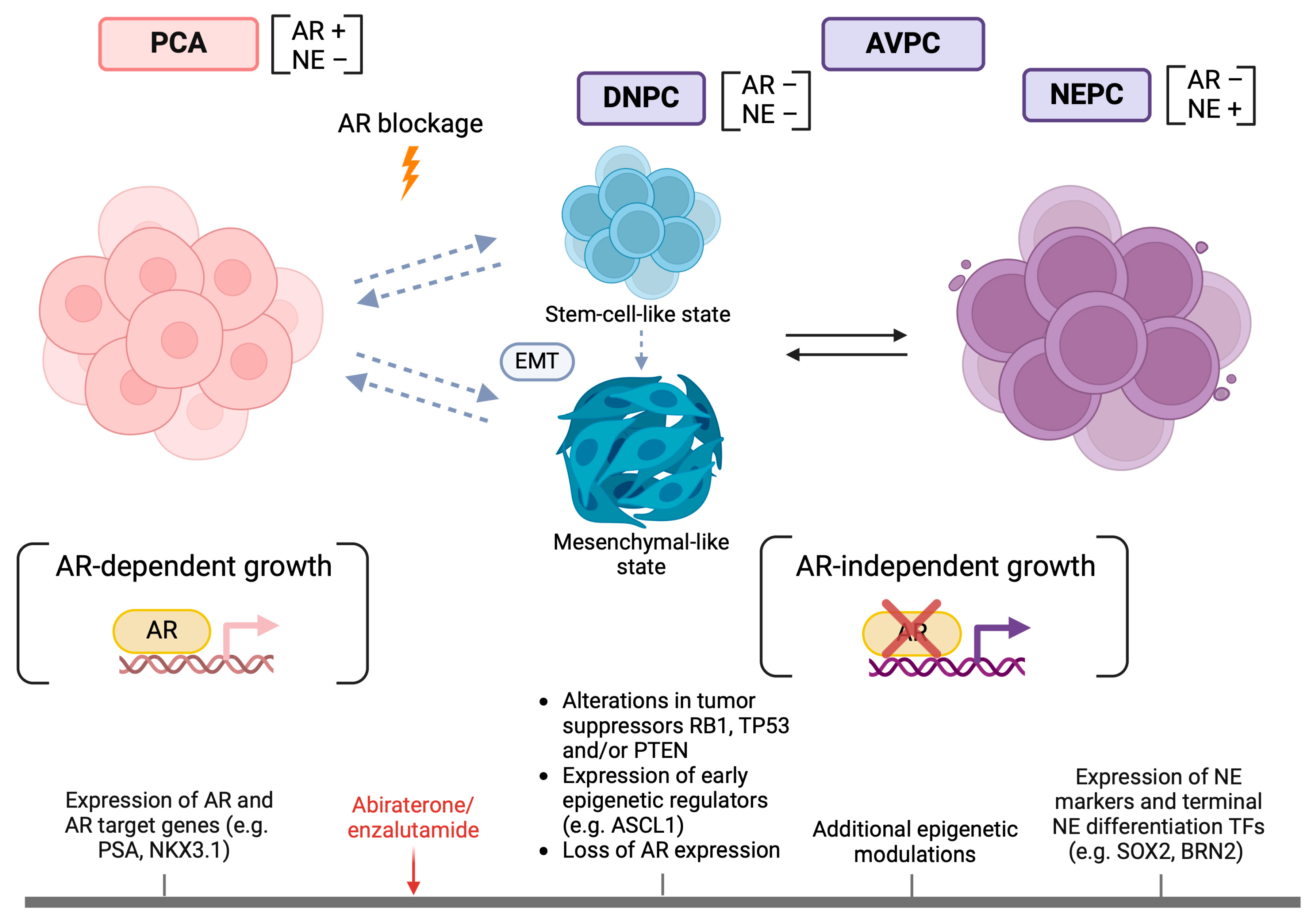
3. NE and AR-Signaling Markers
4. Tumor Suppressors RB1, TP53, and PTEN
5. Oncogenes MYCN and AURKA
6. DNA Damage Repair (DDR) Pathway
7. Gene Expression Profiles, Epigenetic Regulators and Transcription Factors
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef]
- Oczkowski, M.; Dziendzikowska, K. Dietary Factors and Prostate Cancer Development, Progression, and Reduction. Nutrients 2021, 13, 496. [Google Scholar] [CrossRef]
- Keogh, J.W.; MacLeod, R.D. Body composition, physical fitness, functional performance, quality of life, and fatigue benefits of exercise for prostate cancer patients: A systematic review. J. Pain Symptom Manag. 2012, 43, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.C.; Howard, L.E. Obesity increases the risk for high-grade prostate cancer: Results from the REDUCE study. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2936–2942. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, G. Impact of obesity upon prostate cancer-associated mortality: A meta-analysis of 17 cohort studies. Oncol. Lett. 2015, 9, 1307–1312. [Google Scholar] [CrossRef]
- Harrison, S.; Tilling, K. Systematic review and meta-analysis of the associations between body mass index, prostate cancer, advanced prostate cancer, and prostate-specific antigen. Cancer Cases Control 2015, 31, 431–449. [Google Scholar] [CrossRef]
- Langlais, C.S.; Cowan, J.E. Obesity at Diagnosis and Prostate Cancer Prognosis and Recurrence Risk Following Primary Treatment by Radical Prostatectomy. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1917–1925. [Google Scholar] [CrossRef] [PubMed]
- Sarafanov, A.G.; Todorov, T.I. Prostate cancer outcome and tissue levels of metal ions. Prostate 2011, 71, 1231–1238. [Google Scholar] [CrossRef]
- Weischenfeldt, J.; Simon, R. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell 2013, 23, 159–170. [Google Scholar] [CrossRef]
- Scher, H.I.; Halabi, S. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: Recommendations of the Prostate Cancer Clinical Trials Working Group. J. Clin. Oncol. 2008, 26, 1148–1159. [Google Scholar] [CrossRef]
- Chandrasekar, T.; Yang, J.C. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl. Androl. Urol. 2015, 4, 365–380. [Google Scholar] [CrossRef]
- Berchuck, J.E.; Viscuse, P.V. Clinical considerations for the management of androgen indifferent prostate cancer. Prostate Cancer Prostatic Dis. 2021, 24, 623–637. [Google Scholar] [CrossRef]
- Laudato, S.; Aparicio, A. Clonal Evolution and Epithelial Plasticity in the Emergence of AR-Independent Prostate Carcinoma. Trends Cancer 2019, 5, 440–455. [Google Scholar] [CrossRef]
- Terry, S.; Beltran, H. The many faces of neuroendocrine differentiation in prostate cancer progression. Front. Oncol. 2014, 4, 60. [Google Scholar] [CrossRef]
- Aggarwal, R.; Zhang, T. Neuroendocrine prostate cancer: Subtypes, biology, and clinical outcomes. J. Natl. Compr. Cancer Netw. 2014, 12, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Bluemn, E.G.; Coleman, I.M. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell 2017, 32, 474–489.e6. [Google Scholar] [CrossRef]
- Han, H.; Lee, H.H. Prostate epithelial genes define therapy-relevant prostate cancer molecular subtype. Prostate Cancer Prostatic Dis. 2021, 24, 1080–1092. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Sun, Y. p53 Mutation Directs AURKA Overexpression via miR-25 and FBXW7 in Prostatic Small Cell Neuroendocrine Carcinoma. Mol. Cancer. Res. 2015, 13, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, S.; Inoue, T. Clinical and molecular features of treatment-related neuroendocrine prostate cancer. Int. J. Urol. 2018, 25, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, A.M.; Harzstark, A.L. Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin. Cancer Res. 2013, 19, 3621–3630. [Google Scholar] [CrossRef]
- Corn, P.G.; Heath, E.I. Cabazitaxel plus carboplatin for the treatment of men with metastatic castration-resistant prostate cancers: A randomised, open-label, phase 1-2 trial. Lancet. Oncol. 2019, 20, 1432–1443. [Google Scholar] [CrossRef]
- Epstein, J.I.; Amin, M.B. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am. J. Surg. Pathol. 2014, 38, 756–767. [Google Scholar] [CrossRef]
- Slootbeek, P.H.J.; Duizer, M.L. Impact of DNA damage repair defects and aggressive variant features on response to carboplatin-based chemotherapy in metastatic castration-resistant prostate cancer. Int. J. Cancer 2021, 148, 385–395. [Google Scholar] [CrossRef]
- Aparicio, A.M.; Shen, L. Combined Tumor Suppressor Defects Characterize Clinically Defined Aggressive Variant Prostate Cancers. Clin. Cancer Res. 2016, 22, 1520–1530. [Google Scholar] [CrossRef]
- Manucha, V.; Henegan, J. Clinicopathologic Diagnostic Approach to Aggressive Variant Prostate Cancer. Arch. Pathol. Lab. Med. 2020, 144, 18–23. [Google Scholar] [CrossRef]
- Montironi, R.; Cimadamore, A. Morphologic, Molecular and Clinical Features of Aggressive Variant Prostate Cancer. Cells 2020, 9, 1073. [Google Scholar] [CrossRef]
- Taplin, M.E.; George, D.J. Prognostic significance of plasma chromogranin a levels in patients with hormone-refractory prostate cancer treated in Cancer and Leukemia Group B 9480 study. Urology 2015, 66, 386–391. [Google Scholar] [CrossRef]
- Berruti, A.; Mosca, A. Independent prognostic role of circulating chromogranin A in prostate cancer patients with hormone-refractory disease. Endocr. Relat. Cancer 2005, 12, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Speights, V.O., Jr.; Cohen, M.K. Neuroendocrine stains and proliferative indices of prostatic adenocarcinomas in transurethral resection samples. Br. J. Urol. 1997, 80, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Pruneri, G.; Galli, S. Chromogranin A and B and secretogranin II in prostatic adenocarcinomas: Neuroendocrine expression in patients untreated and treated with androgen deprivation therapy. Prostate 1998, 34, 113–120. [Google Scholar] [CrossRef]
- Bonkhoff, H.; Wernert, N. Relation of endocrine-paracrine cells to cell proliferation in normal, hyperplastic, and neoplastic human prostate. Prostate 1991, 19, 91–98. [Google Scholar] [CrossRef]
- Hirano, D.; Okada, Y. Neuroendocrine differentiation in hormone refractory prostate cancer following androgen deprivation therapy. Eur. Urol. 2014, 45, 586–592. [Google Scholar] [CrossRef]
- Steineck, G.; Reuter, V. Cytotoxic treatment of aggressive prostate tumors with or without neuroendocrine elements. Acta Oncol. 2002, 41, 668–674. [Google Scholar] [CrossRef]
- Culine, S.; El Demery, M. Docetaxel and cisplatin in patients with metastatic androgen independent prostate cancer and circulating neuroendocrine markers. J. Urol. 2007, 178, 844–848. [Google Scholar] [CrossRef]
- Loriot, Y.; Massard, C. Combining carboplatin and etoposide in docetaxel-pretreated patients with castration-resistant prostate cancer: A prospective study evaluating also neuroendocrine features. Ann. Oncol. 2009, 20, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Fléchon, A.; Pouessel, D. Phase II study of carboplatin and etoposide in patients with anaplastic progressive metastatic castration-resistant prostate cancer (mCRPC) with or without neuroendocrine differentiation: Results of the French Genito-Urinary Tumor Group (GETUG) P01 trial. Ann. Oncol. 2011, 22, 2476–2481. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Rickman, D.S. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011, 1, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Tzelepi, V.; Zhang, J. Modeling a lethal prostate cancer variant with small-cell carcinoma features. Clin. Cancer. Res. 2012, 18, 666–677. [Google Scholar] [CrossRef]
- Wang, W.; Epstein, J.I. Small cell carcinoma of the prostate. A morphologic and immunohistochemical study of 95 cases. Am. J. Surg. Pathol. 2008, 32, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, R.; Huang, J. Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J. Clin. Oncol. 2018, 36, 2492–2503. [Google Scholar] [CrossRef]
- Tsai, H.K.; Lehrer, J. Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma. BMC Cancer 2017, 17, 759. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.B.; Mehra, R. Androgen-independent prostate cancer is a heterogeneous group of diseases: Lessons from a rapid autopsy program. Cancer Res. 2004, 64, 9209–9216. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Prandi, D. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016, 22, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Abida, W.; Cytra, J. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 11428–11436. [Google Scholar] [CrossRef] [PubMed]
- Labrecque, M.P.; Coleman, I.M. Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J. Clin. Investig. 2019, 129, 4492–4505. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, E.M.; Srinivas, S. Prostate Cancer, Version 4.2023, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2023, 21, 1067–1096. [Google Scholar] [CrossRef]
- Soundararajan, R.; Viscuse, P. Genotype-to-Phenotype Associations in the Aggressive Variant Prostate Cancer Molecular Profile (AVPC-m) Components. Cancers 2022, 14, 3233. [Google Scholar] [CrossRef]
- Guedes, L.B.; Almutairi, F. Analytic, Preanalytic, and Clinical Validation of p53 IHC for Detection of TP53 Missense Mutation in Prostate Cancer. Clin. Canc. Res. 2017, 23, 4693–4703. [Google Scholar] [CrossRef]
- Tan, H.L.; Sood, A. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin. Cancer Res. 2014, 20, 890–903. [Google Scholar] [CrossRef]
- Sharma, A.; Yeow, W.S. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J. Clin. Investig. 2010, 120, 4478–4492. [Google Scholar] [CrossRef]
- Williamson, S.R.; Zhang, S. ERG-TMPRSS2 rearrangement is shared by concurrent prostatic adenocarcinoma and prostatic small cell carcinoma and absent in small cell carcinoma of the urinary bladder: Evidence supporting monoclonal origin. Mod. Pathol. 2011, 24, 1120–1127. [Google Scholar] [CrossRef]
- Hansel, D.E.; Nakayama, M. Shared TP53 gene mutation in morphologically and phenotypically distinct concurrent primary small cell neuroendocrine carcinoma and adenocarcinoma of the prostate. Prostate 2009, 69, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Toivanen, R. Transdifferentiation as a Mechanism of Treatment Resistance in a Mouse Model of Castration-Resistant Prostate Cancer. Cancer Discov. 2017, 7, 736–749. [Google Scholar] [CrossRef] [PubMed]
- Frolov, M.V.; Dyson, N.J. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 2004, 117, 2173–2181. [Google Scholar] [CrossRef] [PubMed]
- Mu, P.; Zhang, Z. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017, 355, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Rickman, D.S.; Beltran, H. Biology and evolution of poorly differentiated neuroendocrine tumors. Nat. Med. 2017, 23, 664–673. [Google Scholar] [CrossRef]
- Meuwissen, R.; Linn, S.C. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 2003, 4, 181–189. [Google Scholar] [CrossRef]
- Maddison, L.A.; Sutherland, B.W. Conditional deletion of Rb causes early stage prostate cancer. Cancer Res. 2004, 64, 6018–6025. [Google Scholar] [CrossRef]
- Elgavish, A.; Wood, P.A. Transgenic mouse with human mutant p53 expression in the prostate epithelium. Prostate 2004, 61, 26–34. [Google Scholar] [CrossRef]
- Chen, Z.; Trotman, L.C. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005, 436, 725–730. [Google Scholar] [CrossRef]
- Zhou, Z.; Flesken-Nikitin, A. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 2006, 66, 7889–7898. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.Y.; Rosario, S. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017, 355, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Masumori, N.; Thomas, T.Z. A probasin-large T antigen transgenic mouse line develops prostate adenocarcinoma and neuroendocrine carcinoma with metastatic potential. Cancer Res. 2011, 61, 2239–2249. [Google Scholar]
- Park, J.W.; Lee, J.K. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science 2018, 362, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Calo, E.; Quintero-Estades, J.A. Rb regulates fate choice and lineage commitment in vivo. Nature 2010, 466, 1110–1114. [Google Scholar] [CrossRef]
- Tschaharganeh, D.F.; Xue, W. p53-dependent Nestin regulation links tumor suppression to cellular plasticity in liver cancer. Cell 2014, 158, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, T.; Suzuki, J. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2019, 460, 1140–1144. [Google Scholar] [CrossRef]
- Yi, L.; Lu, C. Multiple roles of p53-related pathways in somatic cell reprogramming and stem cell differentiation. Cancer Res. 2012, 72, 5635–5645. [Google Scholar] [CrossRef]
- Marión, R.M.; Strati, K. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 2009, 460, 1149–1153. [Google Scholar] [CrossRef]
- Picanço-Castro, V.; Russo-Carbolante, E. Pluripotent reprogramming of fibroblasts by lentiviral mediated insertion of SOX2, C-MYC, and TCL-1A. Stem Cells Dev. 2011, 20, 169–180. [Google Scholar] [CrossRef]
- Chen, H.; Sun, Y. Pathogenesis of prostatic small cell carcinoma involves the inactivation of the P53 pathway. Endocr. Relat. Cancer 2012, 19, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Bookstein, R.; MacGrogan, D. p53 is mutated in a subset of advanced-stage prostate cancers. Cancer Res. 1993, 53, 3369–3373. [Google Scholar]
- Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef]
- Grasso, C.S.; Wu, Y.M. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef]
- Beltran, H.; Yelensky, R. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur. Urol. 2013, 63, 920–926. [Google Scholar] [CrossRef]
- Maughan, B.L.; Guedes, L.B. p53 status in the primary tumor predicts efficacy of subsequent abiraterone and enzalutamide in castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2018, 21, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Annala, M.; Vandekerkhove, G. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov. 2018, 8, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Cairns, P.; Okami, K. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 1997, 57, 4997–5000. [Google Scholar]
- Li, J.; Yen, C. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997, 275, 1943–1947. [Google Scholar] [CrossRef]
- Wang, S.; Garcia, A.J. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc. Natl. Acad. Sci. USA 2006, 103, 1480–1485. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Gao, J. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 2003, 4, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Backman, S.A.; Ghazarian, D. Early onset of neoplasia in the prostate and skin of mice with tissue-specific deletion of Pten. Proc. Natl. Acad. Sci. USA 2004, 101, 1725–1730. [Google Scholar] [CrossRef] [PubMed]
- Feilotter, H.E.; Nagai, M.A. Analysis of PTEN and the 10q23 region in primary prostate carcinomas. Oncogene 1998, 16, 1743–1748. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Rink, K. PTEN/MMAC1 mutations in prostate cancer. Prostate Cancer Prostatic Dis. 2000, 3, S32. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alimonti, A.; Carracedo, A. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 2010, 42, 454–458. [Google Scholar] [CrossRef]
- Alimonti, A.; Nardella, C. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J. Clin. Investig. 2010, 120, 681–693. [Google Scholar] [CrossRef]
- Lee, Y.R.; Chen, M. The functions and regulation of the PTEN tumour suppressor: New modes and prospects. Nat. Rev. Mol. Cell Biol. 2018, 19, 547–562. [Google Scholar] [CrossRef]
- Sircar, K.; Yoshimoto, M. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J. Pathol. 2009, 218, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; Liu, Y.N. Prostate epithelial Pten/TP53 loss leads to transformation of multipotential progenitors and epithelial to mesenchymal transition. Am. J. Pathol. 2011, 179, 422–435. [Google Scholar] [CrossRef] [PubMed]
- Velez, M.G.; Kosiorek, H.E. Differential impact of tumor suppressor gene (TP53, PTEN, RB1) alterations and treatment outcomes in metastatic, hormone-sensitive prostate cancer. Prostate Cancer Prostatic Dis. 2022, 25, 479–483. [Google Scholar] [CrossRef]
- Young, F.P.; Becker, T.M. Biomarkers of Castrate Resistance in Prostate Cancer: Androgen Receptor Amplification and T877A Mutation Detection by Multiplex Droplet Digital PCR. J. Clin. Med. 2022, 11, 257. [Google Scholar] [CrossRef]
- Beltran, H.; Oromendia, C. A Phase II Trial of the Aurora Kinase A Inhibitor Alisertib for Patients with Castration-resistant and Neuroendocrine Prostate Cancer: Efficacy and Biomarkers. Clin. Cancer Res. 2019, 25, 43–51. [Google Scholar] [CrossRef]
- Strieder, V.; Lutz, W. Regulation of N-myc expression in development and disease. Cancer Lett. 2002, 180, 107–119. [Google Scholar] [CrossRef]
- Dardenne, E.; Beltran, H. N-Myc Induces an EZH2-Mediated Transcriptional Program Driving Neuroendocrine Prostate Cancer. Cancer Cell 2016, 30, 563–577. [Google Scholar] [CrossRef]
- Rickman, D.S.; Schulte, J.H. The Expanding World of N-MYC-Driven Tumors. Cancer Discov. 2018, 8, 150–163. [Google Scholar] [CrossRef]
- Petrylak, D.P.; Tangen, C.M. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N. Engl. J. Med. 2004, 351, 1513–1520. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.; Brady, N.J. N-Myc-mediated epigenetic reprogramming drives lineage plasticity in advanced prostate cancer. J. Clin. Investig. 2019, 129, 3924–3940. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Phillips, J.W. N-Myc Drives Neuroendocrine Prostate Cancer Initiated from Human Prostate Epithelial Cells. Cancer Cell 2016, 29, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Brady, N.J.; Bagadion, A.M. Temporal evolution of cellular heterogeneity during the progression to advanced AR-negative prostate cancer. Nat. Commun. 2021, 12, 3372. [Google Scholar] [CrossRef] [PubMed]
- Nikonova, A.S.; Astsaturov, I. Aurora A kinase (AURKA) in normal and pathological cell division. Cell. Mol. Life Sci. 2013, 70, 661–687. [Google Scholar] [CrossRef]
- Buschhorn, H.M.; Klein, R.R. Aurora-A over-expression in high-grade PIN lesions and prostate cancer. Prostate 2005, 64, 341–346. [Google Scholar] [CrossRef]
- Jones, D.; Noble, M. Aurora A regulates expression of AR-V7 in models of castrate resistant prostate cancer. Sci. Rep. 2017, 7, 40957. [Google Scholar] [CrossRef] [PubMed]
- Otto, T.; Horn, S. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell 2009, 15, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Mosquera, J.M.; Beltran, H. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment-related neuroendocrine prostate cancer. Neoplasia 2013, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Meulenbeld, H.J.; Bleuse, J.P. Randomized phase II study of danusertib in patients with metastatic castration-resistant prostate cancer after docetaxel failure. BJU Int. 2013, 111, 44–52. [Google Scholar] [CrossRef]
- Kumar, D.; Sharma, N. Therapeutic Interventions of Cancers Using Intrinsically Disordered Proteins as Drug Targets: C-Myc as Model System. Cancer Inform. 2017, 16, 1176935117699408. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, W.C.; Meyerowitz, J.G. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell 2014, 26, 414–427. [Google Scholar] [CrossRef]
- Richards, M.W.; Burgess, S.G. Structural basis of N-Myc binding by Aurora-A and its destabilization by kinase inhibitors. Proc. Natl. Acad. Sci. USA 2016, 113, 13726–13731. [Google Scholar] [CrossRef]
- Hallett, R.M.; Seong, A.B. Transcript signatures that predict outcome and identify targetable pathways in MYCN-amplified neuroblastoma. Mol. Oncol. 2016, 10, 1461–1472. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, B. Targeting the MYCN-PARP-DNA Damage Response Pathway in Neuroendocrine Prostate Cancer. Clin. Cancer Res. 2018, 24, 696–707. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, S.S. Targeting MYCN in Pediatric and Adult Cancers. Front. Oncol. 2021, 10, 623679. [Google Scholar] [CrossRef]
- Carabet, L.A.; Rennie, P.S. Therapeutic Inhibition of Myc in Cancer. Structural Bases and Computer-Aided Drug Discovery Approaches. Int. J. Mol. Sci. 2018, 20, 120. [Google Scholar] [CrossRef]
- Ton, A.T.; Singh, K. Dual-Inhibitors of N-Myc and AURKA as Potential Therapy for Neuroendocrine Prostate Cancer. Int. J. Mol. Sci. 2020, 21, 8277. [Google Scholar] [CrossRef]
- Santiago, T.; Tarek, N. Correlation Between MYCN Gene Status and MYCN Protein Expression in Neuroblastoma: A Pilot Study To Propose the Use of MYCN Immunohistochemistry in Limited-Resource Areas. J. Glob. Oncol. 2019, 5, 1–7. [Google Scholar] [CrossRef]
- Narod, S.A.; Neuhausen, S. Rapid progression of prostate cancer in men with a BRCA2 mutation. Br. J. Cancer 2018, 99, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.; Goh, C. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J. Clin. Oncol. 2013, 31, 1748–1757. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.; Goh, C. Effect of BRCA Mutations on Metastatic Relapse and Cause-specific Survival After Radical Treatment for Localised Prostate Cancer. Eur. Urol. 2015, 68, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Nombela, P.; Lozano, R. BRCA2 and Other DDR Genes in Prostate Cancer. Cancers 2019, 11, 352. [Google Scholar] [CrossRef]
- Gottipati, P.; Vischioni, B. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 2010, 70, 5389–5398. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Tutt, A.N. Synthetic lethality and cancer therapy: Lessons learned from the development of PARP inhibitors. Annu. Rev. Med. 2015, 66, 455–470. [Google Scholar] [CrossRef]
- de Bono, J.; Mateo, J. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N. Eng. J. Med. 2020, 382, 2091–2102. [Google Scholar] [CrossRef]
- Mateo, J.; Porta, N. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020, 21, 162–174. [Google Scholar] [CrossRef]
- Nizialek, E.; Antonarakis, E.S. PARP Inhibitors in Metastatic Prostate Cancer: Evidence to Date. Cancer Manag. Res. 2020, 12, 8105–8114. [Google Scholar] [CrossRef]
- Zafeiriou, Z.; Bianchini, D. Genomic Analysis of Three Metastatic Prostate Cancer Patients with Exceptional Responses to Carboplatin Indicating Different Types of DNA Repair Deficiency. Eur. Urol. 2019, 75, 184–192. [Google Scholar] [CrossRef]
- Mota, J.M.; Barnett, E. Platinum-Based Chemotherapy in Metastatic Prostate Cancer with DNA Repair Gene Alterations. JCO Precis. Oncol. 2020, 4, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, M.M.; Spisák, S. The association between germline BRCA2 variants and sensitivity to platinum-based chemotherapy among men with metastatic prostate cancer. Cancer 2017, 123, 3532–3539. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.H.; Pritchard, C.C. Biallelic Inactivation of BRCA2 in Platinum-sensitive Metastatic Castration-resistant Prostate Cancer. Eur. Urol. 2016, 69, 992–995. [Google Scholar] [CrossRef] [PubMed]
- Schiewer, M.J.; Knudsen, K.E. DNA Damage Response in Prostate Cancer. Cold Spring Harb. Perspect. Med. 2019, 9, a030486. [Google Scholar] [CrossRef]
- Abida, W.; Cheng, M.L. Analysis of the Prevalence of Microsatellite Instability in Prostate Cancer and Response to Immune Checkpoint Blockade. JAMA Oncol. 2019, 5, 471–478. [Google Scholar] [CrossRef]
- Kumar, A.; White, T.A. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc. Natl. Acad. Sci. USA 2011, 108, 17087–17092. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Morrissey, C. MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat. Commun. 2014, 5, 4988. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Valentin, M.; Joost, P. Frequent mismatch-repair defects link prostate cancer to Lynch syndrome. BMC Urol. 2016, 16, 15. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Piulats, J.M. Pembrolizumab for Treatment-Refractory Metastatic Castration-Resistant Prostate Cancer: Multicohort, Open-Label Phase II KEYNOTE-199 Study. J. Clin. Oncol. 2020, 38, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.J.; Hall, M.C. Deficient nucleotide excision repair capacity enhances human prostate cancer risk. Cancer Res. 2014, 64, 1197–1201. [Google Scholar] [CrossRef]
- Goodwin, J.F.; Kothari, V. DNA-PKcs-Mediated Transcriptional Regulation Drives Prostate Cancer Progression and Metastasis. Cancer Cell 2015, 28, 97–113. [Google Scholar] [CrossRef]
- Chao, O.S.; Goodman, O.B., Jr. DNA-PKc inhibition overcomes taxane resistance by promoting taxane-induced DNA damage in prostate cancer cells. Prostate 2021, 81, 1032–1048. [Google Scholar] [CrossRef]
- Adamson, B.; Brittain, N. The catalytic subunit of DNA-PK regulates transcription and splicing of AR in advanced prostate cancer. J. Clin. Investig. 2023, 133, e169200. [Google Scholar] [CrossRef]
- Hahm, J.Y.; Park, J. 8-Oxoguanine: From oxidative damage to epigenetic and epitranscriptional modification. Exp. Mol. Med. 2022, 54, 1626–1642. [Google Scholar] [CrossRef]
- Trzeciak, A.R.; Nyaga, S.G. Cellular repair of oxidatively induced DNA base lesions is defective in prostate cancer cell lines, PC-3 and DU-145. Carcinogenesis 2004, 25, 1359–1370. [Google Scholar] [CrossRef]
- Yun, S.J.; Ha, Y.S. The hOGG1 mutant genotype is associated with prostate cancer susceptibility and aggressive clinicopathological characteristics in the Korean population. Ann. Oncol. 2012, 23, 401–405. [Google Scholar] [CrossRef]
- Guedes, L.B.; Antonarakis, E.S. MSH2 Loss in Primary Prostate Cancer. Clin. Cancer Res. 2017, 23, 6863–6874. [Google Scholar] [CrossRef]
- Han, H.; Park, C.K. Characteristics of BRCA2 Mutated Prostate Cancer at Presentation. Int. J. Mol. Sci. 2022, 23, 13426. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, G.; Armenia, J. Significance of BRCA2 and RB1 Co-loss in Aggressive Prostate Cancer Progression. Clin. Cancer Res. 2020, 26, 2047–2064. [Google Scholar] [CrossRef] [PubMed]
- Mansour, W.Y.; Tennstedt, P. Loss of PTEN-assisted G2/M checkpoint impedes homologous recombination repair and enhances radio-curability and PARP inhibitor treatment response in prostate cancer. Sci. Rep. 2018, 8, 3947. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, H. Aurora-A: A potential DNA repair modulator. Tumour Biol. 2014, 35, 2831–2836. [Google Scholar] [CrossRef]
- Lapuk, A.V.; Wu, C. From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J. Pathol. 2012, 227, 286–297. [Google Scholar] [CrossRef]
- Cheng, S.; Yu, X. Bioinformatics analyses of publicly available NEPCa datasets. Am. J. Clin. Exp. Urol. 2019, 7, 327–340. [Google Scholar]
- Ostano, P.; Mello-Grand, M. Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 1078. [Google Scholar] [CrossRef] [PubMed]
- Alshalalfa, M.; Liu, Y. Characterization of transcriptomic signature of primary prostate cancer analogous to prostatic small cell neuroendocrine carcinoma. Int. J. Cancer 2019, 145, 3453–3461. [Google Scholar] [CrossRef]
- Akamatsu, S.; Wyatt, A.W. The Placental Gene PEG10 Promotes Progression of Neuroendocrine Prostate Cancer. Cell Rep. 2015, 12, 922–936. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Jenner, R.G. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2016, 125, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Flora, P.; Dalal, G. Polycomb Repressive Complex(es) and Their Role in Adult Stem Cells. Genes 2021, 12, 1485. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yu, J. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007, 67, 10657–10663. [Google Scholar] [CrossRef] [PubMed]
- Karanikolas, B.D.; Figueiredo, M.L. Comprehensive evaluation of the role of EZH2 in the growth, invasion, and aggression of a panel of prostate cancer cell lines. Prostate 2010, 70, 675–688. [Google Scholar] [CrossRef]
- Shan, J.; Al-Muftah, M.A. Targeting Wnt/EZH2/microRNA-708 signaling pathway inhibits neuroendocrine differentiation in prostate cancer. Cell Death Discov. 2019, 5, 139. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, D. Androgen deprivation promotes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nat. Commun. 2018, 9, 4080. [Google Scholar] [CrossRef]
- Kirk, J.S.; Schaarschuch, K. Top2a identifies and provides epigenetic rationale for novel combination therapeutic strategies for aggressive prostate cancer. Oncotarget 2015, 6, 3136–3146. [Google Scholar] [CrossRef]
- Wee, Z.N.; Li, Z. EZH2-mediated inactivation of IFN-γ-JAK-STAT1 signaling is an effective therapeutic target in MYC-driven prostate cancer. Cell Rep. 2014, 8, 204–216. [Google Scholar] [CrossRef]
- Schade, A.E.; Kuzmickas, R. Combating castration-resistant prostate cancer by co-targeting the epigenetic regulators EZH2 and HDAC. PLoS Biol. 2023, 21, e3002038. [Google Scholar] [CrossRef]
- Crea, F.; Hurt, E.M. Pharmacologic disruption of Polycomb Repressive Complex 2 inhibits tumorigenicity and tumor progression in prostate cancer. Mol. Cancer 2011, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Clermont, P.L.; Lin, D. Polycomb-mediated silencing in neuroendocrine prostate cancer. Clin. Epigenet. 2015, 7, 40. [Google Scholar] [CrossRef]
- Su, W.; Han, H.H. The Polycomb Repressor Complex 1 Drives Double-Negative Prostate Cancer Metastasis by Coordinating Stemness and Immune Suppression. Cancer Cell 2019, 36, 139–155.e10. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.A.; Balanis, N.G. A Human Adult Stem Cell Signature Marks Aggressive Variants across Epithelial Cancers. Cell Rep. 2018, 24, 3353–3366.e5. [Google Scholar] [CrossRef] [PubMed]
- Xiang, S.; Zou, P. HOTAIR-mediated reciprocal regulation of EZH2 and DNMT1 contribute to polyphyllin I-inhibited growth of castration-resistant prostate cancer cells in vitro and in vivo. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Ohm, J.E.; McGarvey, K.M. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007, 39, 237–242. [Google Scholar] [CrossRef]
- Gravina, G.L.; Festuccia, C. Chronic azacitidine treatment results in differentiating effects, sensitizes against bicalutamide in androgen-independent prostate cancer cells. Prostate 2008, 68, 793–801. [Google Scholar] [CrossRef]
- Lin, D.; Dong, X. Identification of DEK as a potential therapeutic target for neuroendocrine prostate cancer. Oncotarget 2015, 6, 1806–1820. [Google Scholar] [CrossRef]
- Ci, X.; Hao, J. Heterochromatin Protein 1α Mediates Development and Aggressiveness of Neuroendocrine Prostate Cancer. Cancer Res. 2018, 78, 2691–2704. [Google Scholar] [CrossRef]
- Clermont, P.L.; Ci, X. Treatment-emergent neuroendocrine prostate cancer: Molecularly driven clinical guidelines. Int. J. Encodr. Oncol. 2019, 6, IJE20. [Google Scholar] [CrossRef]
- Dang, Q.; Li, L. Anti-androgen enzalutamide enhances prostate cancer neuroendocrine (NE) differentiation via altering the infiltrated mast cells→androgen receptor (AR)→miRNA32 signals. Mol. Oncol. 2015, 9, 1241–1251. [Google Scholar] [CrossRef]
- Ding, M.; Lin, B. A dual yet opposite growth-regulating function of miR-204 and its target XRN1 in prostate adenocarcinoma cells and neuroendocrine-like prostate cancer cells. Oncotarget 2015, 6, 7686–7700. [Google Scholar] [CrossRef]
- Nam, R.K.; Benatar, T. MicroRNA-652 induces NED in LNCaP and EMT in PC3 prostate cancer cells. Oncotarget 2018, 9, 19159–19176. [Google Scholar] [CrossRef]
- Dankert, J.T.; Wiesehöfer, M. The deregulation of miR-17/CCND1 axis during neuroendocrine transdifferentiation of LNCaP prostate cancer cells. PLoS ONE 2018, 13, e0200472. [Google Scholar] [CrossRef]
- Bhagirath, D.; Liston, M. MicroRNA determinants of neuroendocrine differentiation in metastatic castration-resistant prostate cancer. Oncogene 2020, 39, 7209–7223. [Google Scholar] [CrossRef]
- Ramnarine, V.R.; Alshalalfa, M. The long noncoding RNA landscape of neuroendocrine prostate cancer and its clinical implications. Gigascience 2018, 7, giy050. [Google Scholar] [CrossRef]
- Singh, N.; Ramnarine, V.R. The long noncoding RNA H19 regulates tumor plasticity in neuroendocrine prostate cancer. Nat. Commun. 2021, 12, 7349. [Google Scholar] [CrossRef]
- Li, Y.; Donmez, N. SRRM4 Drives Neuroendocrine Transdifferentiation of Prostate Adenocarcinoma Under Androgen Receptor Pathway Inhibition. Eur. Urol. 2017, 71, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Coleman, I.M. SRRM4 Expression and the Loss of REST Activity May Promote the Emergence of the Neuroendocrine Phenotype in Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2015, 21, 4698–4708. [Google Scholar] [CrossRef] [PubMed]
- Svensson, C.; Ceder, J. REST mediates androgen receptor actions on gene repression and predicts early recurrence of prostate cancer. Nucleic Acids Res. 2014, 42, 999–1015. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, A.; Tzelepi, V. Neuroendocrine prostate cancer xenografts with large-cell and small-cell features derived from a single patient’s tumor: Morphological, immunohistochemical, and gene expression profiles. Prostate 2011, 71, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Nouruzi, S.; Ganguli, D. ASCL1 activates neuronal stem cell-like lineage programming through remodeling of the chromatin landscape in prostate cancer. Nat. Commun. 2022, 13, 2282. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.L.; Thaper, D. The Master Neural Transcription Factor BRN2 Is an Androgen Receptor-Suppressed Driver of Neuroendocrine Differentiation in Prostate Cancer. Cancer Discov. 2017, 7, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, D.; Yang, T.L. BRN4 Is a Novel Driver of Neuroendocrine Differentiation in Castration-Resistant Prostate Cancer and Is Selectively Released in Extracellular Vesicles with BRN2. Clin. Cancer Res. 2019, 25, 6532–6545. [Google Scholar] [CrossRef]
- Baca, S.C.; Takeda, D.Y. Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nat. Commun. 2021, 12, 1979. [Google Scholar] [CrossRef]
- Asrani, K.; Torres, A.F. Reciprocal YAP1 loss and INSM1 expression in neuroendocrine prostate cancer. J. Pathol. 2021, 255, 425–437. [Google Scholar] [CrossRef]
- Cejas, P.; Xie, Y. Subtype heterogeneity and epigenetic convergence in neuroendocrine prostate cancer. Nat. Commun. 2021, 12, 5775. [Google Scholar] [CrossRef]
- Vias, M.; Massie, C.E. Pro-neural transcription factors as cancer markers. BMC Med. Genom. 2018, 1, 17. [Google Scholar] [CrossRef]
- Xin, Z.; Zhang, Y. Insulinoma-associated protein 1 is a novel sensitive and specific marker for small cell carcinoma of the prostate. Hum. Pathol. 2018, 79, 151–159. [Google Scholar] [CrossRef]
- Chen, J.F.; Yang, C. Expression of novel neuroendocrine marker insulinoma-associated protein 1 (INSM1) in genitourinary high-grade neuroendocrine carcinomas: An immunohistochemical study with specificity analysis and comparison to chromogranin, synaptophysin, and CD56. Pathol. Res. Pract. 2020, 216, 152993. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, T. Single-cell transcriptional regulation and genetic evolution of neuroendocrine prostate cancer. iScience 2022, 25, 104576. [Google Scholar] [CrossRef] [PubMed]
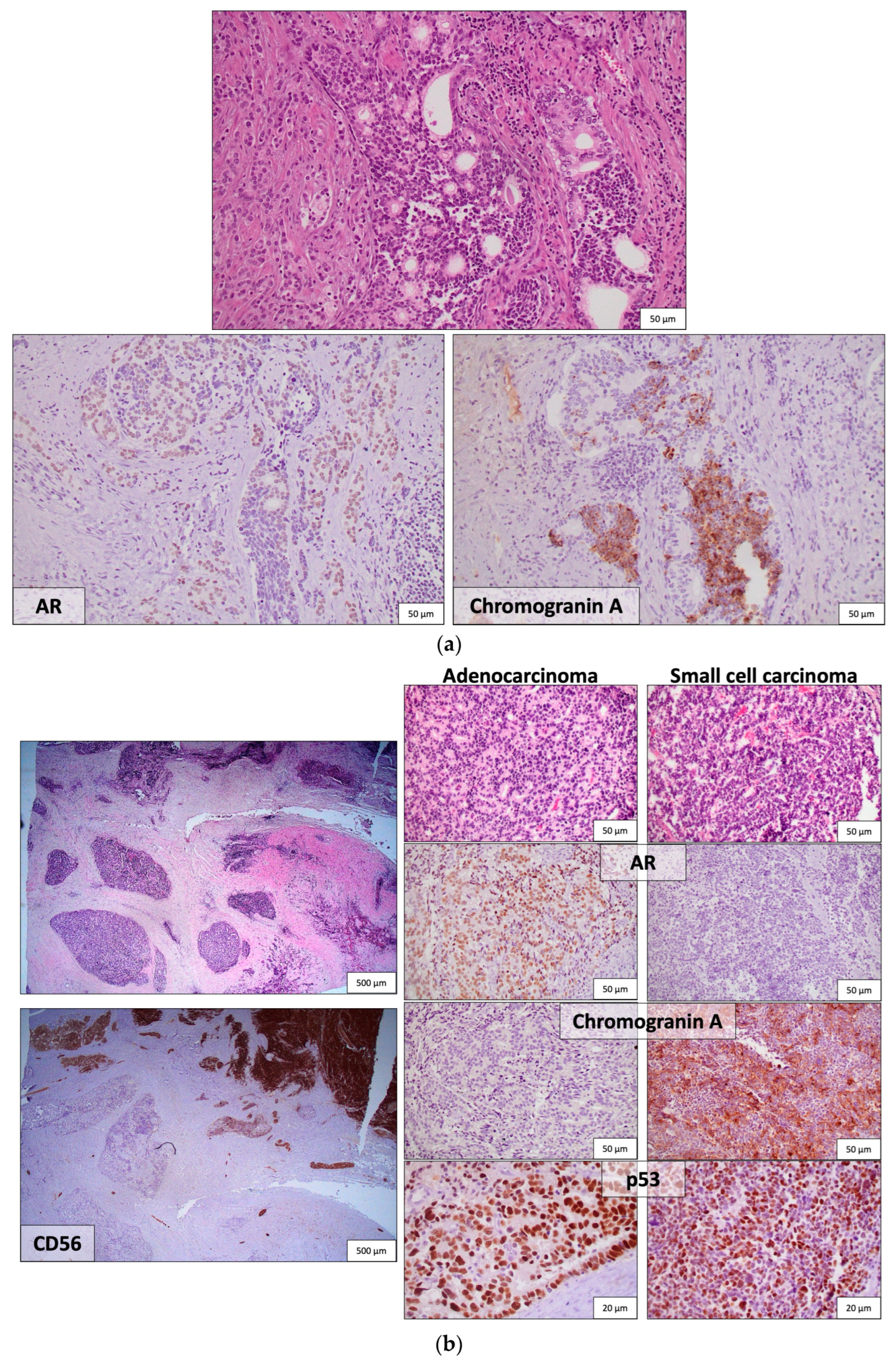
| AVPC | Conventional PCA |
|---|---|
| Adenocarcinoma histology |
| Mostly bone metastases |
| Predominantly osteoblastic bone metastases |
| Variably bulky disease and variable GS |
| Variable PSA levels at presentation and usually higher PSA levels (PSA > 10 ng/mL) at disease progression |
| Focal/no immunohistochemical expression of NE markers and absence of abnormally elevated serum NE markers and non-otherwise explained elevation of serum LDH, CEA or calcium |
| Longer interval time (>6 months) between ADT initiation and progression |
| Candidate Tissue-Based AVPC Biomarkers | Value |
|---|---|
| NE markers | Limited value in the absence of NE morphology |
| AR and AR-regulated genes (PSA, TMRSS2, NKX3.1) | Loss of expression supports AVPC diagnosis but retained expression and transcriptional activity are not preclusive of AVPC |
| Combined NE and AR expression | AR−/NE− and AR−/NE+ phenotypes are consistent with AVPC |
| RB1 | RB1 alterations associated with poor prognosis Predictive of transition to AVPC |
| PTEN | Limited value in the absence of concurrent RB1 or TP53 alterations |
| TP53 | TP53 alterations associated with poor prognosis, but are not specific for AVPC |
| Combined alterations in RB1, PTEN and/or TP53 (≥2/3) | Highly suggestive of AVPC (molecular signature of AVPC) |
| MYCN | Amplification predictive of AVPC/poor response to chemotherapy (docetaxel)/response to alternative treatments (i.e., AURKA inhibitors) |
| AURKA | Amplification predictive of AVPC/response to alternative treatments (i.e., AURKA inhibitors) |
| BRCA2 | BRCA2 defects predictive of response to platinum-based chemotherapy/response to PARP inhibitors |
| MMR proteins | Loss of MMR expression predictive of response to anti-PD-1 therapy |
| PEG10 | Predictive of NEPC |
| NEPC gene expression classifiers | Diagnostic of NEPC and possibly of AVPC in general |
| DNA methylation profile | Probable diagnostic value of AVPC, irrespective of morphology |
| EZH2 | Predictive of transition to NEPC/response to EZH2 inhibitors |
| DEK | Associated with poor prognosis Predictive of transition to NEPC |
| HP1a | Predictive of transition to NEPC |
| ncRNA classifiers | Diagnostic of NEPC and possibly of AVPC in general |
| Proneural TFs (SOX2, ASCL1, BRN2, BRN4, FOXA1, INSM1, NEUROD1) | Predictive of NEPC (taking into account their differential pattern of expression during transition to NEPC) |
| Candidate Tissue-Based AVPC Biomarkers | Method | Evaluation | Sensitivity | Specificity |
| Chromogranin and/or synaptophysin | IHC | Any extent of positive staining | 57% [20] | 0–90% [22] 1 |
| AR | IHC | Reduced (<10%) or weak (1+) staining | 36% [24] | 87% [40] 2 |
| Copy number analysis | Absence of copy number gain | 80% [24] | 30–50% [92] 3 | |
| RB1 | IHC | Reduced (<10%) staining | 61% [24] | 26–93% [49,50] 4 |
| Copy number analysis | Copy number loss | 54% [24] | 72% [76] 5 | |
| p53 | IHC | ≥10% staining | 41% [24] | ≈60% 6 |
| PTEN | Copy number analysis | Copy number loss | 48% [24] | 23% [89] 7 |
| RB1, TP53 and/or PTEN (≥2/3) | DNA sequencing 8 | Combined alterations | 48% [24] | 74% [24] 9 |
| MYCN | Copy number analysis | Copy number gain | 20% [24] | 96% [37] 10 |
| AURKA | Copy number analysis | Copy number gain | 25% [24] | 95% [37] 11 |
| BRCA2 | DNA sequencing | Mutation or deletion | 29% [93] | 87% [74] 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kouroukli, O.; Bravou, V.; Giannitsas, K.; Tzelepi, V. Tissue-Based Diagnostic Biomarkers of Aggressive Variant Prostate Cancer: A Narrative Review. Cancers 2024, 16, 805. https://doi.org/10.3390/cancers16040805
Kouroukli O, Bravou V, Giannitsas K, Tzelepi V. Tissue-Based Diagnostic Biomarkers of Aggressive Variant Prostate Cancer: A Narrative Review. Cancers. 2024; 16(4):805. https://doi.org/10.3390/cancers16040805
Chicago/Turabian StyleKouroukli, Olga, Vasiliki Bravou, Konstantinos Giannitsas, and Vasiliki Tzelepi. 2024. "Tissue-Based Diagnostic Biomarkers of Aggressive Variant Prostate Cancer: A Narrative Review" Cancers 16, no. 4: 805. https://doi.org/10.3390/cancers16040805
APA StyleKouroukli, O., Bravou, V., Giannitsas, K., & Tzelepi, V. (2024). Tissue-Based Diagnostic Biomarkers of Aggressive Variant Prostate Cancer: A Narrative Review. Cancers, 16(4), 805. https://doi.org/10.3390/cancers16040805





