Prediction of a Multi-Gene Assay (Oncotype DX and Mammaprint) Recurrence Risk Group Using Machine Learning in Estrogen Receptor-Positive, HER2-Negative Breast Cancer—The BRAIN Study
Abstract
Simple Summary
Abstract
1. Introduction
Related Work
2. Methods
2.1. Study Population and Data
2.2. Statistical Analysis
2.3. Data Preprocessing and Feature Selection
2.4. AI Modeling and Evaluation
3. Results
3.1. Patient and Tumor Characteristics
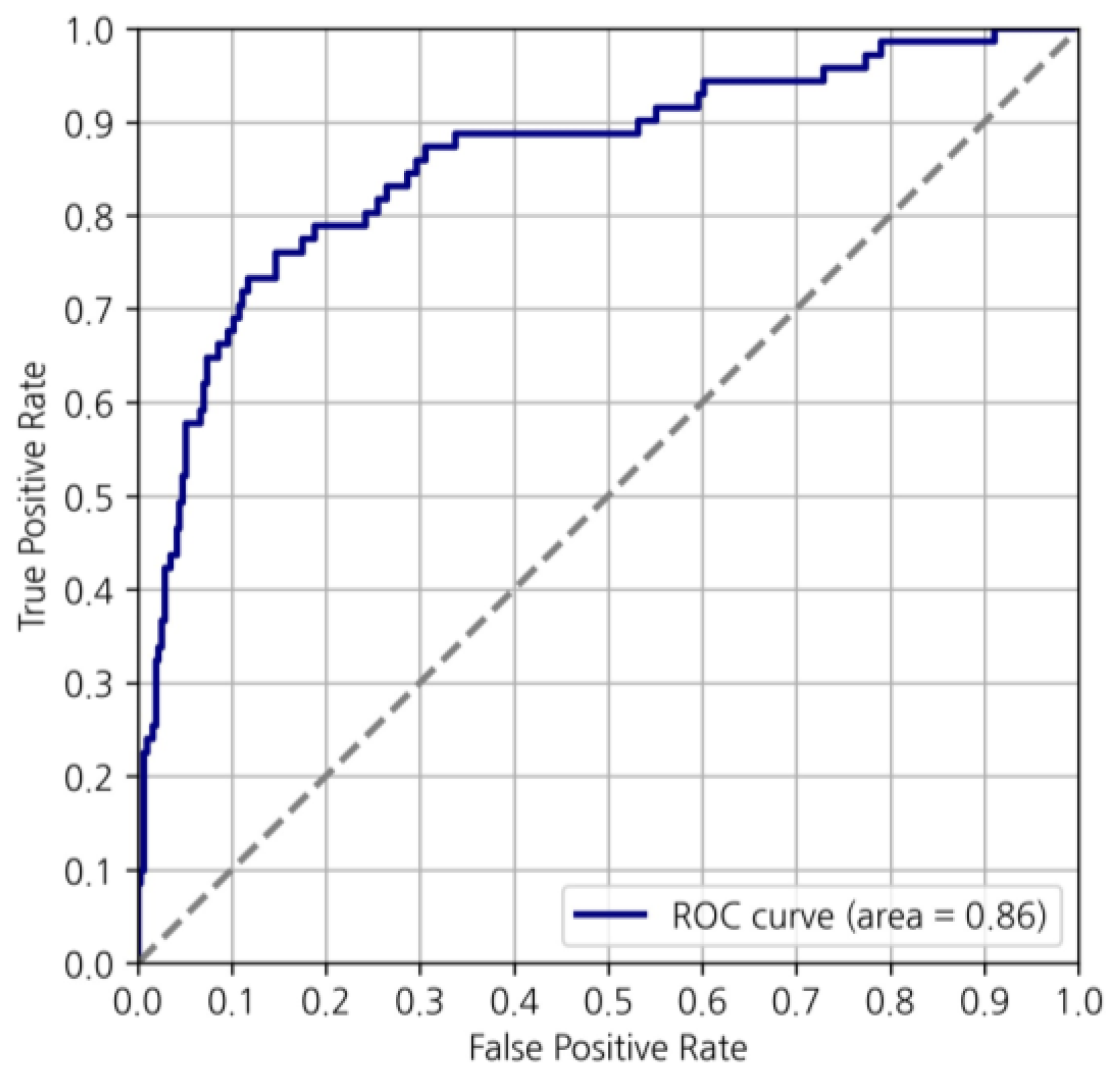
3.2. Machine Learning Model Prediction of MGA Risk Category
3.3. Cross-Tab Analysis and Subgroup Analysis
3.4. Analysis of Model-Underpredicted (Discordant) Cases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Goetz, M.P.; Olson, J.A., Jr.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.Y.; Brain, E.; Causeret, S.; DeLorenzi, M.; et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016, 375, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, A.E.; Connolly, J.L.; Edge, S.B.; Mittendorf, E.A.; Rugo, H.S.; Solin, L.J.; Weaver, D.L.; Winchester, D.J.; Hortobagyi, G.N. Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Ovcaricek, T.; Takac, I.; Matos, E. Multigene expression signatures in early hormone receptor positive HER 2 negative breast cancer. Radiol. Oncol. 2019, 53, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Cognetti, F.; Biganzoli, L.; De Placido, S.; Del Mastro, L.; Masetti, R.; Naso, G.; Pruneri, G.; Santini, D.; Tondini, C.A.; Tinterri, C.; et al. Multigene tests for breast cancer: The physician’s perspective. Oncotarget 2021, 12, 936–947. [Google Scholar] [CrossRef]
- Guth, A.A.; Fineberg, S.; Fei, K.; Franco, R.; Bickell, N.A. Utilization of Oncotype DX in an Inner City Population: Race or Place? Int. J. Breast Cancer 2013, 2013, 653805. [Google Scholar] [CrossRef]
- Lund, M.J.; Mosunjac, M.; Davis, K.M.; Gabram-Mendola, S.; Rizzo, M.; Bumpers, H.L.; Hearn, S.; Zelnak, A.; Styblo, T.; O’Regan, R.M. 21-Gene recurrence scores: Racial differences in testing, scores, treatment, and outcome. Cancer 2012, 118, 788–796. [Google Scholar] [CrossRef]
- Orucevic, A.; Heidel, R.E.; Bell, J.L. Utilization and impact of 21-gene recurrence score assay for breast cancer in clinical practice across the United States: Lessons learned from the 2010 to 2012 National Cancer Data Base analysis. Breast Cancer Res. Treat. 2016, 157, 427–435. [Google Scholar] [CrossRef]
- Harowicz, M.R.; Robinson, T.J.; Dinan, M.A.; Saha, A.; Marks, J.R.; Marcom, P.K.; Mazurowski, M.A. Algorithms for prediction of the Oncotype DX recurrence score using clinicopathologic data: A review and comparison using an independent dataset. Breast Cancer Res. Treat. 2017, 162, 1–10. [Google Scholar] [CrossRef]
- Hou, Y.; Tozbikian, G.; Zynger, D.L.; Li, Z. Using the Modified Magee Equation to Identify Patients Unlikely to Benefit from the 21-Gene Recurrence Score Assay (Oncotype DX Assay). Am. J. Clin. Pathol. 2017, 147, 541–548. [Google Scholar] [CrossRef]
- Sughayer, M.; Alaaraj, R.; Alsughayer, A. Applying new Magee equations for predicting the Oncotype DX recurrence score. Breast Cancer 2018, 25, 597–604. [Google Scholar] [CrossRef]
- Yeo, B.; Zabaglo, L.; Hills, M.; Dodson, A.; Smith, I.; Dowsett, M. Clinical utility of the IHC4+C score in oestrogen receptor-positive early breast cancer: A prospective decision impact study. Br. J. Cancer 2015, 113, 390–395. [Google Scholar] [CrossRef]
- Li, H.; Zhu, Y.; Burnside, E.S.; Drukker, K.; Hoadley, K.A.; Fan, C.; Conzen, S.D.; Whitman, G.J.; Sutton, E.J.; Net, J.M.; et al. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology 2016, 281, 382–391. [Google Scholar] [CrossRef]
- Baltres, A.; Al Masry, Z.; Zemouri, R.; Valmary-Degano, S.; Arnould, L.; Zerhouni, N.; Devalland, C. Prediction of Oncotype DX recurrence score using deep multi-layer perceptrons in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer 2020, 27, 1007–1016. [Google Scholar] [CrossRef]
- Kim, I.; Choi, H.J.; Ryu, J.M.; Lee, S.K.; Yu, J.H.; Kim, S.W.; Nam, S.J.; Lee, J.E. A predictive model for high/low risk group according to oncotype DX recurrence score using machine learning. Eur. J. Surg. Oncol. 2019, 45, 134–140. [Google Scholar] [CrossRef]
- Pawloski, K.R.; Gonen, M.; Wen, H.Y.; Tadros, A.B.; Thompson, D.; Abbate, K.; Morrow, M.; El-Tamer, M. Supervised machine learning model to predict oncotype DX risk category in patients over age 50. Breast Cancer Res. Treat. 2022, 191, 423–430. [Google Scholar] [CrossRef]
- Li, H.; Wang, J.; Li, Z.; Dababneh, M.; Wang, F.; Zhao, P.; Smith, G.H.; Teodoro, G.; Li, M.; Kong, J.; et al. Deep Learning-Based Pathology Image Analysis Enhances Magee Feature Correlation with Oncotype DX Breast Recurrence Score. Front. Med. 2022, 9, 886763. [Google Scholar] [CrossRef]
- Romeo, V.; Cuocolo, R.; Sanduzzi, L.; Carpentiero, V.; Caruso, M.; Lama, B.; Garifalos, D.; Stanzione, A.; Maurea, S.; Brunetti, A. MRI Radiomics and Machine Learning for the Prediction of Oncotype DX Recurrence Score in Invasive Breast Cancer. Cancers 2023, 15, 1840. [Google Scholar] [CrossRef] [PubMed]
- Orucevic, A.; Bell, J.L.; King, M.; McNabb, A.P.; Heidel, R.E. Nomogram update based on TAILORx clinical trial results—Oncotype DX breast cancer recurrence score can be predicted using clinicopathologic data. Breast 2019, 46, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Ismaila, N.; Henry, N.L.; Somerfield, M.R.; Bast, R.C.; Barlow, W.; Collyar, D.E.; Hammond, M.E.; Kuderer, N.M.; Liu, M.C.; et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women with Early-Stage Invasive Breast Cancer: ASCO Clinical Practice Guideline Update-Integration of Results From TAILORx. J. Clin. Oncol. 2019, 37, 1956–1964. [Google Scholar] [CrossRef] [PubMed]
- Henry, N.L.; Somerfield, M.R.; Abramson, V.G.; Allison, K.H.; Anders, C.K.; Chingos, D.T.; Hurria, A.; Openshaw, T.H.; Krop, I.E. Role of Patient and Disease Factors in Adjuvant Systemic Therapy Decision Making for Early-Stage, Operable Breast Cancer: American Society of Clinical Oncology Endorsement of Cancer Care Ontario Guideline Recommendations. J. Clin. Oncol. 2016, 34, 2303–2311. [Google Scholar] [CrossRef]
- Győrffy, B.; Hatzis, C.; Sanft, T.; Hofstatter, E.; Aktas, B.; Pusztai, L. Multigene prognostic tests in breast cancer: Past, present, future. Breast Cancer Res. 2015, 17, 11. [Google Scholar] [CrossRef]
- Prat, A.; Parker, J.S.; Fan, C.; Cheang, M.C.U.; Miller, L.D.; Bergh, J.; Chia, S.K.L.; Bernard, P.S.; Nielsen, T.O.; Ellis, M.J.; et al. Concordance among gene expression-based predictors for ER-positive breast cancer treated with adjuvant tamoxifen. Ann. Oncol. 2012, 23, 2866–2873. [Google Scholar] [CrossRef]
- Iwamoto, T.; Lee, J.S.; Bianchini, G.; Hubbard, R.E.; Young, E.; Matsuoka, J.; Kim, S.B.; Symmans, W.F.; Hortobagyi, G.N.; Pusztai, L. First generation prognostic gene signatures for breast cancer predict both survival and chemotherapy sensitivity and identify overlapping patient populations. Breast Cancer Res. Treat. 2011, 130, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Mazo, C.; Kearns, C.; Mooney, C.; Gallagher, W.M. Clinical Decision Support Systems in Breast Cancer: A Systematic Review. Cancers 2020, 12, 369. [Google Scholar] [CrossRef] [PubMed]
- Hall, P.S.; McCabe, C.; Stein, R.C.; Cameron, D. Economic evaluation of genomic test-directed chemotherapy for early-stage lymph node-positive breast cancer. J. Natl. Cancer Inst. 2012, 104, 56–66. [Google Scholar] [CrossRef]
- Lyman, G.H.; Cosler, L.E.; Kuderer, N.M.; Hornberger, J. Impact of a 21-gene RT-PCR assay on treatment decisions in early-stage breast cancer: An economic analysis based on prognostic and predictive validation studies. Cancer 2007, 109, 1011–1018. [Google Scholar] [CrossRef]
- Kondo, M.; Hoshi, S.L.; Yamanaka, T.; Ishiguro, H.; Toi, M. Economic evaluation of the 21-gene signature (Oncotype DX) in lymph node-negative/positive, hormone receptor-positive early-stage breast cancer based on Japanese validation study (JBCRG-TR03). Breast Cancer Res. Treat. 2011, 127, 739–749. [Google Scholar] [CrossRef]
- Rouzier, R.; Pronzato, P.; Chéreau, E.; Carlson, J.; Hunt, B.; Valentine, W.J. Multigene assays and molecular markers in breast cancer: Systematic review of health economic analyses. Breast Cancer Res. Treat. 2013, 139, 621–637. [Google Scholar] [CrossRef]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Perez, E.A.; Olson, J.A., Jr.; et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2015, 373, 2005–2014. [Google Scholar] [CrossRef] [PubMed]




| Variables | MMP (n = 526) | ||
|---|---|---|---|
| Low (n = 335) | High (n = 191) | p-Value | |
| Age (mean ± SD) | 53.27 ± 9.07 | 52.62 ± 10.44 | 0.473 |
| Height (mean ± SD) | 159.23 ± 5.41 | 158.81 ± 5.06 | 0.380 |
| Weight (mean ± SD) | 60.54 ± 10.09 | 60.62 ± 8.09 | 0.920 |
| BMI (mean ± SD) | 23.87 ± 3.78 | 24.05 ± 3.10 | 0.590 |
| Menarche age (mean ± SD) | 14.41 ± 1.76 | 14.42 ± 1.57 | 0.933 |
| Menopausal status | 0.656 | ||
| Premenopausal | 176 (52.5%) | 96 (50.3%) | |
| Postmenopausal | 158 (47.2%) | 95 (49.7%) | |
| Unknown | 1 (0.3%) | 0 (0.0%) | |
| Preoperative E2 (mean ± SD) | 73.15 ± 98.06 | 77.01 ± 98.34 | 0.676 |
| Preoperative FSH (mean ± SD) | 40.93 ± 36.29 | 38.24 ± 34.68 | 0.422 |
| Tumor size (mean ± SD) | 1.62 ± 0.65 | 1.97 ± 0.82 | <0.001 |
| Multiple lesions | 0.213 | ||
| No | 251 (74.9%) | 153 (80.1%) | |
| Yes | 84 (25.1%) | 38 (19.9%) | |
| HG | <0.001 | ||
| 1 | 123 (36.7%) | 30 (15.7%) | |
| 2 | 197 (58.8%) | 101 (52.9%) | |
| 3 | 12 (3.6%) | 59 (30.9%) | |
| Unknown | 3 (0.9%) | 1 (0.5%) | |
| NG | <0.001 | ||
| 1 | 32 (9.6%) | 3 (1.6%) | |
| 2 | 274 (81.7%) | 134 (70.2%) | |
| 3 | 28 (8.4%) | 53 (27.7%) | |
| Unknown | 1 (0.3%) | 1 (0.5%) | |
| EIC | 0.131 | ||
| No | 218 (65.1%) | 138 (72.2%) | |
| Yes | 108 (32.2%) | 46 (24.1%) | |
| Unknown | 9 (2.7%) | 7 (3.7%) | |
| LVI | 0.192 | ||
| No | 285 (85.1%) | 153 (80.1%) | |
| Yes | 49 (14.6%) | 37 (19.4%) | |
| Unknown | 1 (0.3%) | 1 (0.5%) | |
| SLN (mean ± SD) | 2.55±1.60 | 2.80±2.50 | 0.172 |
| Lymph node metastasis | 0.352 | ||
| No | 182 (54.3%) | 93 (48.7%) | |
| Yes | 148 (44.2%) | 93 (48.7%) | |
| Unknown | 5 (1.5%) | 5 (2.6%) | |
| Perinodal extension | 0.545 | ||
| No | 288 (86.0%) | 169 (88.5%) | |
| Yes | 46 (13.7%) | 22 (11.5%) | |
| Unknown | 1 (0.3%) | 0 (0.0%) | |
| Estrogen receptor | 0.271 | ||
| Low (0~5) | 11 (3.3%) | 10 (5.2%) | |
| High (6~8) | 324 (96.7%) | 181 (94.8%) | |
| Progesterone receptor | 0.112 | ||
| Negative | 39 (11.6%) | 34 (17.8%) | |
| Positive | 296 (88.4%) | 157 (82.2%) | |
| HER2 receptor | 0.062 | ||
| 0 | 77 (23.0%) | 50 (26.2%) | |
| 1+ | 156 (46.6%) | 69 (36.1%) | |
| 2+ | 102 (30.4%) | 72 (37.7%) | |
| Ki-67 (mean ± SD) | 13.17 ± 11.73 | 27.96 ± 18.89 | <0.001 |
| Variables | ODX (n = 2039) | ||
|---|---|---|---|
| Low (n = 1742) | High (n = 297) | p-Value | |
| Age (mean ± SD) | 50.21 ± 9.52 | 52.76 ± 9.81 | <0.001 |
| Height (mean ± SD) | 159.31 ± 5.34 | 158.88 ± 5.35 | 0.201 |
| Weight (mean ± SD) | 58.77 ± 8.31 | 58.25 ± 8.22 | 0.315 |
| BMI (mean ± SD) | 23.15 ± 3.27 | 23.01 ± 3.21 | 0.516 |
| Menarche age (mean ± SD) | 14.18 ± 1.56 | 14.60 ± 1.70 | <0.001 |
| Menopausal status | <0.001 | ||
| Premenopausal | 1061 (60.9%) | 122 (41.1%) | |
| Postmenopausal | 671 (38.5%) | 174 (58.6%) | |
| Unknown | 10 (0.6%) | 1 (0.3%) | |
| Preoperative E2 (mean ± SD) | 107.12 ± 149.39 | 72.06 ± 103.40 | <0.001 |
| Preoperative FSH (mean ± SD) | 30.68 ± 33.55 | 44.22 ± 37.24 | <0.001 |
| Tumor size (mean ± SD) | 1.62 ± 0.72 | 1.77 ± 0.67 | 0.001 |
| Multiple lesions | 0.191 | ||
| No | 1320 (75.8%) | 236 (79.5%) | |
| Yes | 422 (24.2%) | 61 (20.5%) | |
| HG | <0.001 | ||
| 1 | 522 (30.0%) | 29 (9.8%) | |
| 2 | 1124 (64.5%) | 195 (65.7%) | |
| 3 | 92 (5.3%) | 72 (24.2%) | |
| Unknown | 4 (0.2%) | 1 (0.3%) | |
| NG | <0.001 | ||
| 1 | 134 (7.6%) | 7 (2.4%) | |
| 2 | 1419 (81.5%) | 187 (63.0%) | |
| 3 | 184 (10.6%) | 102 (34.3%) | |
| Unknown | 5 (0.3%) | 1 (0.3%) | |
| EIC | 0.293 | ||
| No | 934 (53.6%) | 175 (58.9%) | |
| Yes | 568 (32.6%) | 91 (30.7%) | |
| Unknown | 240 (13.8%) | 31 (10.4%) | |
| LVI | 0.559 | ||
| No | 1498 (86.0%) | 260 (87.5%) | |
| Yes | 236 (13.5%) | 36 (12.1%) | |
| Unknown | 8 (0.5%) | 1 (0.3%) | |
| SLN (mean ± SD) | 2.81±2.05 | 2.64±1.67 | 0.161 |
| Lymph node metastasis | 0.093 | ||
| No | 1531 (87.9%) | 273 (91.9%) | |
| Yes | 189 (10.8%) | 23 (7.8%) | |
| Unknown | 22 (1.3%) | 1 (0.3%) | |
| Perinodal extension | 0.430 | ||
| No | 1571 (90.2%) | 273 (91.9%) | |
| Yes | 39 (2.2%) | 4 (1.3%) | |
| Unknown | 132 (7.6%) | 20 (6.8%) | |
| Estrogen receptor | 0.004 | ||
| Low (0~5) | 82 (4.7%) | 26 (8.8%) | |
| High (6~8) | 1660 (95.3%) | 271 (91.2%) | |
| Progesterone receptor | <0.001 | ||
| Negative | 193 (11.1%) | 109 (36.7%) | |
| Positive | 1549 (88.9%) | 188 (63.3%) | |
| HER2 receptor | <0.001 | ||
| 0 | 506 (29.0%) | 78 (26.3%) | |
| 1+ | 805 (46.2%) | 113 (38.0%) | |
| 2+ | 431 (24.8%) | 106 (35.7%) | |
| Ki-67 (mean ± SD) | 13.91 ± 11.64 | 24.93 ± 16.68 | <0.001 |
| Cohort | Modeling/Test | Accuracy | F1 Score |
|---|---|---|---|
| MMP | 447/79 | 84.8% | 0.8889 |
| ODX | 1733/306 | 87.9% | 0.9287 |
| Ensemble (MMP + ODX) | 2180/385 | 86.8% | 0.9184 |
| MGA Cohort | Sensitivity | Specificity | Precision | F1 Score | Accuracy |
|---|---|---|---|---|---|
| MMP | 0.8571 | 0.8261 | 0.9231 | 0.8889 | 0.848 |
| ODX | 0.9341 | 0.5833 | 0.9234 | 0.9287 | 0.879 |
| Ensemble | 0.9140 | 0.6620 | 0.9228 | 0.9184 | 0.868 |
| Modeling Dataset | Test Dataset | Modeling/Test | Accuracy | F1 Score |
|---|---|---|---|---|
| MMP data | ODX data | 447/2039 | 77.0% | 0.8584 |
| ODX data | MMP data | 1733/526 | 71.5% | 0.8077 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, J.-H.; Ahn, S.G.; Yoo, Y.; Park, S.-Y.; Kim, J.-H.; Jeong, J.-Y.; Park, S.; Lee, I. Prediction of a Multi-Gene Assay (Oncotype DX and Mammaprint) Recurrence Risk Group Using Machine Learning in Estrogen Receptor-Positive, HER2-Negative Breast Cancer—The BRAIN Study. Cancers 2024, 16, 774. https://doi.org/10.3390/cancers16040774
Ji J-H, Ahn SG, Yoo Y, Park S-Y, Kim J-H, Jeong J-Y, Park S, Lee I. Prediction of a Multi-Gene Assay (Oncotype DX and Mammaprint) Recurrence Risk Group Using Machine Learning in Estrogen Receptor-Positive, HER2-Negative Breast Cancer—The BRAIN Study. Cancers. 2024; 16(4):774. https://doi.org/10.3390/cancers16040774
Chicago/Turabian StyleJi, Jung-Hwan, Sung Gwe Ahn, Youngbum Yoo, Shin-Young Park, Joo-Heung Kim, Ji-Yeong Jeong, Seho Park, and Ilkyun Lee. 2024. "Prediction of a Multi-Gene Assay (Oncotype DX and Mammaprint) Recurrence Risk Group Using Machine Learning in Estrogen Receptor-Positive, HER2-Negative Breast Cancer—The BRAIN Study" Cancers 16, no. 4: 774. https://doi.org/10.3390/cancers16040774
APA StyleJi, J.-H., Ahn, S. G., Yoo, Y., Park, S.-Y., Kim, J.-H., Jeong, J.-Y., Park, S., & Lee, I. (2024). Prediction of a Multi-Gene Assay (Oncotype DX and Mammaprint) Recurrence Risk Group Using Machine Learning in Estrogen Receptor-Positive, HER2-Negative Breast Cancer—The BRAIN Study. Cancers, 16(4), 774. https://doi.org/10.3390/cancers16040774









