Simple Summary
In our glioblastoma patients treated with standard therapy, the TERTp C250T mutation occurred less frequently than the C228T mutation. Patients with the C250T mutation had better prognosis than those with either TERTp-wt or TERTp C228T mutations, even when adjusted for key glioblastoma prognostic factors. This may be due to the lesser involvement of the C250T mutation in telomere- and chromosome-related pathways, as evidenced by the results of a gene enrichment analysis adjusted for MGMTp methylation status: TERTp C250T was differentially enriched compared to TERTp-wt and C228T. There were no differences according to TERTp mutation status in the mutations or copy number variants of other genes commonly present in glioblastoma. The biological pathways by which TERTp and MGMTp exert their effects are independent.
Abstract
The aim of this study was to determine how TERTp mutations impact glioblastoma prognosis. Materials and Methods: TERTp mutations were assessed in a retrospective cohort of 258 uniformly treated glioblastoma patients. RNA-sequencing and whole exome sequencing results were available in a subset of patients. Results: Overall, there were no differences in outcomes between patients with mutated TERTp-wt or TERTp. However, we found significant differences according to the type of TERTp mutation. Progression-free survival (mPFS) was 9.1 months for those with the C250T mutation and 7 months for those with either the C228T mutation or TERTp-wt (p = 0.016). Overall survival (mOS) was 21.9 and 15 months, respectively (p = 0.026). This differential effect was more pronounced in patients with MGMTp methylation (mPFS: p = 0.008; mOS: p = 0.021). Multivariate analysis identified the C250T mutation as an independent prognostic factor for longer mOS (HR 0.69; p = 0.044). We found no differences according to TERTp mutation status in molecular alterations common in glioblastoma, nor in copy number variants in genes related to alternative lengthening of telomeres. Nevertheless, in the gene enrichment analysis adjusted for MGMTp methylation status, some Reactome gene sets were differentially enriched, suggesting that the C250T mutation may exert a lesser effect on telomeres or chromosomes. Conclusions: In our series, patients exhibiting the C250T mutation had a more favorable prognosis compared to those with either TERPp-wt or TERTp C228T mutations. Additionally, our findings suggest a reduced involvement of the C250T mutation in the underlying biological mechanisms related to telomeres.
1. Introduction
Glioblastoma is currently defined by the WHO 2021 classification as the highest tumor grade in the astrocyte lineage, without mutations in the isocitrate dehydrogenase (IDH) genes. It is the most frequent malignant CNS tumor in adults, with few therapeutic options, and progress in the search for new treatment strategies has been very slow. Median survival for patients receiving the standard treatment of maximal resection followed by post-operative chemoradiotherapy with temozolomide is around 15 months, while it is only 8 months for those who are ineligible for this treatment [1]. Recognized prognostic factors used for therapeutic decisions include age, extent of surgery, KPS, and methyl-guanine-methyl-transferase promoter (MGMTp) methylation [2,3].
Telomeres are highly repetitive non-coding DNA regions located at the terminal end of eukaryotic chromosomes; they prevent chromosome recombination, end-to-end fusion, and DNA-damage recognition. They control the replicative capacity of human cells, as they shorten with each cell division until they reach a certain limit, after which they cannot continue to function, leading to cell apoptosis or senescence [4]. Telomerase reactivation may occur via multiple genetic and epigenetic mechanisms, including TERT and TERC (the RNA template component of the telomerase structure) amplification; genomic rearrangements; somatic mutations within TERTp; and epigenetic modifications through TERTp methylation [4,5,6,7]. In addition, telomeres may be maintained by alternative lengthening of telomeres (ALT), which involves mutations in the genes encoding for the α-thalassemia/mental retardation syndrome X-linked protein (ATRX); the death domain-associated protein (DAXX); and the SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 1 (SMARCA1) [8,9]. Telomerase activation is estimated to occur in 85–90% of cancers; ALT pathway is observed in approximately 10–15% of them [10].
Telomerase reverse transcriptase promoter (TERTp, HGNC:11730) mutations are frequent in multiple tumor types [11], and are present in 70–80% of glioblastomas and around 80% of oligodendrogliomas, where they always co-occur with IDH1/2 mutations [11,12,13]. The presence of a TERTp mutation in a morphologically low-grade IDH-wild-type (wt) glioma reclassifies it as glioblastoma, as these tumors will behave similarly to glioblastoma in terms of outcome [14]. However, TERTp mutations do not constitute a molecular grading factor among oligodendrogliomas, which have a much better prognosis [15,16]. Several TERTp mutations have been described but they primarily occur at one of two hotspots as a C-to-T transition in the coding strand. The most frequent transitions—g.1295228C>T (chr5, hg19) (C228T) and g.1295250C>T (chr5, hg19) (C250T)—are located at −124 and −146 bp, respectively, upstream of the start codon, and are mutually exclusive [11]. C228T is by far the most common TERTp mutation except in skin cancer, where C250T occurs almost as frequently [17].
Contradictory results have been reported on the prognostic impact of TERTp mutations in glioblastoma. Some studies have found that TERTp mutations are a negative prognostic factor, both in gliomas in general [18] and in glioblastomas in particular [12,19,20,21], while others have found no differences in outcomes associated with TERTp mutations [22,23,24,25,26] if other known prognostic factors are taken into account [22]. The potential interaction between MGMTp methylation and TERTp mutations on clinical results has also been extensively investigated, though without definitive results at the biological level [22,25,26,27,28].
We have analyzed the prognostic value of TERTp mutations in a retrospective cohort of 258 uniformly treated glioblastoma patients. In addition to data on TERTp mutation status, we have collected information on all other known prognostic factors, including MGMTp methylation status. For further analyses of other molecular alterations potentially related to TERTp mutations and their effect on prognosis, we had RNA-sequencing (RNA-Seq) and whole exome sequencing analysis (WES) results available in a subset of the patients.
2. Material and Methods
2.1. Patients
This was a retrospective multicenter study including 258 patients diagnosed and treated at six Spanish institutions from May 2005 to November 2022. All patients were diagnosed with glioblastoma IDH wt and were uniformly treated with the standard treatment of surgery followed by concomitant radiation plus temozolomidec, and adjuvant temozolomide. The patients were included in two previous studies by the GLIOCAT group. Clinical data were collected from hospital records. The primary endpoint of the study was to determine the impact of TERTp mutation status on patient outcome, taking into account other known prognostic markers.
2.2. Molecular Analyses
Diffuse midline glioma, H3 p.K28M (K27M)/p.K28I (K27I) and diffuse hemispheric glioma H3.3 p.G35R (G34R)/p.G35V (G34V) were ruled out via immunohistochemical analysis as previously described [29] and WES. MGMTp methylation was assessed via methylation-specific PCR as previously described [30]. The presence of IDH mutations was ruled out via immunohistochemistry, and/or via PCR in patients younger than 56 years or via WES. TERTp mutation status was determined using PCR/Sanger sequencing in 240 patients (93%); the Oncomine Comprehensive Assay and the IonChef-S5 Sequence Platform (Thermo Fisher, Waltham, MA, USA) followed by analysis with Ion Reporter Software (Thermo Fisher, https://ionreporter.thermofisher.com/ir/, last accessed on 21 November 2022) in 12 (4.7%); and with FoundationOne CDx (Roche Foundation, Cambridge, MA, USA) in 6 (2.3%). The lack of detection of TERTp mutations on these last two tests did not automatically classify the tumor as TERTp-wt unless the wt status was confirmed through alternative methods.
For PCR/Sanger sequencing, TERTp was amplified with the forward 5′-GCACCCGTCCTGCCCCTTCACC-3′ and reverse 5′-GGCTTCCCACGTGCGCAGCAGGA-3′ primers using DNA obtained from macrodissected formalin-fixed paraffin-embedded (FFPE) tumor tissue. The PCR conditions were standard, with an annealing temperature of 62 °C, 2 mM MgCl2 and 5% DMSO. PCR bands were cleaned up with IlustraTMExoProStarTM 1-Step (GE HealthCare, Chicago, IL, USA) and sequenced with the forward primer and the BigDye® Terminator v1.1 Cycle sequencing kit (Applied Biosystems, Waltham, MA, USA). Sequences were processed on SeqStudio Genetic Analyzer (Applied Biosystems) and analyzed with SeqScanner2 software 2 (version 2.0).
Results of the TERTp mutation analyses were unified, and tumors were classified as having the C228T or the C250T mutation or as TERTp-wt.
RNA-Seq had been performed on FFPE samples from 85 patients, as previously described [31]. Differential gene expression analysis comparing samples with the C228T or C250T mutation, or TERTp-wt, was performed with limma-voom [32] using the RSEM [33] raw counts as input and adjusting the model for age, sex and, in a second analysis, also by MGMTp methylation status. Data were analyzed with the Kruskal–Wallis test with TERT log2cpm. Gene set enrichment analysis (GSEA) was done with the R fgsea package [34,35], using the output of the differential expression analysis. Genes were ranked by the limma-moderated t-statistic (Preranked-GSEA) and Reactome gene sets were used as the predefined gene list. The direction of the enrichment was provided by the normalized enrichment score (NES), a metric that corrects for differences in the enrichment score between gene sets due to differences in gene set size. Reactome pathways with an adjusted p-value < 0.05 were considered significant.
WES had been performed on DNA from the FFPE samples from 92 patients. SureSelect Human All Exon V5 (Agilent Technologies, Santa Clara, CA, USA) capture was used for whole exome enrichment with the modified protocol for FFPE samples. The KAPA Hyper Prep Kit (Roche, Basel, Switzerland) was used for DNA pre-capture library preparation. The starting material was 0.2–0.5 μg of FFPE-extracted genomic DNA (gDNA). The gDNA was sheared on a Covaris E220 Plus (Covaris, Woburn, MA, USA), end-repaired and adenylated. Illumina platform-compatible adaptors with unique dual indexes and unique molecular identifiers (Integrated DNA Technologies, Coralville, IA, USA) were ligated. The adaptor-modified end library was amplified with 8–18 PCR cycles using the KAPA HiFi HotStart ReadyMix (2X) BARBB (Roche, Basel, Switzerland), varying in function of the initial FFPE gDNA quality metrics. The quality of the PCR product was controlled on the Agilent 2100 Bioanalyzer 7500 chip (Agilent Technologies) to confirm size range and quantity of the library. It was then hybridized for 24 h at 65 °C with the 2720 Thermal Cycler (Applied Biosystems, Thermo Fisher), The hybridization mix was washed and the eluate PCR-amplified for 12 cycles using KAPA HiFi HotStart ReadyMix (2X). The final library size and the concentration were determined on the 2100 Bioanalyzer 7500 (Agilent Technologies).
The captured libraries were sequenced on NovaSeq 6000 (Illumina, San Diego, CA, USA) in paired-end mode with a read length of 2 × 101 bp following the manufacturer’s protocol for dual indexing. Image analysis, base calling and quality scoring of the run were processed using the manufacturer’s software, Real Time Analysis (RTA 3.4.4), followed by generation of FASTQ sequence files.
After sequencing, reads were mapped to the human genome (hs37d5) using the BWA-mem [36] with default parameters. Alignment files (BAM format) containing only properly paired, unique mapping reads were processed using Picard tools version 1.110 (http://broadinstitute.github.io/picard/ (accessed on 31 August 2017)) to add read groups and remove duplicates. The Genome Analysis Tool Kit was used for local indel realignment and base recalibration (https://www.broadinstitute.org/gatk/about/citing (accessed on 31 August 2017)) [37].
Processed BAM files were used to perform somatic variant calling of single nucleotide variants, and small insertions and deletions with Mutect2 (GATK v4.0.8.1). Since matched control samples were not available, Mutect2 was run in tumor-only mode. To further discriminate somatic from germline variants, two sets of variants were provided to Mutect2: (1) the aggregate variants from a “panel of normals” of 400 individuals; and (2) a set of human variants from gnomAD (https://gnomad.broadinstitute.org (accessed on 31 August 2017)), as included in the GATK suite. Only the variants considered as PASS by Mutect2, using FilterMutectCalls (GATK v4.0.8.1), were considered for the study. To further eliminate potential artefacts, variants had to have a minimum depth of coverage of ten reads, at least three reads supporting the alternative allele, and an alternative allele frequency >0.05.
Functional annotations were added to the resulting VCF file using snpEff [38] with the gene annotation obtained from ENSEMBL version 75 (http://www.ensembl.org/ (accessed on 31 August 2017)).
Copy number variants (CNV) were predicted using Control-FREEC [39]. A baseline created with the panel of normals was used as a control.
2.3. Statistical Analyses
Categorical variables were compared with the χ2 or Fisher’s exact test, as appropriate, and continuous variables with the Kruskal–Wallis test. Progression free survival (mPFS) was calculated from the date of surgery to the date of progression, death, last follow-up, or administrative censoring (15 March 2023). Overall survival (mOS) was calculated from the date of surgery to the date of death from any cause, last follow-up, or administrative censoring. Median PFS and OS, with their 95% confidence intervals (CIs), were estimated with the Kaplan–Meier method and compared with the log-rank test. Multivariate Cox proportional hazards models were used to assess the prognostic value of TERTp, adjusted by MGMTp methylation status, KPS, age and extent of surgery. Hazard ratios (HRs) and their 95% CIs were calculated. All reported p-values were two-sided. Analyses were performed with R software v3.4.2 and SPSS v24 (IBM).
2.4. Ethics Statement
This study was approved by the Institutional Review Board of the Hospital Germans Trias i Pujol (PI-14-016 and PI-18-259), and by the Ethics Committees of all the participating institutions and their biobanks. The study was conducted in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments. All patients or their representatives gave their written informed consent to participate in this study.
3. Results
3.1. Patients
Patient characteristics are shown in Table 1. Ninety patients were over 65 years of age. Surgery consisted of gross total resection in 103 patients, partial resection in 122, and biopsy only in 32. KPS was <80% in 59 patients. MGMTp methylation status was known in 253 patients (98.4%), 135 (52.3%) of whom had MGMTp methylation. At recurrence, patients were treated with second surgery, re-irradiation, or systemic treatment (including nitrosoureas or temozolomide), or were included in clinical trials. Bevacizumab was administered to 156 patients (60.7%) as a second or further line of treatment.

Table 1.
Patient characteristics.
3.2. TERTp Mutations
TERTp mutations were detected in 202 patients (78.7%). The C228T mutation was detected in 56.2% of patients and the C250T mutation in 22.1%, while 21.3% had TERTp-wt. In addition, one patient had the C229T mutation but was excluded from the analysis. This patient was a 53-year-old woman with a gross total resection and no MGMTp methylation; she was progression free for 11.2 months and survived for 19.7 months (Table 1).
3.3. Outcomes
Progression or death occurred in 229 patients (89.1%); 107 (76.7%) had died at the time of analysis. Median PFS for all patients was 8 months (95% CI 7.4–91) and mOS was 16.3 months (95% CI 14.2–18).
Age ≤ 65 years, KPS ≥ 80%, gross total resection, and MGMTp methylation improved prognosis in the univariate analysis. No significant differences in mPFS or mOS were observed according to TERTp mutation status (Table 2 and Supplementary Figure S1A,B). However, we found significant differences in both mPFS and mOS according to the type of TERTp mutation. Median OS was 21.9 months (95% CI 15.1–32.7) for patients with the C250T mutation, compared to 16 months (95% CI 14–17.1) for those with the C228T mutation, and 14.2 months (95% CI 11.9–22.4) for those with TERTp-wt (p = 0.047) (Table 2 and Supplementary Figure S2A). When patients with the C250T mutation were compared to all other patients (C228T and TERTp-wt), the significance increased: mOS for those with the C250T mutation was 21.9 months (95% CI 15.1–32.7), while it was 15 months (95% CI 14–17.1) for those with either the C228T mutation or TERTp-wt (p = 0.016) (Table 2 and Supplementary Figure S2B). Differences in mPFS were similar (Table 2 and Supplementary Figure S3A,B).
As expected, patients with methylated MGMTp had better prognosis than those with unmethylated MGMTp, regardless of TERTp mutation status (Supplementary Table S1). However, the greatest benefit in both mPFS and mOS was for patients with both MGMTp methylation and the TERTp C250T mutation: mPFS for these patients was 12.1 months (95% CI 9.8–22.3) (p = 0.002) and mOS was 24.8 months (95% CI 21.7–not reached) (p = 0.007) (Table 2 and Figure 1A,B).
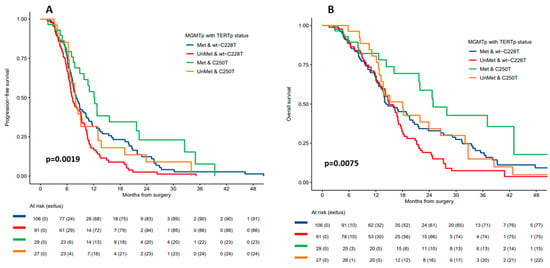
Figure 1.
(A) Progression-free survival and (B) overall survival according to MGMTp methylation status and TERTp mutation status (C250T versus C228T mutation + TERTp-wt).

Table 2.
Median progression-free survival (mPFS) and median overall survival (mOS) according to TERTp mutation status and MGMTp methylation status.
Table 2.
Median progression-free survival (mPFS) and median overall survival (mOS) according to TERTp mutation status and MGMTp methylation status.
| Comparisons | mPFS | mOS | |||
|---|---|---|---|---|---|
| Months (95% CI) | p | Months (95% CI) | p | ||
| All patients | 8 (7.4–9.1) | 16.3 (14.2–18) | |||
| TERTp wt vs. mut | wt | 7 (6.3–9) | 0.103 | 14.2 (11.9–22.4) | 0.801 |
| C228T + C250T | 8.4 (7.6–9.8) | 16.7 (14.7–18.4) | |||
| TERTp C250T vs. C228T vs. wt | C250T | 9.1 (7.8–12.8) | 0.048 | 21.9 (15.1–32.7) | 0.047 |
| C228T | 8.1 (7.3–9.4) | 16 (14–17.1) | |||
| wt | 7 (6.3–9) | 14.2 (11.9–22.4) | |||
| TERTp C250T vs. Wt + C228T | C250T | 9.1 (7.8–12.8) | 0.026 | 21.9 (15.1–32.7) | 0.016 |
| wt + C228T | 7.7 (7.1–8.9) | 15 (14–17.1) | |||
| MGMTp status & TERTp C250T vs. Wt + C228T | Met & C250T | 12.1 (9.8–22.3) | 0.002 | 24.8 (21.7–NR) | 0.007 |
| Met & wt + C228T | 8.1 (7.3–1.3) | 14.2 (13.4–2.7) | |||
| UnMet & wt + C228T | 7.1 (6.7–9.2) | 15 (14–17.1) | |||
| UnMet & C250T | 7.9 (6.6–13) | 18 (13.2–32.7) | |||
wt, wild-type; mut, mutated; Met, methylated; UnMet, unmethylated; NR: not reached; When TERTp mutation status was included in multivariate analyses together with age, extent of surgery, KPS and MGMTp methylation, the C250T mutation—but not the C228T mutation—emerged as a prognostic factor for longer mOS (HR = 0.69, 95% CI 0.48–0.99; p = 0.04) (Table 2 and Figure 2A,B).
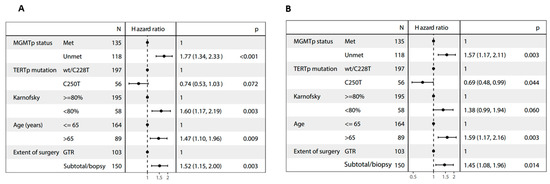
Figure 2.
Forest plots of multivariate analyses of (A) progression-free survival and (B) overall survival.
Of the 92 patients with informative WES results, three harbored TERT variants, all of which were protein-coding missense mutations: one with c.2935C>T; one with c.2572C>T; and one with both c.1457G>A and c.470C>T. The first three of these mutations are possibly pathogenic, while the fourth is considered benign. All three patients also had the TERTp C228T mutation. We detected no CNV of the TERT gene.
The analysis of genes related to glioblastoma, ALT or TERT transcription: EGFR amplification; mutations in EGFR, TP53, PTEN, BRAF, PI3K, MYC, DAXX, SMARCA, and ATRX; CNV of CDKN2A/B; and RNA expression of the LncRNA TERC showed no differences in the distribution of these alterations depending on the TERTp mutation status (Table 3). Similarly, there were no significant distinctions when patients were grouped based on C250 or C228T mutations and wild-type status.

Table 3.
Molecular alterations according to TERTp mutation status, as detected via RNA-Seq and/or WES in tumor samples.
Additionally, there were no fusions of another gene with TERT and no differences in TERT RNA expression associated with TERTp mutation status (Supplementary Figure S4).
RNA-Seq comparing samples according to TERTp mutation status, using a false discovery rate (FDR) threshold of <5%, did not reveal any significant differences associated with TERTp mutation status. However, the gene set enrichment analysis showed that some Reactome gene sets were differentially enriched, with significant differences in the NES (Figure 3). These results were maintained even when adjusting for MGMTp methylation status. Compared to TERTp-wt samples, those with TERTp mutations were enriched in pathways involved with telomeres (packaging of telomere ends; telomere maintenance; inhibition of DNA recombination at telomere pathways; DNA damage/telomere stress-induced senescence; and extension of telomeres) and chromosomes (prophase/prometaphase chromosome condensation; chromosome maintenance; chromatin-modifying enzymes; and chromatin organization).
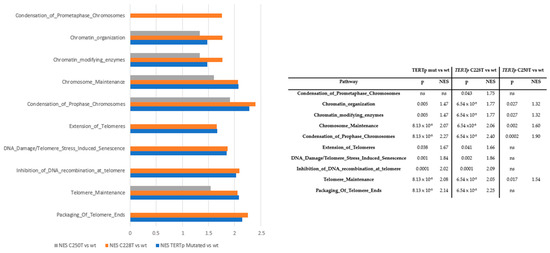
Figure 3.
Gene enrichment analysis according to TERTp mutation status adjusted for age, sex and MGMTp methylation status. NES: normalized enrichment score.
Interestingly, the greatest enrichment of these pathways was seen in the comparison between samples with the C228T mutation and those with TERTp-wt, particularly in pathways involved in prophase/prometaphase chromosome condensation. There was also some enrichment observed in the comparison between samples with the C250T mutation and those with TERTp-wt, but the significance was maintained only in pathways involved in telomere maintenance; prophase/metaphase chromosome condensation; chromosome maintenance; chromatin-modifying enzymes; and chromatin organization. No significant differences in these pathways were found between samples with the C228T or C250T mutations.
4. Discussion
In the present study, TERTp mutations in general did not affect prognosis, but the TERTp C250T mutation was associated with longer mPFS (p = 0.026) and mOS (p = 0.016) compared to the C228T mutation or TERTp-wt. The observed effect persisted in terms of mOS during multivariate analyses, which incorporated established prognostic factors such as age, extent of surgery, KPS, and MGMTp methylation status (p = 0.04). Specifically, the C250T mutation exhibited a favorable impact on prognosis, even among patients with MGMTp unmethylation. The mOS was 24.8 months for individuals with MGMTp methylation and 18 months for those without MGMTp methylation, surpassing the outcomes associated with the C228T mutation (16 and 14.8 months, respectively).
Notably, TERTp mutations, particularly C228T, have been linked to poorer prognosis in head and neck cancer patients [40]. Conversely, a study involving 887 gliomas of various grades reported a positive association between the C250T mutation and improved prognosis [41].
The adverse prognostic implications of TERTp mutations in lower-grade astrocytomas prompted numerous investigations into their potential impact on glioblastoma prognosis, particularly their potential interaction with MGMTp methylation [19,22,25,26,27,42]. However, inconsistencies arose from variations in study methodologies, including the incorporation of recognized prognostic factors, diverse treatment approaches, and limited examination of differences in prognosis based on the specific type of TERTp mutation.
An analysis by Nguyen et al., involving 303 glioblastoma patients, failed to identify significant differences in mPFS or mOS based on TERTp mutation status. Interestingly, they observed that MGMTp methylation only improved prognosis in the presence of TERTp mutations, while the mutations worsened prognosis in patients without MGMTp methylation [25]. A separate study by Arita et al., encompassing 260 IDH-wt glioblastoma patients, validated in a larger cohort, suggested that patients with both MGMTp methylation and TERTp mutations exhibited the most favorable prognosis, followed by those with TERTp wt and MGMTp methylation [22].
Despite these findings, data on the specific impact of the type of TERTp mutation were lacking. Furthermore, a meta-analysis indicated that the influence of TERTp mutations might be modulated by MGMTp methylation, suggesting that not all patients with methylated MGMTp would benefit from temozolomide, but only those with concurrent TERTp mutations [43].
Supporting this notion, a methylome analysis incorporating TERTp mutation status revealed a distinct methylation profile between mutated and wild-type TERTp tumors, implying potential differences in sensitivity to temozolomide [28].
In contrast, other studies have reported a lack of interaction between MGMTp methylation and TERTp mutation status [26,27]. Gramatzki et al. [26] included 298 patients from the German Glioma Network (GGN), 205 of whom received the standard treatment, and 302 patients from an independent retrospective cohort, 238 of whom received the standard treatment. Each cohort was analyzed separately, and neither showed variations in mPFS or mOS based on the combination of MGMTp methylation and TERTp mutations. In the GGN cohort, there were 42 patients with the C250T mutation, while the retrospective cohort included 45 patients with this mutation. Although slight variations in mPFS and mOS were observed initially between these two mutations, these distinctions ceased to hold statistical significance in the subsequent multivariate analyses (supplementary data in [26]).
Our findings in the present study indicate that both MGMTp methylation and TERTp mutations seem to affect prognosis independently, and that the TERTp C250T mutation improved the prognosis of patients regardless of their MGMTp methylation status.
The differential prognosis based on TERTp mutational status observed within our own patient cohort could stem from genomic disparities linked to TERT RNA transcription and ALT, both of which have been associated with TERTp mutation status [11]. Transcription factors are essential both to the regulation of TERT transcription and to the accessibility of DNA [44]. Somatic TERTp mutations have been shown to create an E-twenty-six (ETS) transcription factor binding site, enhancing the transcriptional activity of TERTp [17,45].
These mutations are also associated with increased telomerase activity and TERT upregulation in gliomas. [11,46] Specifically, gliomas with the C228T mutation exhibited a 14-fold increase in mRNA expression compared to TERTp-wild-type tumors, while those with the C250T mutation had a 7-fold increase (p < 0.001). This differential expression was hypothesized to result from varying access to transcription factors due to chromatin remodeling [24]. Notably, while both mutations generate the ETS binding site, only the C250T mutation appears to be influenced by non-canonical NF-kappa B signaling, and ETS binding to the mutant TERTp may not be sufficient to drive its transcription [47].
Contrary to these previous findings, our study did not identify differences in TERT expression based on TERTp mutation status. Several factors could contribute to this lack of association, including the small sample size; potential variations in tumor representation among samples; different combinations of TERT alpha and beta transcripts [48,49]; and the possibility of alternative biological mechanisms influencing TERT expression. The study by Salgado et al. [50] in melanoma, which similarly failed to establish a clear association between TERT expression levels and mutational status, emphasized the role of promoter methylation in TERT expression. This underscores the complexity of regulatory mechanisms governing TERT expression and highlights the need for further exploration to unravel these intricate relationships.
Nevertheless, in contrast to TERTp-wt tumors, we found that those with TERTp mutations had enrichment of the pathways involved in the biological mechanisms of telomeres and chromosomes. Differences between mutated and wt tumors were highly significant, and we observed more differential enrichment between tumors with the C228T mutation and TERTp-wt tumors than between those with the C250T mutation and TERTp-wt tumors. This leads us to suggest that the C250T mutation is less efficient in activating telomere lengthening than the C228T, which could explain both the deleterious impact of C228T and the beneficial impact of the C250T mutation on patient prognosis. These differences were maintained even when adjusted for MGMTp methylation status, suggesting that the effect of TERTp mutations and MGMTp methylation follow different—and not necessarily related—biological pathways, so that their effect on prognosis could be additive or restrictive.
In addition, we found that mutations in genes involved in ALT (ATRX, DAXX and SMARCA1), or in those indirectly regulating TERT transcription—including the MYC oncogene, which acts through the Myc/Max/Mad protein family—and the PIK3 gene family [9,51,52], were not universally or exclusively present in TERTp-wt tumors, which leads us to surmise that these tumors do not have telomeres as their main oncogenic pathway [53]. There were no rearrangements of TERT in our series, and the only three patients with TERT mutations also had the TERTp C228T mutation. Finally, our study did not reveal any special enrichment of a specific pathway in TERTp-wt patients. Consequently, we cannot posit this as an alternative explanation of our findings regarding the impact of TERTp mutations on outcome.
Our study has several limitations, including its retrospective nature and the relatively low number of patients with tumors with TERTp-wt or the C250T mutation, which is a characteristic shared by several previous reports [22,25,26]. In addition, we had no data on TERTp methylation and no functional data on the effect of differences in genomic expression on telomere lengthening or telomerase levels in tumors with different TERTp mutational status. This potential interaction between genomic expression and TERTp mutations warrants further investigation in future studies, especially in trials of TERT-targeted therapies.
5. Conclusions
In contrast to other studies, our univariate analyses showed a significant association between the C250T mutation and better prognosis, which remained significant in the multivariate analyses even when other prognostic factors were included. These differences with previous studies could be due to the highly curated clinical data we collected, as well as to the fact that we included a homogeneously treated series of patients.
In summary, our findings strongly indicate that patients carrying the TERTp C250T mutation exhibit a more favorable prognosis compared to their counterparts, irrespective of their MGMTp methylation status. This improved prognosis associated with the C250T mutation may be attributed to its lesser involvement in telomere activity compared to the C228T mutation, thereby limiting its potentially deleterious effects on patient outcomes.
Notably, our investigation did not reveal significant differences in molecular alterations related to other glioblastoma-associated genes, or in genes associated with the ALT system. This suggests that the prognostic impact of the TERTp C250T mutation may be specifically linked to its modulation of telomere activity rather than broader alterations in the molecular landscape of glioblastomas. Further research and comprehensive analyses are warranted to delve into the intricate molecular mechanisms underlying the observed prognostic differences associated with TERTp mutations, and to unravel potential therapeutic implications.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers16040735/s1, Table S1: Progression-free survival (PFS) and overall survival (OS) according to MGMTp methylation status and MGMTp and TERTp status; Figure S1: (A) Progression-free survival and (B) overall survival according to TERTp mutation status (wild-type vs. mutated); Figure S2: Overall survival according to TERTp mutation status (A) C250T mutation vs C228T mutation vs wild-type and (B) C250 mutation vs. C228T mutation and wild-type; Figure S3: Progression-free survival according to TERTp mutation status (A) C250T mutation vs C228T mutation vs wild-type and (B) C250 mutation vs. C228T mutation and wild-type; Figure S4: TERT RNA expression was assessed in relation to TERTp mutation status using the Kruskal-Wallis test. The results indicated no statistically significant differences across all comparative statuses (C2228T vs. C250T vs. wt: p = 0.25, C228T vs. wt: p = 0.1, C228T vs. C250T: p = 0.71, C250T vs. wt: p = 0.35), as all p-values exceeded the threshold of 0.05.
Author Contributions
Conceptualization, E.P., B.M., F.A. and C.B.; Methodology, A.M.M.-M., C.C. (Cristina Carrato) and C.B.; Software, A.M.M.-M., A.E.-C., G.P. and M.M.; Validation, C.S., B.M., M.G. and A.E.; Formal analysis, T.G., C.C. (Carme Crous), E.P., A.H., M.D., C.S., P.J., B.M., A.E., G.P., F.A., C.C. (Cristina Carrato), I.A. and N.d.l.I.; Investigation, C.C. (Carme Crous), E.P., A.H., M.D., C.S., P.J., A.M.M.-M., O.A.-L., M.G., A.E.-C., G.P., F.A. and I.A.; Data curation, T.G., C.C. (Carme Crous), M.G., A.E., A.E.-C., C.C. (Cristina Carrato) and C.B.; Writing—original draft, C.B.; Writing—review & editing, C.B.; Visualization, A.E.-C. and G.P.; Supervision, E.P. and C.B.; Funding acquisition, F.A., N.d.l.I. and C.B. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Fundació La Marató TV3 (665/C/2013) (http://www.ccma.cat/tv3/marato/projectes-financats/2012/231/ (accessed on 1 February 2024)) and the Instituto de la Salud Carlos III, project PI118/01062, Ministerio de Economía y Competitividad.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the Hospital Universitari Germans Trias I Pujol (code protocol PI-14-016 data 4 March 2014 and code protocol PI-18-259 data 14 December 2018) for studies involving humans.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study or their representatives.
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request. Molecular data underlying the findings described in the manuscript are fully available without restriction from the Bioproject Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/PRJNA833243; http://www.ncbi.nlm.nih.gov/bioproject/PRJNA613395; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1073422).
Acknowledgments
The authors thank the biobanks of the participating institutions: Fundació Institut Mar d’Investigacions Mèdiques; Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS); Institut d’Investigació en Ciències de la Salut Germans Trias i Pujol (IGTP); Fundació Institut de Recerca de l’Hospital de la Santa Creu i St. Pau (IIB Sant Pau); and the Xarxa de Bancs de Tumors sponsored by Pla Director d’Oncologia de Catalunya (XBTC).
Conflicts of Interest
The authors declare no conflicts of interest for the data they are presenting.
References
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013-2017. Neuro Oncol. 2020, 22, iv1–iv96. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186. [Google Scholar] [CrossRef] [PubMed]
- Mohile, N.A.; Messersmith, H.; Gatson, N.T.; Hottinger, A.F.; Lassman, A.; Morton, J.; Ney, D.; Nghiemphu, P.L.; Olar, A.; Olson, J.; et al. Therapy for Diffuse Astrocytic and Oligodendroglial Tumors in Adults: ASCO-SNO Guideline. J. Clin. Oncol. 2022, 40, 403–426. [Google Scholar] [CrossRef] [PubMed]
- Dratwa, M.; Wysoczanska, B.; Lacina, P.; Kubik, T.; Bogunia-Kubik, K. TERT-Regulation and Roles in Cancer Formation. Front. Immunol. 2020, 11, 589929. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, N.; Rachakonda, S.; Kumar, R. Telomeres and Telomere Length: A General Overview. Cancers 2020, 12, 558. [Google Scholar] [CrossRef] [PubMed]
- Chebly, A.; Ropio, J.; Peloponese, J.M.; Poglio, S.; Prochazkova-Carlotti, M.; Cherrier, F.; Ferrer, J.; Idrissi, Y.; Segal-Bendirdjian, E.; Chouery, E.; et al. Exploring hTERT promoter methylation in cutaneous T-cell lymphomas. Mol. Oncol. 2022, 16, 1931–1946. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.D.; Komosa, M.; Sudhaman, S.; Leao, R.; Zhang, C.H.; Apolonio, J.D.; Hermanns, T.; Wild, P.J.; Klocker, H.; Nassiri, F.; et al. Dual role of allele-specific DNA hypermethylation within the TERT promoter in cancer. J. Clin. Invest. 2021, 131, e146915. [Google Scholar] [CrossRef]
- Heaphy, C.M.; Subhawong, A.P.; Hong, S.M.; Goggins, M.G.; Montgomery, E.A.; Gabrielson, E.; Netto, G.J.; Epstein, J.I.; Lotan, T.L.; Westra, W.H.; et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011, 179, 1608–1615. [Google Scholar] [CrossRef]
- Diplas, B.H.; He, X.; Brosnan-Cashman, J.A.; Liu, H.; Chen, L.H.; Wang, Z.; Moure, C.J.; Killela, P.J.; Loriaux, D.B.; Lipp, E.S.; et al. The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat. Commun. 2018, 9, 2087. [Google Scholar] [CrossRef]
- Shay, J.W.; Wright, W.E. Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 2019, 20, 299–309. [Google Scholar] [CrossRef]
- Huang, D.S.; Wang, Z.; He, X.J.; Diplas, B.H.; Yang, R.; Killela, P.J.; Meng, Q.; Ye, Z.Y.; Wang, W.; Jiang, X.T.; et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur. J. Cancer 2015, 51, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Killela, P.J.; Pirozzi, C.J.; Healy, P.; Reitman, Z.J.; Lipp, E.; Rasheed, B.A.; Yang, R.; Diplas, B.H.; Wang, Z.; Greer, P.K.; et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget 2014, 5, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, K. TERT promoter mutation as a diagnostic marker for diffuse gliomas. Neuro Oncol. 2019, 21, 417–418. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Arita, H.; Satomi, K.; Yamasaki, K.; Matsushita, Y.; Nakamura, T.; Miyakita, Y.; Umehara, T.; Kobayashi, K.; Tamura, K.; et al. TERT promoter mutation status is necessary and sufficient to diagnose IDH-wildtype diffuse astrocytic glioma with molecular features of glioblastoma. Acta Neuropathol. 2021, 142, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Stichel, D.; Ebrahimi, A.; Reuss, D.; Schrimpf, D.; Ono, T.; Shirahata, M.; Reifenberger, G.; Weller, M.; Hanggi, D.; Wick, W.; et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018, 136, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Berzero, G.; Di Stefano, A.L.; Ronchi, S.; Bielle, F.; Villa, C.; Guillerm, E.; Capelle, L.; Mathon, B.; Laurenge, A.; Giry, M.; et al. IDH-wildtype lower-grade diffuse gliomas: The importance of histological grade and molecular assessment for prognostic stratification. Neuro Oncol. 2021, 23, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly recurrent TERT promoter mutations in human melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef] [PubMed]
- Geng, P.; Zhao, X.; Ou, J.; Li, J.; Sa, R.; Liang, H. TERT Genetic Mutations as Prognostic Marker in Glioma. Mol. Neurobiol. 2017, 54, 3665–3669. [Google Scholar] [CrossRef]
- Malkki, H. Neuro-oncology: TERT promoter mutations could indicate poor prognosis in glioblastoma. Nat. Rev. Neurol. 2014, 10, 546. [Google Scholar]
- Simon, M.; Hosen, I.; Gousias, K.; Rachakonda, S.; Heidenreich, B.; Gessi, M.; Schramm, J.; Hemminki, K.; Waha, A.; Kumar, R. TERT promoter mutations: A novel independent prognostic factor in primary glioblastomas. Neuro Oncol. 2015, 17, 45–52. [Google Scholar] [CrossRef]
- Arita, H.; Ichimura, K. Prognostic significance of TERT promoter mutations in adult-type diffuse gliomas. Brain Tumor Pathol. 2022, 39, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Arita, H.; Yamasaki, K.; Matsushita, Y.; Nakamura, T.; Shimokawa, A.; Takami, H.; Tanaka, S.; Mukasa, A.; Shirahata, M.; Shimizu, S.; et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol. Commun. 2016, 4, 79. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, B.; Rachakonda, P.S.; Hosen, I.; Volz, F.; Hemminki, K.; Weyerbrock, A.; Kumar, R. TERT promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget 2015, 6, 10617–10633. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Lie, A.; Li, T.; Chowdhury, R.; Liu, F.; Ozer, B.; Wei, B.; Green, R.M.; Ellingson, B.M.; Wang, H.J.; et al. Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro Oncol. 2017, 19, 394–404. [Google Scholar] [PubMed]
- Gramatzki, D.; Felsberg, J.; Hentschel, B.; Wolter, M.; Schackert, G.; Westphal, M.; Regli, L.; Thon, N.; Tatagiba, M.; Wick, W.; et al. Telomerase reverse transcriptase promoter mutation– and O6-methylguanine DNA methyltransferase promoter methylation–mediated sensitivity to temozolomide in isocitrate dehydrogenase–wild-type glioblastoma: Is there a link? Eur. J. Cancer 2021, 147, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Giunco, S.; Padovan, M.; Angelini, C.; Cavallin, F.; Cerretti, G.; Morello, M.; Caccese, M.; Rizzo, B.; d’Avella, D.; Puppa, A.D.; et al. Prognostic role and interaction of TERT promoter status, telomere length and MGMT promoter methylation in newly diagnosed IDH wild-type glioblastoma patients. ESMO Open 2023, 8, 101570. [Google Scholar] [CrossRef]
- Kessler, T.; Berberich, A.; Sadik, A.; Sahm, F.; Gorlia, T.; Meisner, C.; Hoffmann, D.C.; Wick, A.; Kickingereder, P.; Rubmann, P.; et al. Methylome analyses of three glioblastoma cohorts reveal chemotherapy sensitivity markers within DDR genes. Cancer Med. 2020, 9, 8373–8385. [Google Scholar] [CrossRef]
- Carrato, C.; Alameda, F.; Esteve-Codina, A.; Pineda, E.; Arpi, O.; Martinez-Garcia, M.; Mallo, M.; Gut, M.; Lopez-Martos, R.; Barco, S.D.; et al. Glioblastoma TCGA Mesenchymal and IGS 23 Tumors are Identifiable by IHC and have an Immune-phenotype Indicating a Potential Benefit from Immunotherapy. Clin. Cancer Res. 2020, 26, 6600–6609. [Google Scholar] [CrossRef]
- Estival, A.; Sanz, C.; Ramirez, J.-L.; Maria Velarde, J.; Domenech, M.; Carrato, C.; de las Penas, R.; Gil-Gil, M.; Sepulveda, J.; Armengol, R.; et al. Pyrosequencing versus methylation-specific PCR for assessment of MGMT methylation in tumor and blood samples of glioblastoma patients. Sci. Rep. 2019, 9, 11125. [Google Scholar] [CrossRef]
- Esteve-Codina, A.; Alameda, F.; Carrato, C.; Pineda, E.; Arpi, O.; Martinez Garcia, M.; Mallo, M.; Gut, M.; Dabad, M.; Tortosa, A.; et al. RNA-Sequencing and immunohistochemistry reveal ZFN7 as a stronger marker of survival than molecular subtypes in G-CIMP-negative glioblastoma. Clin. Cancer Res. 2020, 27, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Sergushichev, A.A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv 2016, 60012, 1–9. [Google Scholar]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Boeva, V.; Popova, T.; Bleakley, K.; Chiche, P.; Cappo, J.; Schleiermacher, G.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2012, 28, 423–425. [Google Scholar] [CrossRef]
- Boscolo-Rizzo, P.; Tirelli, G.; Polesel, J.; Sia, E.; Phillips, V.; Borsetto, D.; De Rossi, A.; Giunco, S. TERT promoter mutations in head and neck squamous cell carcinoma: A systematic review and meta-analysis on prevalence and prognostic significance. Oral. Oncol. 2023, 140, 106398. [Google Scholar] [CrossRef]
- You, H.; Wu, Y.; Chang, K.; Shi, X.; Chen, X.D.; Yan, W.; Li, R. Paradoxical prognostic impact of TERT promoter mutations in gliomas depends on different histological and genetic backgrounds. CNS Neurosci. Ther. 2017, 23, 790–797. [Google Scholar] [CrossRef]
- Huse, J.T. TERT promoter mutation designates biologically aggressive primary glioblastoma. Neuro Oncol. 2015, 17, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.G.; Nguyen, T.Q.; Ngo, T.N.M.; Nguyen, H.C.; Fung, K.M.; Dunn, I.F. The interaction between TERT promoter mutation and MGMT promoter methylation on overall survival of glioma patients: A meta-analysis. BMC Cancer 2020, 20, 897. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 2019, 38, 6172–6183. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, B.; Kumar, R. TERT promoter mutations in telomere biology. Mutat. Res. Rev. Mutat. Res. 2017, 771, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.S.; Wen, J.; Bacchetti, S. The human telomerase catalytic subunit hTERT: Organization of the gene and characterization of the promoter. Hum. Mol. Genet. 1999, 8, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, Q.L.; Sun, W.; Chandrasekharan, P.; Cheng, H.S.; Ying, Z.; Lakshmanan, M.; Raju, A.; Tenen, D.G.; Cheng, S.Y.; et al. Non-canonical NF-kappaB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat. Cell Biol. 2015, 17, 1327–1338. [Google Scholar] [CrossRef]
- Ropio, J.; Prochazkova-Carlotti, M.; Batista, R.; Pestana, A.; Chebly, A.; Ferrer, J.; Idrissi, Y.; Cappellen, D.; Duraes, C.; Boaventura, P.; et al. Spotlight on hTERT Complex Regulation in Cutaneous T-Cell Lymphomas. Genes 2023, 14, 439. [Google Scholar] [CrossRef]
- Mavrogiannou, E.; Strati, A.; Stathopoulou, A.; Tsaroucha, E.G.; Kaklamanis, L.; Lianidou, E.S. Real-time RT-PCR quantification of human telomerase reverse transcriptase splice variants in tumor cell lines and non-small cell lung cancer. Clin. Chem. 2007, 53, 53–61. [Google Scholar] [CrossRef]
- Salgado, C.; Roelse, C.; Nell, R.; Gruis, N.; van Doorn, R.; van der Velden, P. Interplay between TERT promoter mutations and methylation culminates in chromatin accessibility and TERT expression. PLoS ONE 2020, 15, e0231418. [Google Scholar] [CrossRef]
- Ohba, S.; Kuwahara, K.; Yamada, S.; Abe, M.; Hirose, Y. Correlation between IDH, ATRX, and TERT promoter mutations in glioma. Brain Tumor Pathol. 2020, 37, 33–40. [Google Scholar] [CrossRef]
- Pierini, T.; Nardelli, C.; Lema Fernandez, A.G.; Pierini, V.; Pellanera, F.; Nofrini, V.; Gorello, P.; Moretti, M.; Arniani, S.; Roti, G.; et al. New somatic TERT promoter variants enhance the Telomerase activity in Glioblastoma. Acta Neuropathol. Commun. 2020, 8, 145. [Google Scholar] [CrossRef]
- Williams, E.A.; Miller, J.J.; Tummala, S.S.; Penson, T.; Iafrate, A.J.; Juratli, T.A.; Cahill, D.P. TERT promoter wild-type glioblastomas show distinct clinical features and frequent PI3K pathway mutations. Acta Neuropathol. Commun. 2018, 6, 106. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).