Machine-Learning-Based Classification Model to Address Diagnostic Challenges in Transbronchial Lung Biopsy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
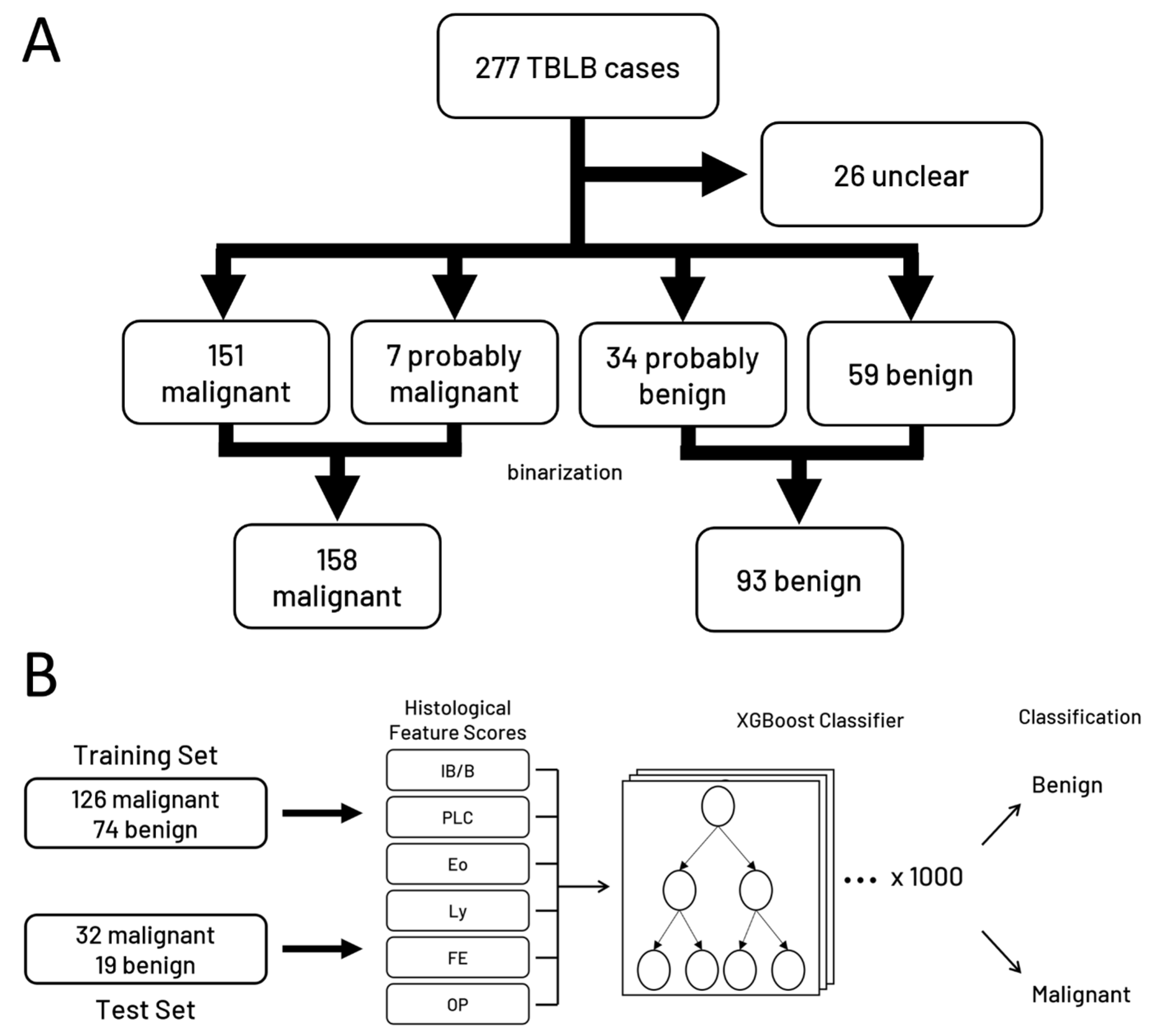
2.1. Patient Cohort
2.2. Pathological Findings and Scoring
2.3. Ground Truth
2.4. Machine Learning Model Development
2.5. Statistical Analysis
3. Results
3.1. Clinicopathological Characteristics of Patients
3.2. Pathological Findings
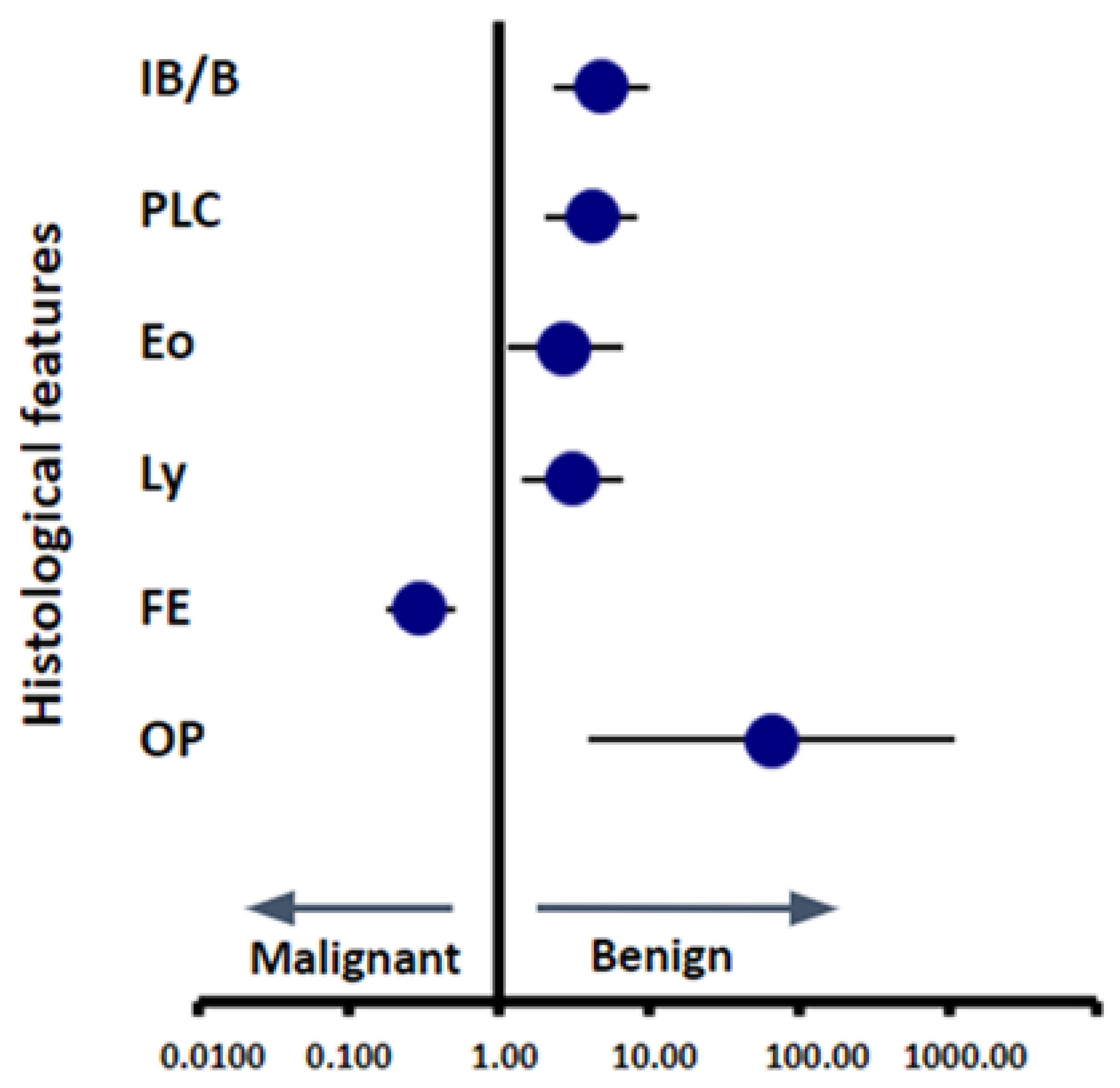
3.3. Scoring of Pathological Findings
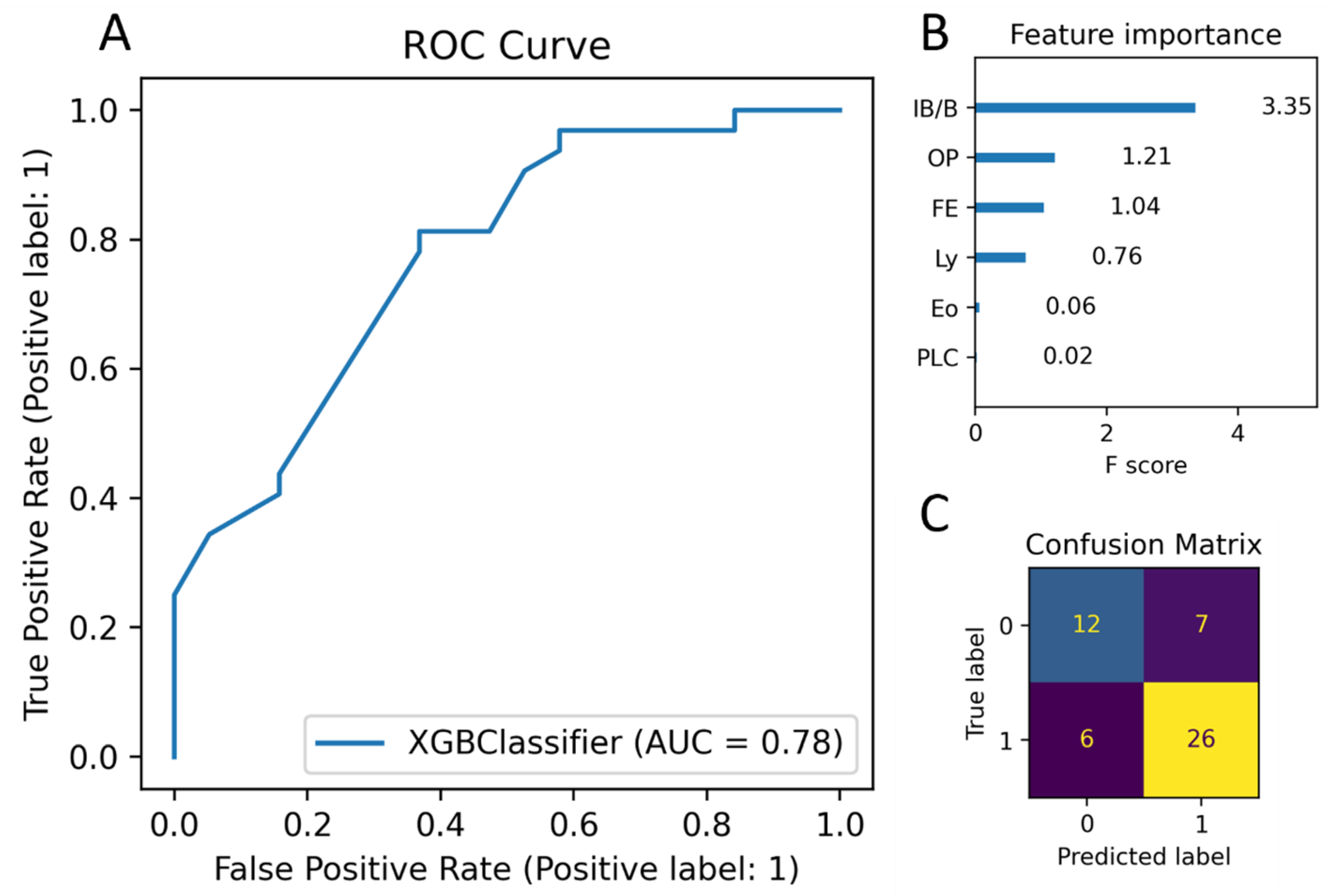
3.4. Building a Machine Learning Model to Classify Cases based on Scoring Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mazzone, P.J.; Silvestri, G.A.; Souter, L.H.; Caverly, T.J.; Kanne, J.P.; Katki, H.A.; Wiener, R.S.; Detterbeck, F.C. Screening for Lung Cancer: CHEST Guideline and Expert Panel Report. Chest 2021, 160, e427. [Google Scholar] [CrossRef]
- Oudkerk, M.; Devaraj, A.; Vliegenthart, R.; Henzler, T.; Prosch, H.; Heussel, C.P.; Bastarrika, G.; Sverzellati, N.; Mascalchi, M.; Delorme, S.; et al. European position statement on lung cancer screening. Lancet Oncol. 2017, 18, e754–e766. [Google Scholar] [CrossRef]
- Usman Ali, M.; Miller, J.; Peirson, L.; Fitzpatrick-Lewis, D.; Kenny, M.; Sherifali, D.; Raina, P. Screening for lung cancer: A systematic review and meta-analysis. Prev. Med. 2016, 89, 301–314. [Google Scholar] [CrossRef]
- Chen, F.F.; Zhang, D.; Wang, Y.L.; Xiong, B. Video-assisted thoracoscopic surgery lobectomy versus open lobectomy in patients with clinical stage Ⅰ non-small cell lung cancer: A meta-analysis. Eur. J. Surg. Oncol. 2013, 39, 957–963. [Google Scholar] [CrossRef]
- Zeng, W.; Zhang, W.; Zhang, J.; You, G.; Mao, Y.; Xu, J.; Yu, D.; Peng, J.; Wei, Y. Systematic review and meta-analysis of video-assisted thoracoscopic surgery segmentectomy versus lobectomy for stage I non-small cell lung cancer. World J. Surg. Oncol. 2020, 18, 44. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Rivera, M.P.; Mehta, A.C.; Wahidi, M.M. Establishing the diagnosis of lung cancer: Diagnosis and management of lung cancer, 3rd ed.: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013, 143 (Suppl. 5), E142S–E165S. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Du, Y.; Yang, H.F.; Yu, J.H.; Xu, X.X. CT-guided percutaneous core needle biopsy for small (≤20 mm) pulmonary lesions. Clin. Radiol. 2013, 68, e43–e48. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Park, C.M.; Kim, H.; Lee, J.H.; Goo, J.M. Non-specific benign pathological results on transthoracic core-needle biopsy: How to differentiate false-negatives? Eur. Radiol. 2017, 27, 3888–3895. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, Y.; Wang, W.; Yuan, Q.; Hu, H.d.; Li, L. Improved diagnostic yield of transbronchial lung biopsy in peripheral pulmonary lesions using a combination of endobronchial ultrasound and rapid on-site evaluation. J. Int. Med. Res. 2021, 49, 0300060521999535. [Google Scholar] [CrossRef] [PubMed]
- Bremnes, R.M.; Dønnem, T.; Al-Saad, S.; AI-Shibli, K.; Anderson, S.; Sirera, R.; Camps, C.; Marinez, I.; Busund, L.T. The Role of Tumor Stroma in Cancer Progression and Prognosis: Emphasis on Carcinoma-Associated Fibroblasts and Non-small Cell Lung Cancer. J. Thorac. Oncol. 2011, 6, 209–217. [Google Scholar] [CrossRef]
- De Wever, O.; Mareel, M. Role of tissue stroma in cancer cell invasion. J. Pathol. 2003, 200, 429–447. [Google Scholar] [CrossRef] [PubMed]
- Kradin, R.L.; Digumarthy, S. The pathology of pulmonary bacterial infection. Semin. Diagn. Pathol. 2017, 34, 498–509. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, X.L.; Feng, K.; Liu, X.Q.; Yang, J. Benign pathologies results from lung nodule percutaneous biopsies: How to differentiate true and false benign? J. Cancer Res. Ther. 2021, 17, 658–663. [Google Scholar] [CrossRef]
- Fu, Y.F.; Jiang, L.H.; Wang, T.; Li, G.C.; Cao, W.; Shi, Y.B. Nonspecific benign pathological results on computed tomography-guided lung biopsy: A predictive model of true negatives. J. Cancer Res. Ther. 2019, 15, 1464–1470. [Google Scholar] [CrossRef]
- Rui, Y.; Han, M.; Zhou, W.; He, Q.; Li, H.; Li, P.; Zhang, F.; Shi, Y.; Su, X. Non-malignant pathological results on transthoracic CT guided core-needle biopsy: When is benign really benign? Clin. Radiol. 2018, 73, e1–e757. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.; Bhushan, M. Machine learning and deep learning approach for medical image analysis: Diagnosis to detection. Multimed. Tools Appl. 2022, 82, 26731–26769. [Google Scholar] [CrossRef]
- Eswara Rao, G.V.; Rajitha, B.; Srinivasu, P.N.; Ijaz, M.F.; Woźniak, M. Hybrid framework for respiratory lung diseases detection based on classical CNN and quantum classifiers from chest X-rays. Biomed. Signal Process. Control 2024, 88, 105567. [Google Scholar] [CrossRef]
- Ahmad, I.; Merla, A.; Ali, F.; Shah, B.; AlZubi, A.A.; AlZubi, M.A. A deep transfer learning approach for COVID-19 detection and exploring a sense of belonging with Diabetes. Front. Public Health 2023, 11, 1308404. [Google Scholar] [CrossRef]
- Liu, X.L.; Li, W.; Yang, W.X.; Rui, M.P.; Li, Z.; Lv, L.; Yang, L.P. Computed tomography-guided biopsy of small lung nodules: Diagnosticaccuracy and analysis for true negatives. J. Int. Med. Res. 2020, 48, 0300060519879006. [Google Scholar] [CrossRef]
- Hui, H.; Ma, G.L.; Yin, H.T.; Zhou, Y.; Xie, X.M.; Gao, Y.G. Computed tomography-guided cutting needle biopsy for lung nodules: When the biopsy-based benign results are real benign. World J. Surg. Oncol. 2022, 20, 180. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Lim, K.Y.; Suh, Y.J.; Hur, J.; Han, D.H.; Kang, M.J.; Choo, J.Y.; Kim, C.; Kim, J.I.; Yoon, S.H.; et al. Nondiagnostic Percutaneous Transthoracic Needle Biopsy of Lung Lesions: A Multicenter Study of Malignancy Risk. Radiology 2019, 290, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Visscher, D.W.; Myers, J.L. Bronchiolitis: The pathologist’s perspective. Proc. Am. Thorac. Soc. 2006, 3, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Colby, T.V. Bronchiolitis: Pathologic Considerations. Am. J. Clin. Pathol. 1998, 109, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; King, T.E.; Bateman, E.D.; Lynch, D.A.; Capron, F.; Center, D.; Colby, T.V.; Cordier, J.F.; Dubois, R.M.; Galvin, J.; et al. American Thoracic Society/European Respiratory Society International Multidisciplinary Consensus Classification of the Idiopathic Interstitial Pneumonias. Am. J. Respir. Crit. Care Med. 2002, 165, 277–304. [Google Scholar] [CrossRef]
- Talmadge, E.; King, J.; Lee, J.S. Cryptogenic Organizing Pneumonia. N. Engl. J. Med. 2022, 386, 1058–1069. [Google Scholar] [CrossRef]
- Radzikowska, E.; Nowicka, U.; Wiatr, E.; Jakubowska, L.; Langfort, R.; Chabowski, M.; Roszkowski, K. Organising pneumonia and lung cancer—Case report and review of the literature. Pneumol. Alergol. Pol. 2007, 75, 394–397. [Google Scholar] [CrossRef]
- Mao, R.; Zhang, L.; Hou, J.; Zou, Y.; Zhu, L.; Chen, Z. Organizing pneumonia secondary to lung cancer of unknown primary site. Respir. Med. Case Rep. 2019, 28, 100892. [Google Scholar] [CrossRef]
- Romero, S.; Barroso, E.; Rodriguez-Paniagua, M.; Aranda, F.I. Organizing pneumonia adjacent to lung cancer: Frequency and clinico-pathologic features. Lung Cancer 2002, 35, 195–201. [Google Scholar] [CrossRef]
- Fukushima, M.; Fukuda, Y.; Kawamoto, M.; Yamanaka, N. Elastosis in lung carcinoma: Immunohistochemical, ultrastructural and clinical studies. Pathol. Int. 2000, 50, 626–635. [Google Scholar] [CrossRef]
- Eto, T.; Suzuki, H.; Honda, A.; Nagashima, Y. The changes of the stromal elastotic framework in the growth of peripheral lung adenocarcinomas. Cancer 1996, 77, 646–656. [Google Scholar] [CrossRef]
- Zaizen, Y.; Tachibana, Y.; Kashima, Y.; Bychkov, A.; Tabata, K.; Otani, K.; Kinoshita, Y.; Yamano, Y.; Kataoka, K.; Ichikado, K.; et al. Alveolar Epithelial Denudation Is a Major Factor in the Pathogenesis of Pleuroparenchymal Fibroelastosis. J. Clin. Med. 2021, 10, 895. [Google Scholar] [CrossRef] [PubMed]
- Ricoy, J.; Suárez-Antelo, J.; Antúnez, J.; Algería, A.M.; Ferreiro, L.; Toubes, M.E.; Casal, A.; Valdés, L. Pleuroparenchymal fibroelastosis: Clinical, radiological and histopathological features. Respir. Med. 2022, 191, 106437. [Google Scholar] [CrossRef] [PubMed]
- Kian, S.; Cheng, H.; Khoon, C.; Chuah, L. Pleuroparenchymal Fibroelastosis of the Lung: A Review. Arch. Pathol. Lab. Med. 2016, 140, 849–853. [Google Scholar] [CrossRef]



| Variable | Malignant (n = 158) | Benign (n = 93) | p-Value |
|---|---|---|---|
| Age | 73.7 ± 8.0 (74) | 68.5 ± 13.0 (71) | <0.01 |
| Gender M/F | 93/65 | 41/52 | 0.02 |
| Smoking Index | 746 ± 660 (662) | 341 ± 528 (0) | <0.01 |
| Number of specimens | 7.8 ± 3.3 (7) | 5.7 ± 2.6 (5) | <0.01 |
| Location (lobe) | >0.05 | ||
| Right upper | 54 [21%] | 29 [11%] | |
| Right middle | 10 [4%] | 14 [6%] | |
| Right lower | 38 [15%] | 17 [7%] | |
| Left upper | 36 [14%] | 19 [8%] | |
| Left lower | 20 [8%] | 14 [6%] |
| Judgement | Definition | N |
|---|---|---|
| Malignant | Pathologically confirmed malignancy at the time of biopsy, follow-up biopsy, cytology, or in subsequent surgical materials | 151 |
| Probably Malignant | Clinically diagnosed and treated as malignant without pathological evidence of malignancy | 7 |
| Unclear | No pathological evidence of malignancy, with a clinical diagnosis of difficulty in determining malignancy and ongoing follow-up | 26 |
| Probably Benign | No pathological evidence of malignancy, with a clinical diagnosis of benign disease and ongoing follow-up | 34 |
| Benign | No pathological evidence of malignancy, clinically diagnosed as benign, and treated (e.g., antibiotics for infection) or discharged | 59 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sano, H.; Okoshi, E.N.; Tachibana, Y.; Tanaka, T.; Lami, K.; Uegami, W.; Ohta, Y.; Brcic, L.; Bychkov, A.; Fukuoka, J. Machine-Learning-Based Classification Model to Address Diagnostic Challenges in Transbronchial Lung Biopsy. Cancers 2024, 16, 731. https://doi.org/10.3390/cancers16040731
Sano H, Okoshi EN, Tachibana Y, Tanaka T, Lami K, Uegami W, Ohta Y, Brcic L, Bychkov A, Fukuoka J. Machine-Learning-Based Classification Model to Address Diagnostic Challenges in Transbronchial Lung Biopsy. Cancers. 2024; 16(4):731. https://doi.org/10.3390/cancers16040731
Chicago/Turabian StyleSano, Hisao, Ethan N. Okoshi, Yuri Tachibana, Tomonori Tanaka, Kris Lami, Wataru Uegami, Yoshio Ohta, Luka Brcic, Andrey Bychkov, and Junya Fukuoka. 2024. "Machine-Learning-Based Classification Model to Address Diagnostic Challenges in Transbronchial Lung Biopsy" Cancers 16, no. 4: 731. https://doi.org/10.3390/cancers16040731
APA StyleSano, H., Okoshi, E. N., Tachibana, Y., Tanaka, T., Lami, K., Uegami, W., Ohta, Y., Brcic, L., Bychkov, A., & Fukuoka, J. (2024). Machine-Learning-Based Classification Model to Address Diagnostic Challenges in Transbronchial Lung Biopsy. Cancers, 16(4), 731. https://doi.org/10.3390/cancers16040731








