Exploring Novel Frontiers: Leveraging STAT3 Signaling for Advanced Cancer Therapeutics
Abstract
Simple Summary
Abstract
1. Introduction
2. Mechanism of the STAT3 Signaling Pathway (Figure 1 and Figure 2)
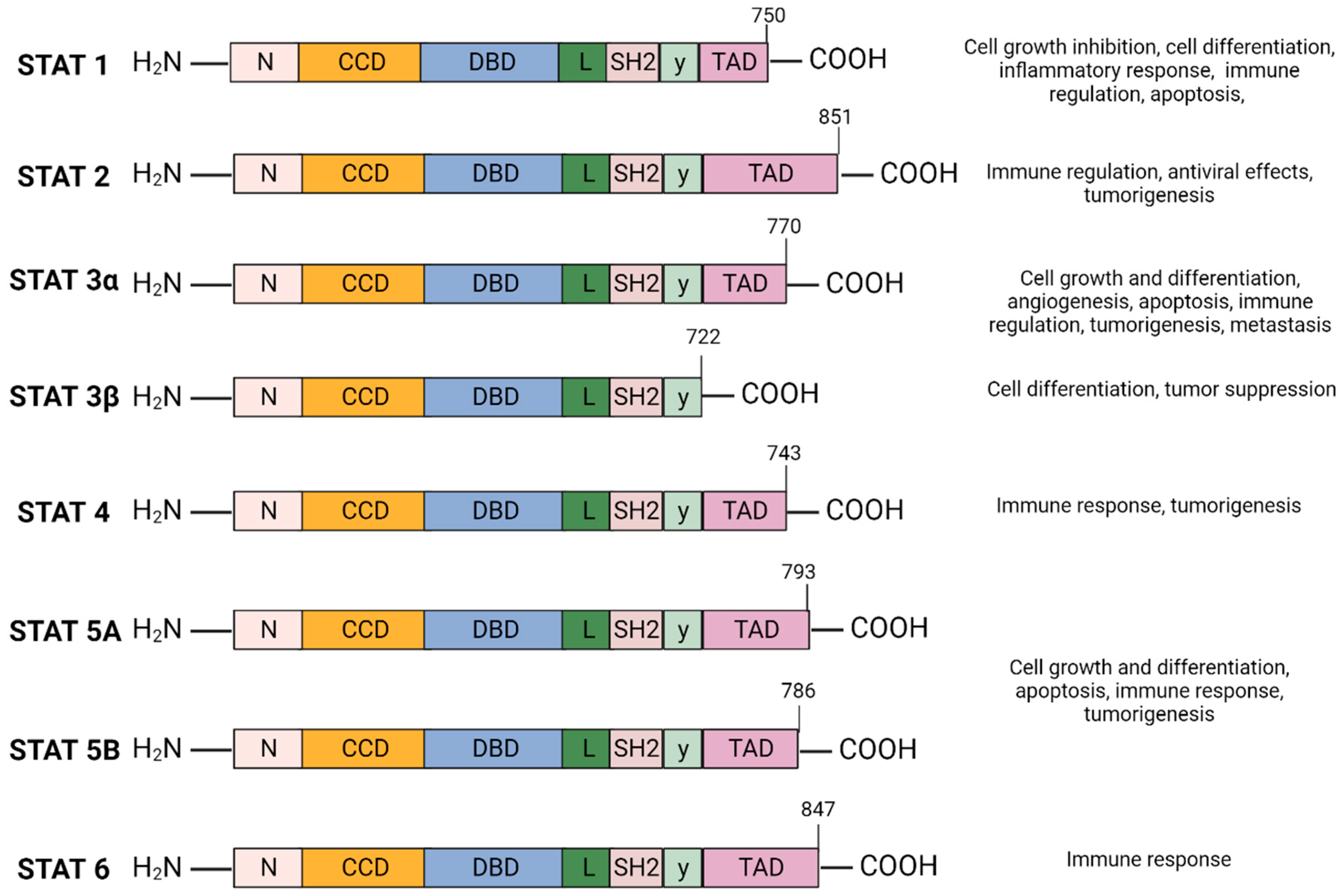
2.1. STAT3 Structure and Isoforms (Figure 1)
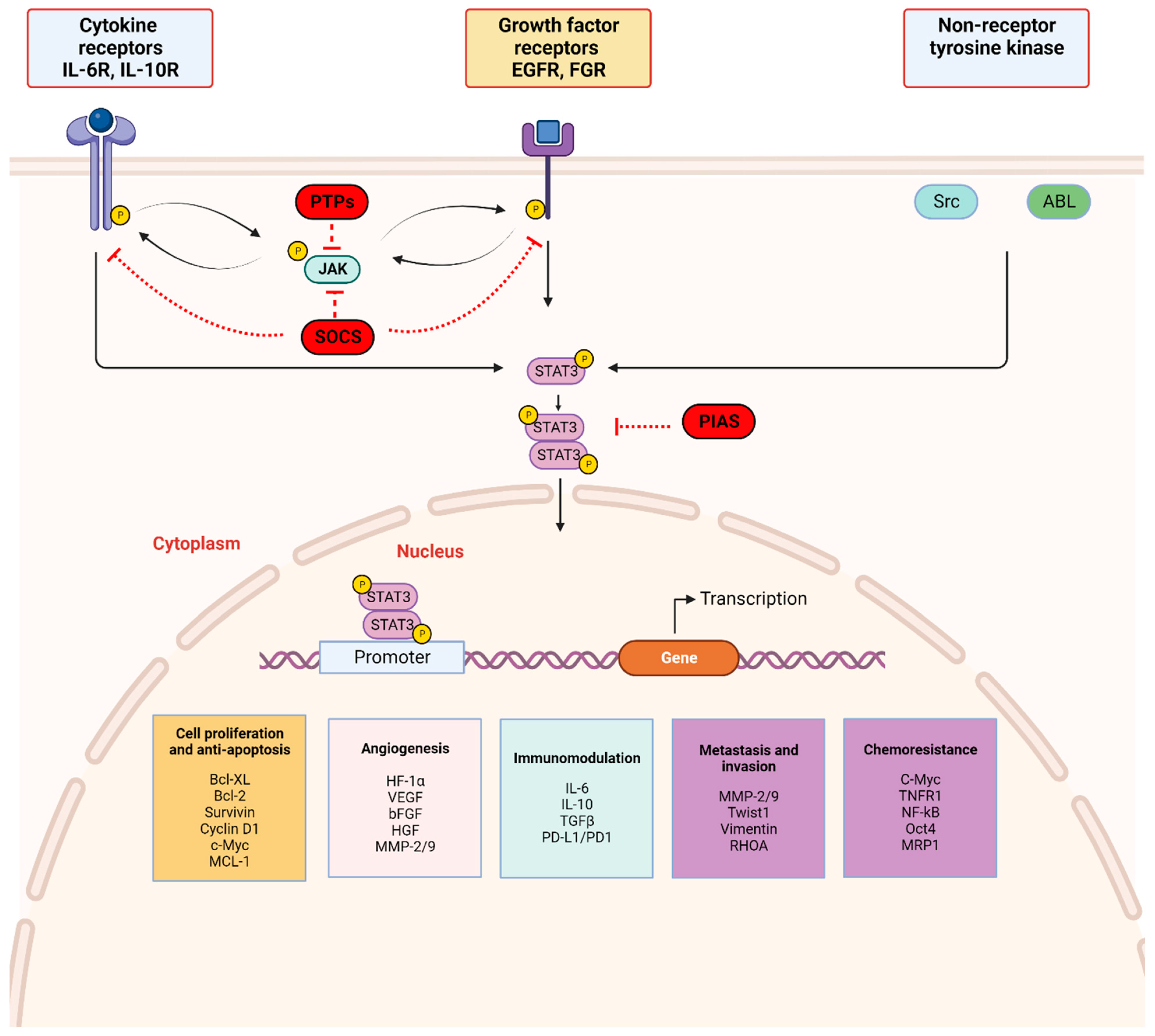
2.2. STAT3 Signaling and Activation (Figure 2)
3. STAT3 Activation in Cancer
3.1. STAT3 and Tumorigenesis
3.2. STAT3 as a Tumor Suppressor
4. Strategies for Targeting STAT3 Signaling
4.1. Indirect Inhibitors of STAT3
4.2. Direct Inhibitors of STAT3: Targeting the Various STAT3 Domain Structures to Block the Transcriptional Activity of STAT3 (Figure 3)
4.2.1. Inhibitors of the SH2 Domain
4.2.2. Inhibitors of the DNA Binding Domain
4.2.3. Inhibitors of the N-Terminal Domain (NTD)
4.3. Other Mechanisms of STAT3 Inhibition
5. Current Challenges and Future Direction
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| STAT3α | Signal Transducer and Activator of Transcription 3α |
| STAT3β | Signal Transducer and Activator of Transcription 3β |
| STAT1 | Signal Transducer and Activator of Transcription 1 |
| STAT2 | Signal Transducer and Activator of Transcription 2 |
| STAT4 | Signal Transducer and Activator of Transcription 4 |
| STAT5a | Signal Transducer and Activator of Transcription 5a |
| STAT5b | Signal Transducer and Activator of Transcription 5b |
| STAT6 | Signal Transducer and Activator of Transcription 6 |
| NTD | N-terminal domain |
| CCD | Coiled-coil domain |
| DBD | DNA-binding domain |
| SH2 | SRC-homology-2 domain |
| TAD | Carboxyl-terminal transactivation domain |
| IFN | Interferon |
| IL | Interleukin |
| JAK1 | Janus kinase 1 |
| JAK2 | Janus kinase 2 |
| Gp130 | Glycoprotein 130 |
| BCL6 | B cell lymphoma protein 6 |
| PI3K | Phosphoinositide 3 kinase pathways |
| MAPK | Mitogen activated protein kinase pathways |
| ERK | Extracellular receptor kinase pathways |
| EGF | Epidermal growth factor |
| FGF | Fibroblast growth factor |
| IGF | Insulin-like growth factor |
| Gp130 | Glycoprotein 130 |
| Tyr705 | Tyrosine 705 |
| PTPs | Protein tyrosine phosphatases |
| PIAS | Protein inhibitors of activated STAT |
| SOCS | Suppressors of cytokine signaling |
| SHP-1/2 | Src homology region 2 domain-containing phosphatase 1 and 2 |
| NSCLC | Non-small cell lung cancer |
| siRNA | Small interfering RNA |
| Rb | Retinoblastoma protein |
| EMT | Epithelial-to-mesenchymal transition |
| Twist1 | Twist-related protein 1 |
| bHLH | Basic helix-loop-helix |
| E-cadherin | Epithelial cadherin |
| N-cadherin | Neural cadherin |
| MMPs | Matrix metalloproteinases |
| HIF1α | Hypoxia-inducible factor-1α |
| VEGF | Vascular endothelial growth factor |
| PD-L1 | Programmed death-ligand 1 |
| NPCs | Neural precursor cells |
| PTC | Papillary thyroid carcinoma |
| TPO | Thyroid peroxidase |
| Snail1 | Snail Family Transcriptional Repressor 1 |
| GSK | Glycogen synthase kinase |
| HNSCCs | Head and neck squamous cell cancer cell lines |
| DLTs | Dose limiting toxicities |
| FDA | U.S. Food and Drug Administration |
| TUNEL | Transferase-mediated dUTP nick-end labeling |
| AML | Acute Myelogenous Leukemia |
| GVHD | Graft versus host disease |
| ORR | Objective response rate |
| OSCC | Oral squamous cell carcinoma |
| RT-PCR | Reverse transcription-polymerase chain reaction |
| ELISA | Enzyme-linked immunosorbent assay |
| TKIs | Tyrosine kinase inhibitors |
| HCC | Human hepatocellular carcinoma |
| HepPten− | Hepatocyte-specific deletion of Pten |
| cPR | Confirmed partial response |
| RECIST | Response Evaluation Criteria in Solid Tumors |
| CLL | Chronic Lymphocytic Lymphoma |
| BBI608 | Napabucasin |
| SH2 | Src homology region 2 |
| Shp1 | Domain-containing phosphatase 1 |
| TKI258 | Dovitinib |
| FGFR1 | Fibroblast growth factor receptor 1 |
| FGFR2 | Fibroblast growth factor receptor 2 |
| FGFR3 | Fibroblast growth factor receptor 3 |
| HBCx2 | Xenograft model |
| siRNAs | Small interfering RNAs |
| shRNA | Short hairpin RNA |
| RNAi | RNA interference |
| dsRNA | Double stranded RNA |
| Gint4.T-STAT3 | Glioblastoma a novel aptamer-siRNA chimera |
| BTK | Bruton tyrosine kinase |
| 3′-UTR | 3′-untranslated region |
| NHL | Non-Hodgkin’s Lymphoma |
| IDH | Isocitrate dehydrogenase enzyme |
| IDH1 | Isocitrate dehydrogenase enzyme 1 |
| IDH2 | Isocitrate dehydrogenase enzyme 2 |
References
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Tong, Q.; Liu, B.; Huang, W.; Tian, Y.; Fu, X. Targeting STAT3 in Cancer Immunotherapy. Mol. Cancer 2020, 19, 145. [Google Scholar] [CrossRef] [PubMed]
- Dimberg, A.; Karlberg, I.; Nilsson, K.; Oberg, F. Ser727/Tyr701-phosphorylated Stat1 is required for the regulation of c-Myc, cyclins, and p27Kip1 associated with ATRA-induced G0/G1 arrest of U-937 cells. Blood 2003, 102, 254–261. [Google Scholar] [CrossRef]
- Wang, Y.; Song, Q.; Huang, W.; Lin, Y.; Wang, X.; Wang, C.; Willard, B.; Zhao, C.; Nan, J.; Holvey-Bates, E.; et al. A virus-induced conformational switch of STAT1-STAT2 dimers boosts antiviral defenses. Cell Res. 2021, 31, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Li, S.; Cha, E.; Schindler, C. Immune response in Stat2 knockout mice. Immunity 2000, 13, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Thierfelder, W.E.; van Deursen, J.M.; Yamamoto, K.; Tripp, R.A.; Sarawar, S.R.; Carson, R.T.; Sangster, M.Y.; Vignali, D.A.; Doherty, P.C.; Grosveld, G.C.; et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature 1996, 382, 171–174. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.H.; Sun, Y.L.; Hoey, T.; Grusby, M.J. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature 1996, 382, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.H.; Schindler, U.; Smiley, S.T.; Grusby, M.J. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity 1996, 4, 313–319. [Google Scholar] [CrossRef]
- Shimoda, K.; van Deursen, J.; Sangster, M.Y.; Sarawar, S.R.; Carson, R.T.; Tripp, R.A.; Chu, C.; Quelle, F.W.; Nosaka, T.; Vignali, D.A.; et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature 1996, 380, 630–633. [Google Scholar] [CrossRef]
- Liu, X.; Robinson, G.W.; Wagner, K.U.; Garrett, L.; Wynshaw-Boris, A.; Hennighausen, L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997, 11, 179–186. [Google Scholar] [CrossRef]
- Neilson, L.M.; Zhu, J.; Xie, J.; Malabarba, M.G.; Sakamoto, K.; Wagner, K.U.; Kirken, R.A.; Rui, H. Coactivation of janus tyrosine kinase (Jak)1 positively modulates prolactin-Jak2 signaling in breast cancer: Recruitment of ERK and signal transducer and activator of transcription (Stat)3 and enhancement of Akt and Stat5a/b pathways. Mol. Endocrinol. 2007, 21, 2218–2232. [Google Scholar] [CrossRef] [PubMed]
- Catlett-Falcone, R.; Landowski, T.H.; Oshiro, M.M.; Turkson, J.; Levitzki, A.; Savino, R.; Ciliberto, G.; Moscinski, L.; Fernández-Luna, J.L.; Nuñez, G.; et al. Constitutive Activation of Stat3 Signaling Confers Resistance to Apoptosis in Human U266 Myeloma Cells. Immunity 1999, 10, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Konnikova, L.; Kotecki, M.; Kruger, M.M.; Cochran, B.H. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer 2003, 3, 23. [Google Scholar] [CrossRef] [PubMed]
- Sen, M.; Pollock, N.I.; Black, J.; DeGrave, K.A.; Wheeler, S.; Freilino, M.L.; Joyce, S.; Lui, V.W.; Zeng, Y.; Chiosea, S.I.; et al. JAK kinase inhibition abrogates STAT3 activation and head and neck squamous cell carcinoma tumor growth. Neoplasia 2015, 17, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.K.; Yagiz, K.; Pradhan, D.; Luthringer, D.J.; Amin, M.B.; Alkan, S.; Cinar, B. STAT3 and STAT5A are potential therapeutic targets in castration-resistant prostate cancer. Oncotarget 2017, 8, 85997–86010. [Google Scholar] [CrossRef] [PubMed]
- Mirtti, T.; Leiby, B.E.; Abdulghani, J.; Aaltonen, E.; Pavela, M.; Mamtani, A.; Alanen, K.; Egevad, L.; Granfors, T.; Josefsson, A.; et al. Nuclear Stat5a/b predicts early recurrence and prostate cancer-specific death in patients treated by radical prostatectomy. Hum. Pathol. 2013, 44, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Klupp, F.; Diers, J.; Kahlert, C.; Neumann, L.; Halama, N.; Franz, C.; Schmidt, T.; Lasitschka, F.; Warth, A.; Weitz, J.; et al. Expressional STAT3/STAT5 Ratio is an Independent Prognostic Marker in Colon Carcinoma. Ann. Surg. Oncol. 2015, 22 (Suppl. S3), S1548–S1555. [Google Scholar] [CrossRef]
- Takeda, K.; Noguchi, K.; Shi, W.; Tanaka, T.; Matsumoto, M.; Yoshida, N.; Kishimoto, T.; Akira, S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 1997, 94, 3801–3804. [Google Scholar] [CrossRef]
- Kiu, H.; Nicholson, S.E. Biology and significance of the JAK/STAT signalling pathways. Growth Factors 2012, 30, 88–106. [Google Scholar] [CrossRef]
- Akira, S. Roles of STAT3 defined by tissue-specific gene targeting. Oncogene 2000, 19, 2607–2611. [Google Scholar] [CrossRef]
- Bromberg, J.F.; Wrzeszczynska, M.H.; Devgan, G.; Zhao, Y.; Pestell, R.G.; Albanese, C.; Darnell, J.E., Jr. Stat3 as an oncogene. Cell 1999, 98, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Nishio, Y.; Inoue, M.; Wang, X.-J.; We, S.; Matsusaka, T.; Yoshida, K.; Sudo, T.; Naruto, M.; Kishimoto, T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 1994, 77, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Abroun, S.; Saki, N.; Ahmadvand, M.; Asghari, F.; Salari, F.; Rahim, F. STATs: An Old Story, Yet Mesmerizing. Cell J. 2015, 17, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Calò, V.; Migliavacca, M.; Bazan, V.; Macaluso, M.; Buscemi, M.; Gebbia, N.; Russo, A. STAT proteins: From normal control of cellular events to tumorigenesis. J. Cell. Physiol. 2003, 197, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Mohammad, I.S.; Liu, Z. Overview of the STAT-3 signaling pathway in cancer and the development of specific inhibitors (Review). Oncol. Lett. 2020, 19, 2585–2594. [Google Scholar] [CrossRef] [PubMed]
- Wake, M.S.; Watson, C.J. STAT3 the oncogene—Still eluding therapy? FEBS J. 2015, 282, 2600–2611. [Google Scholar] [CrossRef] [PubMed]
- Della Pietra, L.; Bressan, A.; Pezzotti, A.R.; Serlupi-Crescenzi, O. Highly conserved amino-acid sequence between murine STAT3 and a revised human STAT3 sequence. Gene 1998, 213, 119–124. [Google Scholar] [CrossRef]
- Ilaria, R.L., Jr. STAT isoforms: Mediators of STAT specificity or leukemogenesis? Leuk. Res. 2001, 25, 483–484. [Google Scholar] [CrossRef]
- Caldenhoven, E.; van Dijk, T.B.; Solari, R.; Armstrong, J.; Raaijmakers, J.A.; Lammers, J.W.; Koenderman, L.; de Groot, R.P. STAT3beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J. Biol. Chem. 1996, 271, 13221–13227. [Google Scholar] [CrossRef]
- Huang, Y.; Qiu, J.; Dong, S.; Redell, M.S.; Poli, V.; Mancini, M.A.; Tweardy, D.J. Stat3 isoforms, alpha and beta, demonstrate distinct intracellular dynamics with prolonged nuclear retention of Stat3beta mapping to its unique C-terminal end. J. Biol. Chem. 2007, 282, 34958–34967. [Google Scholar] [CrossRef]
- Chakraborty, A.; Tweardy, D.J. Stat3 and G-CSF-induced myeloid differentiation. Leuk. Lymphoma 1998, 30, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-F.; Chen, Y.; Wu, C.; Wu, Z.-Y.; Tweardy, D.J.; Alshareef, A.; Liao, L.-D.; Xue, Y.-J.; Wu, J.-Y.; Chen, B.; et al. The Opposing Function of STAT3 as an Oncoprotein and Tumor Suppressor Is Dictated by the Expression Status of STAT3β in Esophageal Squamous Cell Carcinoma. Clin. Cancer Res. 2016, 22, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Dyer, K.F.; Cascio, M.; Mietzner, T.A.; Tweardy, D.J. Identification of a novel Stat3 recruitment and activation motif within the granulocyte colony-stimulating factor receptor. Blood 1999, 93, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Bowman, T.; Garcia, R.; Turkson, J.; Jove, R. STATs in oncogenesis. Oncogene 2000, 19, 2474–2488. [Google Scholar] [CrossRef]
- Silver, D.L.; Naora, H.; Liu, J.; Cheng, W.; Montell, D.J. Activated Signal Transducer and Activator of Transcription (STAT) 3: Localization in Focal Adhesions and Function in Ovarian Cancer Cell Motility. Cancer Res. 2004, 64, 3550–3558. [Google Scholar] [CrossRef]
- Niu, G.; Wright, K.L.; Huang, M.; Song, L.; Haura, E.; Turkson, J.; Zhang, S.; Wang, T.; Sinibaldi, D.; Coppola, D.; et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene 2002, 21, 2000–2008. [Google Scholar] [CrossRef]
- Benekli, M.; Baer, M.R.; Baumann, H.; Wetzler, M. Signal transducer and activator of transcription proteins in leukemias. Blood 2003, 101, 2940–2954. [Google Scholar] [CrossRef]
- Turkson, J.; Jove, R. STAT proteins: Novel molecular targets for cancer drug discovery. Oncogene 2000, 19, 6613–6626. [Google Scholar] [CrossRef]
- Wingelhofer, B.; Neubauer, H.A.; Valent, P.; Han, X.; Constantinescu, S.N.; Gunning, P.T.; Müller, M.; Moriggl, R. Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia 2018, 32, 1713–1726. [Google Scholar] [CrossRef]
- Li, W.X. Canonical and non-canonical JAK–STAT signaling. Trends Cell Biol. 2008, 18, 545–551. [Google Scholar] [CrossRef]
- Benekli, M.; Baumann, H.; Wetzler, M. Targeting signal transducer and activator of transcription signaling pathway in leukemias. J. Clin. Oncol. 2009, 27, 4422–4432. [Google Scholar] [CrossRef] [PubMed]
- Ilaria, R.L., Jr.; Van Etten, R.A. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J. Biol. Chem. 1996, 271, 31704–31710. [Google Scholar] [CrossRef] [PubMed]
- Reich, N.C. STATs get their move on. Jakstat 2013, 2, e27080. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, U.; Kasembeli, M.M.; Tweardy, D.J. STAT3 Inhibitors in Cancer: A Comprehensive Update. In STAT Inhibitors in Cancer; Ward, A.C., Ed.; Springer International Publishing: Cham, Swizerland, 2016; pp. 95–161. [Google Scholar] [CrossRef]
- Sgrignani, J.; Garofalo, M.; Matkovic, M.; Merulla, J.; Catapano, C.V.; Cavalli, A. Structural Biology of STAT3 and Its Implications for Anticancer Therapies Development. Int. J. Mol. Sci. 2018, 19, 1591. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, C.; Susini, C.; Melmed, S. Inhibitory roles for SHP-1 and SOCS-3 following pituitary proopiomelanocortin induction by leukemia inhibitory factor. J. Clin. Investig. 1999, 104, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Baumann, H. Dual signaling role of the protein tyrosine phosphatase SHP-2 in regulating expression of acute-phase plasma proteins by interleukin-6 cytokine receptors in hepatic cells. Mol. Cell. Biol. 1999, 19, 5326–5338. [Google Scholar] [CrossRef]
- Resemann, H.K.; Watson, C.J.; Lloyd-Lewis, B. The Stat3 paradox: A killer and an oncogene. Mol. Cell. Endocrinol. 2014, 382, 603–611. [Google Scholar] [CrossRef]
- Wang, H.-Q.; Man, Q.-W.; Huo, F.-Y.; Gao, X.; Lin, H.; Li, S.-R.; Wang, J.; Su, F.-C.; Cai, L.; Shi, Y.; et al. STAT3 pathway in cancers: Past, present, and future. MedComm 2022, 3, e124. [Google Scholar] [CrossRef]
- Kortylewski, M.; Jove, R.; Yu, H. Targeting STAT3 affects melanoma on multiple fronts. Cancer Metastasis Rev. 2005, 24, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.L.; Meyer, D.J.; Campbell, G.S.; Larner, A.C.; Carter-Su, C.; Schwartz, J.; Jove, R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science 1995, 269, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Grandis, J.R.; Drenning, S.D.; Chakraborty, A.; Zhou, M.Y.; Zeng, Q.; Pitt, A.S.; Tweardy, D.J. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor- mediated cell growth In vitro. J. Clin. Investig. 1998, 102, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Du, H.; Qin, Y.; Roberts, J.; Cummings, O.W.; Yan, C. Activation of the signal transducers and activators of the transcription 3 pathway in alveolar epithelial cells induces inflammation and adenocarcinomas in mouse lung. Cancer Res. 2007, 67, 8494–8503. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.V.; Greulich, H.; Sellers, W.R.; Meyerson, M.; Frank, D.A. Signal transducer and activator of transcription 3 is required for the oncogenic effects of non-small-cell lung cancer-associated mutations of the epidermal growth factor receptor. Cancer Res. 2006, 66, 3162–3168. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Zhang, Y.; Li, Y.; Lv, T.; Liu, J.; Wang, X. Prognostic significance of STAT3 expression and its correlation with chemoresistance of non-small cell lung cancer cells. Acta Histochem. 2012, 114, 151–158. [Google Scholar] [CrossRef] [PubMed]
- D’Arcangelo, M.; Cappuzzo, F. K-Ras Mutations in Non-Small-Cell Lung Cancer: Prognostic and Predictive Value. ISRN Mol. Biol. 2012, 2012, 837306. [Google Scholar] [CrossRef]
- Grabner, B.; Schramek, D.; Mueller, K.M.; Moll, H.P.; Svinka, J.; Hoffmann, T.; Bauer, E.; Blaas, L.; Hruschka, N.; Zboray, K.; et al. Disruption of STAT3 signalling promotes KRAS-induced lung tumorigenesis. Nat. Commun. 2015, 6, 6285. [Google Scholar] [CrossRef]
- Gritsko, T.; Williams, A.; Turkson, J.; Kaneko, S.; Bowman, T.; Huang, M.; Nam, S.; Eweis, I.; Diaz, N.; Sullivan, D.; et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin. Cancer Res. 2006, 12, 11–19. [Google Scholar] [CrossRef]
- Garcia, R.; Yu, C.L.; Hudnall, A.; Catlett, R.; Nelson, K.L.; Smithgall, T.; Fujita, D.J.; Ethier, S.P.; Jove, R. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ. 1997, 8, 1267–1276. [Google Scholar]
- Watson, C.J.; Miller, W.R. Elevated levels of members of the STAT family of transcription factors in breast carcinoma nuclear extracts. Br. J. Cancer 1995, 71, 840–844. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.W.; Hsu, S.C.; Ali-Seyed, M.; Gunduz, M.; Xia, W.; Wei, Y.; Bartholomeusz, G.; Shih, J.Y.; Hung, M.C. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell 2005, 7, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Pectasides, E.; Egloff, A.M.; Sasaki, C.; Kountourakis, P.; Burtness, B.; Fountzilas, G.; Dafni, U.; Zaramboukas, T.; Rampias, T.; Rimm, D.; et al. Nuclear localization of signal transducer and activator of transcription 3 in head and neck squamous cell carcinoma is associated with a better prognosis. Clin. Cancer Res. 2010, 16, 2427–2434. [Google Scholar] [CrossRef] [PubMed]
- Shinagawa, K.; Yanamoto, S.; Naruse, T.; Kawakita, A.; Morishita, K.; Sakamoto, Y.; Rokutanda, S.; Umeda, M. Clinical Roles of Interleukin-6 and STAT3 in Oral Squamous Cell Carcinoma. Pathol. Oncol. Res. 2017, 23, 425–431. [Google Scholar] [CrossRef] [PubMed]
- de la Iglesia, N.; Puram, S.V.; Bonni, A. STAT3 regulation of glioblastoma pathogenesis. Curr. Mol. Med. 2009, 9, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Couto, J.P.; Daly, L.; Almeida, A.; Knauf, J.A.; Fagin, J.A.; Sobrinho-Simões, M.; Lima, J.; Máximo, V.; Soares, P.; Lyden, D.; et al. STAT3 negatively regulates thyroid tumorigenesis. Proc. Natl. Acad. Sci. USA 2012, 109, E2361–E2370. [Google Scholar] [CrossRef] [PubMed]
- Pencik, J.; Schlederer, M.; Gruber, W.; Unger, C.; Walker, S.M.; Chalaris, A.; Marié, I.J.; Hassler, M.R.; Javaheri, T.; Aksoy, O.; et al. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat. Commun. 2015, 6, 7736, Erratum in Nat. Commun. 2015, 6, 8802. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, R.B.; Contino, G.; Deshpande, V.; Tzatsos, A.; Conrad, C.; Benes, C.H.; Levy, D.E.; Settleman, J.; Engelman, J.A.; Bardeesy, N. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011, 71, 5020–5029. [Google Scholar] [CrossRef]
- Lee, J.; Kim, J.C.; Lee, S.E.; Quinley, C.; Kim, H.; Herdman, S.; Corr, M.; Raz, E. Signal transducer and activator of transcription 3 (STAT3) protein suppresses adenoma-to-carcinoma transition in Apcmin/+ mice via regulation of Snail-1 (SNAI) protein stability. J. Biol. Chem. 2012, 287, 18182–18189. [Google Scholar] [CrossRef]
- Benekli, M.; Xia, Z.; Donohue, K.A.; Ford, L.A.; Pixley, L.A.; Baer, M.R.; Baumann, H.; Wetzler, M. Constitutive activity of signal transducer and activator of transcription 3 protein in acute myeloid leukemia blasts is associated with short disease-free survival. Blood 2002, 99, 252–257. [Google Scholar] [CrossRef]
- Mencalha, A.L.; Binato, R.; Ferreira, G.M.; Du Rocher, B.; Abdelhay, E. Forkhead box M1 (FoxM1) gene is a new STAT3 transcriptional factor target and is essential for proliferation, survival and DNA repair of K562 cell line. PLoS ONE 2012, 7, e48160. [Google Scholar] [CrossRef] [PubMed]
- Adamaki, M.; Tsotra, M.; Vlahopoulos, S.; Zampogiannis, A.; Papavassiliou, A.G.; Moschovi, M. STAT transcript levels in childhood acute lymphoblastic leukemia: STAT1 and STAT3 transcript correlations. Leuk. Res. 2015, 39, 1285–1291. [Google Scholar] [CrossRef] [PubMed]
- Turton, K.B.; Annis, D.S.; Rui, L.; Esnault, S.; Mosher, D.F. Ratios of Four STAT3 Splice Variants in Human Eosinophils and Diffuse Large B Cell Lymphoma Cells. PLoS ONE 2015, 10, e0127243. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zong, C.S.; Zeng, L.; Jiang, Y.; Sadowski, H.B.; Wang, L.H. Stat3 plays an important role in oncogenic Ros- and insulin-like growth factor I receptor-induced anchorage-independent growth. J. Biol. Chem. 1998, 273, 28065–28072. [Google Scholar] [CrossRef] [PubMed]
- Danial, N.N.; Pernis, A.; Rothman, P.B. Jak-STAT signaling induced by the v-abl oncogene. Science 1995, 269, 1875–1877. [Google Scholar] [CrossRef] [PubMed]
- Migone, T.S.; Lin, J.X.; Cereseto, A.; Mulloy, J.C.; O’Shea, J.J.; Franchini, G.; Leonard, W.J. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I. Science 1995, 269, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Weber-Nordt, R.M.; Egen, C.; Wehinger, J.; Ludwig, W.; Gouilleux-Gruart, V.; Mertelsmann, R.; Finke, J. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood 1996, 88, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Zhang, Q.; Goradia, A.; Raghunath, P.N.; Liu, X.; Paessler, M.; Wang, H.Y.; Wysocka, M.; Cheng, M.; Ruggeri, B.A.; et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc. Natl. Acad. Sci. USA 2008, 105, 20852–20857. [Google Scholar] [CrossRef]
- Lee, T.; Yeh, J.; Van Waes, C.; Chen, Z. Bcl-xL is regulated by NF-KappaB and STAT3 through p53-dependent control in head and neck squamous cell carcinoma. Cancer Res. 2004, 64, 1115. [Google Scholar]
- Kunigal, S.; Lakka, S.S.; Sodadasu, P.K.; Estes, N.; Rao, J.S. Stat3-siRNA induces Fas-mediated apoptosis in vitro and in vivo in breast cancer. Int. J. Oncol. 2009, 34, 1209–1220. [Google Scholar]
- Bhattacharya, S.; Ray, R.M.; Johnson, L.R. STAT3-mediated transcription of Bcl-2, Mcl-1 and c-IAP2 prevents apoptosis in polyamine-depleted cells. Biochem. J. 2005, 392, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Du, Y.; Song, Q.; Nan, J.; Guan, P.; Guo, J.; Wang, X.; Yang, J.; Zhao, C. E2F is required for STAT3-mediated upregulation of cyclin B1 and Cdc2 expressions and contributes to G2-M phase transition. Acta Biochim. Biophys. Sin. 2019, 51, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Zhang, Z.-G.; Tian, X.-Q.; Sun, D.-F.; Liang, Q.-C.; Zhang, Y.-J.; Lu, R.; Chen, Y.-X.; Fang, J.-Y. Inhibition of JAK1, 2/STAT3 Signaling Induces Apoptosis, Cell Cycle Arrest, and Reduces Tumor Cell Invasion in Colorectal Cancer Cells. Neoplasia 2008, 10, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Alle, K.M.; Henshall, S.M.; Field, A.S.; Sutherland, R.L. Cyclin D1 protein is overexpressed in hyperplasia and intraductal carcinoma of the breast. Clin. Cancer Res. 1998, 4, 847–854. [Google Scholar] [PubMed]
- Cheng, F.; Wang, H.-W.; Cuenca, A.; Huang, M.; Ghansah, T.; Brayer, J.; Kerr, W.G.; Takeda, K.; Akira, S.; Schoenberger, S.P.; et al. A Critical Role for Stat3 Signaling in Immune Tolerance. Immunity 2003, 19, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Xiong, H.; Hong, J.; Du, W.; Lin, Y.W.; Ren, L.L.; Wang, Y.C.; Su, W.Y.; Wang, J.L.; Cui, Y.; Wang, Z.H.; et al. Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J. Biol. Chem. 2012, 287, 5819–5832. [Google Scholar] [CrossRef]
- Pensa, S.; Regis, G.; Boselli, D.; Novelli, F.; Poli, V. STAT1 and STAT3 in tumorigenesis: Two sides of the same coin. In Madame Curie Bioscience Database; Landes Bioscience: Austin, TX, USA, 2000. [Google Scholar]
- Lee, H.J.; Zhuang, G.; Cao, Y.; Du, P.; Kim, H.J.; Settleman, J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 2014, 26, 207–221. [Google Scholar] [CrossRef]
- Szakács, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [CrossRef]
- Kettner, N.M.; Vijayaraghavan, S.; Durak, M.G.; Bui, T.; Kohansal, M.; Ha, M.J.; Liu, B.; Rao, X.; Wang, J.; Yi, M.; et al. Combined Inhibition of STAT3 and DNA Repair in Palbociclib-Resistant ER-Positive Breast Cancer. Clin. Cancer Res. 2019, 25, 3996–4013. [Google Scholar] [CrossRef]
- Lo, H.W.; Hsu, S.C.; Xia, W.; Cao, X.; Shih, J.Y.; Wei, Y.; Abbruzzese, J.L.; Hortobagyi, G.N.; Hung, M.C. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007, 67, 9066–9076. [Google Scholar] [CrossRef] [PubMed]
- El Ghouzzi, V.; Legeai-Mallet, L.; Aresta, S.; Benoist, C.; Munnich, A.; de Gunzburg, J.; Bonaventure, J. Saethre-Chotzen mutations cause TWIST protein degradation or impaired nuclear location. Hum. Mol. Genet. 2000, 9, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Puisieux, A.; Valsesia-Wittmann, S.; Ansieau, S. A twist for survival and cancer progression. Br. J. Cancer 2006, 94, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Heimann, R.; Lan, F.; McBride, R.; Hellman, S. Separating favorable from unfavorable prognostic markers in breast cancer: The role of E-cadherin. Cancer Res. 2000, 60, 298–304. [Google Scholar] [PubMed]
- Lee, T.K.; Poon, R.T.; Yuen, A.P.; Ling, M.T.; Kwok, W.K.; Wang, X.H.; Wong, Y.C.; Guan, X.Y.; Man, K.; Chau, K.L.; et al. Twist overexpression correlates with hepatocellular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clin. Cancer Res. 2006, 12, 5369–5376. [Google Scholar] [CrossRef] [PubMed]
- Ohuchida, K.; Mizumoto, K.; Ohhashi, S.; Yamaguchi, H.; Konomi, H.; Nagai, E.; Yamaguchi, K.; Tsuneyoshi, M.; Tanaka, M. Twist, a novel oncogene, is upregulated in pancreatic cancer: Clinical implication of Twist expression in pancreatic juice. Int. J. Cancer 2007, 120, 1634–1640. [Google Scholar] [CrossRef]
- Cheng, G.Z.; Chan, J.; Wang, Q.; Zhang, W.; Sun, C.D.; Wang, L.H. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007, 67, 1979–1987. [Google Scholar] [CrossRef]
- Cheng, G.Z.; Zhang, W.Z.; Sun, M.; Wang, Q.; Coppola, D.; Mansour, M.; Xu, L.M.; Costanzo, C.; Cheng, J.Q.; Wang, L.H. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J. Biol. Chem. 2008, 283, 14665–14673. [Google Scholar] [CrossRef]
- Friedl, P.; Alexander, S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef]
- Pan, Y.R.; Chen, C.C.; Chan, Y.T.; Wang, H.J.; Chien, F.T.; Chen, Y.L.; Liu, J.L.; Yang, M.H. STAT3-coordinated migration facilitates the dissemination of diffuse large B-cell lymphomas. Nat. Commun. 2018, 9, 3696. [Google Scholar] [CrossRef]
- Xie, T.X.; Wei, D.; Liu, M.; Gao, A.C.; Ali-Osman, F.; Sawaya, R.; Huang, S. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene 2004, 23, 3550–3560. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huang, C.; Huang, K.; Wu, W.; Jiang, T.; Cao, J.; Feng, Z.; Qiu, Z. STAT3 knockdown reduces pancreatic cancer cell invasiveness and matrix metalloproteinase-7 expression in nude mice. PLoS ONE 2011, 6, e25941. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Heller, R.; Catlett-Falcone, R.; Coppola, D.; Jaroszeski, M.; Dalton, W.; Jove, R.; Yu, H. Gene therapy with dominant-negative Stat3 suppresses growth of the murine melanoma B16 tumor in vivo. Cancer Res. 1999, 59, 5059–5063. [Google Scholar] [PubMed]
- Wang, T.; Niu, G.; Kortylewski, M.; Burdelya, L.; Shain, K.; Zhang, S.; Bhattacharya, R.; Gabrilovich, D.; Heller, R.; Coppola, D.; et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 2004, 10, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, Y.; Huang, M.; Kusmartsev, S.; Bhattacharya, R.; Cheng, P.; Salup, R.; Jove, R.; Gabrilovich, D. Hyperactivation of STAT3 Is Involved in Abnormal Differentiation of Dendritic Cells in Cancer. J. Immunol. 2004, 172, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, Y.; Nagaraj, S.; Rosenbauer, A.; Muro-Cacho, C.; Sebti, S.M.; Gabrilovich, D.I. Regulation of dendritic cell differentiation and antitumor immune response in cancer by pharmacologic-selective inhibition of the janus-activated kinase 2/signal transducers and activators of transcription 3 pathway. Cancer Res. 2005, 65, 9525–9535. [Google Scholar] [CrossRef] [PubMed]
- Attili, I.; Karachaliou, N.; Bonanno, L.; Berenguer, J.; Bracht, J.; Codony-Servat, J.; Codony-Servat, C.; Ito, M.; Rosell, R. STAT3 as a potential immunotherapy biomarker in oncogene-addicted non-small cell lung cancer. Ther. Adv. Med. Oncol. 2018, 10, 1758835918763744. [Google Scholar] [CrossRef]
- Lu, C.; Talukder, A.; Savage, N.M.; Singh, N.; Liu, K. JAK-STAT-mediated chronic inflammation impairs cytotoxic T lymphocyte activation to decrease anti-PD-1 immunotherapy efficacy in pancreatic cancer. Oncoimmunology 2017, 6, e1291106. [Google Scholar] [CrossRef]
- Zerdes, I.; Wallerius, M.; Sifakis, E.G.; Wallmann, T.; Betts, S.; Bartish, M.; Tsesmetzis, N.; Tobin, N.P.; Coucoravas, C.; Bergh, J.; et al. STAT3 Activity Promotes Programmed-Death Ligand 1 Expression and Suppresses Immune Responses in Breast Cancer. Cancers 2019, 11, 1479. [Google Scholar] [CrossRef]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef]
- Klein, S.L.; Morgan, R. The impact of sex and gender on immunotherapy outcomes. Biol. Sex Differ. 2020, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Real, P.J.; Sierra, A.; De Juan, A.; Segovia, J.C.; Lopez-Vega, J.M.; Fernandez-Luna, J.L. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene 2002, 21, 7611–7618. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.K.; Yang, Z.F.; Lam, S.P.; Lam, C.T.; Ngai, P.; Tam, K.H.; Poon, R.T.; Fan, S.T. Inhibition of Stat3 activity by YC-1 enhances chemo-sensitivity in hepatocellular carcinoma. Cancer Biol. Ther. 2007, 6, 1900–1907. [Google Scholar] [CrossRef] [PubMed]
- Yoshimatsu, T.; Kawaguchi, D.; Oishi, K.; Takeda, K.; Akira, S.; Masuyama, N.; Gotoh, Y. Non-cell-autonomous action of STAT3 in maintenance of neural precursor cells in the mouse neocortex. Development 2006, 133, 2553–2563. [Google Scholar] [CrossRef] [PubMed]
- Rajan, P.; McKay, R.D. Multiple routes to astrocytic differentiation in the CNS. J. Neurosci. 1998, 18, 3620–3629. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H.; Zhang, W.; Huang, H.J.; Liao, W.S.L.; Fuller, G.N. Analysis of the activation status of Akt, NFκB, and Stat3 in human diffuse gliomas. Lab. Investig. 2004, 84, 941–951. [Google Scholar] [CrossRef] [PubMed]
- de la Iglesia, N.; Konopka, G.; Puram, S.V.; Chan, J.A.; Bachoo, R.M.; You, M.J.; Levy, D.E.; Depinho, R.A.; Bonni, A. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008, 22, 449–462. [Google Scholar] [CrossRef]
- Dolled-Filhart, M.; Camp, R.L.; Kowalski, D.P.; Smith, B.L.; Rimm, D.L. Tissue microarray analysis of signal transducers and activators of transcription 3 (Stat3) and phospho-Stat3 (Tyr705) in node-negative breast cancer shows nuclear localization is associated with a better prognosis. Clin. Cancer Res. 2003, 9, 594–600. [Google Scholar]
- Yates, L.R.; Knappskog, S.; Wedge, D.; Farmery, J.H.R.; Gonzalez, S.; Martincorena, I.; Alexandrov, L.B.; Van Loo, P.; Haugland, H.K.; Lilleng, P.K.; et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 2017, 32, 169–184.e167. [Google Scholar] [CrossRef]
- Coleman, N.; Rodon, J. Taking Aim at the Undruggable; American Society of Clinical Oncology Educational Book; American Society of Clinical Oncology: Alexandria, VA, USA, 2021; pp. e145–e152. [Google Scholar] [CrossRef]
- Wong, A.L.A.; Hirpara, J.L.; Pervaiz, S.; Eu, J.-Q.; Sethi, G.; Goh, B.-C. Do STAT3 inhibitors have potential in the future for cancer therapy? Expert Opin. Investig. Drugs 2017, 26, 883–887. [Google Scholar] [CrossRef]
- Fragoulis, G.E.; McInnes, I.B.; Siebert, S. JAK-inhibitors. New players in the field of immune-mediated diseases, beyond rheumatoid arthritis. Rheumatology 2019, 58, i43–i54. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Hu, J.; Guo, Q.H.; Zhao, Y.; Cheng, J.J.; Zhang, D.S.; Fei, Q.; Li, J.; Sun, Y.M. AZD1480, a JAK inhibitor, inhibits cell growth and survival of colorectal cancer via modulating the JAK2/STAT3 signaling pathway. Oncol. Rep. 2014, 32, 1991–1998. [Google Scholar] [CrossRef][Green Version]
- Stuart, E.; Buchert, M.; Putoczki, T.; Thiem, S.; Farid, R.; Elzer, J.; Huszar, D.; Waring, P.M.; Phesse, T.J.; Ernst, M. Therapeutic inhibition of Jak activity inhibits progression of gastrointestinal tumors in mice. Mol. Cancer Ther. 2014, 13, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Plimack, E.R.; Lorusso, P.M.; McCoon, P.; Tang, W.; Krebs, A.D.; Curt, G.; Eckhardt, S.G. AZD1480: A phase I study of a novel JAK2 inhibitor in solid tumors. Oncologist 2013, 18, 819–820. [Google Scholar] [CrossRef] [PubMed]
- Przepiorka, D.; Luo, L.; Subramaniam, S.; Qiu, J.; Gudi, R.; Cunningham, L.C.; Nie, L.; Leong, R.; Ma, L.; Sheth, C.; et al. FDA Approval Summary: Ruxolitinib for Treatment of Steroid-Refractory Acute Graft-Versus-Host Disease. Oncologist 2020, 25, e328–e334. [Google Scholar] [CrossRef] [PubMed]
- Ganetsky, A. Ruxolitinib: A new treatment option for myelofibrosis. Pharmacotherapy 2013, 33, 84–92. [Google Scholar] [CrossRef]
- Tefferi, A. Primary myelofibrosis: 2021 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2021, 96, 145–162. [Google Scholar] [CrossRef]
- Quintás-Cardama, A.; Vaddi, K.; Liu, P.; Manshouri, T.; Li, J.; Scherle, P.A.; Caulder, E.; Wen, X.; Li, Y.; Waeltz, P.; et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: Therapeutic implications for the treatment of myeloproliferative neoplasms. Blood 2010, 115, 3109–3117. [Google Scholar] [CrossRef]
- Benoy, I.; Salgado, R.; Colpaert, C.; Weytjens, R.; Vermeulen, P.B.; Dirix, L.Y. Serum interleukin 6, plasma VEGF, serum VEGF, and VEGF platelet load in breast cancer patients. Clin. Breast Cancer 2002, 2, 311–315. [Google Scholar] [CrossRef]
- Zhang, G.J.; Adachi, I. Serum interleukin-6 levels correlate to tumor progression and prognosis in metastatic breast carcinoma. Anticancer Res. 1999, 19, 1427–1432. [Google Scholar]
- DeMichele, A.; Gray, R.; Horn, M.; Chen, J.; Aplenc, R.; Vaughan, W.P.; Tallman, M.S. Host Genetic Variants in the Interleukin-6 Promoter Predict Poor Outcome in Patients with Estrogen Receptor-Positive, Node-Positive Breast Cancer. Cancer Res. 2009, 69, 4184–4191. [Google Scholar] [CrossRef] [PubMed]
- Yokoe, T.; Iino, Y.; Morishita, Y. Trends of IL-6 and IL-8 levels in patients with recurrent breast cancer: Preliminary report. Breast Cancer 2000, 7, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, R.; Nagao, K.; Miyayama, H.; Matsuda, M.; Baba, K.; Matsuoka, Y.; Yamashita, H.; Fukuda, M.; Mizumoto, T.; Hamamoto, R. An analysis of serum interleukin-6 levels to predict benefits of medroxyprogesterone acetate in advanced or recurrent breast cancer. Oncology 2000, 59, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Conze, D.; Weiss, L.; Regen, P.S.; Bhushan, A.; Weaver, D.; Johnson, P.; Rincón, M. Autocrine production of interleukin 6 causes multidrug resistance in breast cancer cells. Cancer Res. 2001, 61, 8851–8858. [Google Scholar] [PubMed]
- Makhlin, I.; McAndrew, N.P.; Wileyto, E.P.; Clark, A.S.; Holmes, R.; Bottalico, L.N.; Mesaros, C.; Blair, I.A.; Jeschke, G.R.; Fox, K.R.; et al. Ruxolitinib and exemestane for estrogen receptor positive, aromatase inhibitor resistant advanced breast cancer. NPJ Breast Cancer 2022, 8, 122. [Google Scholar] [CrossRef]
- Ferrajoli, A.; Faderl, S.; Van, Q.; Koch, P.; Harris, D.; Liu, Z.; Hazan-Halevy, I.; Wang, Y.; Kantarjian, H.M.; Priebe, W.; et al. WP1066 disrupts Janus kinase-2 and induces caspase-dependent apoptosis in acute myelogenous leukemia cells. Cancer Res. 2007, 67, 11291–11299. [Google Scholar] [CrossRef]
- Tsujita, Y.; Horiguchi, A.; Tasaki, S.; Isono, M.; Asano, T.; Ito, K.; Asano, T.; Mayumi, Y.; Kushibiki, T. STAT3 inhibition by WP1066 suppresses the growth and invasiveness of bladder cancer cells. Oncol. Rep. 2017, 38, 2197–2204. [Google Scholar] [CrossRef]
- Iwamaru, A.; Szymanski, S.; Iwado, E.; Aoki, H.; Yokoyama, T.; Fokt, I.; Hess, K.; Conrad, C.; Madden, T.; Sawaya, R.; et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene 2007, 26, 2435–2444. [Google Scholar] [CrossRef]
- Zhou, X.; Ren, Y.; Liu, A.; Jin, R.; Jiang, Q.; Huang, Y.; Kong, L.; Wang, X.; Zhang, L. WP1066 sensitizes oral squamous cell carcinoma cells to cisplatin by targeting STAT3/miR-21 axis. Sci. Rep. 2014, 4, 7461. [Google Scholar] [CrossRef]
- Sau, S.; Mondal, S.K.; Kashaw, S.K.; Iyer, A.K.; Banerjee, R. Combination of cationic dexamethasone derivative and STAT3 inhibitor (WP1066) for aggressive melanoma: A strategy for repurposing a phase I clinical trial drug. Mol. Cell. Biochem. 2017, 436, 119–136. [Google Scholar] [CrossRef]
- Schroeder, M.A.; Khoury, H.J.; Jagasia, M.; Ali, H.; Schiller, G.J.; Staser, K.; Choi, J.; Gehrs, L.; Arbushites, M.C.; Yan, Y.; et al. A phase 1 trial of itacitinib, a selective JAK1 inhibitor, in patients with acute graft-versus-host disease. Blood Adv. 2020, 4, 1656–1669. [Google Scholar] [CrossRef] [PubMed]
- Zeiser, R.; Socié, G.; Schroeder, M.A.; Abhyankar, S.; Vaz, C.P.; Kwon, M.; Clausen, J.; Volodin, L.; Giebel, S.; Chacon, M.J.; et al. Efficacy and safety of itacitinib versus placebo in combination with corticosteroids for initial treatment of acute graft-versus-host disease (GRAVITAS-301): A randomised, multicentre, double-blind, phase 3 trial. Lancet Haematol. 2022, 9, e14–e25. [Google Scholar] [CrossRef] [PubMed]
- Pemmaraju, N.; Harrison, C.; Gupta, V.; Verstovsek, S.; Scott, B.; Oh, S.T.; Palandri, F.; Al-Ali, H.K.; Sobas, M.; McMullin, M.F.; et al. Risk-adjusted safety analysis of the oral JAK2/IRAK1 inhibitor pacritinib in patients with myelofibrosis. EJHaem 2022, 3, 1346–1351. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, J.; Hoffman, R.; Talpaz, M.; Gerds, A.T.; Stein, B.; Gupta, V.; Szoke, A.; Drummond, M.; Pristupa, A.; Granston, T.; et al. Pacritinib vs. Best Available Therapy, Including Ruxolitinib, in Patients with Myelofibrosis: A Randomized Clinical Trial. JAMA Oncol. 2018, 4, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Lippitz, B.E.; Harris, R.A. Cytokine patterns in cancer patients: A review of the correlation between interleukin 6 and prognosis. Oncoimmunology 2016, 5, e1093722. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Chen, B. Siltuximab (CNTO 328): A promising option for human malignancies. Drug Des. Dev. Ther. 2015, 9, 3455–3458. [Google Scholar] [CrossRef]
- Mihara, M.; Kasutani, K.; Okazaki, M.; Nakamura, A.; Kawai, S.; Sugimoto, M.; Matsumoto, Y.; Ohsugi, Y. Tocilizumab inhibits signal transduction mediated by both mIL-6R and sIL-6R, but not by the receptors of other members of IL-6 cytokine family. Int. Immunopharmacol. 2005, 5, 1731–1740. [Google Scholar] [CrossRef]
- Deisseroth, A.; Ko, C.-W.; Nie, L.; Zirkelbach, J.F.; Zhao, L.; Bullock, J.; Mehrotra, N.; Del Valle, P.; Saber, H.; Sheth, C.; et al. FDA Approval: Siltuximab for the Treatment of Patients with Multicentric Castleman Disease. Clin. Cancer Res. 2015, 21, 950–954. [Google Scholar] [CrossRef]
- Angevin, E.; Tabernero, J.; Elez, E.; Cohen, S.J.; Bahleda, R.; van Laethem, J.-L.; Ottensmeier, C.; Lopez-Martin, J.A.; Clive, S.; Joly, F.; et al. A Phase I/II, Multiple-Dose, Dose-Escalation Study of Siltuximab, an Anti-Interleukin-6 Monoclonal Antibody, in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2014, 20, 2192–2204. [Google Scholar] [CrossRef]
- Lu, Z.Y.; Brailly, H.; Wijdenes, J.; Bataille, R.; Rossi, J.F.; Klein, B. Measurement of whole body interleukin-6 (IL-6) production: Prediction of the efficacy of anti-IL-6 treatments. Blood 1995, 86, 3123–3131. [Google Scholar] [CrossRef]
- Shinriki, S.; Jono, H.; Ota, K.; Ueda, M.; Kudo, M.; Ota, T.; Oike, Y.; Endo, M.; Ibusuki, M.; Hiraki, A.; et al. Humanized Anti-Interleukin-6 Receptor Antibody Suppresses Tumor Angiogenesis and In Vivo Growth of Human Oral Squamous Cell Carcinoma. Clin. Cancer Res. 2009, 15, 5426–5434. [Google Scholar] [CrossRef] [PubMed]
- Bei, R.; Budillon, A.; Masuelli, L.; Cereda, V.; Vitolo, D.; Di Gennaro, E.; Ripavecchia, V.; Palumbo, C.; Ionna, F.; Losito, S.; et al. Frequent overexpression of multiple ErbB receptors by head and neck squamous cell carcinoma contrasts with rare antibody immunity in patients. J. Pathol. 2004, 204, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Soulieres, D.; Senzer, N.N.; Vokes, E.E.; Hidalgo, M.; Agarwala, S.S.; Siu, L.L. Multicenter phase II study of erlotinib, an oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with recurrent or metastatic squamous cell cancer of the head and neck. J. Clin. Oncol. 2004, 22, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Stanam, A.; Love-Homan, L.; Joseph, T.S.; Espinosa-Cotton, M.; Simons, A.L. Upregulated interleukin-6 expression contributes to erlotinib resistance in head and neck squamous cell carcinoma. Mol. Oncol. 2015, 9, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
- Quintás-Cardama, A.; Cortes, J.E.; O’Brien, S.; Ravandi, F.; Borthakur, G.; Liu, D.; Bleickardt, E.; Chen, T.T.; Kantarjian, H.M. Dasatinib early intervention after cytogenetic or hematologic resistance to imatinib in patients with chronic myeloid leukemia. Cancer 2009, 115, 2912–2921. [Google Scholar] [CrossRef] [PubMed]
- Johnson, F.M.; Agrawal, S.; Burris, H.; Rosen, L.; Dhillon, N.; Hong, D.; Blackwood-Chirchir, A.; Luo, F.R.; Sy, O.; Kaul, S.; et al. Phase 1 pharmacokinetic and drug-interaction study of dasatinib in patients with advanced solid tumors. Cancer 2010, 116, 1582–1591. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.P.; Tran, C.; Lee, F.Y.; Chen, P.; Norris, D.; Sawyers, C.L. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science 2004, 305, 399–401. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2020 update on diagnosis, therapy and monitoring. Am. J. Hematol. 2020, 95, 691–709. [Google Scholar] [CrossRef]
- Beebe, J.D.; Liu, J.Y.; Zhang, J.T. Two decades of research in discovery of anticancer drugs targeting STAT3, how close are we? Pharmacol. Ther. 2018, 191, 74–91. [Google Scholar] [CrossRef]
- Heppler, L.N.; Frank, D.A. Targeting Oncogenic Transcription Factors: Therapeutic Implications of Endogenous STAT Inhibitors. Trends Cancer 2017, 3, 816–827. [Google Scholar] [CrossRef]
- Zhang, T.; Kee, W.H.; Seow, K.T.; Fung, W.; Cao, X. The coiled-coil domain of Stat3 is essential for its SH2 domain-mediated receptor binding and subsequent activation induced by epidermal growth factor and interleukin-6. Mol. Cell. Biol. 2000, 20, 7132–7139. [Google Scholar] [CrossRef] [PubMed]
- Kraskouskaya, D.; Duodu, E.; Arpin, C.C.; Gunning, P.T. Progress towards the development of SH2 domain inhibitors. Chem. Soc. Rev. 2013, 42, 3337–3370. [Google Scholar] [CrossRef] [PubMed]
- Uddalak, B.; Moses, M.K.; Prema, R.; David, J.T. Targeting Janus Kinases and Signal Transducer and Activator of Transcription 3 To Treat Inflammation, Fibrosis, and Cancer: Rationale, Progress, and Caution. Pharmacol. Rev. 2020, 72, 486. [Google Scholar] [CrossRef]
- Turkson, J.; Ryan, D.; Kim, J.S.; Zhang, Y.; Chen, Z.; Haura, E.; Laudano, A.; Sebti, S.; Hamilton, A.D.; Jove, R. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J. Biol. Chem. 2001, 276, 45443–45455. [Google Scholar] [CrossRef] [PubMed]
- Siddiquee, K.A.; Gunning, P.T.; Glenn, M.; Katt, W.P.; Zhang, S.; Schrock, C.; Sebti, S.M.; Jove, R.; Hamilton, A.D.; Turkson, J. An oxazole-based small-molecule Stat3 inhibitor modulates Stat3 stability and processing and induces antitumor cell effects. ACS Chem. Biol. 2007, 2, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, U.; Eckols, T.K.; Xu, X.; Kasembeli, M.M.; Chen, Y.; Adachi, M.; Song, Y.; Mo, Q.; Lai, S.Y.; Tweardy, D.J. Small-molecule inhibition of STAT3 in radioresistant head and neck squamous cell carcinoma. Oncotarget 2016, 7, 26307–26330. [Google Scholar] [CrossRef] [PubMed]
- Kasembeli, M.M.; Singhmar, P.; Ma, J.; Edralin, J.; Tang, Y.; Adams, C., 3rd; Heijnen, C.J.; Kavelaars, A.; Tweardy, D.J. TTI-101: A competitive inhibitor of STAT3 that spares oxidative phosphorylation and reverses mechanical allodynia in mouse models of neuropathic pain. Biochem. Pharmacol. 2021, 192, 114688. [Google Scholar] [CrossRef]
- Redell, M.S.; Ruiz, M.J.; Alonzo, T.A.; Gerbing, R.B.; Tweardy, D.J. Stat3 signaling in acute myeloid leukemia: Ligand-dependent and-independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood J. Am. Soc. Hematol. 2011, 117, 5701–5709. [Google Scholar] [CrossRef]
- Jung, K.H.; Yoo, W.; Stevenson, H.L.; Deshpande, D.; Shen, H.; Gagea, M.; Yoo, S.-Y.; Wang, J.; Eckols, T.K.; Bharadwaj, U.; et al. Multifunctional Effects of a Small-Molecule STAT3 Inhibitor on NASH and Hepatocellular Carcinoma in Mice. Clin. Cancer Res. 2017, 23, 5537–5546. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Vining, D.J.; Arora, S.P.; de Achaval, S.; Larson, J.; Cartwright, C.; Avritscher, R.; Alibhai, I.; Kaseb, A.O. Phase 1 trial evaluating TTI-101, a first-in-class, orally bioavailable, small molecule, inhibitor of STAT3, in patients with advanced solid tumors. J. Clin. Oncol. 2023, 41, 3018. [Google Scholar] [CrossRef]
- Tvardi Therapeutics Announces First Patients Dosed in Its Phase 2 Liver Cancer Trial Using TTI-101, a Novel STAT3 Inhibitor. News Release. Tvardi Therapeutics, Inc. 22 June 2023. Available online: https://tinyurl.com/5n6c5zbs (accessed on 17 September 2023).
- NCT05384119, Study of TTI-101 in Combination for Participants With Metastatic Hormone Receptor Positive and Human Epithelial Receptor 2 (HER2)-Negative Breast Cancer. 20 May 2022. Available online: https://ClinicalTrials.gov/show/NCT05384119 (accessed on 11 November 2022).
- A Study of TTI-101 as Monotherapy and in Combination in Participants With Locally Advanced or Metastatic, and Unresectable Hepatocellular Carcinoma. ClinicalTrials.gov Identifier: NCT05440708. Available online: https://clinicaltrials.gov/study/NCT05440708 (accessed on 12 September 2023).
- Adachi, M.; Cui, C.; Dodge, C.T.; Bhayani, M.K.; Lai, S.Y. Targeting STAT3 inhibits growth and enhances radiosensitivity in head and neck squamous cell carcinoma. Oral Oncol. 2012, 48, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, H.; Li, X.; Han, L.; Xu, N.; Shi, A. Signaling pathway inhibitors target breast cancer stem cells in triple-negative breast cancer. Oncol. Rep. 2019, 41, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Schust, J.; Sperl, B.; Hollis, A.; Mayer, T.U.; Berg, T. Stattic: A small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 2006, 13, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Bijak, M. Silybin, a Major Bioactive Component of Milk Thistle (Silybum marianum L. Gaernt.)-Chemistry, Bioavailability, and Metabolism. Molecules 2017, 22, 1942. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, C.; Tyagi, A.; Kaur, M.; Agarwal, R. Silibinin inhibits constitutive activation of Stat3, and causes caspase activation and apoptotic death of human prostate carcinoma DU145 cells. Carcinogenesis 2007, 28, 1463–1470. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-X.; Cai, H.; Jiang, G.; Zhou, T.-B.; Wu, H. Silibinin inhibits proliferation, induces apoptosis and causes cell cycle arrest in human gastric cancer MGC803 cells via STAT3 pathway inhibition. Asian Pac. J. Cancer Prev. 2014, 15, 6791–6798. [Google Scholar] [CrossRef]
- Priego, N.; Zhu, L.; Monteiro, C.; Mulders, M.; Wasilewski, D.; Bindeman, W.; Doglio, L.; Martínez, L.; Martínez-Saez, E.; Ramón, Y.C.S.; et al. STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nat. Med. 2018, 24, 1024–1035. [Google Scholar] [CrossRef]
- Pérez-Sánchez, A.; Cuyàs, E.; Ruiz-Torres, V.; Agulló-Chazarra, L.; Verdura, S.; González-Álvarez, I.; Bermejo, M.; Joven, J.; Micol, V.; Bosch-Barrera, J.; et al. Intestinal Permeability Study of Clinically Relevant Formulations of Silibinin in Caco-2 Cell Monolayers. Int. J. Mol. Sci. 2019, 20, 1606. [Google Scholar] [CrossRef]
- Addeo, R. Silibinin: A New Opportunity for the Treatment of Brain Metastasis from Lung Cancer. J. Exp. Pharmacol. 2021, 13, 901–903. [Google Scholar] [CrossRef]
- Brown, J.R.; Walker, S.R.; Heppler, L.N.; Tyekucheva, S.; Nelson, E.A.; Klitgaard, J.; Nicolais, M.; Kroll, Y.; Xiang, M.; Yeh, J.E.; et al. Targeting constitutively active STAT3 in chronic lymphocytic leukemia: A clinical trial of the STAT3 inhibitor pyrimethamine with pharmacodynamic analyses. Am. J. Hematol. 2021, 96, E95–E98. [Google Scholar] [CrossRef]
- Bielinska, A.; Shivdasani, R.A.; Zhang, L.Q.; Nabel, G.J. Regulation of gene expression with double-stranded phosphorothioate oligonucleotides. Science 1990, 250, 997–1000. [Google Scholar] [CrossRef] [PubMed]
- Leong, P.L.; Andrews, G.A.; Johnson, D.E.; Dyer, K.F.; Xi, S.; Mai, J.C.; Robbins, P.D.; Gadiparthi, S.; Burke, N.A.; Watkins, S.F.; et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc. Natl. Acad. Sci. USA 2003, 100, 4138–4143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, P.; Zhang, B.; Mao, H.; Shen, L.; Ma, Y. Inhibitory effects of STAT3 decoy oligodeoxynucleotides on human epithelial ovarian cancer cell growth in vivo. Int. J. Mol. Med. 2013, 32, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Lewis, H.D.; Winter, A.; Murphy, T.F.; Tripathi, S.; Pandey, V.N.; Barton, B.E. STAT3 inhibition in prostate and pancreatic cancer lines by STAT3 binding sequence oligonucleotides: Differential activity between 5′ and 3′ ends. Mol. Cancer Ther. 2008, 7, 1543–1550. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sun, X.; Zhang, J.; Wang, L.; Tian, Z. Growth inhibition of human hepatocellular carcinoma cells by blocking STAT3 activation with decoy-ODN. Cancer Lett. 2008, 262, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Sen, M.; Paul, K.; Freilino, M.L.; Li, H.; Li, C.; Johnson, D.E.; Wang, L.; Eiseman, J.; Grandis, J.R. Systemic Administration of a Cyclic Signal Transducer and Activator of Transcription 3 (STAT3) Decoy Oligonucleotide Inhibits Tumor Growth without Inducing Toxicological Effects. Mol. Med. 2014, 20, 46–56. [Google Scholar] [CrossRef]
- Sen, M.; Thomas, S.M.; Kim, S.; Yeh, J.I.; Ferris, R.L.; Johnson, J.T.; Duvvuri, U.; Lee, J.; Sahu, N.; Joyce, S.; et al. First-in-Human Trial of a STAT3 Decoy Oligonucleotide in Head and Neck Tumors: Implications for Cancer Therapy. Cancer Discov. 2012, 2, 694–705. [Google Scholar] [CrossRef]
- Li, Y.; Rogoff, H.A.; Keates, S.; Gao, Y.; Murikipudi, S.; Mikule, K.; Leggett, D.; Li, W.; Pardee, A.B.; Li, C.J. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc. Natl. Acad. Sci. USA 2015, 112, 1839–1844. [Google Scholar] [CrossRef]
- Jonker, D.J.; Nott, L.; Yoshino, T.; Gill, S.; Shapiro, J.; Ohtsu, A.; Zalcberg, J.; Vickers, M.M.; Wei, A.C.; Gao, Y.; et al. Napabucasin versus placebo in refractory advanced colorectal cancer: A randomised phase 3 trial. Lancet Gastroenterol. Hepatol. 2018, 3, 263–270. [Google Scholar] [CrossRef]
- Vogt, M.; Domoszlai, T.; Kleshchanok, D.; Lehmann, S.; Schmitt, A.; Poli, V.; Richtering, W.; Müller-Newen, G. The role of the N-terminal domain in dimerization and nucleocytoplasmic shuttling of latent STAT3. J. Cell Sci. 2011, 124, 900–909. [Google Scholar] [CrossRef]
- Zhang, X.; Darnell, J.E., Jr. Functional importance of Stat3 tetramerization in activation of the alpha 2-macroglobulin gene. J. Biol. Chem. 2001, 276, 33576–33581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sif, S.; DeWille, J. The mouse C/EBPdelta gene promoter is regulated by STAT3 and Sp1 transcriptional activators, chromatin remodeling and c-Myc repression. J. Cell. Biochem. 2007, 102, 1256–1270. [Google Scholar] [CrossRef] [PubMed]
- Cimica, V.; Chen, H.C.; Iyer, J.K.; Reich, N.C. Dynamics of the STAT3 transcription factor: Nuclear import dependent on Ran and importin-β1. PLoS ONE 2011, 6, e20188. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, O.A.; Tarasova, N.I.; Zhang, X.; Chasovskikh, S.; Cheema, A.K.; Wang, H.; Brown, M.L.; Dritschilo, A. STAT3 suppresses transcription of proapoptotic genes in cancer cells with the involvement of its N-terminal domain. Proc. Natl. Acad. Sci. USA 2013, 110, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Varone, A.; Spano, D.; Corda, D. Shp1 in Solid Cancers and Their Therapy. Front. Oncol. 2020, 10, 935. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Sethi, G.; Ahn, K.S.; Sandur, S.K.; Pandey, M.K.; Kunnumakkara, A.B.; Sung, B.; Ichikawa, H. Targeting signal-transducer-and-activator-of-transcription-3 for prevention and therapy of cancer: Modern target but ancient solution. Ann. N. Y. Acad. Sci. 2006, 1091, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Kannappan, R.; Reuter, S.; Kim, J.H.; Aggarwal, B.B. Chemosensitization of tumors by resveratrol. Ann. N. Y. Acad. Sci. 2011, 1215, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.-T.; Cheng, A.-L.; Shiau, C.-W.; Huang, H.-P.; Huang, J.-W.; Chen, P.-J.; Chen, K.-F. Signal transducer and activator of transcription 3 is a major kinase-independent target of sorafenib in hepatocellular carcinoma. J. Hepatol. 2011, 55, 1041–1048. [Google Scholar] [CrossRef]
- André, F.; Bachelot, T.; Campone, M.; Dalenc, F.; Perez-Garcia, J.M.; Hurvitz, S.A.; Turner, N.; Rugo, H.; Smith, J.W.; Deudon, S.; et al. Targeting FGFR with Dovitinib (TKI258): Preclinical and Clinical Data in Breast Cancer. Clin. Cancer Res. 2013, 19, 3693–3702. [Google Scholar] [CrossRef]
- Huang, T.T.; Su, J.C.; Liu, C.Y.; Shiau, C.W.; Chen, K.F. Alteration of SHP-1/p-STAT3 Signaling: A Potential Target for Anticancer Therapy. Int. J. Mol. Sci. 2017, 18, 1234. [Google Scholar] [CrossRef]
- Musolino, A.; Campone, M.; Neven, P.; Denduluri, N.; Barrios, C.H.; Cortes, J.; Blackwell, K.; Soliman, H.; Kahan, Z.; Bonnefoi, H.; et al. Phase II, randomized, placebo-controlled study of dovitinib in combination with fulvestrant in postmenopausal patients with HR+, HER2− breast cancer that had progressed during or after prior endocrine therapy. Breast Cancer Res. 2017, 19, 18. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Tong, X.; Fu, L.; Zhang, R. Knockdown of STAT3 by shRNA inhibits the growth of CAOV3 ovarian cancer cell line in vitro and in vivo. Acta Biochim. Et Biophys. Sin. 2008, 40, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Cai, J.H.; Xie, S.J.; Li, G.X.; Song, W.Q.; Yan, Q.H.; Yan, L.; Zhang, F. Therapeutic effects of signal transducer and activator of transcription 3 siRNA on human breast cancer in xenograft mice. Chin. Med. J. (Engl.) 2011, 124, 1854–1861. [Google Scholar] [PubMed]
- Lu, Y.M.; Zhang, S.L.; Meng, L.R.; Zhao, Y.Y. Influence of human epidermal growth factor receptor-2 siRNA on chemosensitivity to cisplatin of human ovarian carcinoma cells: An in vitro experiment. Zhonghua Yi Xue Za Zhi 2008, 88, 909–913. [Google Scholar] [PubMed]
- Ma, Y.; Kowolik, C.M.; Swiderski, P.M.; Kortylewski, M.; Yu, H.; Horne, D.A.; Jove, R.; Caballero, O.L.; Simpson, A.J.; Lee, F.T.; et al. Humanized Lewis-Y specific antibody based delivery of STAT3 siRNA. ACS Chem. Biol. 2011, 6, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.D.; Vorhies, J.S.; Senzer, N.; Nemunaitis, J. siRNA vs. shRNA: Similarities and differences. Adv. Drug Deliv. Rev. 2009, 61, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Esposito, C.L.; Nuzzo, S.; Catuogno, S.; Romano, S.; de Nigris, F.; de Franciscis, V. STAT3 Gene Silencing by Aptamer-siRNA Chimera as Selective Therapeutic for Glioblastoma. Mol. Ther. Nucleic Acids 2018, 10, 398–411. [Google Scholar] [CrossRef]
- Pedroso de Lima, M.C.; Simões, S.; Pires, P.; Faneca, H.; Düzgüneş, N. Cationic lipid-DNA complexes in gene delivery: From biophysics to biological applications. Adv. Drug Deliv. Rev. 2001, 47, 277–294. [Google Scholar] [CrossRef]
- Huang, S.; Chen, M.; Shen, Y.; Shen, W.; Guo, H.; Gao, Q.; Zou, X. Inhibition of activated Stat3 reverses drug resistance to chemotherapeutic agents in gastric cancer cells. Cancer Lett. 2012, 315, 198–205. [Google Scholar] [CrossRef]
- Zhao, C.; Li, H.; Lin, H.J.; Yang, S.; Lin, J.; Liang, G. Feedback Activation of STAT3 as a Cancer Drug-Resistance Mechanism. Trends Pharmacol. Sci. 2016, 37, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Roschewski, M.; Izumi, R.; Hamdy, A.; Patel, M.R.; Arkenau, H.-T.; de Vos, S.; Reagan, P.M.; Zinzani, P.L.; Davies, A.; Pagel, J.M.; et al. PRISM: A Platform Protocol for the Treatment of Relapsed/Refractory Aggressive Non-Hodgkin Lymphoma. Blood 2019, 134, 2869. [Google Scholar] [CrossRef]
- Strati, P.; de Vos, S.; Ruan, J.; Maddocks, K.J.; Flowers, C.R.; Rule, S.; Patel, P.; Xu, Y.; Wei, H.; Frigault, M.M.; et al. Acalabrutinib for treatment of diffuse large B-cell lymphoma: Results from a phase Ib study. Haematologica 2021, 106, 2774–2778. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Kurzrock, R.; Kim, Y.; Woessner, R.; Younes, A.; Nemunaitis, J.; Fowler, N.; Zhou, T.; Schmidt, J.; Jo, M.; et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 2015, 7, 314ra185. [Google Scholar] [CrossRef] [PubMed]
- Odate, S.; Veschi, V.; Yan, S.; Lam, N.; Woessner, R.; Thiele, C.J. Inhibition of STAT3 with the Generation 2.5 Antisense Oligonucleotide, AZD9150, Decreases Neuroblastoma Tumorigenicity and Increases Chemosensitivity. Clin. Cancer Res. 2017, 23, 1771–1784. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, W.; Dong, S.; Li, W.; Zhu, W.; Zhou, W. STAT3 Inhibitors: A Novel Insight for Anticancer Therapy of Pancreatic Cancer. Biomolecules 2022, 12, 1450. [Google Scholar] [CrossRef]
- Reilley, M.J.; McCoon, P.; Cook, C.; Lyne, P.; Kurzrock, R.; Kim, Y.; Woessner, R.; Younes, A.; Nemunaitis, J.; Fowler, N.; et al. STAT3 antisense oligonucleotide AZD9150 in a subset of patients with heavily pretreated lymphoma: Results of a phase 1b trial. J. Immunother. Cancer 2018, 6, 119. [Google Scholar] [CrossRef]
- NCT03421353, AZD9150 Plus Durvalumab Alone or in Combination with Chemotherapy in Patients with Advanced, Solid Tumours and in Patients With Non-Small-Cell Lung Cancer, 22 November. Available online: https://clinicaltrials.gov/ct2/show/NCT03421353 (accessed on 9 December 2022).
- NCT03527147, Platform Study for the Treatment of Relapsed or Refractory Aggressive Non-Hodgkin’s Lymphoma (PRISM Study) (PRISM), 22 November. Available online: https://clinicaltrials.gov/ct2/show/NCT03527147 (accessed on 9 December 2022).
- Bewersdorf, J.P.; Rampal, R.K. Hitting the brakes on accelerated and blast-phase myeloproliferative neoplasms: Current and emerging concepts. Hematol. Am. Soc. Hematol. Educ. Program. 2022, 2022, 218–224. [Google Scholar] [CrossRef]
- Andreozzi, F.; Massaro, F.; Wittnebel, S.; Spilleboudt, C.; Lewalle, P.; Salaroli, A. New Perspectives in Treating Acute Myeloid Leukemia: Driving towards a Patient-Tailored Strategy. Int. J. Mol. Sci. 2022, 23, 3887. [Google Scholar] [CrossRef]
- Rampal, R.; Ahn, J.; Abdel-Wahab, O.; Nahas, M.; Wang, K.; Lipson, D.; Otto, G.A.; Yelensky, R.; Hricik, T.; McKenney, A.S.; et al. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc. Natl. Acad. Sci. USA 2014, 111, E5401–E5410. [Google Scholar] [CrossRef]
- Bai, L.; Zhou, H.; Xu, R.; Zhao, Y.; Chinnaswamy, K.; McEachern, D.; Chen, J.; Yang, C.Y.; Liu, Z.; Wang, M.; et al. A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo. Cancer Cell 2019, 36, 498–511.e417. [Google Scholar] [CrossRef] [PubMed]

| Cancer Type | STAT3 Function | References |
|---|---|---|
| Multiple myeloma | ||
| Cell lines | Oncogene | Catlett-Falcone R et al., 1999 [12] |
| Astrocytoma | ||
| Cell lines | Oncogene | Konnikova L et al., 2003 [13] |
| Breast cancer | ||
| Cell lines | Oncogene | Gritsko T et al., 2006 [60] |
| Cell lines | Oncogene | Garcia R et al., 1997 [61] |
| Tumor | Oncogene | Watson CJ et al., 1995 [62] |
| Tumor | Oncogene | Lo H et al., 2005 [63] |
| Head and neck squamous cell cancer | ||
| Cell lines | Oncogene | Grandis JR et al., 1998 [54] |
| Tumor | Oncogene | Pectasides E et al., 2010 [64] |
| Tumor | Tumor suppressor | Shinagawa K et al., 2017 [65] |
| Non-small cell Lung Cancer | ||
| Cell lines | Onocogene | Alvarez JV et al., 2006 [56] |
| Tumor | Tumor suppressor | Grabner et al., 2015 [59] |
| Glioblastoma | ||
| Cell lines | Tumor suppressor | de la Iglesia N et al., 2009 [66] |
| Papillary thyroid carcinoma | ||
| Cell lines | Tumor suppressor | Couto JP et al., 2012 [67] |
| Prostate | ||
| Patient derived xenografts | Tumor suppressor | Pencik J et al., 2015 [68] |
| Pancreas | ||
| Cell lines | Oncogene | Corcoran RB et al., 2011 [69] |
| Colorectal | ||
| Cell lines | Tumor suppressor | Lee J et al., 2012 [70] |
| Acute Myelogenous Leukemia | ||
| Tumor | Oncogene | Benekli M et al., 2002 [71] |
| Chronic Myelogenous Leukemia | ||
| Cell lines | Oncogene | Mencalha AL., 2012 [72] |
| Acute Lymphocytic Leukemia | ||
| Tumor | Oncogene | Adamaki M et al., 2015 [73] |
| B-cell lymphoma | ||
| Cell lines | Oncogene | Turton KB et al., 2015 [74] |
| Agent | Trial Number | Target | Type | Tumor type | Phase |
|---|---|---|---|---|---|
| Direct inhibitors | |||||
| TTI-101 | NCT03195699 | SH2 domain | Small molecules | Advanced Solid Tumors | I |
| Pyrimethamine | NCT01066663 | SH2 domain | Small molecule | CLL, Small Lymphocytic Lymphoma | I/II |
| Silibinin | NCT05689619 | SH2 domain | Small molecule | NSCLC and BC Patients with Single Brain Metastasis | II |
| Indirect inhibitors | |||||
| AZD4205 | NCT04105010 | JAK 1 | Small molecule | Peripheral T cell lymphoma (PTCL) | I/II |
| Itacitinib | NCT03670069 | JAK 1 | Small molecule | Sarcoma | I |
| Itacitinib | NCT04358185 | JAK1 | Small molecule | Advanced hepatocellular carcinoma | I |
| Ruxolitinib | NCT05592015 | JAK1/2 | Small molecule | T-cell large granular lymphocyte leukemia | II |
| WP1066 | NCT01904123, NCT04334863 | JAK2 | Small molecule | Recurrent malignant Glioma or progressive metastatic melanoma in the brain | I |
| Pacritinib | NCT03645824 | JAK2 | Small molecule | Myelofibrosis | II |
| Combination strategies | |||||
| TTI-101 + palbociclib and aromatase inhibitor | NCT05384119 | SH2 domain | Small molecules | Metastatic Hormone Receptor (HR)-Positive and Human Epidermal Receptor 2 (HER2)-Negative Breast Cancer | I/II |
| AZD9150 + Acalabrutinib | NCT03527147 | STAT3 mRNA | Antisense oligonucleotides | Relapsed/refractory aggressive Non-Hodgkin’s Lymphoma (NHL) | I |
| Fedratinib + Ivosidenib/Enasidenib | NCT04955938 | JAK2 | Small molecule | Blood cancers that show Isocitrate dehydrogenase (IDH) | I |
| Ruxolitinib + Decitabine | NCT04282187 | JAK1/2 | small molecule | Accelerated/blast phase myeloproliferative neoplasms | II |
| Ruxolitinib and Venetoclax | NCT03874052 | JAK1/2 | Small molecule | Relapsed or Refractory Acute Myeloid Leukemia | I |
| Ruxolitinib, Carfilzomib, and Low Dose Dexamethasone | NCT03773107 | JAK1/2 | Small molecule | Carfilzomib-Refractory Multiple Myeloma | I/II |
| Ruxolitinib and Pelabresib (CPI-0610) | NCT04603495 | JAK1/2 | Small molecule | Myelofibrosis | III |
| Ruxolitinib and CPX-351 | NCT03878199 | JAK1/2 | Small molecule | Accelerated phase or blast phase myeloproliferative neoplasm | I/II |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adesoye, T.; Tripathy, D.; Hunt, K.K.; Keyomarsi, K. Exploring Novel Frontiers: Leveraging STAT3 Signaling for Advanced Cancer Therapeutics. Cancers 2024, 16, 492. https://doi.org/10.3390/cancers16030492
Adesoye T, Tripathy D, Hunt KK, Keyomarsi K. Exploring Novel Frontiers: Leveraging STAT3 Signaling for Advanced Cancer Therapeutics. Cancers. 2024; 16(3):492. https://doi.org/10.3390/cancers16030492
Chicago/Turabian StyleAdesoye, Taiwo, Debasish Tripathy, Kelly K. Hunt, and Khandan Keyomarsi. 2024. "Exploring Novel Frontiers: Leveraging STAT3 Signaling for Advanced Cancer Therapeutics" Cancers 16, no. 3: 492. https://doi.org/10.3390/cancers16030492
APA StyleAdesoye, T., Tripathy, D., Hunt, K. K., & Keyomarsi, K. (2024). Exploring Novel Frontiers: Leveraging STAT3 Signaling for Advanced Cancer Therapeutics. Cancers, 16(3), 492. https://doi.org/10.3390/cancers16030492






