The Role of SOX2 and SOX9 in Radioresistance and Tumor Recurrence
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression and Clinical Data Sets
2.2. Differential Gene Expression Analysis and Co-Expression
2.3. Images Analysis
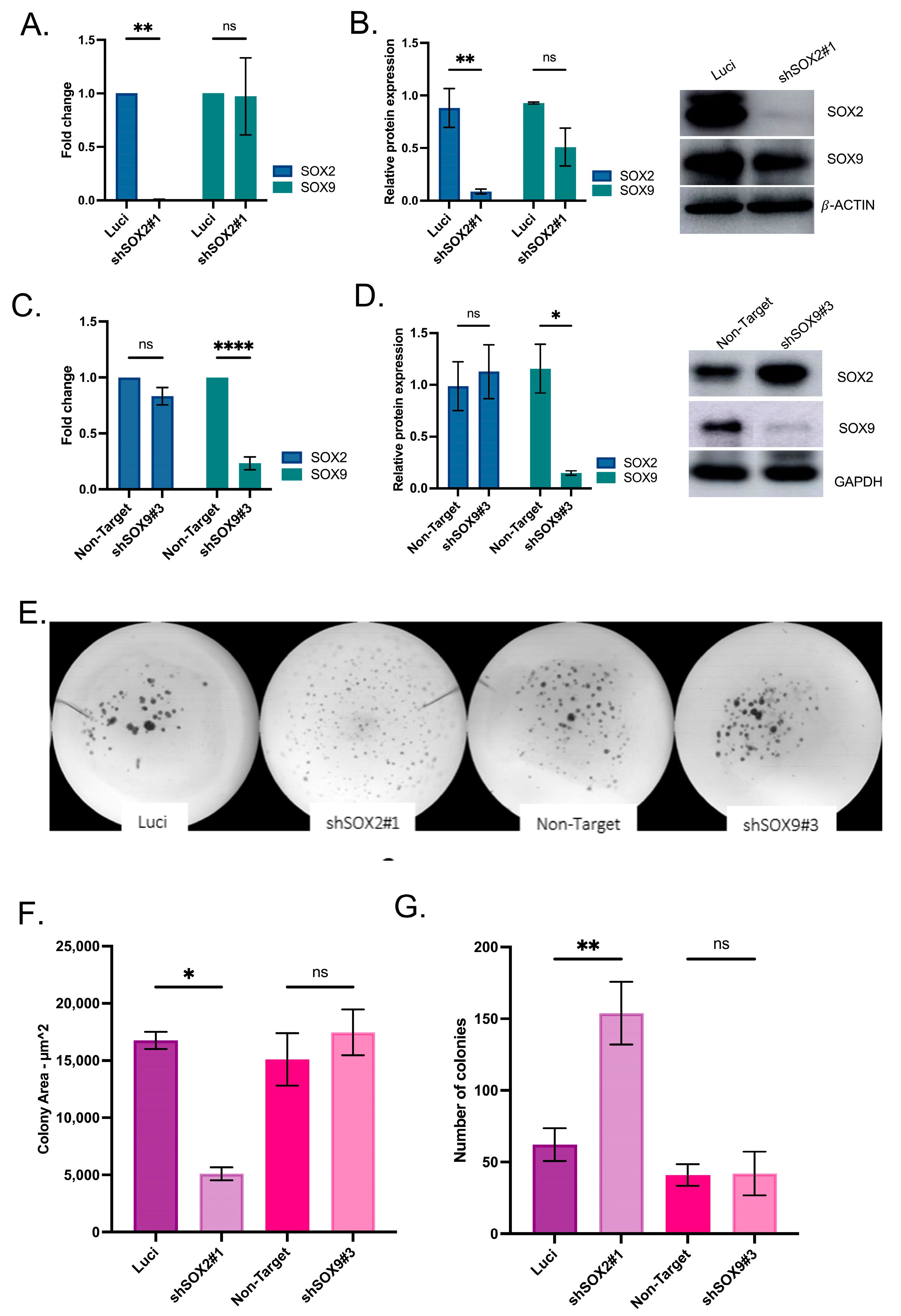
2.4. Cellular Culture and Gene Silencing
2.5. Colony Formation Assay and Irradiation Assay
2.6. mRNA Expression Analysis
2.7. RNA Sequencing
2.8. Western Blotting Analysis
2.9. Immunohistochemistry (IHC)
2.10. Statistical Analysis
3. Results
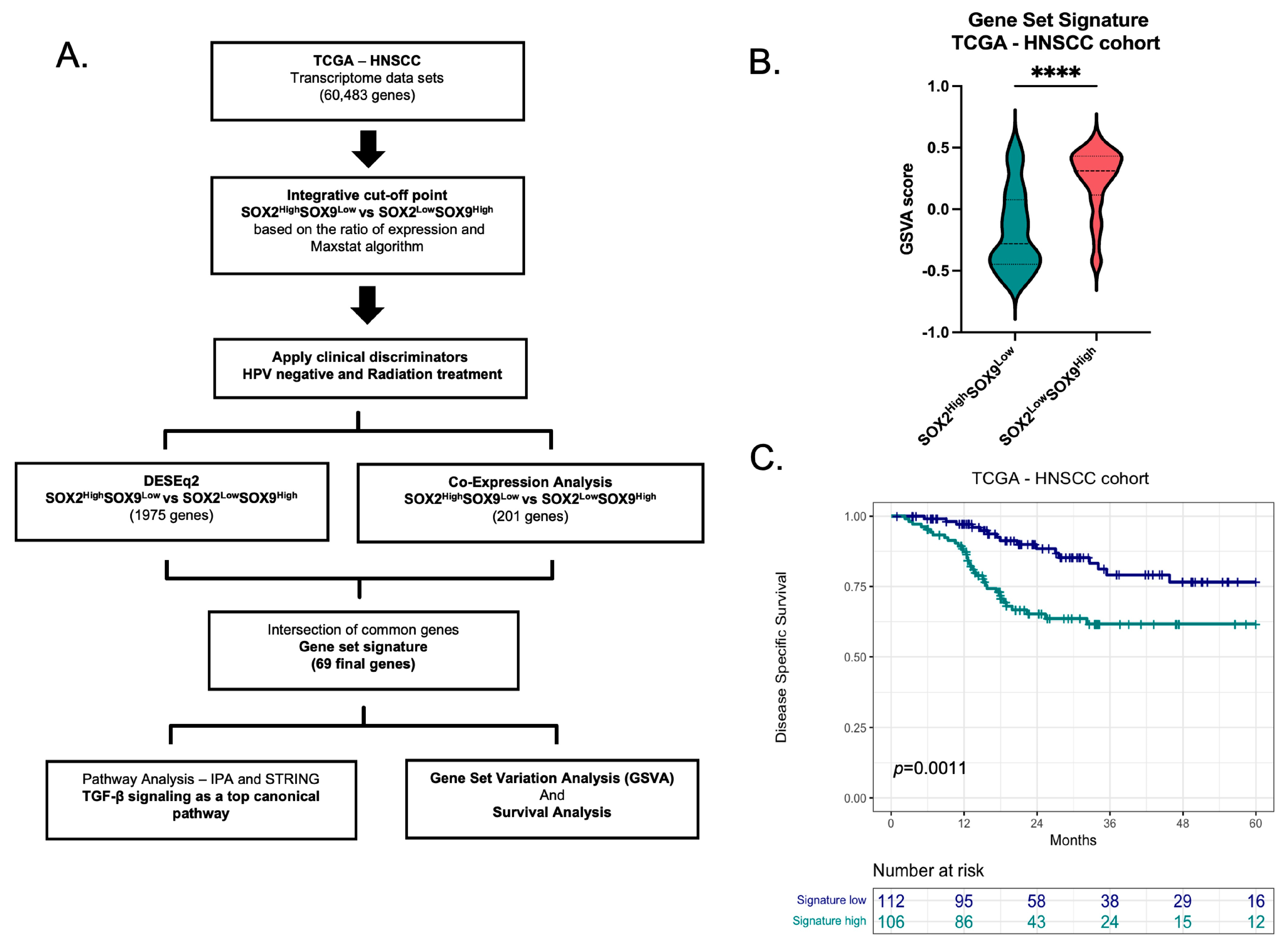
3.1. Inverse SOX2 and SOX9 Expression Correlates with Disease Specific Survival in HNSCC
3.2. Gene signature Based in the Inverse Expression of SOX2 and SOX9
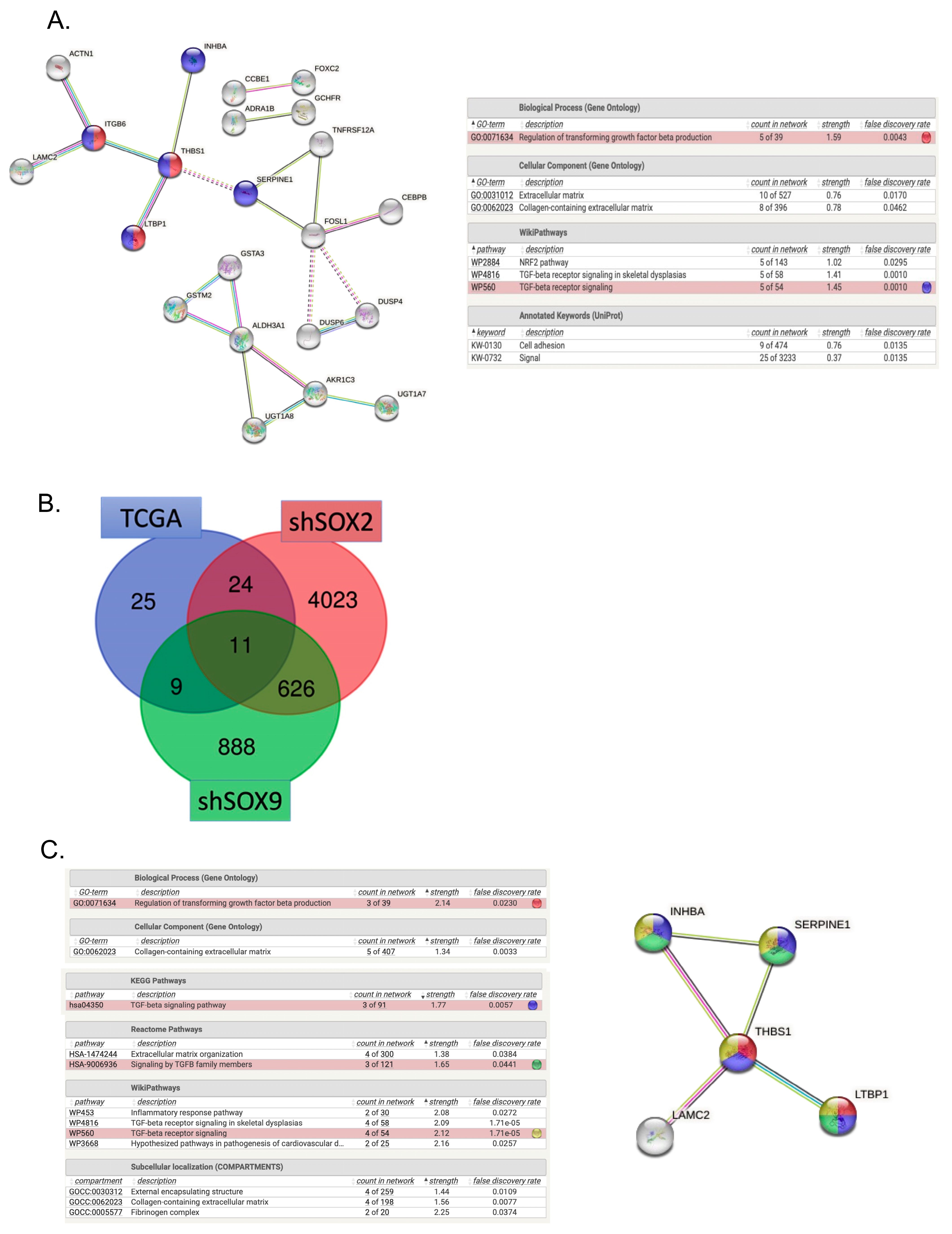
3.3. TGF-β Signaling as One of the Top Canonical Pathways Regulated by Inverse SOX2/SOX9 Expression
3.4. SOX2 and SOX9 Are Independent in Gene Expression
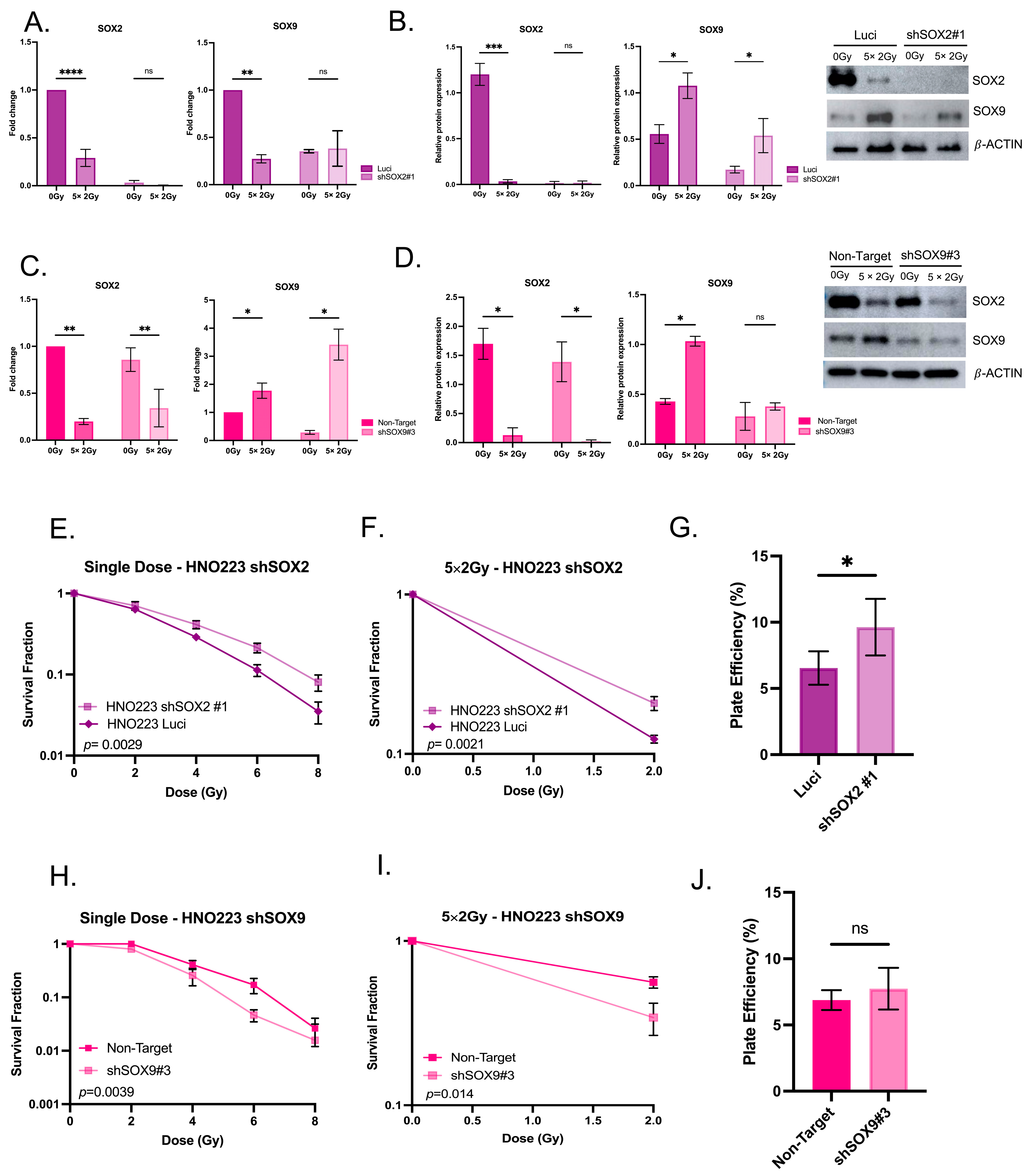
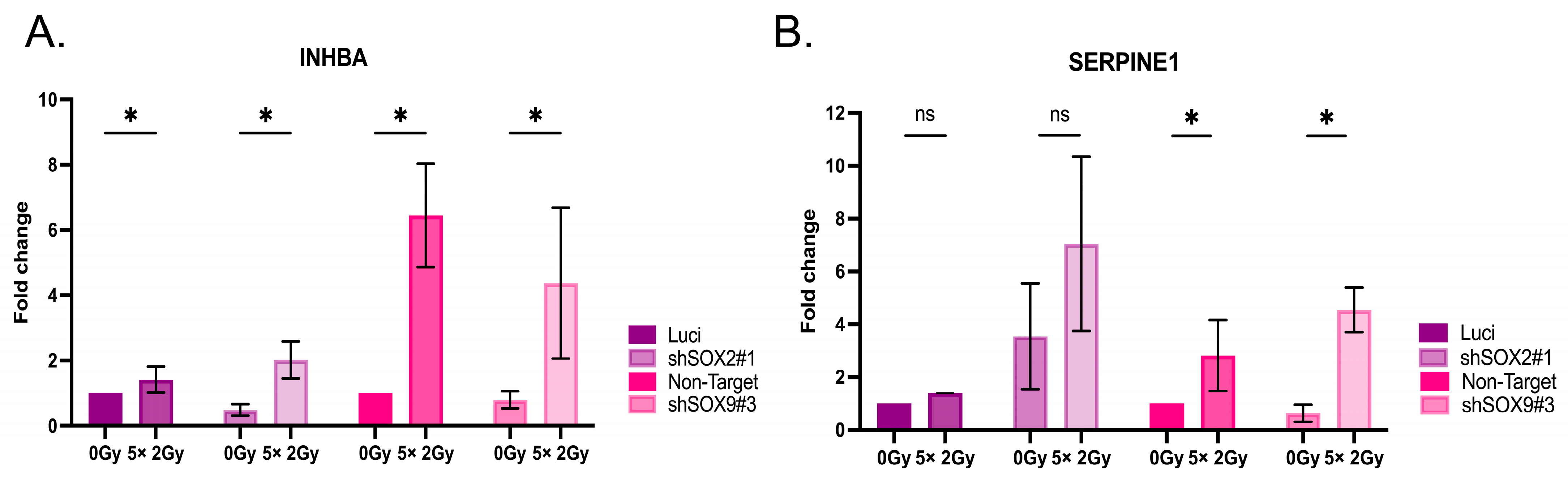
3.5. Radiation Impacts SOX2 and SOX9 Expression in Head and Neck Cell Lines
3.6. Radiation Impacts Genes in the TGF-β Signaling Pathway
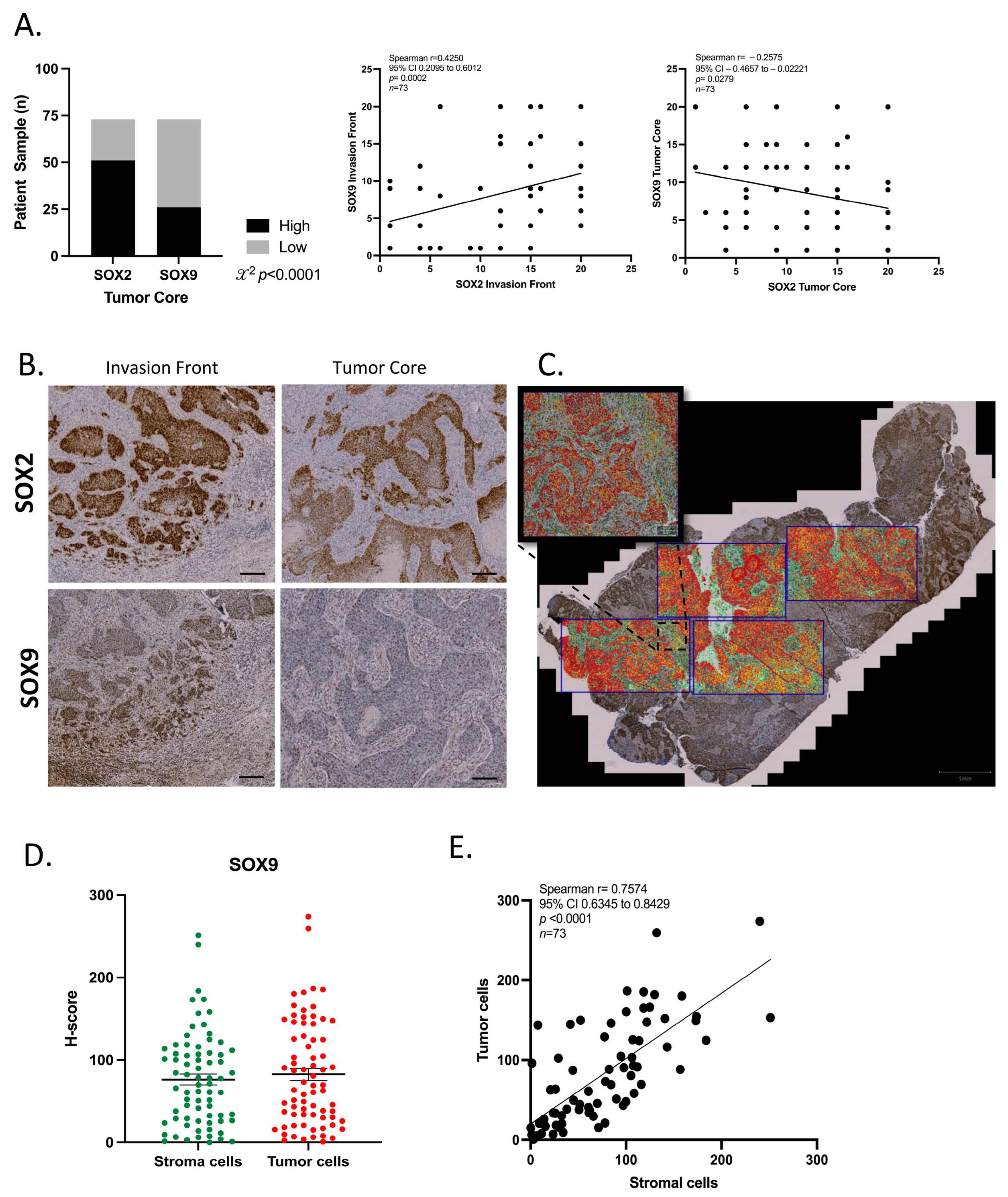
3.7. SOX2 and SOX9 Present Difference Expression in Tumor Compartments
3.8. SOX9 Is Expressed in the Tumor Microenvironment
4. Discussion
4.1. Inverse SOX2 and SOX9 Expression Correlates with Disease Specific Survival in HNSCC
4.2. Gene Signature Based on the Inverse Expression of SOX2 and SOX9
4.3. TGF-β Signaling as One of the Top Canonical Pathways Regulated by Inverse SOX2/SOX9 Expression
4.4. SOX9 in the Tumor Microenvironment
4.5. Cancer Stem Cells, the Tumor Microenvironment and Radiation Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Braakhuis, B.J.; Brakenhoff, R.H. The Molecular Biology of Head and Neck Cancer. Nat. Rev. Cancer 2011, 11, 9–22. [Google Scholar] [CrossRef]
- Cogliano, V.J.; Baan, R.; Straif, K.; Grosse, Y.; Lauby-Secretan, B.; El Ghissassi, F.; Bouvard, V.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; et al. Preventable Exposures Associated with Human Cancers. J. Natl. Cancer Inst. 2011, 103, 1827–1839. [Google Scholar] [CrossRef] [PubMed]
- Shamseddine, A.A.; Burman, B.; Lee, N.Y.; Zamarin, D.; Riaz, N. Tumor Immunity and Immunotherapy for Hpv-Related Cancers. Cancer Discov. 2021, 11, 1896–1912. [Google Scholar] [CrossRef]
- Seliger, B.; Massa, C.; Yang, B.; Bethmann, D.; Kappler, M.; Eckert, A.W.; Wickenhauser, C. Immune Escape Mechanisms and Their Clinical Relevance in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2020, 21, 7032. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, N.; Yadav, J.; Thakur, K.; Bibban, R.; Chhokar, A.; Tripathi, T.; Bhat, A.; Singh, T.; Jadli, M.; Singh, U.; et al. Human Papillomavirus Infection in Head and Neck Squamous Cell Carcinomas: Transcriptional Triggers and Changed Disease Patterns. Front. Cell. Infect. Microbiol. 2020, 10, 537650. [Google Scholar] [CrossRef]
- Kostareli, E.; Holzinger, D.; Bogatyrova, O.; Hielscher, T.; Wichmann, G.; Keck, M.; Lahrmann, B.; Grabe, N.; Flechtenmacher, C.; Schmidt, C.R.; et al. Hpv-Related Methylation Signature Predicts Survival in Oropharyngeal Squamous Cell Carcinomas. J. Clin. Investig. 2013, 123, 2488–2501. [Google Scholar] [CrossRef]
- Gillison, M.L.; Chaturvedi, A.K.; Anderson, W.F.; Fakhry, C. Epidemiology of Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma. J. Clin. Oncol. 2015, 33, 3235–3242. [Google Scholar] [CrossRef]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef]
- Malladi, S.; Macalinao, D.G.; Jin, X.; He, L.; Basnet, H.; Zou, Y.; de Stanchina, E.; Massague, J. Metastatic Latency and Immune Evasion through Autocrine Inhibition of Wnt. Cell 2016, 165, 45–60. [Google Scholar] [CrossRef]
- Fan, Y.; Li, Y.; Yao, X.; Jin, J.; Scott, A.; Liu, B.; Wang, S.; Huo, L.; Wang, Y.; Wang, R.; et al. Epithelial Sox9 Drives Progression and Metastases of Gastric Adenocarcinoma by Promoting Immunosuppressive Tumour Microenvironment. Gut 2023, 72, 624–637. [Google Scholar] [CrossRef]
- Angelozzi, M.; Lefebvre, V. Soxopathies: Growing Family of Developmental Disorders Due to Sox Mutations. Trends Genet 2019, 35, 658–671. [Google Scholar] [CrossRef]
- Sarkar, A.; Huebner, A.J.; Sulahian, R.; Anselmo, A.; Xu, X.; Flattery, K.; Desai, N.; Sebastian, C.; Yram, M.A.; Arnold, K.; et al. Sox2 Suppresses Gastric Tumorigenesis in Mice. Cell Rep. 2016, 16, 1929–1941. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, D.; Ramkairsingh, M.; Lin, X.; Kapoor, A.; Major, P.; Tang, D. The Central Contributions of Breast Cancer Stem Cells in Developing Resistance to Endocrine Therapy in Estrogen Receptor (Er)-Positive Breast Cancer. Cancers 2019, 11, 1028. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.-H.; Lee, A.-C.; Hsiao, S.-H.; Lin, S.-E.; Chiu, Y.-F.; Yang, L.-H.; Yu, C.-C.; Chiou, S.-H.; Huang, H.-N.; Ko, J.-C.; et al. Crosstalk between Sox2 and Tgf-Β Signaling Regulates Egfr-Tki Tolerance and Lung Cancer Dissemination. Cancer Res. 2020, 80, 4426–4438. [Google Scholar] [CrossRef] [PubMed]
- Bayo, P.; Jou, A.; Stenzinger, A.; Shao, C.; Gross, M.; Jensen, A.; Grabe, N.; Mende, C.H.; Rados, P.V.; Debus, J.; et al. Loss of Sox2 Expression Induces Cell Motility Via Vimentin up-Regulation and Is an Unfavorable Risk Factor for Survival of Head and Neck Squamous Cell Carcinoma. Mol. Oncol. 2015, 9, 1704–1719. [Google Scholar] [CrossRef]
- Baumeister, P.; Hollmann, A.; Kitz, J.; Afthonidou, A.; Simon, F.; Shakhtour, J.; Mack, B.; Kranz, G.; Libl, D.; Leu, M.; et al. High Expression of Epcam and Sox2 Is a Positive Prognosticator of Clinical Outcome for Head and Neck Carcinoma. Sci. Rep. 2018, 8, 14582. [Google Scholar] [CrossRef]
- Aguilar-Medina, M.; Avendano-Felix, M.; Lizarraga-Verdugo, E.; Bermudez, M.; Romero-Quintana, J.G.; Ramos-Payan, R.; Ruiz-Garcia, E.; Lopez-Camarillo, C. Sox9 Stem-Cell Factor: Clinical and Functional Relevance in Cancer. J. Oncol. 2019, 2019, 6754040. [Google Scholar] [CrossRef]
- Castillo, S.D.; Sanchez-Cespedes, M. The Sox Family of Genes in Cancer Development: Biological Relevance and Opportunities for Therapy. Expert Opin. Ther. Targets 2012, 16, 903–919. [Google Scholar] [CrossRef]
- Huang, J.Q.; Wei, F.K.; Xu, X.L.; Ye, S.X.; Song, J.W.; Ding, P.K.; Zhu, J.; Li, H.F.; Luo, X.P.; Gong, H.; et al. Sox9 Drives the Epithelial-Mesenchymal Transition in Non-Small-Cell Lung Cancer through the Wnt/Β-Catenin Pathway. J. Transl. Med. 2019, 17, 143. [Google Scholar] [CrossRef]
- Li, T.; Huang, H.; Shi, G.; Zhao, L.; Li, T.; Zhang, Z.; Liu, R.; Hu, Y.; Liu, H.; Yu, J.; et al. Tgf-Β1-Sox9 Axis-Inducible Col10a1 Promotes Invasion and Metastasis in Gastric Cancer Via Epithelial-to-Mesenchymal Transition. Cell Death Dis. 2018, 9, 849. [Google Scholar] [CrossRef]
- Kamachi, Y.; Kondoh, H. Sox Proteins: Regulators of Cell Fate Specification and Differentiation. Development 2013, 140, 4129–4144. [Google Scholar] [CrossRef]
- Lin, S.C.; Chou, Y.T.; Jiang, S.S.; Chang, J.L.; Chung, C.H.; Kao, Y.R.; Chang, I.S.; Wu, C.W. Epigenetic Switch between Sox2 and Sox9 Regulates Cancer Cell Plasticity. Cancer Res. 2016, 76, 7036–7048. [Google Scholar] [CrossRef] [PubMed]
- Laughney, A.M.; Hu, J.; Campbell, N.R.; Bakhoum, S.F.; Setty, M.; Lavallée, V.P.; Xie, Y.; Masilionis, I.; Carr, A.J.; Kottapalli, S.; et al. Regenerative Lineages and Immune-Mediated Pruning in Lung Cancer Metastasis. Nat. Med. 2020, 26, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, K.; Molfenter, B.; Laureano, N.K.; Tawk, B.; Bieg, M.; Hostench, X.P.; Weichenhan, D.; Ullrich, N.D.; Shang, V.; Richter, D.; et al. Somatic Mutations and Promotor Methylation of the Ryanodine Receptor 2 Is a Common Event in the Pathogenesis of Head and Neck Cancer. Int. J. Cancer 2019, 145, 3299–3310. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for Rna-Seq Data with Deseq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The Cbio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci. Signal. 2013, 6, 269. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. Qupath: Open Source Software for Digital Pathology Image Analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed]
- Kurth, I.; Hein, L.; Mäbert, K.; Peitzsch, C.; Koi, L.; Cojoc, M.; Kunz-Schughart, L.; Baumann, M.; Dubrovska, A. Cancer Stem Cell Related Markers of Radioresistance in Head and Neck Squamous Cell Carcinoma. Oncotarget 2015, 6, 34494. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-Tailed Prior Distributions for Sequence Count Data: Removing the Noise and Preserving Large Differences. Bioinformatics 2019, 35, 2048–2092. [Google Scholar] [CrossRef]
- Mamun, M.A.; Mannoor, K.; Cao, J.; Qadri, F.; Song, X. Sox2 in Cancer Stemness: Tumor Malignancy and Therapeutic Potentials. J. Mol. Cell Biol. 2018, 12, 85–98. [Google Scholar] [CrossRef]
- Boumahdi, S.; Driessens, G.; Lapouge, G.; Rorive, S.; Nassar, D.; Le Mercier, M.; Delatte, B.; Caauwe, A.; Lenglez, S.; Nkusi, E.; et al. Sox2 Controls Tumour Initiation and Cancer Stem-Cell Functions in Squamous-Cell Carcinoma. Nature 2014, 511, 246–250. [Google Scholar] [CrossRef]
- Jo, A.; Denduluri, S.; Zhang, B.; Wang, Z.; Yin, L.; Yan, Z.; Kang, R.; Shi, L.L.; Mok, J.; Lee, M.J.; et al. The Versatile Functions of Sox9 in Development, Stem Cells, and Human Diseases. Genes Dis. 2014, 1, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Aleman, A.; Adrien, L.; Lopez-Serra, L.; Cordon-Cardo, C.; Esteller, M.; Belbin, T.J.; Sanchez-Carbayo, M. Identification of DNA Hypermethylation of Sox9 in Association with Bladder Cancer Progression Using Cpg Microarrays. Br. J. Cancer 2007, 98, 466–473. [Google Scholar] [CrossRef]
- Athwal, V.S.; Pritchett, J.; Martin, K.; Llewellyn, J.; Scott, J.; Harvey, E.; Zaitoun, A.M.; Mullan, A.F.; Zeef, L.A.H.; Friedman, S.L.; et al. Sox9 Regulated Matrix Proteins Are Increased in Patients Serum and Correlate with Severity of Liver Fibrosis. Sci. Rep. 2018, 8, 17905. [Google Scholar] [CrossRef]
- Panda, M.; Tripathi, S.K.; Biswal, B.K. Sox9: An Emerging Driving Factor from Cancer Progression to Drug Resistance. Biochim. Et Biophys. Acta. Rev. Cancer 2021, 1875, 188517. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Khorani, K.; Schwaerzler, J.; Burkart, S.; Kurth, I.; Holzinger, D.; Flechtenmacher, C.; Plinkert, P.K.; Zaoui, K.; Hess, J. Establishment of a Plasticity-Associated Risk Model Based on a Sox2- and Sox9-Related Gene Set in Head and Neck Squamous Cell Carcinoma. Mol. Cancer Res. MCR 2021, 19, 1676–1687. [Google Scholar] [CrossRef] [PubMed]
- Domenici, G.; Aurrekoetxea-Rodríguez, I.; Simões, B.M.; Rábano, M.; Lee, S.Y.; Millán, J.S.; Comaills, V.; Oliemuller, E.; López-Ruiz, J.A.; Zabalza, I.; et al. A Sox2-Sox9 Signalling Axis Maintains Human Breast Luminal Progenitor and Breast Cancer Stem Cells. Oncogene 2019, 38, 3151–3169. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Cao, E.Y.; Kumar, V.; Zhang, X.; Leong, H.S.; Wong, A.M.L.; Ramakrishnan, N.; Hakimullah, M.; Teo, H.M.V.; Chong, F.T.; et al. Longitudinal Single-Cell Rna Sequencing of Patient-Derived Primary Cells Reveals Drug-Induced Infidelity in Stem Cell Hierarchy. Nat. Commun. 2018, 9, 4931. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef]
- Patil, S.; Linge, A.; Grosser, M.; Lohaus, F.; Gudziol, V.; Kemper, M.; Nowak, A.; Haim, D.; Tinhofer, I.; Budach, V.; et al. Development and Validation of a 6-Gene Signature for the Prognosis of Loco-Regional Control in Patients with Hpv-Negative Locally Advanced Hnscc Treated by Postoperative Radio(Chemo)Therapy. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2022, 171, 91–100. [Google Scholar] [CrossRef]
- Bufalino, A.; Cervigne, N.K.; de Oliveira, C.E.; Fonseca, F.P.; Rodrigues, P.C.; Macedo, C.C.; Sobral, L.M.; Miguel, M.C.; Lopes, M.A.; Paes Leme, A.F.; et al. Low Mir-143/Mir-145 Cluster Levels Induce Activin a Overexpression in Oral Squamous Cell Carcinomas, Which Contributes to Poor Prognosis. PLoS ONE 2015, 10, e0136599. [Google Scholar] [CrossRef]
- Wu, Z.-H.; Tang, Y.; Niu, X.; Cheng, Q. Expression and Gene Regulation Network of Inhba in Head and Neck Squamous Cell Carcinoma Based on Data Mining. Sci. Rep. 2019, 9, 14341. [Google Scholar] [CrossRef]
- Schmidt, S.; Linge, A.; Zwanenburg, A.; Leger, S.; Lohaus, F.; Krenn, C.; Appold, S.; Gudziol, V.; Nowak, A.; von Neubeck, C.; et al. Development and Validation of a Gene Signature for Patients with Head and Neck Carcinomas Treated by Postoperative Radio(Chemo)Therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 1364–1374. [Google Scholar] [CrossRef]
- Lee, Y.C.; Yu, C.C.; Lan, C.; Lee, C.H.; Lee, H.T.; Kuo, Y.L.; Wang, P.H.; Chang, W.W. Plasminogen Activator Inhibitor-1 as Regulator of Tumor-Initiating Cell Properties in Head and Neck Cancers. Head Neck 2016, 38 (Suppl. 1), E895–E904. [Google Scholar] [CrossRef]
- Pal, S.K.; Nguyen, C.T.; Morita, K.I.; Miki, Y.; Kayamori, K.; Yamaguchi, A.; Sakamoto, K. Thbs1 Is Induced by Tgfb1 in the Cancer Stroma and Promotes Invasion of Oral Squamous Cell Carcinoma. J. Oral Pathol. Med. Off. Publ. Int. Assoc. Oral Pathol. Am. Acad. Oral Pathol. 2015, 45, 730–739. [Google Scholar] [CrossRef]
- Derynck, R.; Turley, S.J.; Akhurst, R.J. Tgfβ Biology in Cancer Progression and Immunotherapy. Nat. Rev. Clin. Oncol. 2020, 18, 9–34. [Google Scholar] [CrossRef]
- Ikushima, H.; Miyazono, K. Tgfbeta Signalling: A Complex Web in Cancer Progression. Nat. Rev. Cancer 2010, 10, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, M.; Cheng, M.; Wang, X.; Li, K.; Chen, J.; Chen, Z.; Chen, S.; Chen, J.; Xiong, G.; et al. Tumor Microenvironment in Head and Neck Squamous Cell Carcinoma: Functions and Regulatory Mechanisms. Cancer Lett. 2021, 507, 55–69. [Google Scholar] [CrossRef]
- Loomans, H.A.; Andl, C.D. Intertwining of Activin a and Tgfβ Signaling: Dual Roles in Cancer Progression and Cancer Cell Invasion. Cancers 2014, 7, 70–91. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Che, D.; Yang, F.; Chi, C.; Meng, H.; Shen, J.; Qi, L.; Liu, F.; Lv, L.; Li, Y.; et al. Tumor-Associated Macrophages Promote Tumor Metastasis Via the Tgf-Β/Sox9 Axis in Non-Small Cell Lung Cancer. Oncotarget 2017, 8, 99801. [Google Scholar] [CrossRef]
- Loomans, H.A.; Arnold, S.A.; Hebron, K.; Taylor, C.J.; Zijlstra, A.; Andl, C.D. Loss of Acvrib Leads to Increased Squamous Cell Carcinoma Aggressiveness through Alterations in Cell-Cell and Cell-Matrix Adhesion Proteins. Am. J. Cancer Res. 2017, 7, 2422. [Google Scholar] [PubMed]
- Tsai, C.N.; Tsai, C.L.; Yi, J.S.; Kao, H.K.; Huang, Y.; Wang, C.I.; Lee, Y.S.; Chang, K.P. Activin a Regulates the Epidermal Growth Factor Receptor Promoter by Activating the Pi3k/Sp1 Pathway in Oral Squamous Cell Carcinoma Cells. Sci. Rep. 2019, 9, 5197. [Google Scholar] [CrossRef]
- Markwell, S.M.; Weed, S.A. Tumor and Stromal-Based Contributions to Head and Neck Squamous Cell Carcinoma Invasion. Cancers 2015, 7, 382–406. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e24. [Google Scholar] [CrossRef]
- Koontongkaew, S. The Tumor Microenvironment Contribution to Development, Growth, Invasion and Metastasis of Head and Neck Squamous Cell Carcinomas. J. Cancer 2013, 4, 66–83. [Google Scholar] [CrossRef]
- Roche, K.C.; Gracz, A.D.; Liu, X.F.; Newton, V.; Akiyama, H.; Magness, S.T. Sox9 Maintains Reserve Stem Cells and Preserves Radioresistance in Mouse Small Intestine. Gastroenterology 2015, 149, 1553–1563. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yin, W.; Yu, Z.H.; Zhou, Y.J.; Chi, J.R.; Ge, J.; Cao, X.C. Mir-190 Enhances Endocrine Therapy Sensitivity by Regulating Sox9 Expression in Breast Cancer. J. Exp. Clin. Cancer Res. CR 2019, 38, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Pang, W.; Yang, N.; Hao, L.; Wang, L. Microrna-511 Inhibits Malignant Behaviors of Breast Cancer by Directly Targeting Sox9 and Regulating the Pi3k/Akt Pathway. Int. J. Oncol. 2018, 53, 2715–2726. [Google Scholar] [CrossRef]
- Seymour, P.A.; Freude, K.K.; Tran, M.N.; Mayes, E.E.; Jensen, J.; Kist, R.; Scherer, G.; Sander, M. Sox9 Is Required for Maintenance of the Pancreatic Progenitor Cell Pool. Proc. Natl. Acad. Sci. USA 2007, 104, 1865–1870. [Google Scholar] [CrossRef]
- Sumita, Y.; Yamazaki, M.; Maruyama, S.; Abé, T.; Cheng, J.; Takagi, R.; Tanuma, J.I. Cytoplasmic Expression of Sox9 as a Poor Prognostic Factor for Oral Squamous Cell Carcinoma. Oncol. Rep. 2018, 40, 2487–2496. [Google Scholar] [CrossRef]
- Haga, K.; Yamazaki, M.; Maruyama, S.; Kawaharada, M.; Suzuki, A.; Hoshikawa, E.; Chan, N.N.; Funayama, A.; Mikami, T.; Kobayashi, T.; et al. Crosstalk between Oral Squamous Cell Carcinoma Cells and Cancer-Associated Fibroblasts Via the Tgf-Β/Sox9 Axis in Cancer Progression. Transl. Oncol. 2021, 14, 101236. [Google Scholar] [CrossRef] [PubMed]
- Riemenschnitter, C.; Teleki, I.; Tischler, V.; Guo, W.; Varga, Z. Stability and Prognostic Value of Slug, Sox9 and Sox10 Expression in Breast Cancers Treated with Neoadjuvant Chemotherapy. SpringerPlus 2013, 2, 695. [Google Scholar] [CrossRef][Green Version]
- Olivares-Urbano, M.A.; Griñán-Lisón, C.; Marchal, J.A.; Núñez, M.I. Csc Radioresistance: A Therapeutic Challenge to Improve Radiotherapy Effectiveness in Cancer. Cells 2020, 9, 1651. [Google Scholar] [CrossRef]
- De Martino, M.; Daviaud, C.; Diamond, J.M.; Kraynak, J.; Alard, A.; Formenti, S.C.; Miller, L.D.; Demaria, S.; Vanpouille-Box, C. Activin a Promotes Regulatory T-Cell-Mediated Immunosuppression in Irradiated Breast Cancer. Cancer Immunol. Res. 2021, 9, 89–102. [Google Scholar] [CrossRef]
- Carl, C.; Flindt, A.; Hartmann, J.; Dahlke, M.; Rades, D.; Dunst, J.; Lehnert, H.; Gieseler, F.; Ungefroren, H. Ionizing Radiation Induces a Motile Phenotype in Human Carcinoma Cells in Vitro through Hyperactivation of the Tgf-Beta Signaling Pathway. Cell. Mol. Life Sci. CMLS 2016, 73, 427–443. [Google Scholar] [CrossRef]
- Keysar, S.B.; Le, P.N.; Miller, B.; Jackson, B.C.; Eagles, J.R.; Nieto, C.; Kim, J.; Tang, B.; Glogowska, M.J.; Morton, J.J.; et al. Regulation of Head and Neck Squamous Cancer Stem Cells by Pi3k and Sox2. J. Natl. Cancer Inst. 2017, 109, djw189. [Google Scholar] [CrossRef] [PubMed]
- Vlashi, E.; Chen, A.M.; Boyrie, S.; Yu, G.; Nguyen, A.; Brower, P.A.; Hess, C.B.; Pajonk, F. Radiation-Induced Dedifferentiation of Head and Neck Cancer Cells into Cancer Stem Cells Depends on Human Papillomavirus Status. Int. J. Radiat. Oncol. Biol. Phys. 2016, 94, 1198–1206. [Google Scholar] [CrossRef]
- Chung, J.H.; Jung, H.R.; Jung, A.R.; Lee, Y.C.; Kong, M.; Lee, J.S.; Eun, Y.G. Sox2 Activation Predicts Prognosis in Patients with Head and Neck Squamous Cell Carcinoma. Sci. Rep. 2018, 8, 1677. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wei, Y.; Hummel, M.; Hoffmann, T.K.; Gross, M.; Kaufmann, A.M.; Albers, A.E. Evidence for Epithelial-Mesenchymal Transition in Cancer Stem Cells of Head and Neck Squamous Cell Carcinoma. PLoS ONE 2011, 6, e16466. [Google Scholar] [CrossRef] [PubMed]

| Feature | SOX2HighSOX9Low n (%) | SOX2LowSOX9High n (%) | p | |
|---|---|---|---|---|
| Patients (n) | 260 | 214 | ||
| Vital Status | Alive | 157 (60.4) | 124 (57.9) | 0.63 |
| Dead | 103 (39.6) | 90 (42.1) | ||
| Overall Survival 5-year | 0 | 167 (64.2) | 132 (61.7) | 0.56 |
| 1 | 93 (35.7) | 82 (38.3) | ||
| Disease Specific Survival 5-years | 0 | 195 (75) | 155 (72.4) | 0.14 |
| 1 | 65 (25) | 59 (27.6) | ||
| HPV status 1 | Negative | 194 (77) | 196 (98) | 0.0001 |
| Positive | 58 (23) | 4 (2) | ||
| Alcohol | No | 77 (30.3) | 68 (32.4) | 0.68 |
| Yes | 177 (69.7) | 142 (67.6) | ||
| Smoking | No | 52 (20.4) | 55 (26) | 0.15 |
| Yes | 203 (79.6) | 156 (74) | ||
| Gender | Female | 55 (21.2) | 67 (31.3) | 0.01 |
| Male | 205 (78.8) | 147 (68.7) | ||
| Age (median, range) | 60 (19, 84) | 61 (24, 90) | >0.99 | |
| Subsite | Hypopharynx | 4 (1.5) | 6 (2.8) | 0.0001 |
| Larynx | 76 (29.2) | 29 (13.5) | ||
| Oral Cavity | 122 (49.9) | 169 (78.9) | ||
| Oropharynx | 58 (19.4) | 10 (4.8) | ||
| Pathological Grading | G1 | 27 (10.4) | 31 (14.5) | 0.31 |
| G2 | 154 (59.3) | 132 (61.7) | ||
| G3 | 67 (25.7) | 47 (21.9) | ||
| G4 | 2 (0.7) | 0 | ||
| GX | 8 (3) | 4 (1.9) | ||
| Tumor Size | cT1 | 19 (7.3) | 14 (6.5) | 0.74 |
| cT2 | 76 (29.2) | 56 (26.2) | ||
| cT3 | 67 (32.6) | 55 (25.7) | ||
| cT4 | 91 (35) | 81 (37.8) | ||
| cTX | 6 (2.3) | 5 (2.3) | ||
| Lymph Node metastasis | cN0 | 130 (50) | 93 (43.4) | 0.40 |
| cN1-3 | 119 (45.7) | 110 (51.4) | ||
| cNX | 10 (3.9) | 8 (3.8) | ||
| Distant metastasis | M0 | 247 (95) | 198 (92.5) | 0.47 |
| M1 | 1 (0.3) | 3 (1.4) | ||
| MX | 11 (4.2) | 9 (4.2) | ||
| Tumor Size | pT0 | 1 (0.4) | 0 | 0.76 |
| pT1 | 23 (8.8) | 20 (9.3) | ||
| pT2 | 73 (28) | 53 (24.8) | ||
| pT3 | 49 (18.9) | 40 (18.7) | ||
| pT4 | 85 (32.7) | 78 (36.4) | ||
| pTX | 20 (7.7) | 12 (5.6) | ||
| Lymph Node metastasis | pN0 | 90 (34.6) | 71 (33.2) | 0.52 |
| pN1 | 121 (46.6) | 107 (50) | ||
| pNX | 39 (15) | 25 (11.7) | ||
| pM | pM0 | 95 (36.5) | 84 (39.2) | 0.42 |
| pM1 | 1 (0.4) | 0 | ||
| pMX | 36 (13.8) | 24 (11.2) | ||
| Pathological Staging | Stage I | 14 (5.3) | 11 (5.1) | 0.67 |
| Stage II | 38 (14.7) | 28 (13.1) | ||
| Stage III | 43 (16.5) | 30 (14.1) | ||
| Stage IVA | 121 (46.5) | 116 (54.2) | ||
| Stage IVB | 7 (2.7) | 4 (1.8) | ||
| Neoadjuvant Treatment | No | 257 (98.8) | 212 (99) | >0.99 |
| Yes | 3 (1.2) | 2 (1) | ||
| Radiation | No | 72 (32.2) | 71 (36.8) | 0.01 |
| Yes | 152 (67.8) | 122 (63.2) | ||
| Additional Radiation Therapy | No | 54 (58.7) | 56 (70) | 0.15 |
| Yes | 38 (41.3) | 24 (30) |
| Function Name | p-Value | Number of Molecules |
|---|---|---|
| Cellular Development | 4.70 × 10−2–2.02 × 10−6 | 15 |
| Cellular Growth and Proliferation | 4.70 × 10−2–2.02 × 10−6 | 15 |
| Cellular Movement | 3.86 × 10−2–5.65 × 10−4 | 12 |
| Cellular Function and Maintenance | 3.16 × 10−2–9.01 × 10−4 | 4 |
| Cell-To-Cell Signaling and Interaction | 4.30 × 10−2–1.19 × 10−3 | 9 |
| HNO223 shSOX2 | HNO223 shSOX9 | |||
|---|---|---|---|---|
| Gene | log2FoldChange | padj | log2FoldChange | padj |
| INHBA | −1.5583044 | 0.00110584 | −1.1062817 | 6.67 × 10−5 |
| SERPINE1 | 3.09471999 | 4.07 × 10−13 | −1.2736951 | 0.00033041 |
| THBS1 | −1.3113053 | 0.00875671 | −2.3902272 | 0.00427583 |
| LTBP1 | 4.60426832 | 3.43 × 10−28 | 0.91433761 | 0.03975225 |
| GCHFR | −1.1650005 | 0.01166647 | 0.69411435 | 0.04855759 |
| PRDM8 | −1.4274595 | 0.00463216 | 0.39562131 | 1.38 × 10−5 |
| DUSP6 | −3.3943627 | 9.16 × 10−10 | −1.8248729 | 5.81 × 10−7 |
| TNS4 | −9.1657433 | 4.84 × 10−59 | 1.07067784 | 0.00040597 |
| TINAGL1 | −5.9954209 | 8.60 × 10−54 | −0.60474 | 0.04086991 |
| LAMC2 | −3.2760053 | 5.55 × 10−16 | −0.9932665 | 5.14 × 10−5 |
| SERPINB7 | −3.9976105 | 1.03 × 10−15 | −1.0997699 | 0.00022133 |
| SOX2 | −7.6332362 | 9.28 × 10−5 | 1.29689157 | 0.0589497 |
| SOX9 | −2.0600676 | 1.76 × 10−6 | −1.9138004 | 4.31 × 10−10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbosa, S.; Laureano, N.K.; Hadiwikarta, W.W.; Visioli, F.; Bonrouhi, M.; Pajdzik, K.; Conde-Lopez, C.; Herold-Mende, C.; Eidt, G.; Langie, R.; et al. The Role of SOX2 and SOX9 in Radioresistance and Tumor Recurrence. Cancers 2024, 16, 439. https://doi.org/10.3390/cancers16020439
Barbosa S, Laureano NK, Hadiwikarta WW, Visioli F, Bonrouhi M, Pajdzik K, Conde-Lopez C, Herold-Mende C, Eidt G, Langie R, et al. The Role of SOX2 and SOX9 in Radioresistance and Tumor Recurrence. Cancers. 2024; 16(2):439. https://doi.org/10.3390/cancers16020439
Chicago/Turabian StyleBarbosa, Silvia, Natalia Koerich Laureano, Wahyu Wijaya Hadiwikarta, Fernanda Visioli, Mahnaz Bonrouhi, Kinga Pajdzik, Cristina Conde-Lopez, Christel Herold-Mende, Gustavo Eidt, Renan Langie, and et al. 2024. "The Role of SOX2 and SOX9 in Radioresistance and Tumor Recurrence" Cancers 16, no. 2: 439. https://doi.org/10.3390/cancers16020439
APA StyleBarbosa, S., Laureano, N. K., Hadiwikarta, W. W., Visioli, F., Bonrouhi, M., Pajdzik, K., Conde-Lopez, C., Herold-Mende, C., Eidt, G., Langie, R., Lamers, M. L., Stögbauer, F., Hess, J., Kurth, I., & Jou, A. (2024). The Role of SOX2 and SOX9 in Radioresistance and Tumor Recurrence. Cancers, 16(2), 439. https://doi.org/10.3390/cancers16020439








