Clinical and Technical Validation of OncoIndx® Assay—A Comprehensive Genome Profiling Assay for Pan-Cancer Investigations
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
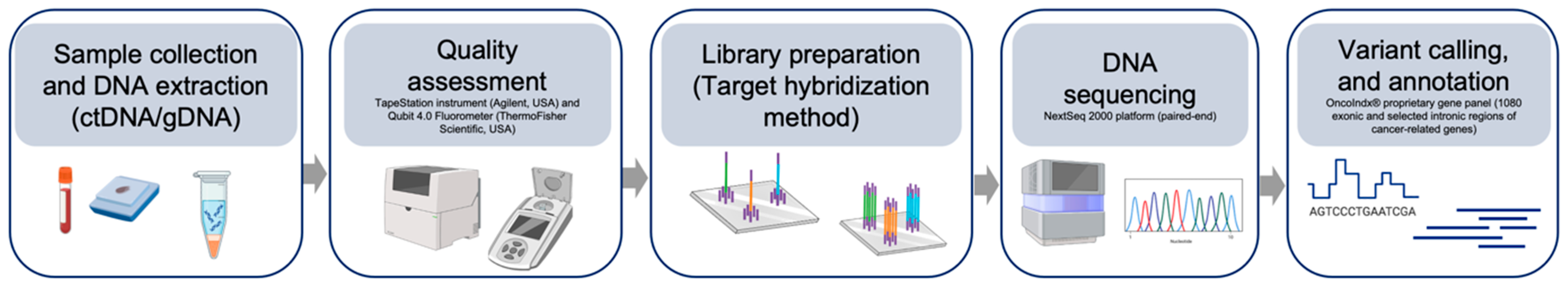
2.1. Sample Collection and Targeted Exon Sequencing
2.2. Bioinformatic Data Processing
2.3. Variant Prioritization and Interpretation
2.4. Validating Test Outcomes
2.4.1. Level 1: Reference Standards
2.4.2. Level 2: Clinical Samples
2.4.3. Level 3: Orthogonal Validation
3. Results and Discussion
3.1. OncoIndx® Detected Genomic Alterations from NGS Standard Reference Samples with High Concordance and Analytical Precision
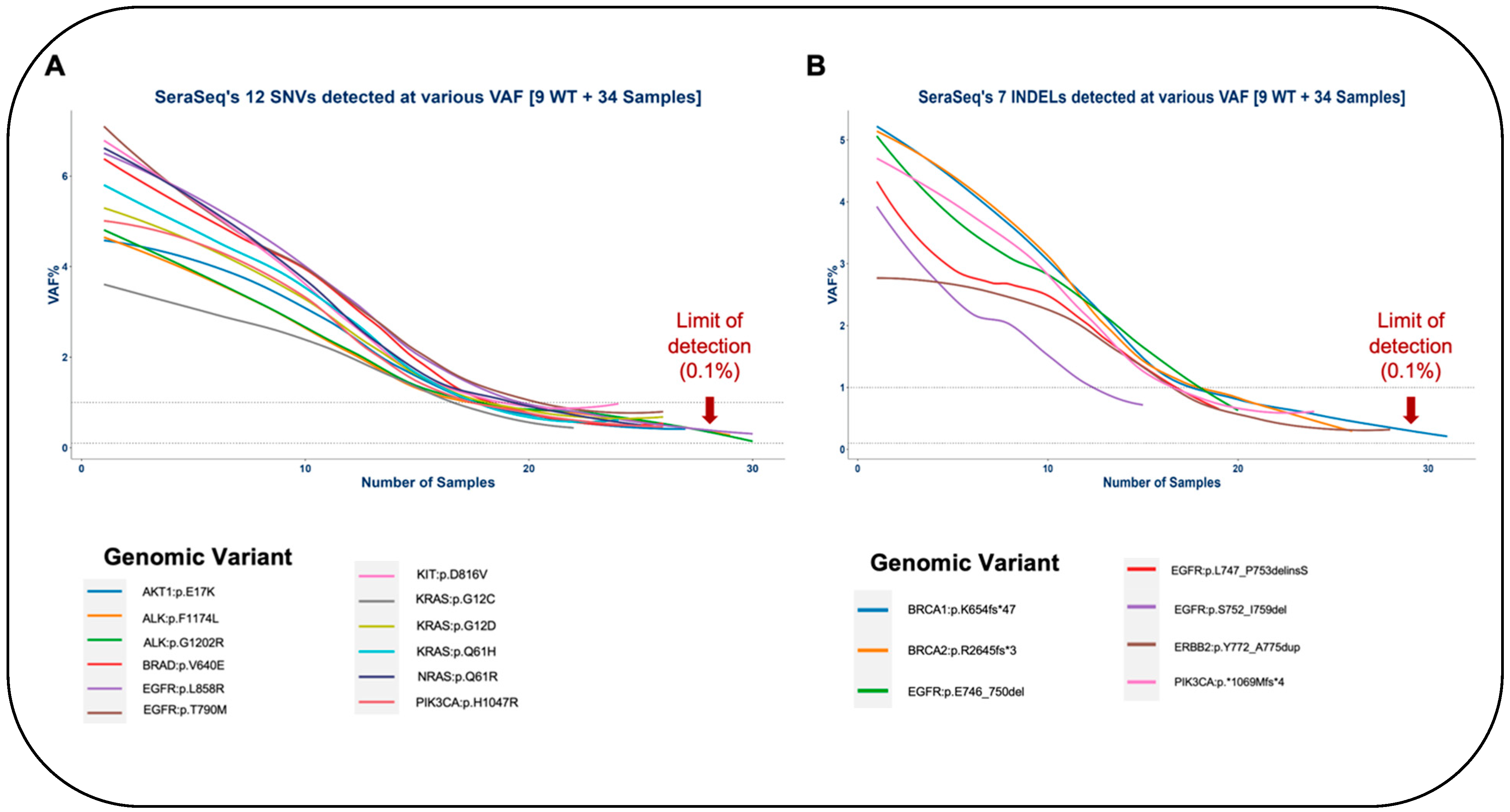
3.1.1. Single Nucleotide Variants and INDELs
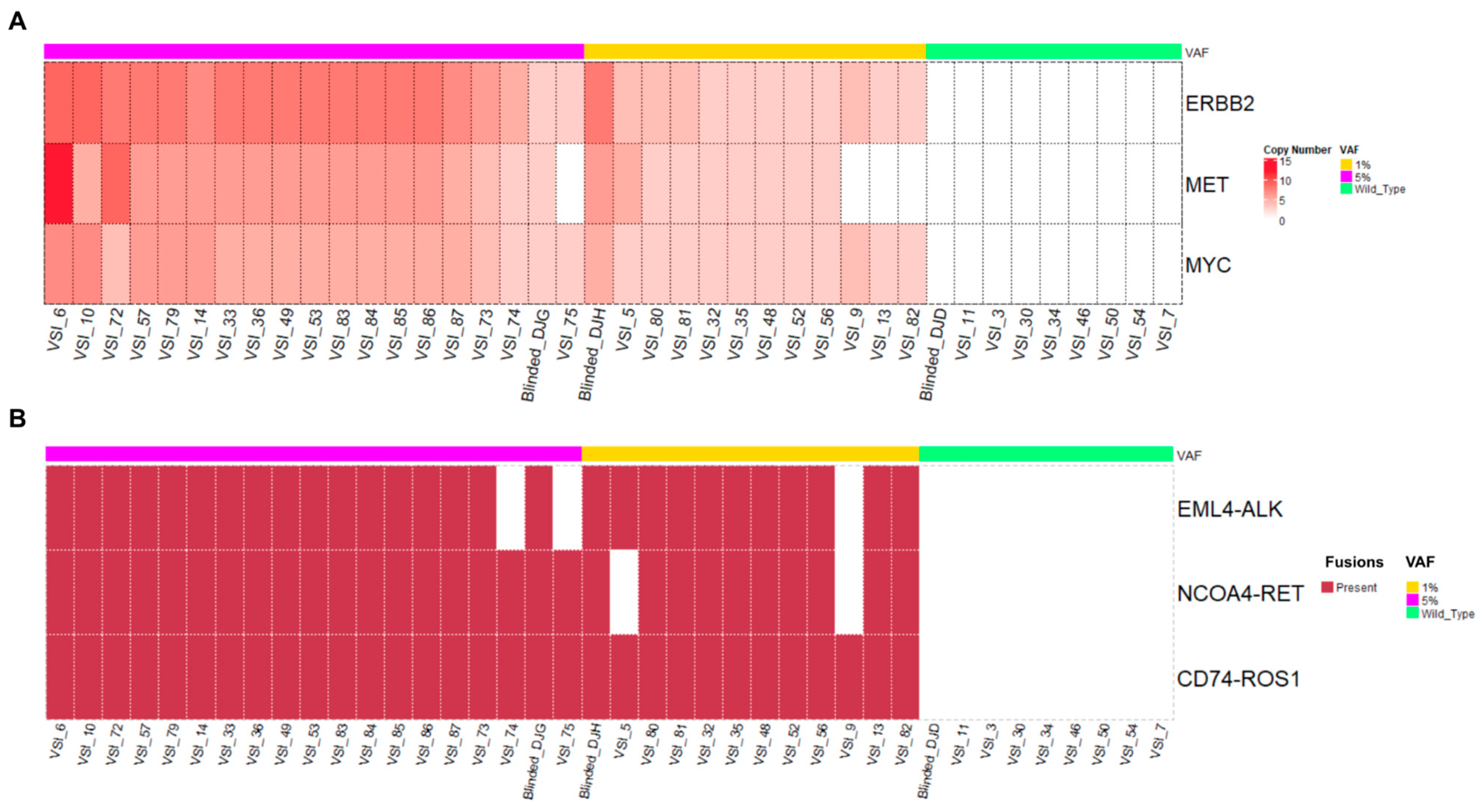
3.1.2. Copy Number Alterations and Fusions
3.1.3. Limit of Detection of OncoIndx®
3.2. High Concordance of Genomic Alterations Obtained from Clinical Samples
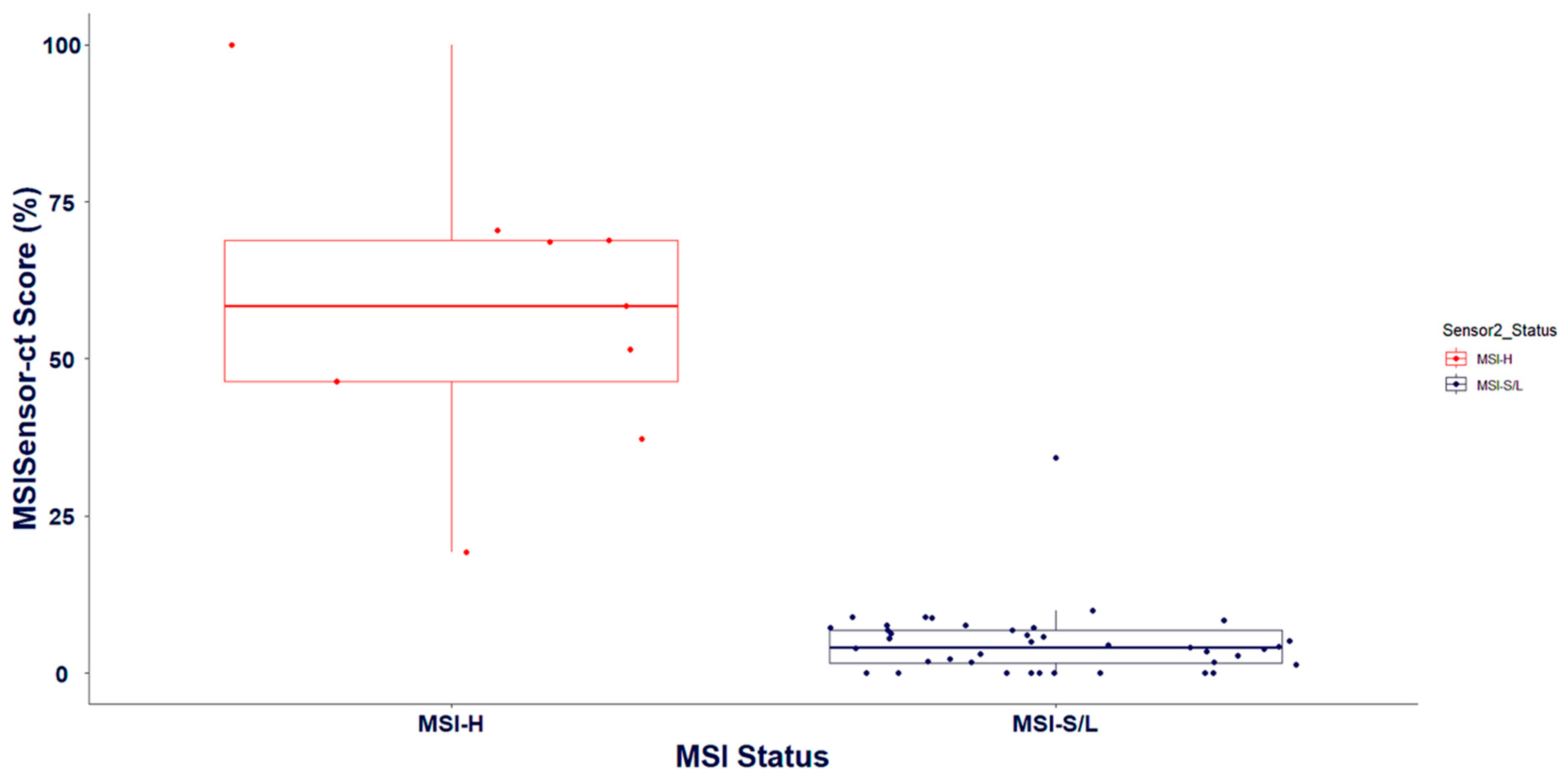
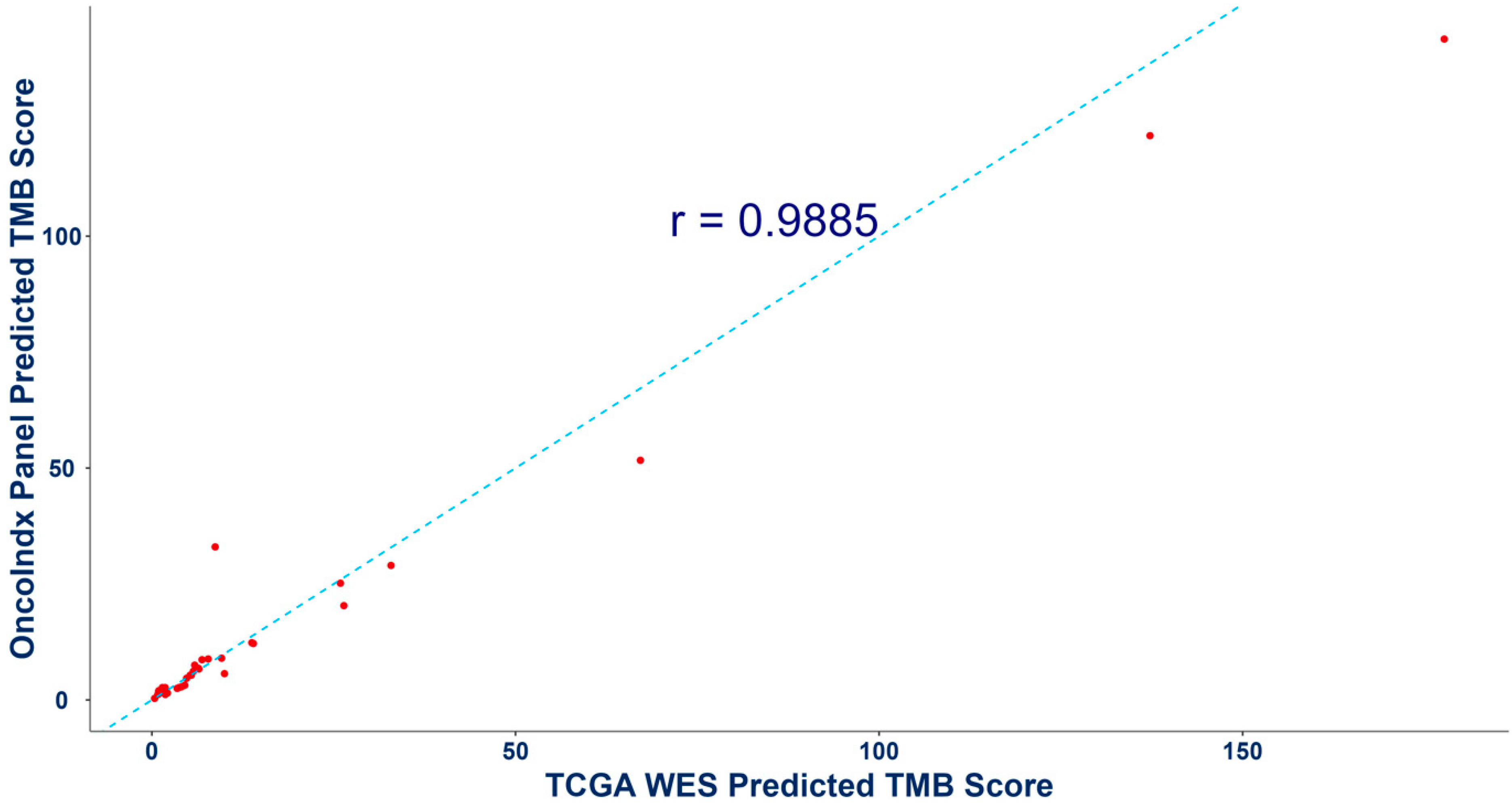
3.3. Validation of Biomarker Signatures against Reference Laboratories: Microsatellite Instability and Tumor Mutation Burden
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Park, Y.; Heider, D.; Hauschild, A.C. Integrative analysis of next-generation sequencing for next-generation cancer research toward artificial intelligence. Cancers 2021, 13, 3148. [Google Scholar] [CrossRef]
- Xu, J.; Yang, P.; Xue, S.; Sharma, B.; Sanchez-Martin, M.; Wang, F.; Beaty, K.A.; Dehan, E.; Parikh, B. Translating cancer genomics into precision medicine with artificial intelligence: Applications, challenges and future perspectives. Hum. Genet. 2019, 138, 109–124. [Google Scholar] [CrossRef]
- Waarts, M.R.; Stonestrom, A.J.; Park, Y.C.; Levine, R.L. Targeting mutations in cancer. J. Clin. Investig. 2022, 132, e154943. [Google Scholar] [CrossRef]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef]
- Morganti, S.; Tarantino, P.; Ferraro, E.; D’Amico, P.; Viale, G.; Trapani, D.; Duso, B.A.; Curigliano, G. Complexity of genome sequencing and reporting: Next generation sequencing (NGS) technologies and implementation of precision medicine in real life. Crit. Rev. Oncol. Hematol. 2019, 133, 171–182. [Google Scholar] [CrossRef]
- Arnedos, M.; Vielh, P.; Soria, J.C.; Andre, F. The genetic complexity of common cancers and the promise of personalized medicine: Is there any hope? J. Pathol. 2014, 232, 274–282. [Google Scholar] [CrossRef]
- Conroy, J.M.; Pabla, S.; Glenn, S.T.; Seager, R.J.; Van Roey, E.; Gao, S.; Burgher, B.; Andreas, J.; Giamo, V.; Mallon, M.; et al. A scalable high-throughput targeted next-generation sequencing assay for comprehensive genomic profiling of solid tumors. PLoS ONE 2021, 16, e0260089. [Google Scholar] [CrossRef]
- Delgado Serrano, L.F. Identification and Characterization of New Complex Patterns of Structural DNA and RNA Alterations in Cancer. Ph.D. Thesis, University of Barcelona, Barcelona, Spain, 2021. [Google Scholar]
- Hiemenz, M.C.; Graf, R.P.; Schiavone, K.; Harries, L.; Oxnard, G.R.; Ross, J.S.; Huang, R.S. Real-world comprehensive genomic profiling success rates in tissue and liquid prostate carcinoma specimens. Oncology 2022, 27, e970–e972. [Google Scholar] [CrossRef]
- Barata, P.C.; Reisinger, R.; Bilen, M.A.; Heath, E.I.; Nandagopal, L.; Swami, U.; Kessel, A.; Jaeger, E.; Wesolowski, S.; Chipman, J.; et al. Differences in the tumor genomic landscape between African Americans (AA) and Caucasians (CA) advanced prostate cancer (aPC) patients (pts) by comprehensive genomic profiling (CGP) of cell-free DNA (cfDNA). J. Clin. Oncol. 2021, 39 (Suppl. 15), 5058. [Google Scholar] [CrossRef]
- Shafi, G.; Dongare, M.; Bharde, A.; Fauzul, M.; Hariramani, K.; D’Souza, A.; Jadhav, B.; Kad, T.; Prajapati, S.; Jadhav, V.; et al. Abstract PR007: Comprehensive ctDNA profiling reveals potential metastatic genomic signatures in treatment-naive early-stage breast cancer patients. Cancer Res. 2023, 83 (Suppl. 2), PR007. [Google Scholar] [CrossRef]
- Mathew, A.; Joseph, S.; Boby, J.; Benny, S.; Veedu, J.; Rajappa, S.; Rohatgi, N.; Sirohi, B.; Jain, R.; Agarwala, V.; et al. Clinical benefit of comprehensive genomic profiling for advanced cancers in India. JCO Glob. Oncol. 2022, 8, e2100421. [Google Scholar] [CrossRef]
- Bharde, A.; Bhonde, M.; Bose, C.; Raut, N.V.; Kothavade, H.; D’Souza, A.; Kad, T.; Jadhav, B.; Prajapati, S.; Jadhav, V.B.L.; et al. Comprehensive ctDNA analysis to identify genome-instability and actionable mutation landscape in gynecologic cancers. J. Clin. Oncol. 2023, 41, e17503. [Google Scholar] [CrossRef]
- Karol, D.; McKinnon, M.; Mukhtar, L.; Awan, A.; Lo, B.; Wheatley-Price, P. The impact of foundation medicine testing on cancer patients: A single academic centre experience. Front. Oncol. 2021, 11, 687730. [Google Scholar] [CrossRef]
- Takeda, M.; Takahama, T.; Sakai, K.; Shimizu, S.; Watanabe, S.; Kawakami, H.; Tanaka, K.; Sato, C.; Hayashi, H.; Nonagase, Y.; et al. Clinical application of the FoundationOne CDx assay to therapeutic decision-making for patients with advanced solid tumors. Oncology 2021, 26, e588–e596. [Google Scholar] [CrossRef]
- Bharde, A.; Noronha, V.; Ratnaparkhi, M.; Patil, V.; Jadhav, B.; Prajapati, S.; Choughule, A.; Chandrani, P.; Haldar, S.; Moubeen, F.; et al. Comprehensive genomic profiling of ctDNA reveals distinct genomic signatures and therapeutic implications for immunotherapy response in advanced head and neck cancer. Cancer Res. 2024, 84 (Suppl. 6), 2408. [Google Scholar] [CrossRef]
- Krzyszczyk, P.; Acevedo, A.; Davidoff, E.J.; Timmins, L.M.; Marrero-Berrios, I.; Patel, M.; White, C.; Lowe, C.; Sherba, J.J.; Hartmanshenn, C.; et al. The growing role of precision and personalized medicine for cancer treatment. Technology 2018, 6, 79–100. [Google Scholar] [CrossRef]
- Mukhopadhyay, D. Mechanistic Role of the Calcium Channel TRPC6 in Driving Aggressive Traits in Triple-Negative Breast Cancer. Ph.D. Thesis, UMass Chan Medical School, Worcester, MA, USA, 2024. [Google Scholar]
- Fabrizio, D.; Cristescu, R.; Albacker, L.; Snyder, A.; Ward, A.; Lunceford, J.; Aurora-Garg, D.; Jin, F.; Hopkins, J.; Rubin, E.; et al. Real-world prevalence across 159 872 patients with cancer supports the clinical utility of TMB-H to define metastatic solid tumors for treatment with pembrolizumab. Ann. Oncol. 2024, 32, 1193–1194. [Google Scholar] [CrossRef]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for validation of next-generation sequencing–based oncology panels: A joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef]
- Roy, S.; Coldren, C.; Karunamurthy, A.; Kip, N.S.; Klee, E.W.; Lincoln, S.E.; Leon, A.; Pullambhatla, M.; Temple-Smolkin, R.L.; Voelkerding, K.V.; et al. Standards and guidelines for validating next-generation sequencing bioinformatics pipelines: A joint recommendation of the Association for Molecular Pathology and the College of American Pathologists. J. Mol. Diagn. 2018, 20, 4–27. [Google Scholar] [CrossRef]
- Pereira, R.; Oliveira, J.; Sousa, M. Bioinformatics and computational tools for next-generation sequencing analysis in clinical genetics. J. Clin. Med. 2020, 9, 132. [Google Scholar] [CrossRef]
- Leclerc, J.; Vermaut, C.; Buisine, M.P. Diagnosis of Lynch syndrome and strategies to distinguish Lynch-related tumors from sporadic MSI/dMMR tumors. Cancers 2021, 13, 467. [Google Scholar] [CrossRef]
- Lorenzi, M.; Amonkar, M.; Zhang, J.; Mehta, S.; Liaw, K.L. Epidemiology of microsatellite instability high (MSI-H) and deficient mismatch repair (dMMR) in solid tumors: A structured literature review. J. Oncol. 2020, 2020, 1807929. [Google Scholar] [CrossRef]
- Poulogiannis, G.; Frayling, I.M.; Arends, M.J. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology 2010, 56, 167–179. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Taniue, K.; Akimitsu, N. Fusion genes and RNAs in cancer development. Non-Coding RNA 2021, 7, 10. [Google Scholar] [CrossRef]
- Wang, L.; Geng, H.; Liu, Y.; Liu, L.; Chen, Y.; Wu, F.; Liu, Z.; Ling, S.; Wang, Y.; Zhou, L. Hot and cold tumors: Immunological features and the therapeutic strategies. MedComm 2023, 4, e343. [Google Scholar] [CrossRef]
- Bai, J.; Chen, H.; Bai, X. Relationship between microsatellite status and immune microenvironment of colorectal cancer and its application to diagnosis and treatment. J. Clin. Lab. Anal. 2021, 35, e23810. [Google Scholar] [CrossRef]
- Zhang, J.; Späth, S.S.; Marjani, S.L.; Zhang, W.; Pan, X. Characterization of cancer genomic heterogeneity by next-generation sequencing advances precision medicine in cancer treatment. Precis. Clin. Med. 2018, 1, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Kadri, S.; Long, B.C.; Mujacic, I.; Zhen, C.J.; Wurst, M.N.; Sharma, S.; McDonald, N.; Niu, N.; Benhamed, S.; Tuteja, J.H.; et al. Clinical validation of a next-generation sequencing genomic oncology panel via cross-platform benchmarking against established amplicon sequencing assays. J. Mol. Diagn. 2017, 19, 43–56. [Google Scholar] [CrossRef] [PubMed]
| Alteration Type | Total Number of Alterations | True Positives | False Positives | True Negatives | False Negatives | * PPV | * NPV | Accuracy | Specificity | Sensitivity |
|---|---|---|---|---|---|---|---|---|---|---|
| SNVs | 264 | 156 | 0 | 108 | 0 | 100 | 100 | 100 | 100 | 100 |
| Small INDELs | 154 | 87 | 0 | 63 | 4 | 100 | 94.03 | 97.40 | 100 | 95.60 |
| CNA | 66 | 39 | 0 | 27 | 0 | 100 | 100 | 100 | 100 | 100 |
| Fusions | 66 | 38 | 0 | 27 | 1 | 100 | 96.43 | 98.48 | 100 | 97.44 |
| List of SNVs and INDELs Validated from the Industrial Samples | |
|---|---|
| AKT1:p.E17K | EGFR:p.T790M |
| ALK:p.F1174L | ERBB2:p.Y772_A775dup |
| ALK:p.G1202R | KIT:p.D816V |
| BRAF:p.V640E | KRAS:p.G12C |
| BRCA1:p.K654fs*47 | KRAS:p.G12D |
| BRCA2:p.R2645fs*3 | KRAS:p.Q61H |
| EGFR:p.E746_A750del | KRAS:p.Q61R |
| EGFR:p.L747_P753delinsS | NRAS:p.Q61R |
| EGFR:p.L858R | PIK3CA:p.*1069Mfs*4 |
| EGFR:p.S752_I759del | PIK3CA:p.H1047R |
| S. No. | Genes | Concordant Genomic Findings from OncoIndx® Assay | Concordance Levels Obtained from OncoIndx® Assay |
|---|---|---|---|
| 1 | EGFR | L858R E746_A750del L747_S752del | 100% |
| 2 | ALK | NPM1-ALK ALK-EML4 Fusion G1202R G1269A | 100% |
| 3 | KRAS | A146T | 100% |
| 4 | PIK3CA | H1047R | 50% |
| 5 | BRCA2 | S636* | 100% |
| Statistics of MSI Detection in OncoIndx® (Percentage %) | |
|---|---|
| Positive Predictive Value (PPV) | 100 |
| Negative Predictive Value (PPV) | 90.91 |
| Sensitivity | 90 |
| Specificity | 100 |
| Accuracy | 95 |
| Sample Type | FDA-Approved Test Prediction | OncoIndx® Test Prediction |
|---|---|---|
| Blood | MSS | 3.2 (MSI-low) |
| Blood | MSS | 1.55 (MSI-low) |
| Blood | MSS | 0.79 (MSI-low) |
| Blood | MSS | 3.07 (MSI-low) |
| Blood | MSS | 3.61 (MSI-low) |
| Biomarker/s | Outcome | Blood/Pleural Effusion | FFPE/RNALater |
|---|---|---|---|
| MSI | MSI-H | ≥20 | ≥20 |
| MSI-I | ≥10 | ≥10 | |
| MSI-L | <10 | <10 | |
| MSI-S | 0 | 0 |
| Sample | Sample Type | True Prediction | OncoIndx® Test Prediction |
|---|---|---|---|
| Control | Healthy control | Negative control | 1.5 |
| Healthy control | Negative control | 1.5 | |
| Healthy control | Negative control | 1.5 | |
| Healthy control | Negative control | 2.5 | |
| Healthy control | Negative control | 1.67 | |
| Healthy control | Negative control | 0 | |
| Healthy control | Negative control | 0 | |
| Healthy control | Negative control | 0.5 | |
| SeraSeqTM reference samples | TMB Mix Score 7 (0%) | 5.8–9.2 | 8.67 |
| TMB Mix Score 7 (0.5%) | 10.5–15.7 (d = 3.5–7.7) | 10.83 | |
| TMB Mix Score 7 (2%) | 16.6–19.2 (d = 3.5–7.7) | 6.67 | |
| TMB Mix Score 20 (0%) | 6.1–8.9 | 6.5 | |
| TMB Mix Score 20 (0.5%) | 23.7–28.3 (d = 15.8–21.2) | 8.17 | |
| TMB Mix Score 20 (2%) | 34.6–36.6 (d = 26.4–29.8) | 5.83 | |
| CAP gDNA samples | gDNA | 9 | 11.5 |
| gDNA | 26 | 19.5 | |
| gDNA | 9 | 9.83 | |
| gDNA | 26 | 5.67 |
| Sample Type | FDA-Approved Test Prediction | OncoIndx® Test Prediction |
|---|---|---|
| Blood | 1 | 3.2 |
| Blood | 7.26 | 1.55 |
| Blood | 6.7 | 0.79 |
| Blood | 3 | 3.07 |
| Blood | 4 | 3.61 |
| Blood | 4.77 | 3.167 |
| Biomarker/s | Outcomes | Blood/Pleural Effusion | FFPE/RNALater |
|---|---|---|---|
| TMB | TMB-H | ≥10 | ≥10 |
| TMB-L | <10 | <10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramesh, A.; Bharde, A.; D’Souza, A.; Jadhav, B.; Prajapati, S.; Hariramani, K.; Basavalingegowda, M.; Iyer, S.; Halder, S.; Deochake, M.; et al. Clinical and Technical Validation of OncoIndx® Assay—A Comprehensive Genome Profiling Assay for Pan-Cancer Investigations. Cancers 2024, 16, 3415. https://doi.org/10.3390/cancers16193415
Ramesh A, Bharde A, D’Souza A, Jadhav B, Prajapati S, Hariramani K, Basavalingegowda M, Iyer S, Halder S, Deochake M, et al. Clinical and Technical Validation of OncoIndx® Assay—A Comprehensive Genome Profiling Assay for Pan-Cancer Investigations. Cancers. 2024; 16(19):3415. https://doi.org/10.3390/cancers16193415
Chicago/Turabian StyleRamesh, Aarthi, Atul Bharde, Alain D’Souza, Bhagwat Jadhav, Sangeeta Prajapati, Kanchan Hariramani, Madhura Basavalingegowda, Sandhya Iyer, Sumit Halder, Mahesh Deochake, and et al. 2024. "Clinical and Technical Validation of OncoIndx® Assay—A Comprehensive Genome Profiling Assay for Pan-Cancer Investigations" Cancers 16, no. 19: 3415. https://doi.org/10.3390/cancers16193415
APA StyleRamesh, A., Bharde, A., D’Souza, A., Jadhav, B., Prajapati, S., Hariramani, K., Basavalingegowda, M., Iyer, S., Halder, S., Deochake, M., Kothavade, H., Vasudevan, A., Uttarwar, M., Khandare, J., & Shafi, G. (2024). Clinical and Technical Validation of OncoIndx® Assay—A Comprehensive Genome Profiling Assay for Pan-Cancer Investigations. Cancers, 16(19), 3415. https://doi.org/10.3390/cancers16193415







