Simple Summary
Histopathological and molecular factors could play a role in determining LNM in colon cancer. Extended lymphadenectomies in right-sided patients have been associated with complications. We demonstrated that histopathological and molecular analysis can be useful in predicting LNM and therefore identify high risk patients which could potentially benefit from D3 lymphadenectomy/CME.
Abstract
Background: Lymphadenectomy plays a central role in the treatment of localized colon cancer. While in left colon cancer the D3 lymphadenectomy/CME is considered the standard of care, lymphatic stations to be removed in right colon cancer are still a matter of discussion. The individuation of LNM risk factors could help in choosing the lymphadenectomy in right-sided tumors. This study aims to analyze the correlation of histopathological and molecular characteristics with lymph node metastasis, both in right- and left-sided colon cancer, and their impact on survival; Methods: We conducted a single-center observational retrospective study. The following data were collected and analyzed for each patient: demographics, histopathological and molecular data, and intraoperative and perioperative data. Statistical analyses were performed, including descriptive statistics, multivariate logistic regression and survival analysis; Results: An association between tumor size (pT, p < 0.001), grading (p = 0.013), budding (p < 0.001), LVI (79,4% p < 0.001) and LNM was observed. A multivariate analysis identified pT4 (OR 5.45, p < 0.001) and LVI+ (OR 10.7, p < 0.001) as significant predictors of LNM. Right-sided patients presented a worse OS when associated with LNM, while no significant difference was observed in N0 patients; Conclusions: histological and molecular analysis can help identify high risk patients, which could benefit from extended lymphadenectomies. These patients could be ideal candidates for the D3 lymphadenectomy/CME.
1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed malignancy, and the second most common cause of cancer-related death [1].
Despite often being considered as a single identity, right- and left-sided colon cancers constitute two distinguished entities in terms of anatomy, vascularization, mutational profile and, when in an advanced stage, also in terms of prognosis [2,3].
Neoplasms of the right colon are, in other words, biologically different from those of the left colon [4,5], and those differences can be taken into consideration when we approach their treatment.
Surgery remains a cornerstone in the treatment of both right- and left-sided localized colon cancer [4,5,6], and lymphadenectomy plays a central role: a correctly executed lymphadenectomy is, in fact, not only important in allowing a correct stadiation of the disease, but it is important also in terms of prognosis and future treatment planning [6,7,8]. However, while in left-sided colon cancer there is a strong consensus on the D3 lymphadenectomy/CME being the standard of care for resectable tumors, lymphatic stations to be removed in patients with right-sided colon cancer are still a matter of discussion [6,7,8], with Asian guidelines advocating extended lymph node removal (D3) on a routine basis in T3/T4 and selected T2 stages [9], while Western guidelines recommend the execution of the D2 lymphadenectomy only [6,7,8].
These differences are strongly connected to the vascular anatomy of these tumors: in the left colon, the possibility of ligating the inferior mesenteric pedicles at the origin makes the D3 lymphadenectomy/CME simple and safe to execute. On the contrary, in the right colon, the impossibility of ligating the superior mesenteric pedicles makes the D3 lymphadenectomy/CME more challenging. The D3 lymphadenectomy in right-sided colon cancer has been associated with a higher risk of vascular complications [10], despite it being associated with some survival advantages [11], and is therefore not only generally not recommended by Western guidelines, but also not routinely executed in high-volume centers in Italy [12].
Histological and molecular factors can have an impact on lymph node metastatization. This relation was first studied in colorectal cancer by Ueno et al. [13], who conducted a study on 292 patients with invasive colon cancer, and more recently confirmed by Yasue et al. [14], who conducted a similar study on 846 cases of T1 stage colorectal cancer.
This study aims to analyze the correlation of histopathological and molecular characteristics with lymph node metastasis, both in right- and left-sided colon cancer (primary endpoint). This study also aims to analyze the survival of patients with colon cancer and with or without lymph node metastasis (secondary endpoint).
2. Materials and Methods
A retrospective single-center study has been conducted on patients diagnosed with resectable colon cancer at our institution who underwent surgical resection between January 2019 and December 2023. These patients were identified from a prospectively maintained institutional database.
2.1. Patients Selection
Patients with benign or non-neoplastic histology were excluded, as well as patients aged less than 18 years old. Other exclusion criteria were pregnancy at the time of the resection and emergency surgeries. Out of the 266 patients initially identified, 91 were excluded because they did not meet the inclusion criteria, resulting in a final population of 175 cases (Figure 1).
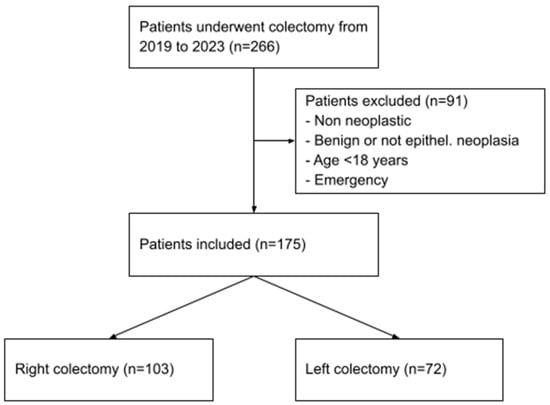
Figure 1.
Patients selection process.
2.2. Analyzed Features and Surgical Technique
Disease was staged according to the indications of the American Joint Committee on Cancer (AJCC) Cancer Staging Manual, 8th edition. Histopathological and molecular data were extracted from histological reports, and the features analyzed were as follows: histotype, tumor T stage (pT), tumor N stage (pN), histological grade of differentiation (G), tumor budding (Bd), lymphovascular invasion (LVI) and microsatellite status (microsatellite instability (MSI) or microsatellite stability (MSS)). Colorectal cancers demonstrating the MSI phenomenon were further divided into two distinct MSI tumor phenotypes: MSI-high (MSH-H) and MSI-low (MSI-L), while tumors that did not present evidence of budding were indicated as Bd0.
Surgical resection was conducted according to the standards adopted by our institution by experienced surgeons. In right colon cancers, a D2 lymphadenectomy was performed, while in left colon cancer, a D3 lymphadenectomy associated with CME was performed.
2.3. Statistical Analysis
Continuous variables are presented as the mean and standard deviation (SD), while categorical variables are expressed as units and percentages. Descriptive statistics were used to summarize information relevant to the study. The differences between groups were analyzed using Student’s t-test for continuous variables, and Pearson’s Chi-squared test or Fisher’s exact test, as appropriate, for categorical variables. A multivariate binomial logistic regression was created to identify independent predictors of outcomes. A survival analysis was conducted according to the Kaplan–Meier method. Differences in survival curves were evaluated by adopting the log-rank test. Significance was accepted at p < 0.05.
All statistical analyses were conducted using R (version 4.3.1; R Foundation for Statistical Computing, Vienna, Austria).
3. Results
3.1. Clinical Characteristics of Patients
Of the 175 patients identified, 105 were males (60%) and 70 females (40%). The mean age was 74 (SD = 11). Most of the patients presented an American Society of Anesthesiologists (ASA) score of II (n = 43, 25%) or greater. Most of the patients in our cohort had a TNM stage of III (n = 36, 21%) or IV (n = 14, 8%). The most frequent localizations were ascending colon (n = 75, 43%) and sigmoid colon (n = 33, 19%). Right-sided colon cancer patients were significantly older (p = 0.048). Baseline characteristics are reported in Table 1.

Table 1.
Patients characteristics.
3.2. Operative Results
The mean operative time was 202 min (SD = 55). The majority of the procedures were conducted video-laparoscopically (n = 121, SD = 55). Left colectomies presented a significantly longer operative time (212.2 vs. 194.9, p = 0.040) and were associated with a higher rate of stoma formation (p = 0.002). The operative results are summarized in Table 2.

Table 2.
Operative results.
3.3. Histopathological and Molecular Features
The mean lymph node yield was 19.3 (SD = 8.5), with a significantly higher number of retrieved lymph nodes in right hemicolectomy specimens (20.8 vs. 17.3, p = 0.007). The R0 rate was 100%. Histological variables (pT, pN, G Bd, LVI) were all comparable except for tumor grading (G, p = 0.028). Left-sided tumors showed a higher incidence of MSS (p < 0.001). Most of the patients had an MSS molecular profile (n = 89, 50.9%), as shown in Table 3.

Table 3.
Histopathological and molecular features.
When comparing the pN0 and pN+ patients, all of the histological features showed statistically significant differences in variable distributions. These differences were also found when analyzing right- and left-sided patients separately, as presented in Table 4 and Figure 2.

Table 4.
Variables distributions in relation to LNM.
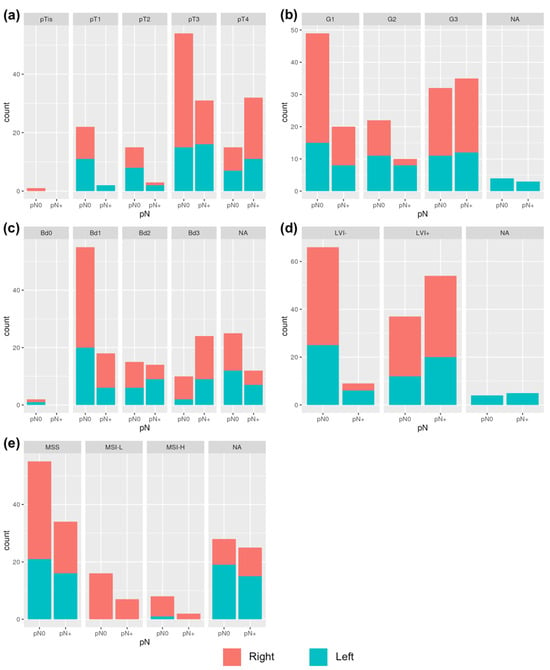
Figure 2.
Variables distributions in relation to LNM in right- and left cancers: (a) correlation between LNM and pT; (b) correlation between LNM and G; (c) correlation between LNM and Bd; (d) correlation between LNM and LVI; (e) correlation between LNM and MSI.
A multivariate analysis showed a significant correlation between lymph node metastasis and lymphovascular invasion (LVI), both in the general population (OR = 20.64, p < 0.001) and in the right-sided and in the left-sided population separately (p = 0.006 and p = 0.041 for right- and left-sided patients, respectively). In right-sided patients, T4 was also found to be a significant predictor of LNM (OR = 8.14, p = 0.019), with near significance in the general population (Table 5).

Table 5.
Multivariate analysis.
3.4. Survival Analysis
A Kaplan–Meier analysis showed no significant differences in terms of overall survival (OS) between right- and left-sided patients (p = 0.35, Figure 3a). However, when the population was grouped according to lymph nodes positivity, right-sided colon cancers with LNM showed a significantly worse prognosis than left-sided colon cancers with LNM (p = 0.012, Figure 3b), while there were no significant differences between right- and left-sided patients without nodes involvement (p = 0.31, Figure 3c).
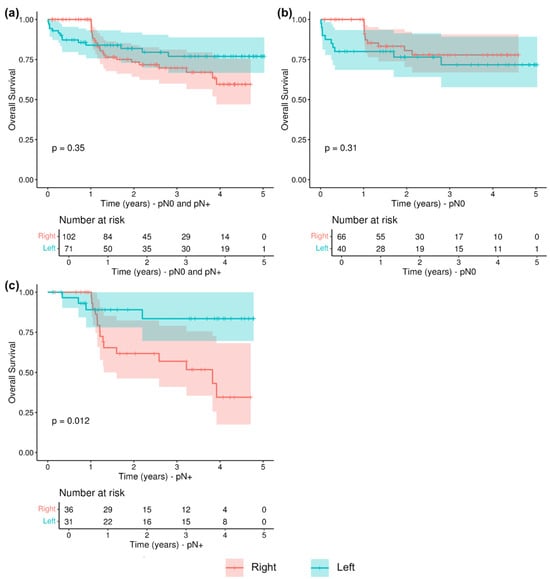
Figure 3.
Overall survival (OS) in relation to LNM and tumor sidedness: (a) OS in pN0 and pN+ patients; (b) OS in pN0 patients; (c) OS in pN+ patients.
4. Discussion
This study showed that lymph node metastases determine a greater prognosis worsening in right-sided colon cancer (RSCC) compared to left-sided colon cancer (LSCC), suggesting the need for a change of perspective in the surgical approach. This investigation of predictive factors has identified specific histopathological features: pT stage, grading, budding and lymphovascular invasion (LVI+).
Despite the absence of any difference in terms of overall survival (OS) between RSCC and LSCC, survival analysis stratified by lymph node staging revealed a significantly worse prognosis of RSCC in the presence of lymph node metastasis (p = 0.012). This interesting finding could be explained by the different approaches of surgical lymphadenectomy. In fact, the standard technique of right hemicolectomy does not involve lymphadenectomy of the mesenteric–celiac axis; on the other hand, left hemicolectomy always includes in these cases the high ligation of the inferior mesenteric artery and thus ensures, if properly performed, local lymph nodal radicality.
A univariate analysis conducted to investigate risk factors for lymph node involvement showed that pT stage, grading, budding and LVI+ had significant correlation with lymph node metastasis, but a multivariate analysis identified pT4 stage and LVI+ as predominant independent predictors, according to ESMO guidelines for stage II that recognize node retrieval and T4 as major risk factors [6]. Moreover, this association appeared to be much stronger in RSCC (OR 8.19 and 25.96, respectively, for pT4 and LVI+).
To our knowledge, the literature on this topic appears limited. The role of histopathologic-molecular factors was studied by Ueno et al. [13] on 292 patients with invasive colon cancer to identify the role of grading, budding and LVI in lymph nodal metastasis. More recently, Yasue et al. [14] conducted a similar analysis on 846 cases of T1-stage colorectal cancer. In this study, however, it was decided to analyze RSCC and LSCC as two separate entities, namely intermediate- and advanced-stage neoplasms.
An analysis of microsatellite stability yielded interesting results. MSI appeared to be a protective factor against lymph nodal invasion (OR < 1), but it did not seem to influence the prognosis in the pN0 patients. However, in the pN+ group, the survival curve of MSS/MSI-Low patients showed a steep drop, suggesting that MSS provides a considerable disadvantage in terms of prognosis in the presence of lymph node metastasis.
Hakki et al. [15] analyzed 1466 cases of colon cancer, of which 361 (25%) were MSI. In these patients, pN+ status was associated with pT stage, LVI+ and MSS.
In view of the above, while in LSCC the current standard surgical technique appears adequate regardless of histologic-molecular arrangement and lymph nodal status, in RSCC a change in perspective to identify two different groups of patients appears needed. Considering the proven importance of proper lymphadenectomy for oncological purposes [16,17], in patients considered “low risk” (MSI and without preoperatory signs of lymph nodal involvement, cN0), the current standard right hemicolectomy technique seems adequate; meanwhile, “high-risk” patients (MSS with cN+) could be appropriate candidates for right hemicolectomy with complete mesocolic excision (CME). Indeed, this procedure is still controversial due to the technical difficulty and the increased complication rate encountered.
In 2022, three criteria for defining CME were established by the Portsmouth Consensus Statements [18]: (1) central vascular ligation, (2) exposure of the superior mesenteric vein (SMV) and (3) excise ion of an intact mesocolon.
Regarding technical feasibility and short-term outcomes, a controlled trial conducted in 2020 on 330 patients [19] showed a greater number of harvested lymph nodes (24 vs. 20, p = 0002) in the CME group, without significant differences in terms of postoperative complications. CME patients also showed a lower rate of recurrence (0% vs. 9.8%, p < 0.001). These positive results were confirmed by a systematic review published in 2021 [20], which included 16 studies. However, the complexity of this technique was confirmed by the results of the CODIG-2 trial published in 2024 [12]: only 37.6% of surgical specimens were type 0 according to the Benz score [21].
In this light, the sensitivity and specificity of the radiological means at our disposal in preoperative staging appear crucial. Current CT techniques facilitate the detection of lymph node metastases with sensitivity and specificity of 71% (CI 59–81%) and 67% (CI 46–83%), respectively, according to the results of a recent meta-analysis [22]. The radiological diagnosis of lymph node invasion still has unresolved critical issues: the most accepted definition of lymph node positivity is the presence of a lymph node larger than 10 mm or a cluster of three or more lymph nodes regardless of size. This approach, however, exposes the risk of false positives (enlarged inflammatory lymph nodes) and false negatives (lymph nodes with micrometastases). The use of radiomics, in this scenario, could be promising to identify lymph node positivity in colorectal cancer. It was first proposed by Huang et al. [23], and in recent studies has been proven to be more precise than conventional techniques [24]; in 2022, Zhao et al. [25] also validated a deep learning model for predicting lymph node metastasis in colorectal cancer with encouraging results. However, stronger data on a larger population are still needed. From a molecular perspective, specific patterns currently studied mainly in metastatic disease (like mutations of KRAS and NRAS genes) [6,26,27] could be a useful tool for selecting high-risk patients. The importance of these factors was confirmed by Roth et al. [28], who analyzed their role in terms of overall survival from 1404 cases in the PETACC3 trial [29].
This study has several limitations: first, its observational and retrospective nature; second, the limited, although well-selected, sample (175 cases), which makes it necessary to confirm these results on a large scale, possibly with a multicenter study which is currently ongoing. We also excluded data regarding perineural invasion, and tumor-infiltrating lymphocytes due to the lack of homogenous reporting in our pathology exams. Finally, the survival analysis was conducted on a population that did not undergo a uniform follow-up period.
5. Conclusions
In conclusion, lymph nodal status in patients with right-sided colon cancer (RSCC) seems to have a stronger impact on survival compared with left-sided (LSCC), with RSCC N+ patients having a dramatic worsening of the prognosis (p = 0.012). In a multivariate analysis, pT4 stage and LVI+ have been shown to be independent predictors for lymph node metastasis in RSCC (OR = 25.96 and 8.19, respectively). Microsatellite instability (MSI) appeared to be a protective factor. Considering the impact of lymph nodal involvement on prognosis, likely due to the absence of a standard and radical lymphadenectomy in right hemicolectomy, this group of patients with pT4 stage, LVI+ and MSS status might be ideal candidates for CME.
Author Contributions
Conceptualization: E.M.M. and F.S.L.C.; methodology: E.M.M., F.S.L.C., A.L.F., A.L. and G.A.; formal analysis: E.M.M. and F.S.L.C.; data curation: F.S.L.C., A.L.F. and L.D.C.; writing—original draft preparation E.M.M., F.S.L.C., A.L.F., A.L., G.A., G.C. and A.S.; writing—review and editing: F.M., G.B. and P.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki. Ethical review and approval were waived for this study due to the nature of the study.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data can be shared up on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sung, H.A.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, M.; Morikawa, T.; Kuchiba, A.; Imamura, Y.; Qian, Z.R.; Nishihara, R.; Liao, X.; Waldron, L.; Hoshida, Y.; Huttenhower, C.; et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut 2012, 61, 847–854. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Loupakis, F.; Yang, D.; Yau, L.; Feng, S.; Cremolini, C.; Zhang, W.; Maus, M.K.; Antoniotti, C.; Langer, C.; Scherer, S.J.; et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J. Natl. Cancer Inst. 2015, 107, dju427. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nawa, T.; Kato, J.; Kawamoto, H.; Okada, H.; Yamamoto, H.; Kohno, H.; Endo, H.; Shiratori, Y. Differences between right- and left-sided colon cancer in patient characteristics, cancer morphology and histology. J. Gastroenterol. Hepatol. 2008, 23, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Baran, B.; Mert Ozupek, N.; Yerli Tetik, N.; Acar, E.; Bekcioglu, O.; Baskin, Y. Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature. Gastroenterol. Res. 2018, 11, 264–273. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Argilés, G.; Tabernero, J.; Labianca, R.; Hochhauser, D.; Salazar, R.; Iveson, T.; Laurent-Puig, P.; Quirke, P.; Yoshino, T.; Taieb, J.; et al. Electronic address: Clinicalguidelines@esmo.org. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2020, 31, 1291–1305. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, A.; Adam, R.; Roselló, S.; Arnold, D.; Normanno, N.; Taïeb, J.; Seligmann, J.; De Baere, T.; Osterlund, P.; Yoshino, T.; et al. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 10–32. [Google Scholar] [CrossRef] [PubMed]
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Arain, M.A.; Chen, Y.J.; Ciombor, K.K.; Cohen, S.; Cooper, H.S.; Deming, D.; Farkas, L.; et al. Colon Cancer, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 329–359. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, Y.; Muro, K.; Saito, Y.; Ito, Y.; Ajioka, Y.; Hamaguchi, T.; Hasegawa, K.; Hotta, K.; Ishida, H.; Ishiguro, M.; et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int. J. Clin. Oncol. 2020, 25, 1–42. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xu, L.; Su, X.; He, Z.; Zhang, C.; Lu, J.; Zhang, G.; Sun, Y.; Du, X.; Chi, P.; Wang, Z.; et al. Short-term outcomes of complete mesocolic excision versus D2 dissection in patients undergoing laparoscopic colectomy for right colon cancer (RELARC): A randomised, controlled, phase 3, superiority trial. Lancet Oncol. 2021, 22, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Bertelsen, C.A.; Neuenschwander, A.U.; Jansen, J.E.; Wilhelmsen, M.; Kirkegaard-Klitbo, A.; Tenma, J.R.; Bols, B.; Ingeholm, P.; Rasmussen, L.A.; Jepsen, L.V.; et al. Disease-free survival after complete mesocolic excision compared with conventional colon cancer surgery: A retrospective, population-based study. Lancet Oncol. 2015, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Anania, G.; Chiozza, M.; Campagnaro, A.; Bagolini, F.; Resta, G.; Azzolina, D.; Silecchia, G.; Cirocchi, R.; Agrusa, A.; Cuccurullo, D.; et al. Laparoscopic right hemicolectomy: A SICE (Società Italiana di Chirurgia Endoscopica e Nuove tecnologie) network prospective study on the approach to right colon lymphadenectomy in Italy: Is there a standard?-CoDIG 2 (ColonDx Italian Group). Surg. Endosc. 2024, 38, 1432–1441. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ueno, H.; Mochizuki, H.; Hashiguchi, Y.; Shimazaki, H.; Aida, S.; Hase, K.; Matsukuma, S.; Kanai, T.; Kurihara, H.; Ozawa, K.; et al. Risk factors for an adverse outcome in early invasive colorectal carcinoma. Gastroenterology 2004, 127, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Yasue, C.; Chino, A.; Takamatsu, M.; Namikawa, K.; Ide, D.; Saito, S.; Igarashi, M.; Fujisaki, J. Pathological risk factors and predictive endoscopic factors for lymph node metastasis of T1 colorectal cancer: A single-center study of 846 lesions. J. Gastroenterol. 2019, 54, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Hakki, L.; Khan, A.; Gonen, M.; Stadler, Z.; Segal, N.H.; Shia, J.; Widmar, M.; Wei, I.H.; Smith, J.J.; Pappou, E.P.; et al. Lymph Node Metastases and Associated Recurrence-Free Survival in Microsatellite Stable and Unstable Colon Cancer. Ann. Surg. Oncol. 2023, 30, 8487–8494. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Marrelli, D.; Piccioni, S.A.; Carbone, L.; Petrioli, R.; Costantini, M.; Malagnino, V.; Bagnacci, G.; Rizzoli, G.; Calomino, N.; Piagnerelli, R.; et al. Posterior and Para-Aortic (D2plus) Lymphadenectomy after Neoadjuvant/Conversion Therapy for Locally Advanced/Oligometastatic Gastric Cancer. Cancers 2024, 16, 1376. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Muttillo, E.M.; La Franca, A.; Stefanelli, S.; Coppola, A.; Li Causi, F.S.; Giannella, R.A.; Pino, E.; Castagnola, G.; Scarinci, A.; Balducci, G.; et al. Oncological Adequacy of Laparoscopic Surgery for Bulky Gastric Cancer: Results of a Western Single-Center Series. Life 2023, 13, 2243. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tejedor, P.; Francis, N.; Jayne, D.; Hohenberger, W.; Khan, J.; on behalf the CME Project Working Group. Consensus statements on complete mesocolic excision for right-sided colon cancer-technical steps and training implications. Surg. Endosc. 2022, 36, 5595–5601. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Wang, C.; Cui, Y.; Shen, Z.; Jiang, K.; Shen, D.; Wang, Y.; Zhan, S.; Guo, P.; Yang, X.; et al. Efficacy and Safety of Complete Mesocolic Excision in Patients With Colon Cancer: Three-year Results From a Prospective, Nonrandomized, Double-blind, Controlled Trial. Ann. Surg. 2020, 271, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Mazzarella, G.; Muttillo, E.M.; Picardi, B.; Rossi, S.; Muttillo, I.A. Complete mesocolic excision and D3 lymphadenectomy with central vascular ligation in right-sided colon cancer: A systematic review of postoperative outcomes, tumor recurrence and overall survival. Surg. Endosc. 2021, 35, 4945–4955. [Google Scholar] [CrossRef] [PubMed]
- Benz, S.; Tannapfel, A.; Tam, Y.; Grünenwald, A.; Vollmer, S.; Stricker, I. Proposal of a new classification system for complete mesocolic excison in right-sided colon cancer. Tech. Coloproctol. 2019, 23, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Nerad, E.; Lahaye, M.J.; Maas, M.; Nelemans, P.; Bakers, F.C.; Beets, G.L.; Beets-Tan, R.G. Diagnostic Accuracy of CT for Local Staging of Colon Cancer: A Systematic Review and Meta-Analysis. AJR Am. J. Roentgenol. 2016, 207, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Q.; Liang, C.H.; He, L.; Tian, J.; Liang, C.S.; Chen, X.; Ma, Z.L.; Liu, Z.Y. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J. Clin. Oncol. 2016, 34, 2157–2164, Erratum in: J. Clin. Oncol. 2016, 34, 2436. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, S.; Koch, V.; Santos, D.P.D.; Ackermann, J.; Grünewald, L.D.; Weitkamp, I.; Yel, I.; Martin, S.S.; Albrecht, M.H.; Scholtz, J.E.; et al. Imaging biomarkers to stratify lymph node metastases in abdominal CT—Is radiomics superior to dual-energy material decomposition? Eur. J. Radiol. Open. 2022, 10, 100459. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, J.; Wang, H.; Zhang, Y.; Wang, R.; Liu, Q.; Li, J.; Li, X.; Huang, H.; Zhang, J.; Zeng, Z.; et al. Deep learning radiomics model related with genomics phenotypes for lymph node metastasis prediction in colorectal cancer. Radiother. Oncol. 2022, 167, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Muttillo, E.M.; Felli, E. Can the mutational status of KRAS drive the treatment of colorectal liver metastases? HPB 2021, 23, 643. [Google Scholar] [CrossRef] [PubMed]
- Formica, V.; Sera, F.; Cremolini, C.; Riondino, S.; Morelli, C.; Arkenau, H.T.; Roselli, M. KRAS and BRAF Mutations in Stage II and III Colon Cancer: A Systematic Review and Meta-Analysis. J. Natl. Cancer Inst. 2022, 114, 517–527. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Roth, A.D.; Delorenzi, M.; Tejpar, S.; Yan, P.; Klingbiel, D.; Fiocca, R.; d’Ario, G.; Cisar, L.; Labianca, R.; Cunningham, D.; et al. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J. Natl. Cancer Inst. 2012, 104, 1635–1646, Erratum in: J. Natl. Cancer Inst. 2013, 105, 151–152. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.D.; Tejpar, S.; Delorenzi, M.; Yan, P.; Fiocca, R.; Klingbiel, D.; Dietrich, D.; Biesmans, B.; Bodoky, G.; Barone, C.; et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: Results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J. Clin. Oncol. 2010, 28, 466–474. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).