The Phenotypical Characterization of Dual-Nature Hybrid Cells in Uveal Melanoma
Abstract
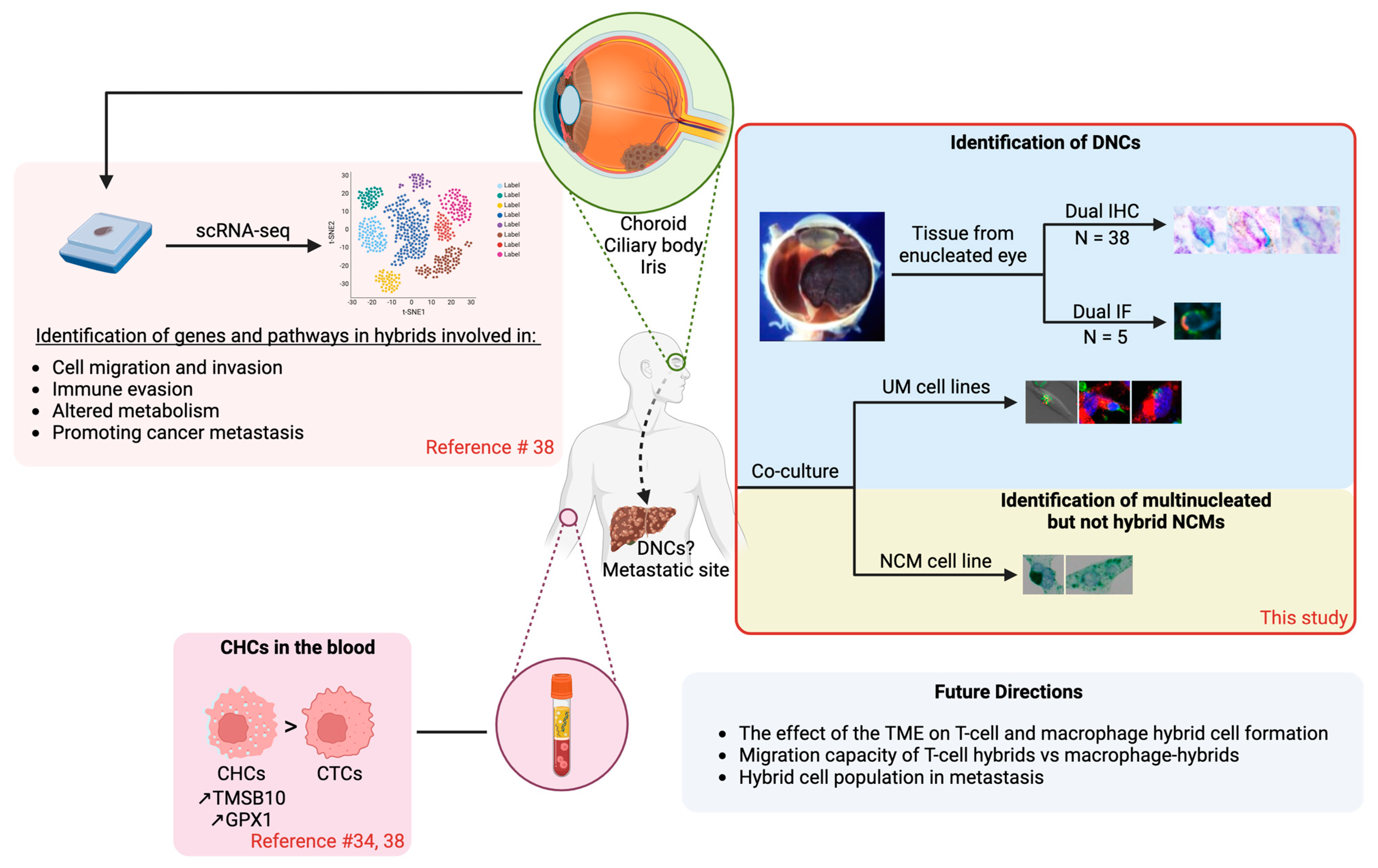
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Specimens
2.2. Histological Staining and Immunohistochemical (IHC) and Immunofluorescence (IF) Analyses
2.3. Cell Culture
2.3.1. Normal Choroidal Melanocyte (NCM) Culture Medium
2.3.2. UM Cell Culture Medium
2.3.3. Isolation of Peripheral Blood Mononuclear Cells (PBMCs)
2.3.4. UM Cells, PBMC Labeling, and Co-Culture
2.4. Phenotypic Determination of Co-Cultured NCM Cells and PBMCs, as Well as UM Cells and PBMCs
2.5. Statistical Analysis
3. Results
3.1. Immunohistochemical (IHC) Analysis of UM Tumors
3.2. Most UM Tumors Contained DNCs with HMB45+/CD45+-, HMB45+/CD8+-, and HMB45+/CD68+-Dominant Phenotypical Profiles
3.3. NCM Cells and PBMCs Are Not Prone to Fusing In Vitro to Form DNCs
3.4. UM Cells and PBMCs Are Prone to Fusing In Vitro to Form DNCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, A.D.; Turell, M.E.; Topham, A.K. Uveal melanoma: Trends in incidence, treatment, and survival. Ophthalmology 2011, 118, 1881–1885. [Google Scholar] [CrossRef] [PubMed]
- Virgili, G.; Gatta, G.; Ciccolallo, L.; Capocaccia, R.; Biggeri, A.; Crocetti, E.; Lutz, J.M.; Paci, E. Incidence of uveal melanoma in Europe. Ophthalmology 2007, 114, 2309–2315. [Google Scholar] [CrossRef]
- Vajdic, C.M.; Kricker, A.; Giblin, M.; McKenzie, J.; Aitken, J.; Giles, G.G.; Armstrong, B.K. Incidence of ocular melanoma in Australia from 1990 to 1998. Int. J. Cancer 2003, 105, 117–122. [Google Scholar] [CrossRef]
- Kaliki, S.; Shields, C.L. Uveal melanoma: Relatively rare but deadly cancer. Eye 2017, 31, 241–257. [Google Scholar] [CrossRef]
- Shields, C.L.; Furuta, M.; Thangappan, A.; Nagori, S.; Mashayekhi, A.; Lally, D.R.; Kelly, C.C.; Rudich, D.S.; Nagori, A.V.; Wakade, O.A.; et al. Metastasis of uveal melanoma millimeter-by-millimeter in 8033 consecutive eyes. Arch. Ophthalmol. 2009, 127, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Damato, B.; Kacperek, A.; Errington, D.; Heimann, H. Proton beAm. radiotherapy of uveal melanoma. Saudi J. Ophthalmol. 2013, 27, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Jensen, O.A. Malignant melanomas of the human uvea: 25-year follow-up of cases in Denmark, 1943–1952. Acta Ophthalmol. 1982, 60, 161–182. [Google Scholar] [CrossRef]
- Kujala, E.; Mäkitie, T.; Kivelä, T. Very long-term prognosis of patients with malignant uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4651–4659. [Google Scholar] [CrossRef]
- Stålhammar, G.; Herrspiegel, C. Long-term relative survival in uveal melanoma: A systematic review and meta-analysis. Commun. Med. 2022, 2, 18. [Google Scholar] [CrossRef]
- Collaborative Ocular Melanoma Study Group. Assessment of metastatic disease status at death in 435 patients with large choroidal melanoma in the Collaborative Ocular Melanoma Study (COMS): COMS report no. 15. Arch. Ophthalmol. 2001, 119, 670–676. [Google Scholar] [CrossRef]
- Diener-West, M.; Reynolds, S.M.; Agugliaro, D.J.; Caldwell, R.; Cumming, K.; Earle, J.D.; Hawkins, B.S.; Hayman, J.A.; Jaiyesimi, I.; Jampol, L.M.; et al. Development of metastatic disease after enrollment in the COMS trials for treatment of choroidal melanoma: Collaborative Ocular Melanoma Study Group Report No. 26. Arch. Ophthalmol. 2005, 123, 1639–1643. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.D.; Shields, C.L.; Shields, J.A. Prognostic factors in uveal melanoma. Melanoma Res. 2001, 11, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Lamas, N.J.; Martel, A.; Nahon-Estève, S.; Goffinet, S.; Macocco, A.; Bertolotto, C.; Lassalle, S.; Hofman, P. Prognostic Biomarkers in Uveal Melanoma: The Status Quo, Recent Advances and Future Directions. Cancers 2021, 14, 96. [Google Scholar] [CrossRef]
- Shields, C.L.; Kaliki, S.; Furuta, M.; Fulco, E.; Alarcon, C.; Shields, J.A. American Joint Committee on Cancer classification of posterior uveal melanoma (tumor size category) predicts prognosis in 7731 patients. Ophthalmology 2013, 120, 2066–2071. [Google Scholar] [CrossRef]
- McLean, I.W.; Foster, W.D.; Zimmerman, L.E. Uveal melanoma: Location, size, cell type, and enucleation as risk factors in metastasis. Hum. Pathol. 1982, 13, 123–132. [Google Scholar] [CrossRef]
- McLean, I.W.; Keefe, K.S.; Burnier, M.N. Uveal melanoma. Comparison of the prognostic value of fibrovascular loops, mean of the ten largest nucleoli, cell type, and tumor size. Ophthalmology 1997, 104, 777–780. [Google Scholar] [CrossRef] [PubMed]
- Folberg, R.; Rummelt, V.; Parys-Van Ginderdeuren, R.; Hwang, T.; Woolson, R.F.; Pe'er, J.; Gruman, L.M. The prognostic value of tumor blood vessel morphology in primary uveal melanoma. Ophthalmology 1993, 100, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- McLean, M.J.; Foster, W.D.; Zimmerman, L.E. Prognostic factors in small malignant melanomas of choroid and ciliary body. Arch. Ophthalmol. 1977, 95, 48–58. [Google Scholar] [CrossRef]
- Affeldt, J.C.; Minckler, D.S.; Azen, S.P.; Yeh, L. Prognosis in uveal melanoma with extrascleral extension. Arch. Ophthalmol. 1980, 98, 1975–1979. [Google Scholar] [CrossRef]
- Jager, M.J.; Ly, L.V.; El Filali, M.; Madigan, M.C. Macrophages in uveal melanoma and in experimental ocular tumor models: Friends or foes? Prog. Retin. Eye Res. 2011, 30, 129–146. [Google Scholar] [CrossRef]
- Herwig, M.C.; Grossniklaus, H.E. Role of macrophages in uveal melanoma. Expert Rev. Ophthalmol. 2011, 6, 405–407. [Google Scholar] [CrossRef] [PubMed]
- Whelchel, J.C.; Farah, S.E.; McLean, I.W.; Burnier, M.N. Immunohistochemistry of infiltrating lymphocytes in uveal malignant melanoma. Investig. Ophthalmol. Vis. Sci. 1993, 34, 2603–2606. [Google Scholar]
- Triozzi, P.L.; Schoenfield, L.; Plesec, T.; Saunthararajah, Y.; Tubbs, R.R.; Singh, A.D. Molecular profiling of primary uveal melanomas with tumor-infiltrating lymphocytes. Oncoimmunology 2019, 8, e947169. [Google Scholar] [CrossRef] [PubMed]
- Gelmi, M.C.; Bas, Z.; Malkani, K.; Ganguly, A.; Shields, C.L.; Jager, M.J. Adding the Cancer Genome Atlas Chromosome Classes to American Joint Committee on Cancer System Offers More Precise Prognostication in Uveal Melanoma. Ophthalmology 2022, 129, 431–437. [Google Scholar] [CrossRef]
- Li, H.; Li, Q.; Dang, K.; Ma, S.; Cotton, J.L.; Yang, S.; Zhu, L.J.; Deng, A.C.; Ip, Y.T.; Johnson, R.L.; et al. YAP/TAZ Activation Drives Uveal Melanoma Initiation and Progression. Cell Rep. 2019, 29, 3200–3211. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.X.; Luo, J.; Mo, J.S.; Liu, G.; Kim, Y.C.; Meng, Z.; Zhao, L.; Peyman, G.; Ouyang, H.; Jiang, W.; et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell 2014, 25, 822–830. [Google Scholar] [CrossRef]
- Boru, G.; Cebulla, C.M.; Sample, K.M.; Massengill, J.B.; Davidorf, F.H.; Abdel-Rahman, M.H. Heterogeneity in Mitogen-Activated Protein Kinase (MAPK) Pathway Activation in Uveal Melanoma With Somatic GNAQ and GNA11 Mutations. Investig. Ophthalmol. Vis. Sci. 2019, 60, 2474–2480. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Huang, H.Y.; Lin, Z.; Ranieri, M.; Li, S.; Sahu, S.; Liu, Y.; Ban, Y.; Guidry, K.; Hu, H.; et al. Genome-Wide CRISPR Screens Identify Multiple Synthetic Lethal Targets That Enhance KRASG12C Inhibitor Efficacy. Cancer Res. 2023, 83, 4095–4111. [Google Scholar] [CrossRef]
- Jin, E.; Burnier, J.V. Liquid Biopsy in Uveal Melanoma: Are We There Yet? Ocul. Oncol. Pathol. 2021, 7, 1–16. [Google Scholar] [CrossRef]
- Callejo, S.A.; Antecka, E.; Blanco, P.L.; Edelstein, C.; Burnier, M.N., Jr. Identification of circulating malignant cells and its correlation with prognostic factors and treatment in uveal melanoma. A prospective longitudinal study. Eye 2007, 21, 752–759. [Google Scholar] [CrossRef]
- Torres, V.; Triozzi, P.; Eng, C.; Tubbs, R.; Schoenfiled, L.; Crabb, J.W.; Saunthararajah, Y.; Singh, A.D. Circulating tumor cells in uveal melanoma. Future Oncol. 2011, 7, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Roszik, J.; Gombos, D.; Upshaw, J.; Sarli, V.; Meas, S.; Lucci, A.; Hall, C.; Patel, S. Pilot Study of Circulating Tumor Cells in Early-Stage and Metastatic Uveal Melanoma. Cancers 2019, 11, 856. [Google Scholar] [CrossRef] [PubMed]
- Logan, P.T.; Fernandes, B.F.; Di Cesare, S.; Marshall, J.C.; Maloney, S.C.; Burnier, M.N., Jr. Single-cell tumor dormancy model of uveal melanoma. Clin. Exp. Metastasis 2008, 25, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Parappilly, M.S.; Chin, Y.; Whalen, R.M.; Anderson, A.N.; Robinson, T.S.; Strgar, L.; Sutton, T.L.; Conley, P.; Klocke, C.; Gibbs, S.L.; et al. Circulating Neoplastic-Immune Hybrid Cells Predict Metastatic Progression in Uveal Melanoma. Cancers 2022, 14, 4617. [Google Scholar] [CrossRef]
- Dietz, M.S.; Sutton, T.L.; Walker, B.S.; Gast, C.E.; Zarour, L.; Sengupta, S.K.; Swain, J.R.; Eng, J.; Parappilly, M.; Limbach, K.; et al. Relevance of circulating hybrid cells as a non-invasive biomarker for myriad solid tumors. Sci. Rep. 2021, 11, 13630. [Google Scholar] [CrossRef]
- Hass, R.; von der Ohe, J.; Dittmar, T. Hybrid Formation and Fusion of Cancer Cells In Vitro and In Vivo. Cancers 2021, 13, 4496. [Google Scholar] [CrossRef]
- Tretyakova, M.S.; Subbalakshmi, A.R.; Menyailo, M.E.; Jolly, M.K.; Denisov, E.V. Tumor Hybrid Cells: Nature and Biological Significance. Front. Cell Dev. Biol. 2022, 10, 814714. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.N.; Conley, P.; Klocke, C.D.; Sengupta, S.K.; Pang, A.; Farley, H.C.; Gillingham, A.R.; Dawson, A.D.; Fan, Y.; Jones, J.A.; et al. Detection of neoplastic-immune hybrid cells with metastatic properties in uveal melanoma. Biomark Res. 2024, 12, 67. [Google Scholar] [CrossRef]
- McLean, I.W.; Foster, W.D.; Zimmerman, L.E.; Gamel, J.W. Modifications of Callender's classification of uveal melanoma at the Armed Forces Institute of Pathology. Am. J. Ophthalmol. 1983, 96, 502–509. [Google Scholar] [CrossRef]
- Valtink, M.; Engelmann, K. Serum-free cultivation of adult normal human choroidal melanocytes. Graefes Arch. Clin. Exp. Ophthalmol. 2007, 245, 1487–1494. [Google Scholar] [CrossRef]
- Jager, M.J.; Magner, J.A.; Ksander, B.R.; Dubovy, S.R. Uveal Melanoma Cell Lines: Where do they come from? (An American Ophthalmological Society Thesis). Trans. Am. Ophthalmol. Soc. 2016, 114, T5. [Google Scholar] [PubMed]
- Finger, P.T.; Kurli, M.; Reddy, S.; Tena, L.B.; Pavlick, A.C. Whole body PET/CT for initial staging of choroidal melanoma. Br. J. Ophthalmol. 2005, 89, 1270–1274. [Google Scholar] [CrossRef] [PubMed]
- Shabo, I.; Midtbö, K.; Andersson, H.; Åkerlund, E.; Olsson, H.; Wegman, P.; Gunnarsson, C.; Lindström, A. Macrophage traits in cancer cells are induced by macrophage-cancer cell fusion and cannot be explained by cellular interaction. BMC Cancer 2015, 15, 922. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.E.; Anderson, E.C.; Davies, P.S.; Silk, A.D.; Pelz, C.; Impey, S.; Wong, M.H. Fusion between Intestinal epithelial cells and macrophages in a cancer context results in nuclear reprogramming. Cancer Res. 2011, 71, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Raskov, H.; Orhan, A.; Christensen, J.P.; Gögenur, I. Cytotoxic CD8(+) T cells in cancer and cancer immunotherapy. Br. J. Cancer 2021, 124, 359–367. [Google Scholar] [CrossRef]
- Gezgin, G.; Visser, M.; Ruano, D.; Santegoets, S.J.; de Miranda, N.; van der Velden, P.A.; Luyten, G.P.M.; van der Burg, S.H.; Verdegaal, E.M.; Jager, M.J. Tumor-Infiltrating T Cells Can Be Expanded Successfully from Primary Uveal Melanoma after Separation from Their Tumor Environment. Ophthalmol. Sci. 2022, 2, 100132. [Google Scholar] [CrossRef]
- Tuthill, R.J.; Unger, J.M.; Liu, P.Y.; Flaherty, L.E.; Sondak, V.K. Risk assessment in localized primary cutaneous melanoma: A Southwest Oncology Group study evaluating nine factors and a test of the Clark logistic regression prediction model. Am. J. Clin. Pathol. 2002, 118, 504–511. [Google Scholar] [CrossRef]
- Bronkhorst, I.H.; Vu, T.H.; Jordanova, E.S.; Luyten, G.P.; Burg, S.H.; Jager, M.J. Different subsets of tumor-infiltrating lymphocytes correlate with macrophage influx and monosomy 3 in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5370–5378. [Google Scholar] [CrossRef]
- Lagouros, E.; Salomao, D.; Thorland, E.; Hodge, D.O.; Vile, R.; Pulido, J.S. Infiltrative T regulatory cells in enucleated uveal melanomas. Trans. Am. Ophthalmol. Soc. 2009, 107, 223–228. [Google Scholar]
- Mougiakakos, D.; Johansson, C.C.; Trocme, E.; All-Ericsson, C.; Economou, M.A.; Larsson, O.; Seregard, S.; Kiessling, R. Intratumoral forkhead box P3-positive regulatory T cells predict poor survival in cyclooxygenase-2-positive uveal melanoma. Cancer 2010, 116, 2224–2233. [Google Scholar] [CrossRef]
- Bronkhorst, I.H.; Jager, M.J. Inflammation in uveal melanoma. Eye 2013, 27, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Niederkorn, J.Y. Immunoregulation of intraocular tumours. Eye 1997, 11 Pt 2, 249–254. [Google Scholar] [CrossRef]
- Staibano, S.; Mascolo, M.; Tranfa, F.; Salvatore, G.; Mignogna, C.; Bufo, P.; Nugnes, L.; Bonavolontà, G.; De Rosa, G. Tumor infiltrating lymphocytes in uveal melanoma: A link with clinical behavior? Int. J. Immunopathol. Pharmacol. 2006, 19, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Irving, B.A.; Weiss, A. The cytoplasmic domain of the T cell receptor zeta chain is sufficient to couple to receptor-associated signal transduction pathways. Cell 1991, 64, 891–901. [Google Scholar] [CrossRef]
- Zhou, R.; Caspi, R.R. Ocular immune privilege. F1000 Biol. Rep. 2010, 2, 1885–1889. [Google Scholar] [CrossRef]
- Bronkhorst, I.H.; Jager, M.J. Uveal melanoma: The inflammatory microenvironment. J. Innate Immun. 2012, 4, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, I.H.; Ly, L.V.; Jordanova, E.S.; Vrolijk, J.; Versluis, M.; Luyten, G.P.; Jager, M.J. Detection of M2-macrophages in uveal melanoma and relation with survival. Investig. Ophthalmol. Vis. Sci. 2011, 52, 643–650. [Google Scholar] [CrossRef]
- Mäkitie, T.; Summanen, P.; Tarkkanen, A.; Kivelä, T. Tumor-infiltrating macrophages (CD68(+) cells) and prognosis in malignant uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2001, 42, 1414–1421. [Google Scholar]
- Li, K.; Sun, L.; Wang, Y.; Cen, Y.; Zhao, J.; Liao, Q.; Wu, W.; Sun, J.; Zhou, M. Single-cell characterization of macrophages in uveal melanoma uncovers transcriptionally heterogeneous subsets conferring poor prognosis and aggressive behavior. Exp. Mol. Med. 2023, 55, 2433–2444. [Google Scholar] [CrossRef]
- Leikam, C.; Hufnagel, A.L.; Otto, C.; Murphy, D.J.; Mühling, B.; Kneitz, S.; Nanda, I.; Schmid, M.; Wagner, T.U.; Haferkamp, S.; et al. In vitro evidence for senescent multinucleated melanocytes as a source for tumor-initiating cells. Cell Death Dis. 2015, 6, e1711. [Google Scholar] [CrossRef]
- Meierjohann, S. Effect of stress-induced polyploidy on melanoma reprogramming and therapy resistance. Semin. Cancer Biol. 2022, 81, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Wakeling, W.F.; Greetham, J.; Bennett, D.C. Efficient spontaneous fusion between some co-cultured cells, especially murine melanoma cells. Cell Biol. Int. 1994, 18, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.; Selby, P.; Southgate, J.; Pittman, K.; Bradley, C.; Blair, G.E. Detection of melanoma cells in peripheral blood by means of reverse transcriptase and polymerase chain reaction. Lancet 1991, 338, 1227–1229. [Google Scholar] [CrossRef] [PubMed]
- Silva-Rodríguez, P.; Fernández-Díaz, D.; Bande, M.; Pardo, M.; Loidi, L.; Blanco-Teijeiro, M.J. GNAQ and GNA11 Genes: A Comprehensive Review on Oncogenesis, Prognosis and Therapeutic Opportunities in Uveal Melanoma. Cancers 2022, 14, 3066. [Google Scholar] [CrossRef]
- Griewank, K.G.; Yu, X.; Khalili, J.; Sozen, M.M.; Stempke-Hale, K.; Bernatchez, C.; Wardell, S.; Bastian, B.C.; Woodman, S.E. Genetic and molecular characterization of uveal melanoma cell lines. Pigment. Cell Melanoma Res. 2012, 25, 182–187. [Google Scholar] [CrossRef]
- Shabo, I.; Svanvik, J.; Lindström, A.; Lechertier, T.; Trabulo, S.; Hulit, J.; Sparey, T.; Pawelek, J. Roles of cell fusion, hybridization and polyploid cell formation in cancer metastasis. World J. Clin. Oncol. 2020, 11, 121–135. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Vinogradov, A.E. Genome multiplication as adaptation to tissue survival: Evidence from gene expression in mammalian heart and liver. Genomics 2007, 89, 70–80. [Google Scholar] [CrossRef]
- Duncan, A.W.; Hickey, R.D.; Paulk, N.K.; Culberson, A.J.; Olson, S.B.; Finegold, M.J.; Grompe, M. Ploidy reductions in murine fusion-derived hepatocytes. PLoS Genet. 2009, 5, e1000385. [Google Scholar] [CrossRef]
- Lindström, A.; Midtbö, K.; Arnesson, L.G.; Garvin, S.; Shabo, I. Fusion between M2-macrophages and cancer cells results in a subpopulation of radioresistant cells with enhanced DNA-repair capacity. Oncotarget 2017, 8, 51370–51386. [Google Scholar] [CrossRef]
- Lu, X.; Kang, Y. Efficient acquisition of dual metastasis organotropism to bone and lung through stable spontaneous fusion between MDA-MB-231 variants. Proc. Natl. Acad. Sci. USA 2009, 106, 9385–9390. [Google Scholar] [CrossRef]
- Pawelek, J.M.; Chakraborty, A.K. The cancer cell--leukocyte fusion theory of metastasis. Adv. Cancer Res. 2008, 101, 397–444. [Google Scholar] [CrossRef] [PubMed]
- Sieler, M.; Weiler, J.; Dittmar, T. Cell-Cell Fusion and the Roads to Novel Properties of Tumor Hybrid Cells. Cells 2021, 10, 1465. [Google Scholar] [CrossRef] [PubMed]
- Gast, C.E.; Silk, A.D.; Zarour, L.; Riegler, L.; Burkhart, J.G.; Gustafson, K.T.; Parappilly, M.S.; Roh-Johnson, M.; Goodman, J.R.; Olson, B.; et al. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci. Adv. 2018, 4, eaat7828. [Google Scholar] [CrossRef] [PubMed]


| Patient | Age | Sex | Location | Size | Cell Type | Vascular Loops | Mitotic Figures | Scleral Invasion |
|---|---|---|---|---|---|---|---|---|
| 1 | 63 | M | Iris—CB | Small | Mixed—SP | No | Yes | No |
| 2 | N/A | M | Choroid | Small | Mixed—EP | No | Yes | Yes |
| 3 | 81 | F | Choroid | Small | Mixed—EP | No | No | No |
| 4 | 51 | M | Choroid | Large | Epithelioid | No | No | No |
| 5 | 76 | F | Choroid | Large | Mixed—EP | Yes | Yes | No |
| 6 | 76 | F | Choroid | Large | Mixed—SP | No | No | No |
| 7 | 63 | F | Choroid | Large | Mixed—EP | No | Yes | No |
| 8 | 55 | M | Choroid | Large | Epithelioid | No | Yes | No |
| 9 | 65 | M | Choroid | Large | Mixed—EP | No | Yes | Yes |
| 10 | 72 | F | Choroid | Large | Spindle | No | Yes | Yes |
| 11 | N/A | N/A | Choroid | Small | Mixed—EP | No | No | Yes |
| 12 | 81 | M | Choroid | Large | Epithelioid | No | No | No |
| 13 | N/A | N/A | Choroid | Large | Epithelioid | No | Yes | Yes |
| 14 | 76 | M | Choroid | Large | Mixed—EP | No | No | No |
| 15 | 76 | M | Choroid—CB | Large | Epithelioid | No | No | No |
| 16 | 59 | F | Choroid | Small | Mixed—SP | Yes | Yes | No |
| 17 | 57 | M | Choroid—CB | Large | Epithelioid | Yes | No | Yes |
| 18 | 77 | M | Choroid | Large | Mixed—EP | No | No | No |
| 19 | 50 | M | Choroid | Small | Mixed—SP | No | No | Yes |
| 20 | 71 | M | Choroid | Small | Mixed—SP | No | No | No |
| 21 | 62 | F | Choroid | Large | Epithelioid | Yes | No | No |
| 22 | 58 | F | Choroid | Small | Epithelioid | No | No | No |
| 23 | 50 | F | Choroid | Large | Spindle | Yes | No | No |
| 24 | N/A | F | Choroid | Large | Epithelioid | Yes | No | No |
| 25 | 67 | F | Choroid—CB | Small | Mixed—SP | Yes | No | No |
| 26 | 57 | M | Choroid | Small | Mixed—SP | No | No | No |
| 27 | 79 | F | Choroid | Small | Mixed—EP | No | Yes | Yes |
| 28 | 93 | M | Choroid | Small | Mixed—EP | Yes | Yes | Yes |
| 29 | 93 | F | Choroid | Small | Mixed—SP | No | No | No |
| 30 | 74 | M | Choroid | Small | Mixed—EP | No | No | No |
| 31 | 51 | F | Choroid | Small | Mixed—SP | No | No | No |
| 32 | 56 | F | Choroid | Small | Mixed—EP | Yes | Yes | No |
| 33 | 34 | M | Choroid | Large | Mixed—SP | Yes | Yes | No |
| 34 | 62 | M | Choroid | Large | Mixed—SP | No | Yes | No |
| 35 | 65 | M | Choroid | Small | Mixed—EP | Yes | Yes | Yes |
| 36 | 60 | F | Choroid | Large | Mixed—SP | No | No | Yes |
| 37 | 78 | M | Choroid | Small | Epithelioid | Yes | Yes | No |
| 38 | 47 | M | Choroid | Large | Mixed—SP | No | No | No |
| DNC Staining | Size | Cell Type | Vascular Loops | Mitotic Figures | Scleral Invasion | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| X2 | FET | X2 | FET | X2 | FET | X2 | FET | X2 | FET | |
| HMB45/CD45 (n = 29) | 0.0583 | 0.0996 | 0.6725 | >0.9999 | 0.2155 | 0.2668 | 0.5844 | >0.9999 | 0.2189 | 0.5320 |
| HMB45/CD8 (n = 28) | >0.9999 | >0.9999 | 0.8322 | >0.9999 | 0.2378 | 0.5053 | 0.9136 | >0.9999 | 0.3968 | 0.4444 |
| HMB45/CD68 (n = 24) | 0.6534 | >0.9999 | 0.0247 * | 0.0686 | 0.5254 | 0.6466 | 0.0577 | 0.0850 | 0.6123 | >0.9999 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcotte, E.; Goyeneche, A.; Abdouh, M.; Burnier, J.V.; Burnier, M.N., Jr. The Phenotypical Characterization of Dual-Nature Hybrid Cells in Uveal Melanoma. Cancers 2024, 16, 3231. https://doi.org/10.3390/cancers16183231
Marcotte E, Goyeneche A, Abdouh M, Burnier JV, Burnier MN Jr. The Phenotypical Characterization of Dual-Nature Hybrid Cells in Uveal Melanoma. Cancers. 2024; 16(18):3231. https://doi.org/10.3390/cancers16183231
Chicago/Turabian StyleMarcotte, Emily, Alicia Goyeneche, Mohamed Abdouh, Julia Valdemarin Burnier, and Miguel Noel Burnier, Jr. 2024. "The Phenotypical Characterization of Dual-Nature Hybrid Cells in Uveal Melanoma" Cancers 16, no. 18: 3231. https://doi.org/10.3390/cancers16183231
APA StyleMarcotte, E., Goyeneche, A., Abdouh, M., Burnier, J. V., & Burnier, M. N., Jr. (2024). The Phenotypical Characterization of Dual-Nature Hybrid Cells in Uveal Melanoma. Cancers, 16(18), 3231. https://doi.org/10.3390/cancers16183231





