MRD in Acute Leukemias: Lessons Learned from Acute Promyelocytic Leukemia
Abstract
Simple Summary
Abstract
1. Introduction
2. Current Treatment Paradigms in Acute Leukemia and the Role of MRD
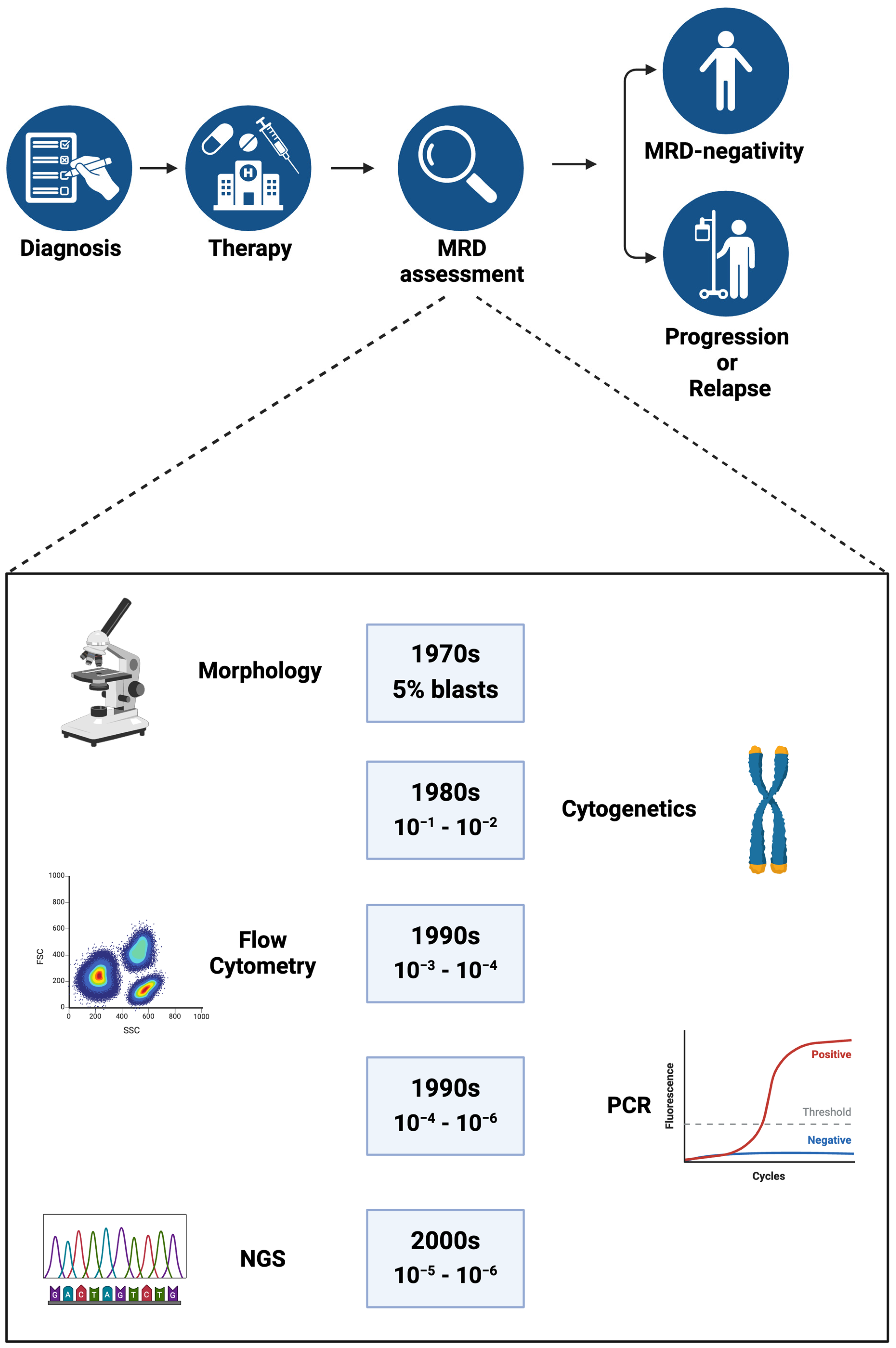
2.1. Traditional Approaches to Determine Treatment Response in Acute Leukemia
2.2. MRD as a Measure to Predict Treatment Response
2.3. Assessing MRD in CR
2.4. MRD-Directed Therapies
3. Lessons Learned from APL
3.1. Pre-ATRA Era: Eliminating MRD, More Is Not Always Better
3.2. The ATRA-Era: From Minimal to Measurable Residual Disease
3.3. Arsenic Trioxide (ATO) as a Single Agent
3.4. ATO and Chemo-Free Regimens
3.5. A Final Lesson from APL: Assessing MRD at the End of Consolidation
4. Does the BM Microenvironment Play a Role in MRD?
4.1. Leukemia Stem Cells as MRD
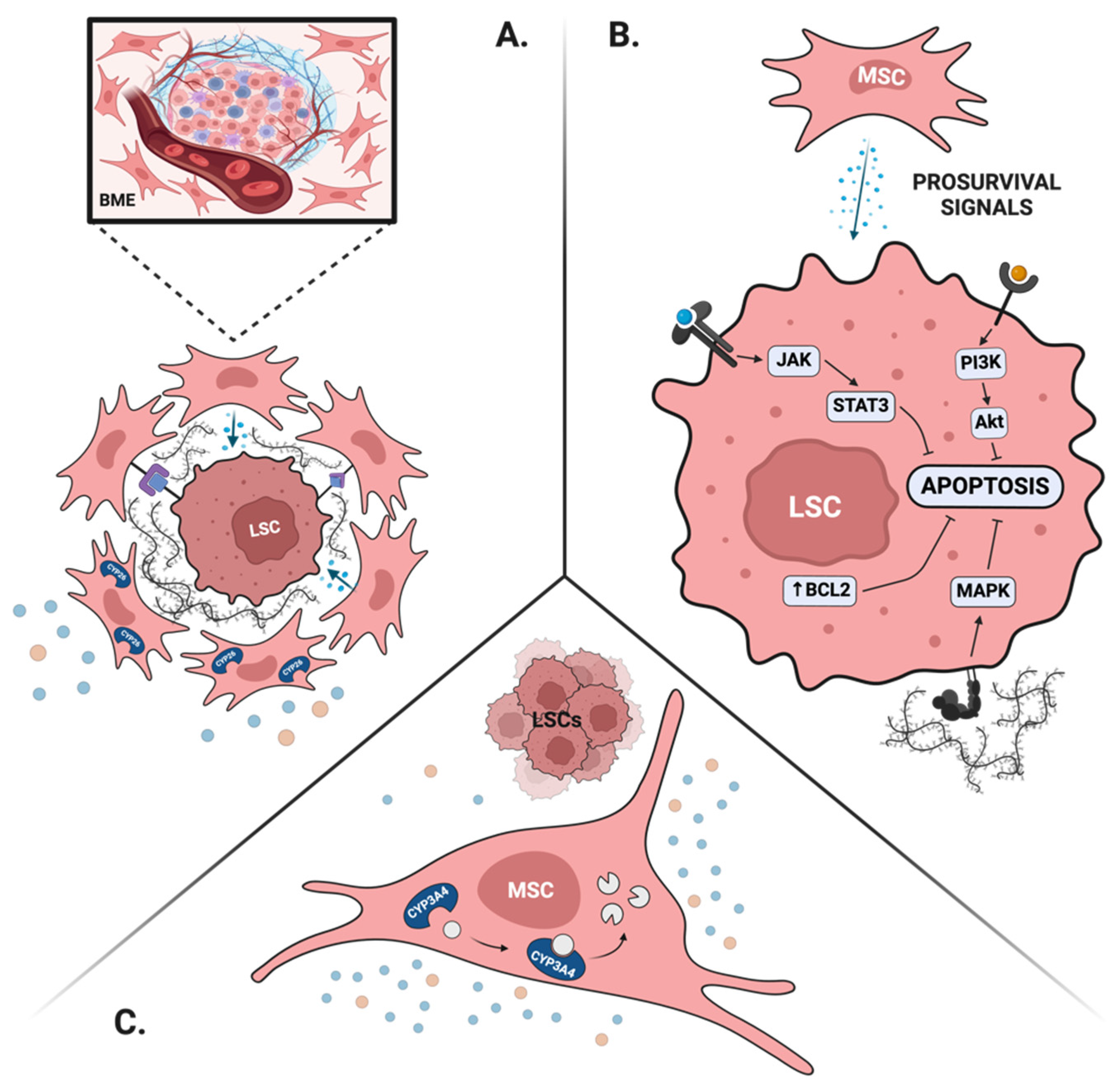
4.2. BME Protects Leukemia Stem Cells and Leads to MRD
4.2.1. BME Maintains LSCs’ Properties
4.2.2. BME Provides Pro-Survival Signals
4.2.3. BME Creates Favorable Drug Pharmacokinetics
4.2.4. BME Promotes Immune Escape of LSCs
4.3. Targeting the BME to Eliminate MRD
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Veith, I. The Yellow Emperor’s Classic of Internal Medicine; University of California Press: Berkeley, CA, USA, 1966. [Google Scholar]
- Avicenna. A Treatise on the Canon of Medicine of Avicenna; Luzac & Co.: London, UK, 1930. [Google Scholar]
- Nutton, V. The Reception of Fracastoro’s Theory of Contagion: The Seed That Fell among Thorns? Osiris 1990, 6, 196–234. [Google Scholar] [CrossRef] [PubMed]
- Ossenkoppele, G.; Schuurhuis, G.J. MRD in AML: Time for redefinition of CR? Blood 2013, 121, 2166–2168. [Google Scholar] [CrossRef] [PubMed]
- Bradstock, K.F.; Janossy, G.; Tidman, N.; Papageorgiou, E.S.; Prentice, H.G.; Willoughby, M.; Hoffbrand, A.V. Immunological Monitoring of Residual Disease in Treated Thymic Acute Lymphoblastic Leukaemia. Leuk. Res. 1981, 5, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Wei, A.; Appelbaum, F.; Craddock, C.; DiNardo, C.; Dombret, H. Diagnosis and Management of AML in Adults: 2022 Recommendations from an International Expert Panel on Behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Gökbuget, N.; Boissel, N.; Chiaretti, S.; Dombret, H.; Doubek, M.; Fielding, A. Management of ALL in Adults: 2024 ELN Recommendations from a European Expert Panel. Blood 2024, 143, 1903–1930. [Google Scholar] [CrossRef]
- Hunger, S.; Mullighan, C. Acute Lymphoblastic Leukemia in Children. N. Engl. J. Med. 2015, 373, 1541–1552. [Google Scholar] [CrossRef]
- Estey, E.; Döhner, H. Acute Myeloid Leukaemia. Lancet 2006, 368, 1894–1907. [Google Scholar] [CrossRef]
- Pui, C.H.; Yang, J.J.; Hunger, S.P.; Pieters, R.; Schrappe, M.; Biondi, A.; Vora, A.; Baruchel, A.; Silverman, L.B.; Schmiegelow, K.; et al. Childhood Acute Lymphoblastic Leukemia: Progress through Collaboration. J. Clin. Oncol. 2015, 33, 2938–2948. [Google Scholar] [CrossRef]
- Basso, G.; Veltroni, M.; Valsecchi, M.G.; Dworzak, M.N.; Ratei, R.; Silvestri, D.; Benetello, A.; Buldini, B.; Maglia, O.; Masera, G.; et al. Risk of Relapse of Childhood Acute Lymphoblastic Leukemia Is Predicted by Flow Cytometric Measurement of Residual Disease on Day 15 Bone Marrow. J. Clin. Oncol. 2009, 27, 5168–5174. [Google Scholar] [CrossRef]
- Wang, H.; Yao, Y.; Mao, L.; Lou, Y.; Ren, Y.; Ye, X.; Yang, M.; Ma, L.; Zhang, Y.; Zhou, Y.; et al. Venetoclax plus “2 + 5” Modified Intensive Chemotherapy with Daunorubicin and Cytarabine in Fit Elderly Patients with Untreated de Novo Acute Myeloid Leukaemia: A Single-Centre Retrospective Analysis. Br. J. Haematol. 2023, 201, 568–572. [Google Scholar] [CrossRef]
- Brüggemann, M.; Schrauder, A.; Raff, T.; Pfeifer, H.; Dworzak, M.; Ottmann, O.G.; Asnafi, V.; Baruchel, A.; Bassan, R.; Benoit, Y.; et al. Standardized MRD Quantification in European ALL Trials: Proceedings of the Second International Symposium on MRD Assessment in Kiel, Germany, 18–20 September 2008. Leukemia 2010, 24, 521–535. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.H.; Pei, D.; Raimondi, S.C.; Coustan-Smith, E.; Jeha, S.; Cheng, C.; Bowman, W.P.; Sandlund, J.T.; Ribeiro, R.C.; Rubnitz, J.E.; et al. Clinical Impact of Minimal Residual Disease in Children with Different Subtypes of Acute Lymphoblastic Leukemia Treated with Response-Adapted Therapy. Leukemia 2017, 31, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Shahkarami, S.; Mehrasa, R.; Younesian, S.; Yaghmaie, M.; Chahardouli, B.; Vaezi, M.; Rezaei, N.; Nikbakht, M.; Alimoghaddam, K.; Ghavamzadeh, A.; et al. Minimal Residual Disease (MRD) Detection Using Rearrangement of Immunoglobulin/T Cell Receptor Genes in Adult Patients with Acute Lymphoblastic Leukemia (ALL). Ann. Hematol. 2018, 97, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.P.; Ross, D.M. Moving Treatment-Free Remission into Mainstream Clinical Practice in CML. Blood 2016, 128, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-H.; Tang, J.-L.; Tien, F.-M.; Kuo, Y.-Y.; Wu, D.-C.; Lin, C.-C.; Tseng, M.-H.; Peng, Y.-L.; Hou, M.-F.; Chuang, Y.-K.; et al. Clinical Implications of Sequential MRD Monitoring by NGS at 2 Time Points after Chemotherapy in Patients with AML. Blood Adv. 2021, 5, 2456–2466. [Google Scholar] [CrossRef] [PubMed]
- Schuurhuis, G.J.; Heuser, M.; Freeman, S.; Béné, M.-C.; Buccisano, F.; Cloos, J.; Grimwade, D.; Haferlach, T.; Hills, R.K.; Hourigan, C.S.; et al. Minimal/Measurable Residual Disease in AML: A Consensus Document from the European LeukemiaNet MRD Working Party. Blood 2018, 131, 1275–1291. [Google Scholar] [CrossRef]
- Gökbuget, N.; Dombret, H.; Bonifacio, M.; Reichle, A.; Graux, C.; Faul, C.; Diedrich, H.; Topp, M.S.; Brüggemann, M.; Horst, H.-A.; et al. Blinatumomab for Minimal Residual Disease in Adults with B-Cell Precursor Acute Lymphoblastic Leukemia. Blood 2018, 131, 1522–1531. [Google Scholar] [CrossRef]
- Litzow, M.R.; Sun, Z.; Mattison, R.J.; Paietta, E.M.; Roberts, K.G.; Zhang, Y.; Racevskis, J.; Lazarus, H.M.; Rowe, J.M.; Arber, D.A.; et al. Blinatumomab for MRD-Negative Acute Lymphoblastic Leukemia in Adults. N. Engl. J. Med. 2024, 391, 320–333. [Google Scholar] [CrossRef]
- Xu, D.; Yang, Y.; Yin, Z.; Tu, S.; Nie, D.; Li, Y.; Huang, Z.; Sun, Q.; Huang, C.; Nie, X.; et al. Risk-Directed Therapy Based on Genetics and MRD Improves the Outcomes of AML1-ETO-Positive AML Patients, a Multi-Center Prospective Cohort Study. Blood Cancer J. 2023, 13, 168. [Google Scholar] [CrossRef]
- Pigazzi, M.; Manara, E.; Buldini, B.; Beqiri, V.; Bisio, V.; Tregnago, C.; Rondelli, R.; Masetti, R.; Putti, M.C.; Fagioli, F.; et al. Minimal Residual Disease Monitored after Induction Therapy by RQ-PCR Can Contribute to Tailor Treatment of Patients with t(8;21) RUNX1-RUNX1T1 Rearrangement. Haematologica 2014, 100, e99–e101. [Google Scholar] [CrossRef]
- Zhu, H.-H.; Zhang, X.-H.; Qin, Y.-Z.; Liu, D.-H.; Jiang, H.; Chen, H.; Jiang, Q.; Xu, L.-P.; Lu, J.; Han, W.; et al. MRD-Directed Risk Stratification Treatment May Improve Outcomes of t(8;21) AML in the First Complete Remission: Results from the AML05 Multicenter Trial. Blood 2013, 121, 4056–4062. [Google Scholar] [CrossRef] [PubMed]
- Boyiadzis, M.; Zhang, M.-J.; Chen, K.; Abdel-Azim, H.; Abid, M.B.; Aljurf, M.; Bacher, U.; Badar, T.; Badawy, S.M.; Battiwalla, M.; et al. Impact of Pre-Transplant Induction and Consolidation Cycles on AML Allogeneic Transplant Outcomes: A CIBMTR Analysis in 3113 AML Patients. Leukemia 2022, 37, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Dillon, L.W.; Gui, G.; Page, K.M.; Ravindra, N.; Wong, Z.C.; Andrew, G.; Mukherjee, D.; Zeger, S.L.; Chaer, F.E.; Spellman, S.; et al. DNA Sequencing to Detect Residual Disease in Adults with Acute Myeloid Leukemia Prior to Hematopoietic Cell Transplant. JAMA 2023, 329, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.-H.; Cheng, Y.-F.; Lu, A.-D.; Wang, Y.; Zuo, Y.-X.; Yan, C.-H.; Wu, J.; Sun, Y.-Q.; Suo, P.; Chen, Y.-H.; et al. Allogeneic Hematopoietic Stem Cell Transplantation Can Improve the Prognosis of High-Risk Pediatric t(8;21) Acute Myeloid Leukemia in First Remission Based on MRD-Guided Treatment. BMC Cancer 2020, 20, 553. [Google Scholar] [CrossRef] [PubMed]
- Perl, A.E.; Larson, R.A.; Podoltsev, N.A.; Strickland, S.; Wang, E.S.; Atallah, E.; Schiller, G.J.; Martinelli, G.; Neubauer, A.; Sierra, J.; et al. Outcomes in Patients with FLT3-Mutated Relapsed/Refractory Acute Myelogenous Leukemia Who Underwent Transplantation in the Phase 3 ADMIRAL Trial of Gilteritinib versus Salvage Chemotherapy. Transplant. Cell. Ther. 2023, 29, 265.e1–265.e10. [Google Scholar] [CrossRef]
- Levis, M.J.; Hamadani, M.; Logan, B.; Jones, R.J.; Singh, A.K.; Litzow, M.; Wingard, J.R.; Papadopoulos, E.B.; Perl, A.E.; Soiffer, R.J.; et al. Gilteritinib as Post-Transplant Maintenance for Acute Myeloid Leukemia with Internal Tandem Duplication Mutation of FLT3. J. Clin. Oncol. 2024, 42, 1766–1775. [Google Scholar] [CrossRef]
- Bercier, P.; De Thé, H. History of Developing Acute Promyelocytic Leukemia Treatment and Role of Promyelocytic Leukemia Bodies. Cancers 2024, 16, 1351. [Google Scholar] [CrossRef]
- Kayser, S.; Conneely, S.E. Management of Acute Promyelocytic Leukemia at Extremes of Age. Cancers 2023, 15, 3637. [Google Scholar] [CrossRef]
- Gill, H.; Raghupathy, R.; Lee, C.Y.Y.; Yung, Y.; Chu, H.-T.; Ni, M.Y.; Xiao, X.; Flores, F.P.; Yim, R.; Lee, P.; et al. Acute Promyelocytic Leukaemia: Population-Based Study of Epidemiology and Outcome with ATRA and Oral-ATO from 1991 to 2021. BMC Cancer 2023, 23, 141. [Google Scholar] [CrossRef]
- Hermsen, J.; Hambley, B. The Coagulopathy of Acute Promyelocytic Leukemia: An Updated Review of Pathophysiology, Risk Stratification, and Clinical Management. Cancers 2023, 15, 3477. [Google Scholar] [CrossRef]
- Fang, H.; Wang, S.A.; Hu, S.; Konoplev, S.N.; Mo, H.; Liu, W.; Zuo, Z.; Xu, J.; Jorgensen, J.L.; Yin, C.C.; et al. Acute Promyelocytic Leukemia: Immunophenotype and Differential Diagnosis by Flow Cytometry. Cytom. Part B Clin. Cytom. 2022, 102, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, J.J.; Chale, R.S.; Abad, A.C.; Schally, A.V. Acute Promyelocytic Leukemia (APL): A Review of the Literature. Oncotarget 2020, 11, 992–1003. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, J.; Xu, H.; Zhao, S.; Wang, L.; Huang, J.; Wang, H.; Tong, H.; Jin, J. Prognosis Influence of Additional Chromosome Abnormalities in Newly Diagnosed Acute Promyelocytic Leukemia with t(15;17)(Q24;Q21). Hematology 2023, 29, 2293513. [Google Scholar] [CrossRef]
- Li, A.Y.; Kashanian, S.M.; Hambley, B.C.; Zacholski, K.; Duong, V.H.; Chaer, F.E.; Holtzman, N.G.; Gojo, I.; Webster, J.A.; Norsworthy, K.J.; et al. FLT3-ITD Allelic Burden and Acute Promyelocytic Leukemia Risk Stratification. Biology 2021, 10, 243. [Google Scholar] [CrossRef]
- Swirsky, D.M.; De Bastos, M.; Parish, S.E.; Rees, J.K.H.; Hayhoe, F.G.J. Features Affecting Outcome during Remission Induction of Acute Myeloid Leukaemia in 619 Adult Patients. Br. J. Haematol. 1986, 64, 435–453. [Google Scholar] [CrossRef]
- Mertelsmann, R.; Tzvi Thaler, H.; To, L.; Gee, T.S.; McKenzie, S.; Schauer, P.; Friedman, A.; Arlin, Z.; Cirrincione, C.; Clarkson, B. Morphological classification, response to therapy, and survival in 263 adult patients with acute nonlymphoblastic leukemia. Blood 1980, 56, 773–781. [Google Scholar] [CrossRef]
- Morrison, F.S.; Kopecky, K.J.; Head, D.R.; Athens, J.W.; Balcerzak, S.P.; Gumbart, C.; Dabich, L.; Costanzi, J.J.; Coltman, C.A.; Saiki, J.H. Late intensification with POMP chemotherapy prolongs survival in acute myelogenous leukemia—Results of a Southwest Oncology Group study of rubidazone versus adriamycin for remission induction, prophylactic intrathecal therapy, late intensification, and levamisole maintenance. Leukemia 1992, 6, 708–714. [Google Scholar]
- Head, D.; Kopecky, K.J.; Weick, J.; Files, J.C.; Ryan, D.; Foucar, K.; Montiel, M.; Bickers, J.; Fishleder, A.; Miller, M. Effect of aggressive daunomycin therapy on survival in acute promyelocytic leukemia. Blood 1995, 86, 1717–1728. [Google Scholar] [CrossRef]
- Nagai, Y.; Ambinder, A.J. The Promise of Retinoids in the Treatment of Cancer: Neither Burnt out nor Fading Away. Cancers 2023, 15, 3535. [Google Scholar] [CrossRef]
- Liang, C.; Qiao, G.; Liu, Y.; Tian, L.; Hui, N.; Li, J.; Ma, Y.; Li, H.; Zhao, Q.; Cao, W.; et al. Overview of All-Trans-Retinoic Acid (ATRA) and Its Analogues: Structures, Activities, and Mechanisms in Acute Promyelocytic Leukaemia. Eur. J. Med. Chem. 2021, 220, 113451. [Google Scholar] [CrossRef]
- Woods, A.C.; Norsworthy, K.J. Differentiation Syndrome in Acute Leukemia: APL and Beyond. Cancers 2023, 15, 4767. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.E.; Ye, Y.C.; Chen, S.R.; Chai, J.R.; Lu, J.X.; Zhoa, L.; Gu, L.J.; Wang, Z.Y. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood 1988, 72, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Tsimberidou, A.-M.; Tirado-Gomez, M.; Andreeff, M.; O’Brien, S.; Kantarjian, H.; Keating, M.; Lopez-Berestein, G.; Estey, E. Single-Agent Liposomal All-Trans Retinoic Acid Can Cure Some Patients with Untreated Acute Promyelocytic Leukemia: An Update of the University of Texas M. D. Anderson Cancer Center Series. Leuk. Lymphoma 2006, 47, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.H.; Kakizuka, A.; Frankel, S.R.; Warrell, R.P.; DeBlasio, A.; Levine, K.; Evans, R.M.; Dmitrovsky, E. Reverse Transcription Polymerase Chain Reaction for the Rearranged Retinoic Acid Receptor Alpha Clarifies Diagnosis and Detects Minimal Residual Disease in Acute Promyelocytic Leukemia. Proc. Natl. Acad. Sci. USA 1992, 89, 2694–2698. [Google Scholar] [CrossRef] [PubMed]
- Lo Coco, F.; Diverio, D.; Avvisati, G.; Arcese, W.; Petti, M.C.; Meloni, G.; Mandelli, F.; Pandolfi, P.P.; Grignani, F.; Pelicci, P.G.; et al. Molecular Evaluation of Residual Disease as a Predictor of Relapse in Acute Promyelocytic Leukaemia. Lancet 1992, 340, 1437–1438. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Chen, Z. Acute Promyelocytic Leukemia: From Highly Fatal to Highly Curable. Blood 2008, 111, 2505–2515. [Google Scholar] [CrossRef]
- Grimwade, D.; Howe, K.; Langabeer, S.; Burnett, A.; Goldstone, A.; Solomon, E. Minimal residual disease detection in acute promyelocytic leukemia by reverse-transcriptase PCR: Evaluation of PML-RAR alpha and RAR alpha-PML assessment in patients who ultimately relapse. Leukemia 1996, 10, 61–66. [Google Scholar]
- Diverio, D.; Rossi, V.; Avvisati, G.; De Santis, S.; Pistilli, A.; Pane, F.; Saglio, G.; Martinelli, G.; Petti, M.C.; Santoro, A.; et al. Early detection of relapse by prospective reverse transcriptase-polymerase chain reaction analysis of the PML/RARalpha fusion gene in patients with acute promyelocytic leukemia enrolled in the GIMEMA-AIEOP multicenter “AIDA” trial. Blood 1998, 92, 784–789. [Google Scholar] [CrossRef]
- Lo Coco, F.; Diverio, D.; Avvisati, G.; Petti, M.C.; Meloni, G.; Pogliani, E.M.; Biondi, A.; Rossi, G.; Carlo-Stella, C.; Selleri, C.; et al. Therapy of molecular relapse in acute promyelocytic leukemia. Blood 1999, 94, 2225–2229. [Google Scholar] [CrossRef]
- Grimwade, D.; Jovanovic, J.V.; Hills, R.K.; Nugent, E.A.; Patel, Y.; Flora, R.; Diverio, D.; Jones, K.; Aslett, H.; Batson, E.; et al. Prospective Minimal Residual Disease Monitoring to Predict Relapse of Acute Promyelocytic Leukemia and to Direct Pre-Emptive Arsenic Trioxide Therapy. J. Clin. Oncol. 2009, 27, 3650–3658. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, S.Y.; Hu, L.H. Studies on the clinical practice and mechanisms of 713 (As2O3) in the treatment of 117 cases of APL. J. Harbin Med. Univ. 1995, 29, 243. [Google Scholar]
- Mathews, V.; George, B.; Lakshmi, K.M.; Viswabandya, A.; Bajel, A.; Balasubramanian, P.; Shaji, R.V.; Srivastava, V.M.; Srivastava, A.; Chandy, M. Single-Agent Arsenic Trioxide in the Treatment of Newly Diagnosed Acute Promyelocytic Leukemia: Durable Remissions with Minimal Toxicity. Blood 2006, 107, 2627–2632. [Google Scholar] [CrossRef] [PubMed]
- Lallemand-Breitenbach, V.; Jeanne, M.; Benhenda, S.; Nasr, R.; Lei, M.; Peres, L.; Zhou, J.; Zhu, J.; Raught, B. Arsenic Degrades PML or PML–RARα through a SUMO-Triggered RNF4/Ubiquitin-Mediated Pathway. Nat. Cell Biol. 2008, 10, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.; Mishra, N.; Shelke, R.; Varma, A.K. Mutations at Proximal Cysteine Residues in PML Impair ATO Binding by Destabilizing the RBCC Domain. FEBS J. 2023, 291, 1422–1438. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Q.; Shi, X.G.; Tang, W.; Xiong, S.M.; Zhu, J.; Cai, X.; Han, Z.G.; Ni, J.H.; Shi, G.Y.; Jia, P.M.; et al. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL): I. As2O3 exerts dose-dependent dual effects on APL cells. Blood 1997, 89, 3345–3353. [Google Scholar]
- Goussetis, D.J.; Altman, J.K.; Glaser, H.; McNeer, J.L.; Tallman, M.S.; Platanias, L.C. Autophagy Is a Critical Mechanism for the Induction of the Antileukemic Effects of Arsenic Trioxide. J. Biol. Chem. 2010, 285, 29989–29997. [Google Scholar] [CrossRef]
- Yan, M.; Wang, H.; Wei, R.; Li, W. Arsenic Trioxide: Applications, Mechanisms of Action, Toxicity and Rescue Strategies to Date. Arch. Pharmacal Res. 2023, 47, 249–271. [Google Scholar] [CrossRef]
- Burnett, A.K.; Russell, N.H.; Hills, R.K.; Bowen, D.; Kell, J.; Knapper, S.; Morgan, Y.G.; Lok, J.; Grech, A.; Jones, G.; et al. Arsenic Trioxide and All-Trans Retinoic Acid Treatment for Acute Promyelocytic Leukaemia in All Risk Groups (AML17): Results of a Randomised, Controlled, Phase 3 Trial. Lancet Oncol. 2015, 16, 1295–1305. [Google Scholar] [CrossRef]
- Ravandi, F.; Estey, E.; Jones, D.; Faderl, S.; O’Brien, S.; Fiorentino, J.; Pierce, S.; Blamble, D.; Estrov, Z.; Wierda, W.; et al. Effective Treatment of Acute Promyelocytic Leukemia with All-Trans-Retinoic Acid, Arsenic Trioxide, and Gemtuzumab Ozogamicin. J. Clin. Oncol. 2009, 27, 504–510. [Google Scholar] [CrossRef]
- Lo-Coco, F.; Avvisati, G.; Vignetti, M.; Thiede, C.; Orlando, S.M.; Iacobelli, S.; Ferrara, F.; Fazi, P.; Cicconi, L.; Di Bona, E.; et al. Retinoic Acid and Arsenic Trioxide for Acute Promyelocytic Leukemia. N. Engl. J. Med. 2013, 369, 111–121. [Google Scholar] [CrossRef]
- Sanz, M.A.; Martín, G.; González, M.; León, A.; Rayón, C.; Rivas, C.; Colomer, D.; Amutio, E.; Capote, F.J.; Milone, G.A.; et al. Risk-Adapted Treatment of Acute Promyelocytic Leukemia with All-Trans-Retinoic Acid and Anthracycline Monochemotherapy: A Multicenter Study by the PETHEMA Group. Blood 2003, 103, 1237–1243. [Google Scholar] [CrossRef] [PubMed]
- Heuser, M.; Freeman, S.D.; Ossenkoppele, G.J.; Buccisano, F.; Hourigan, C.S.; Ngai, L.L.; Tettero, J.M.; Bachas, C.; Baer, C.; Béné, M.-C.; et al. 2021 Update on MRD in Acute Myeloid Leukemia: A Consensus Document from the European LeukemiaNet MRD Working Party. Blood 2021, 138, 2753–2767. [Google Scholar] [CrossRef] [PubMed]
- Fialkow, P.J.; Gartler, S.M.; Yoshida, A. Clonal Origin of Chronic Myelocytic Leukemia in Man. Proc. Natl. Acad. Sci. USA 1967, 58, 1468–1471. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A Cell Initiating Human Acute Myeloid Leukaemia after Transplantation into SCID Mice. Nature 1994, 367, 645–648. [Google Scholar] [CrossRef]
- Boyd, A.L.; Campbell, C.J.V.; Hopkins, C.I.; Fiebig-Comyn, A.; Russell, J.; Ulemek, J.; Foley, R.; Leber, B.; Xenocostas, A.; Collins, T.J.; et al. Niche Displacement of Human Leukemic Stem Cells Uniquely Allows Their Competitive Replacement with Healthy HSPCs. J. Exp. Med. 2014, 211, 1925–1935. [Google Scholar] [CrossRef]
- Gerber, J.M.; Smith, B.D.; Ngwang, B.; Zhang, H.; Vala, M.S.; Morsberger, L.; Galkin, S.; Collector, M.I.; Perkins, B.; Levis, M.J.; et al. A Clinically Relevant Population of Leukemic CD34+CD38− Cells in Acute Myeloid Leukemia. Blood 2012, 119, 3571–3577. [Google Scholar] [CrossRef]
- Jordan, C.; Upchurch, D.; Szilvassy, S.; Guzman, M.; Howard, D.; Pettigrew, A.; Meyerrose, T.; Rossi, R.; Grimes, B.; Rizzieri, D.; et al. The Interleukin-3 Receptor Alpha Chain Is a Unique Marker for Human Acute Myelogenous Leukemia Stem Cells. Leukemia 2000, 14, 1777–1784. [Google Scholar] [CrossRef]
- Jin, L.; Hope, K.J.; Zhai, Q.; Smadja-Joffe, F.; Dick, J.E. Targeting of CD44 Eradicates Human Acute Myeloid Leukemic Stem Cells. Nat. Med. 2006, 12, 1167–1174. [Google Scholar] [CrossRef]
- Hosen, N.; Park, C.Y.; Tatsumi, N.; Oji, Y.; Sugiyama, H.; Gramatzki, M.; Krensky, A.M.; Weissman, I.L. CD96 Is a Leukemic Stem Cell-Specific Marker in Human Acute Myeloid Leukemia. Proc. Natl. Acad. Sci. USA 2007, 104, 11008–11013. [Google Scholar] [CrossRef]
- Van Rhenen, A.; Van Dongen, G.a.M.S.; Kelder, A.; Rombouts, E.J.; Feller, N.; Moshaver, B.; Walsum, M.S.-V.; Zweegman, S.; Ossenkoppele, G.J.; Schuurhuis, G.J. The Novel AML Stem Cell–Associated Antigen CLL-1 Aids in Discrimination between Normal and Leukemic Stem Cells. Blood 2007, 110, 2659–2666. [Google Scholar] [CrossRef]
- Majeti, R.; Chao, M.P.; Alizadeh, A.A.; Pang, W.W.; Jaiswal, S.; Gibbs, K.D.; Van Rooijen, N.; Weissman, I.L. CD47 Is an Adverse Prognostic Factor and Therapeutic Antibody Target on Human Acute Myeloid Leukemia Stem Cells. Cell 2009, 138, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Pabst, C.; Bergeron, A.; Lavallée, V.-P.; Yeh, J.; Gendron, P.; Norddahl, G.L.; Krosl, J.; Boivin, I.; Deneault, E.; Simard, J.; et al. GPR56 Identifies Primary Human Acute Myeloid Leukemia Cells with High Repopulating Potential In Vivo. Blood 2016, 127, 2018–2027. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, B.; Ghiaur, G.; Smith, B.D.; Jones, R.J. Translating Leukemia Stem Cells into the Clinical Setting: Harmonizing the Heterogeneity. Exp. Hematol. 2016, 44, 1130–1137. [Google Scholar] [CrossRef]
- Velten, L.; Story, B.A.; Hernández-Malmierca, P.; Raffel, S.; Leonce, D.R.; Milbank, J.; Paulsen, M.; Demir, A.; Szu-Tu, C.; Frömel, R.; et al. Identification of Leukemic and Pre-Leukemic Stem Cells by Clonal Tracking from Single-Cell Transcriptomics. Nat. Commun. 2021, 12, 1366. [Google Scholar] [CrossRef]
- Han, L.; Qiu, P.; Zeng, Z.; Jorgensen, J.L.; Mak, D.H.; Burks, J.K.; Schober, W.; McQueen, T.J.; Cortes, J.; Tanner, S.D.; et al. Single-cell Mass Cytometry Reveals Intracellular Survival/Proliferative Signaling in FLT3-ITD-mutated AML Stem/Progenitor Cells. Cytom. Part A 2015, 87, 346–356. [Google Scholar] [CrossRef]
- Ghiaur, G.; Gerber, J.; Jones, R.J. Concise Review: Cancer Stem Cells and Minimal Residual Disease. Stem Cells 2011, 30, 89–93. [Google Scholar] [CrossRef]
- Gerber, J.M.; Zeidner, J.F.; Morse, S.; Blackford, A.L.; Perkins, B.; Yanagisawa, B.; Zhang, H.; Morsberger, L.; Karp, J.; Ning, Y.; et al. Association of Acute Myeloid Leukemias Most Immature Phenotype with Risk Groups and Outcomes. Haematologica 2016, 101, 607–616. [Google Scholar] [CrossRef]
- Borowitz, M.J.; Shuster, J.J.; Civin, C.I.; Carroll, A.J.; Look, A.T.; Behm, F.G.; Land, V.J.; Pullen, D.J.; Crist, W.M. Prognostic Significance of CD34 Expression in Childhood B-Precursor Acute Lymphocytic Leukemia: A Pediatric Oncology Group Study. J. Clin. Oncol. 1990, 8, 1389–1398. [Google Scholar] [CrossRef]
- Vergez, F.; Green, A.S.; Tamburini, J.; Sarry, J.-e.; Gaillard, B.; Cornillet-Lefebvre, P.; Pannetier, M.; Neyret, A.; Chapuis, N.; Ifrah, N.; et al. High Levels of CD34+CD38low/−CD123+ Blasts Are Predictive of an Adverse Outcome in Acute Myeloid Leukemia: A Groupe Ouest-Est Des Leucemies Aigues et Maladies Du Sang (GOELAMS) Study. Haematologica 2011, 96, 1792–1798. [Google Scholar] [CrossRef]
- Schepers, K.; Campbell, T.B.; Passegué, E. Normal and Leukemic Stem Cell Niches: Insights and Therapeutic Opportunities. Cell Stem Cell 2015, 16, 254–267. [Google Scholar] [CrossRef]
- Ghiaur, G.; Wroblewski, M.; Loges, S. Acute Myelogenous Leukemia and Its Microenvironment: A Molecular Conversation. Semin. Hematol. 2015, 52, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Ladikou, E.E.; Chevassut, T.; Pepper, C.J.; Pepper, A.G. Dissecting the Role of the CXCL12/CXCR4 Axis in Acute Myeloid Leukaemia. Br. J. Haematol. 2020, 189, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Malfuson, J.; Boutin, L.; Clay, D.; Thépenier, C.; Desterke, C.; Torossian, F.; Guerton, B.; Anginot, A.; De Revel, T.; Lataillade, J.; et al. SP/Drug Efflux Functionality of Hematopoietic Progenitors Is Controlled by Mesenchymal Niche through VLA-4/CD44 Axis. Leukemia 2013, 28, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Kipps, T.J. CXCR4: A Key Receptor in the Crosstalk between Tumor Cells and Their Microenvironment. Blood 2006, 107, 1761–1767. [Google Scholar] [CrossRef] [PubMed]
- Ghiaur, G.; Yegnasubramanian, S.; Perkins, B.; Gucwa, J.L.; Gerber, J.M.; Jones, R.J. Regulation of Human Hematopoietic Stem Cell Self-Renewal by the Microenvironment’s Control of Retinoic Acid Signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 16121–16126. [Google Scholar] [CrossRef]
- Su, M.; Alonso, S.; Jones, J.W.; Yu, J.; Kane, M.A.; Jones, R.J.; Ghiaur, G. All-Trans Retinoic Acid Activity in Acute Myeloid Leukemia: Role of Cytochrome P450 Enzyme Expression by the Microenvironment. PLoS ONE 2015, 10, e0127790. [Google Scholar] [CrossRef]
- Hernandez, D.; Palau, L.; Norsworthy, K.; Anders, N.M.; Alonso, S.; Su, M.; Petkovich, M.; Chandraratna, R.; Rudek, M.A.; Smith, B.D.; et al. Overcoming Microenvironment-Mediated Protection from ATRA Using CYP26-Resistant Retinoids. Leukemia 2020, 34, 3077–3081. [Google Scholar] [CrossRef]
- Konopleva, M.; Konoplev, S.; Hu, W.; Zaritskey, A.; Afanasiev, B.; Andreeff, M. Stromal Cells Prevent Apoptosis of AML Cells by Up-Regulation of Anti-Apoptotic Proteins. Leukemia 2002, 16, 1713–1724. [Google Scholar] [CrossRef]
- Tabe, Y.; Konopleva, M. Resistance to Energy Metabolism—Targeted Therapy of AML Cells Residual in the Bone Marrow Microenvironment. Cancer Drug Resist. 2023, 6, 138–150. [Google Scholar] [CrossRef]
- Alonso, S.; Su, M.; Jones, J.W.; Ganguly, S.; Kane, M.A.; Jones, R.J.; Ghiaur, G. Human Bone Marrow Niche Chemoprotection Mediated by Cytochrome P450 Enzymes. Oncotarget 2015, 6, 14905–14912. [Google Scholar] [CrossRef]
- Chang, Y.-T.; Hernandez, D.; Alonso, S.; Gao, M.; Su, M.; Ghiaur, G.; Levis, M.J.; Jones, R.J. Role of CYP3A4 in Bone Marrow Microenvironment–Mediated Protection of FLT3/ITD AML from Tyrosine Kinase Inhibitors. Blood Adv. 2019, 3, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.-W.; Shi, J.; Chen, J.; Wang, B.; Yu, Y.-H.; Qin, X.; Zhou, X.-C.; Cai, Y.-J.; Li, Z.-Q.; Zhang, F.; et al. Leukemia Propagating Cells Rebuild an Evolving Niche in Response to Therapy. Cancer Cell 2014, 25, 778–793. [Google Scholar] [CrossRef] [PubMed]
- Duarte, D.; Hawkins, E.D.; Lo Celso, C. The Interplay of Leukemia Cells and the Bone Marrow Microenvironment. Blood 2018, 131, 1507–1511. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Chang, Y.; Hernandez, D.; Jones, R.J.; Ghiaur, G. Regulation of Drug Metabolizing Enzymes in the Leukaemic Bone Marrow Microenvironment. J. Cell. Mol. Med. 2019, 23, 4111–4117. [Google Scholar] [CrossRef] [PubMed]
- Vago, L.; Gojo, I. Immune Escape and Immunotherapy of Acute Myeloid Leukemia. J. Clin. Investig. 2020, 130, 1552–1564. [Google Scholar] [CrossRef]
- Riether, C. Regulation of Hematopoietic and Leukemia Stem Cells by Regulatory T Cells. Front. Immunol. 2022, 13, 1049301. [Google Scholar] [CrossRef]
- Cancilla, D.; Rettig, M.P.; DiPersio, J.F. Targeting CXCR4 in AML and ALL. Front. Oncol. 2020, 10, 1672. [Google Scholar] [CrossRef]
- Müller, L.; Tunger, A.; Wobus, M.; Von Bonin, M.; Towers, R.; Bornhäuser, M.; Dazzi, F.; Wehner, R.; Schmitz, M. Immunomodulatory Properties of Mesenchymal Stromal Cells: An Update. Front. Cell Dev. Biol. 2021, 9, 637725. [Google Scholar] [CrossRef]
- Passaro, D.; Di Tullio, A.; Abarrategi, A.; Rouault-Pierre, K.; Foster, K.; Ariza-McNaughton, L.; Montaner, B.; Chakravarty, P.; Bhaw, L.; Diana, G.; et al. Increased Vascular Permeability in the Bone Marrow Microenvironment Contributes to Disease Progression and Drug Response in Acute Myeloid Leukemia. Cancer Cell 2017, 32, 324–341.e6. [Google Scholar] [CrossRef]
- Tettamanti, S.; Pievani, A.; Biondi, A.; Dotti, G.; Serafini, M. Catch Me If You Can: How AML and Its Niche Escape Immunotherapy. Leukemia 2021, 36, 13–22. [Google Scholar] [CrossRef]
- Uy, G.L.; Rettig, M.P.; Motabi, I.H.; McFarland, K.; Trinkaus, K.M.; Hladnik, L.M.; Kulkarni, S.; Abboud, C.N.; Cashen, A.F.; Stockerl-Goldstein, K.E.; et al. A Phase 1/2 Study of Chemosensitization with the CXCR4 Antagonist Plerixafor in Relapsed or Refractory Acute Myeloid Leukemia. Blood 2012, 119, 3917–3924. [Google Scholar] [CrossRef] [PubMed]
- Alonso, S.; Hernandez, D.; Chang, Y.-T.; Gocke, C.D.; McCray, M.; Varadhan, R.; Matsui, W.H.; Jones, R.J.; Ghiaur, G. Hedgehog and Retinoid Signaling Alters Multiple Myeloma Microenvironment and Generates Bortezomib Resistance. J. Clin. Investig. 2016, 126, 4460–4468. [Google Scholar] [CrossRef] [PubMed]
- Menezes, D.L.; See, W.L.; Risueño, A.; Tsai, K.T.; Lee, J.K.; Ma, J.; Khan, R.; Prebet, T.; Skikne, B.; Beach, C.L.; et al. Oral Azacitidine Modulates the Bone Marrow Microenvironment in Patients with Acute Myeloid Leukaemia in Remission: A Subanalysis from the QUAZAR AML-001 Trial. Br. J. Haematol. 2023, 201, 1129–1143. [Google Scholar] [CrossRef] [PubMed]
- Hohtari, H.; Brück, O.; Blom, S.; Turkki, R.; Sinisalo, M.; Kovanen, P.E.; Kallioniemi, O.; Pellinen, T.; Porkka, K.; Mustjoki, S. Immune Cell Constitution in Bone Marrow Microenvironment Predicts Outcome in Adult ALL. Leukemia 2019, 33, 1570–1582. [Google Scholar] [CrossRef] [PubMed]
- Sendker, S.; Reinhardt, D.; Niktoreh, N. Redirecting the Immune Microenvironment in Acute Myeloid Leukemia. Cancers 2021, 13, 1423. [Google Scholar] [CrossRef] [PubMed]
- Perzolli, A.; Koedijk, J.B.; Zwaan, C.M.; Heidenreich, O. Targeting the Innate Immune System in Pediatric and Adult AML. Leukemia 2024, 38, 1191–1201. [Google Scholar] [CrossRef]
- Kenderian, S.S.; Ruella, M.; Shestova, O.; Klichinsky, M.; Aikawa, V.; Morrissette, J.J.D.; Scholler, J.; Song, D.; Porter, D.L.; Carroll, M.; et al. CD33-Specific Chimeric Antigen Receptor T Cells Exhibit Potent Preclinical Activity against Human Acute Myeloid Leukemia. Leukemia 2015, 29, 1637–1647. [Google Scholar] [CrossRef]
- Mardiros, A.; Santos, C.D.; McDonald, T.; Brown, C.E.; Wang, X.; Budde, L.E.; Hoffman, L.; Aguilar, B.; Chang, W.-C.; Bretzlaff, W.; et al. T Cells Expressing CD123-Specific Chimeric Antigen Receptors Exhibit Specific Cytolytic Effector Functions and Antitumor Effects against Human Acute Myeloid Leukemia. Blood 2013, 122, 3138–3148. [Google Scholar] [CrossRef]
- Lemoli, R.M.; Montesinos, P.; Jain, A. Real-World Experience with CPX-351 in High-Risk Acute Myeloid Leukemia. Crit. Rev. Oncol./Hematol. 2023, 185, 103984. [Google Scholar] [CrossRef]
- Ambinder, A.J.; Norsworthy, K.; Hernandez, D.; Palau, L.; Paun, B.; Duffield, A.; Chandraratna, R.; Sanders, M.; Varadhan, R.; Jones, R.J.; et al. A Phase 1 Study of IRX195183, a RARA-Selective CYP26 Resistant Retinoid, in Patients with Relapsed or Refractory AML. Front. Oncol. 2020, 10, 587062. [Google Scholar] [CrossRef]
- De Botton, S.; Cluzeau, T.; Vigil, C.; Cook, R.J.; Rousselot, P.; Rizzieri, D.A.; Liesveld, J.L.; Fenaux, P.; Braun, T.; Banos, A.; et al. Targeting RARA Overexpression with Tamibarotene, a Potent and Selective RARα Agonist, Is a Novel Approach in AML. Blood Adv. 2023, 7, 1858–1870. [Google Scholar] [CrossRef]
- Sanford, D.; Lo-Coco, F.; Sanz, M.A.; Di Bona, E.; Coutre, S.; Altman, J.K.; Wetzler, M.; Allen, S.L.; Ravandi, F.; Kantarjian, H.; et al. Tamibarotene in Patients with Acute Promyelocytic Leukaemia Relapsing after Treatment with All-trans Retinoic Acid and Arsenic Trioxide. Br. J. Haematol. 2015, 171, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Kegyes, D.; Milea, P.A.; Mazga, A.-I.; Tigu, A.-B.; Nistor, M.; Cenariu, D.; Tomai, R.; Buruiana, S.; Einsele, H.; Tănase, A.D.; et al. Looking Ahead to Targeting Macrophages by CAR T- or NK-Cells in Blood Cancers. Expert Opin. Ther. Targets 2024, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wahiduzzaman, M.; Ota, A.; Hosokawa, Y. Novel Mechanistic Insights into the Anti-Cancer Mode of Arsenic Trioxide. Curr. Cancer Drug Targets 2020, 20, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Moosavi, M.A.; Djavaheri-Mergny, M. Autophagy: New Insights into Mechanisms of Action and Resistance of Treatment in Acute Promyelocytic leukemia. Int. J. Mol. Sci. 2019, 20, 3559. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.J.; Kim, J.; Gardner, D.; Beachy, P.A. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc. Natl. Acad. Sci. USA 2010, 107, 13432–13437. [Google Scholar] [CrossRef]
| Mutation | Disease(s) in Which More Prevalent |
|---|---|
| RAS (KRAS, NRAS) | AML, MDS, CHIP |
| SF3B1 | AML, MDS, CHIP |
| SRSF2 | AML, MDS, CHIP |
| ASXL1 | AML, MDS, CHIP |
| DNMT3A | AML, MDS, CHIP |
| TP53 | AML, MDS, CHIP |
| TET2 | AML, MDS, CHIP |
| U2AF1 | AML, MDS, CHIP |
| JAK2 | AML, MDS, CHIP |
| PHF6 | AML, MDS |
| GATA2 | AML, MDS |
| RUNX1-RUNX1T1 | AML, MDS |
| FLT3-ITD | AML, MDS |
| CEBPA | AML, MDS |
| EZH2 | AML, MDS |
| IDH 1 and IDH 2 | AML, MDS |
| ETV6 | AML, MDS |
| PHF6 | AML, MDS |
| ZRSR2 | AML, MDS |
| BCOR | AML, MDS |
| CHEK2 | CHIP |
| ATM | CHIP |
| VAF | CHIP |
| TERT | CHIP |
| SMC4 | CHIP |
| CD164 | CHIP |
| NPAT | CHIP |
| PARP1 | CHIP |
| KDM6A | MDS |
| STAG2 | MDS |
| RAD21 | MDS |
| WT1 | MDS |
| DEK-NUP214 | AML |
| CBFB-MYH11 | AML |
| NPM1 | AML |
| MLLT3-KMT2A | AML |
| MECOM (EVI1) | AML |
| Lessons Learned from APL | Concept | Current Trends in Leukemias |
|---|---|---|
| High-dose cytarabine leads to lower CR and higher relapse rates | More is not always better | Need for MRD-directed randomized clinical trials |
| ATRA induces differentiation syndrome | Inhibiting driver mutations leads to differentiation syndrome | FLT3-, IDH-, and menin-inhibitors induce differentiation syndrome |
| Single-agent ATRA remission without cure | Inhibiting driver mutations alone is not enough to eliminate MRD | FLT3-, IDH-, and menin-inhibitors cannot eliminate MRD as single agents |
| RT-PCR for PML-RARα | Molecular methods to detect MRD | PCR and NGS for somatic mutations (NPM1, FLT3) |
CYP26 protects LSCs from ATRA
| BME creates a biochemical barrier to protect MRD | CYP3A4 protects MRD from chemotherapy and targeted agents
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kegyes, D.; Thiagarajan, P.S.; Ghiaur, G. MRD in Acute Leukemias: Lessons Learned from Acute Promyelocytic Leukemia. Cancers 2024, 16, 3208. https://doi.org/10.3390/cancers16183208
Kegyes D, Thiagarajan PS, Ghiaur G. MRD in Acute Leukemias: Lessons Learned from Acute Promyelocytic Leukemia. Cancers. 2024; 16(18):3208. https://doi.org/10.3390/cancers16183208
Chicago/Turabian StyleKegyes, David, Praveena S. Thiagarajan, and Gabriel Ghiaur. 2024. "MRD in Acute Leukemias: Lessons Learned from Acute Promyelocytic Leukemia" Cancers 16, no. 18: 3208. https://doi.org/10.3390/cancers16183208
APA StyleKegyes, D., Thiagarajan, P. S., & Ghiaur, G. (2024). MRD in Acute Leukemias: Lessons Learned from Acute Promyelocytic Leukemia. Cancers, 16(18), 3208. https://doi.org/10.3390/cancers16183208






