FOXM1 Transcriptionally Co-Upregulates Centrosome Amplification and Clustering Genes and Is a Biomarker for Poor Prognosis in Androgen Receptor-Low Triple-Negative Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Partners in Crime: Centrosome Amplification and Centrosome Clustering Collude to Drive Aggressive Breast Cancer
3. Connecting the Dots: The Chromosome Passenger Complex (CPC) and KIFC1 Are Key Drivers of Centrosome Clustering and Transcriptional Targets of FOXM1
4. The Accomplices: CPC Components AURKB, BIRC5 and CDCA8 Play Distinct Roles in Promoting Mitotic Progression and Genomic Instability in Cancer Cells
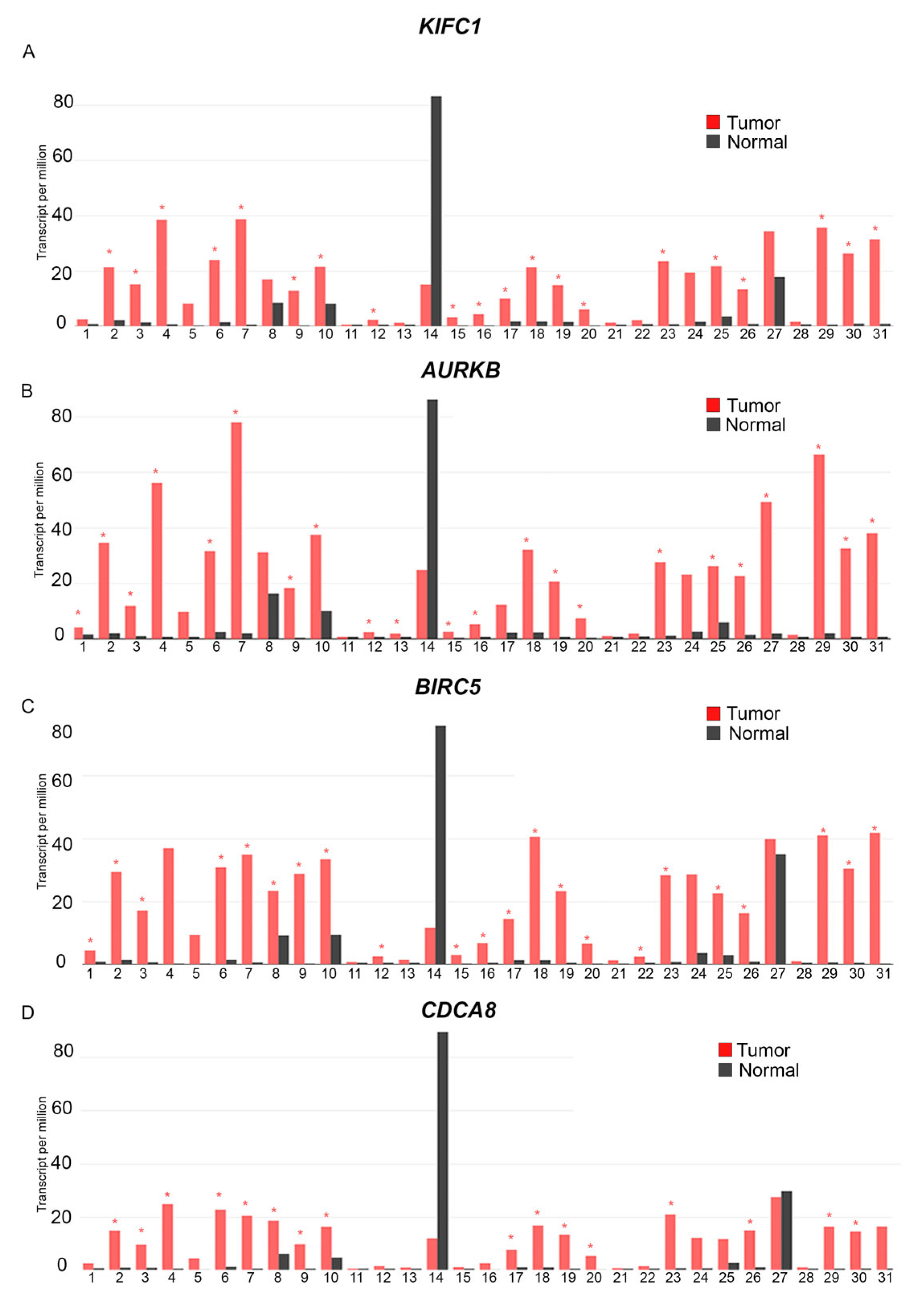
5. Acting in Concert: The Centrosome Clustering Proteins KIFC1, AURKB, BIRC5, and CDCA8 Are Overexpressed in a Variety of Tumor Tissues, Including Breast Cancers
- 1: Adenoid Cystic Carcinoma, Tumor (n = 77), Normal (n = 88).
- 2: Bladder Urothelial Carcinoma, Tumor (n = 404), Normal (n = 28)
- 3: Breast Invasive Carcinoma, Tumor (n = 1085), Normal (n = 291)
- 4: Cervical squamous cell carcinoma and endocervical adenocarcinoma, Tumor (n = 306), Normal (n = 13)
- 5: Cholangiocarcinoma, Tumor (n = 36), Normal (n = 9)
- 6: Colon adenocarcinoma, Tumor (n = 275), Normal (n = 349)
- 7: Lymphoid Neoplasm Diffuse Large B-cell Lymphoma, Tumor (n = 47), Normal (n = 337)
- 8: Esophageal carcinoma) Tumor (n = 182), Normal (n = 286)
- 9: Glioblastoma Multiforme, Tumor (n = 163), Normal (n = 207)
- 10: Head and Neck squamous cell carcinoma, Tumor (n = 519), Normal (n = 44)
- 11: Kidney Chromophobe, Tumor (n = 66), Normal (n =5 3)
- 12: Kidney renal clear cell carcinoma, Tumor (n = 523), Normal (n = 100)
- 13: Kidney renal clear cell carcinoma, Tumor (n = 286), Normal (n = 60)
- 14: Acute Myeloid Leukemia, Tumor (n = 173), Normal (n = 70)
- 15: Brain Lower Grade Glioma, Tumor (n = 518), Normal (n = 207)
- 16: Liver hepatocellular carcinoma, Tumor (n = 369), Normal (n = 160)
- 17: Lung adenocarcinoma, Tumor (n = 485), Normal (n = 347)
- 18: Lung squamous cell carcinoma, Tumor (n = 486), Normal (n = 338)
- 19: Ovarian serous cystadenocarcinoma, Tumor (n = 426), Normal (n = 88)
- 20: Pancreatic adenocarcinoma, Tumor (n = 179), Normal (n = 171)
- 21: Pheochromocytoma and Paraganglioma, Tumor (n = 182), Normal (n = 3)
- 22: Prostate adenocarcinoma, Tumor (n =4 92), Normal (n = 152)
- 23: Rectum adenocarcinoma, Tumor (n = 92), Normal (n = 318)
- 24: Sarcoma, Tumor (n = 261), Normal (n = 2)
- 25: Skin Cutaneous Melanoma, Tumor (n = 461), Normal (n = 558)
- 26: Stomach Adenocarcinoma, Tumor (n = 408), Normal (n = 211)
- 27: Testicular Germ Cell Tumors, Tumor (n = 137) Normal (n = 165)
- 28: Thyroid Cutaneous Carcinoma, Tumor (n = 512), Normal (n = 337)
- 29: Thymoma, Tumor (n = 118), Normal (n = 339)
- 30: Uterine Corpus Endometrial Carcinoma, Tumor (n = 174), Normal (n = 91)
- 31: Uterine Carcinosarcoma, Tumor (n = 57), Normal (n = 78)
6. Aiding and Abetting: Overexpression of Centrosome Clustering Proteins KIFC1, AURKB, BIRC5, and CDCA8 Is Associated with Poor Prognosis, Triple-Negative Status, and TP53 Mutant Status of Breast Cancers
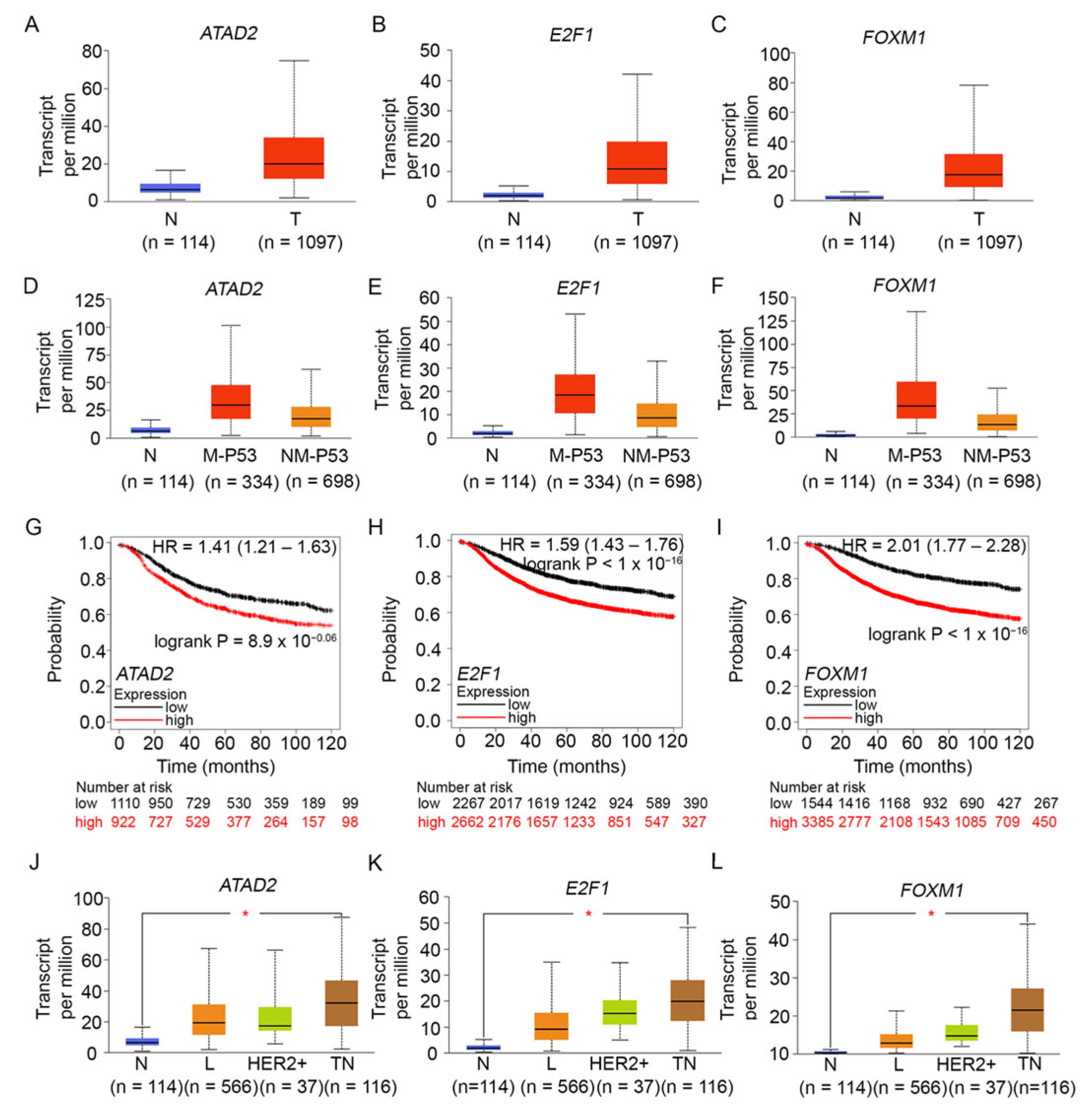
7. Building Alliances: Oncogenic Proteins FOXM1, E2F1, and ATAD2 Are Overexpressed in Breast Tumors, Especially Those with Mutant TP53, and Are Associated with a Poor Prognosis
8. A Team Effort: CPC Genes Interact with p53 in Distinct Ways
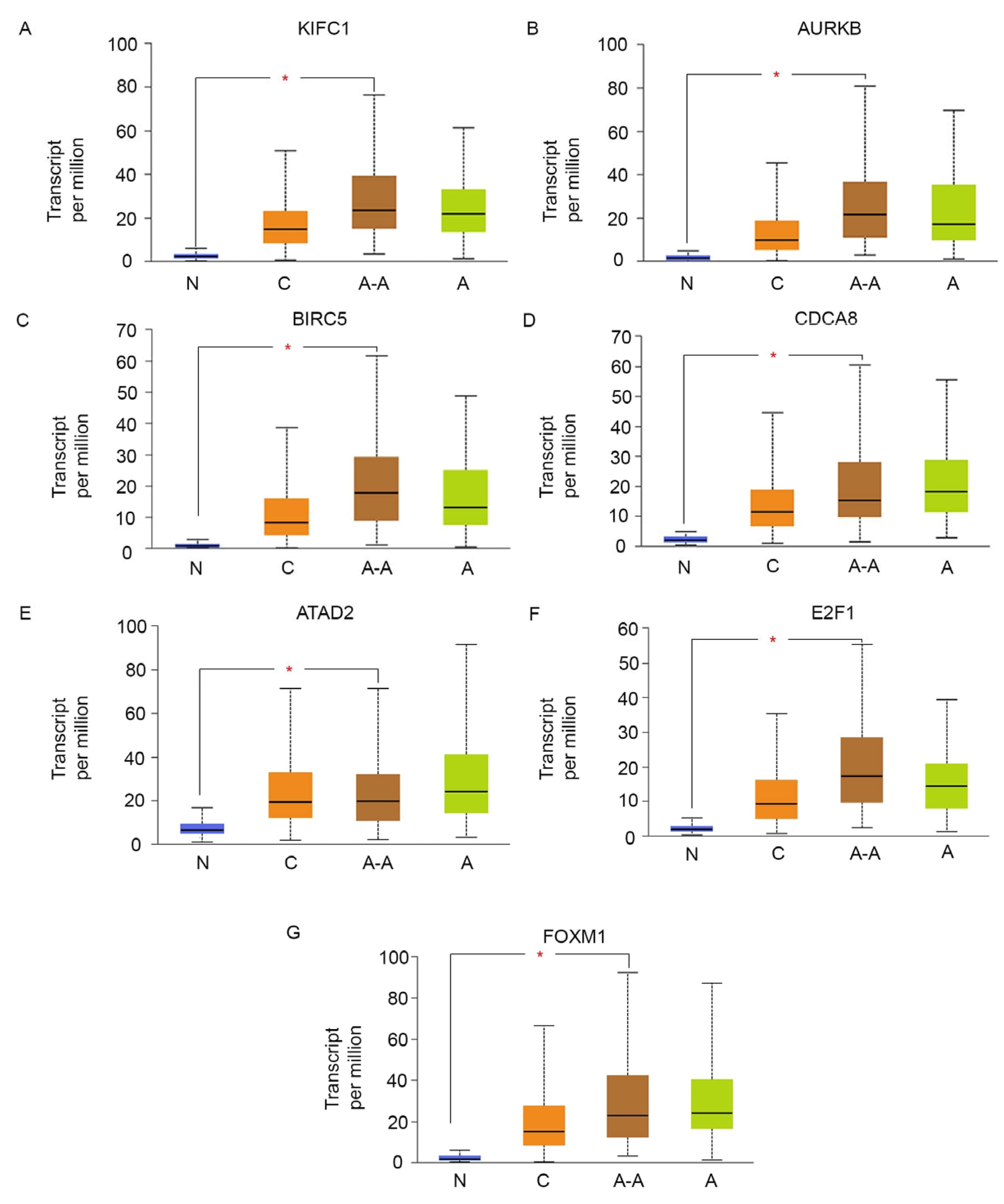
9. Peas in a Pod: FOXM1, ATAD2, E2F1, and Centrosome Clustering Genes Are Co-Upregulated among AR-Low TNBCs
10. Putting the Puzzle Pieces Together: A Core Transcriptional Network Regulates the Expression of Centrosome Clustering Genes
10.1. FOXM1: The Heart and Hub of the Transcriptional Network Controlling G2/M Genes
10.2. DREAM and RB; Engines Controlling Timely Expression of G1/S and G2/M Genes
10.3. MuvB-FOXM1: Coupling the Expression of Centrosome Amplification Genes and Centrosome Clustering Genes
10.4. The TP53-p21-DREAM Pathway Reins in MuvB-FOXM1
10.5. AR and SPDEF: Vital Restraints on Runaway FOXM1 Expression
11. Full Circle: Perspectives on a Novel Actionable Biomarker for AR-Low TNBC
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Feeley, L.P.; Mulligan, A.M.; Pinnaduwage, D.; Bull, S.B.; Andrulis, I.L. Distinguishing luminal breast cancer subtypes by Ki67, progesterone receptor or TP53 status provides prognostic information. Mod. Pathol. 2014, 27, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Isakoff, S.J. Triple-negative breast cancer: Role of specific chemotherapy agents. Cancer J. 2010, 16, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; André, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef]
- Masuda, H.; Baggerly, K.A.; Wang, Y.; Zhang, Y.; Gonzalez-Angulo, A.M.; Meric-Bernstam, F.; Valero, V.; Lehmann, B.D.; Pietenpol, J.A.; Hortobagyi, G.N.; et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin. Cancer Res. 2013, 19, 5533–5540. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Goding Sauer, A.; Newman, L.A.; Jemal, A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J. Clin. 2017, 67, 439–448. [Google Scholar] [CrossRef]
- Burstein, M.D.; Tsimelzon, A.; Poage, G.M. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015, 2, 1688–1698. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanović, B.; Chen, X. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Angajala, A.; Mothershed, E.; Davis, M.B.; Tripathi, S.; He, Q.; Bedi, D.; Dean-Colomb, W.; Yates, C. Quadruple Negative Breast Cancers (QNBC) Demonstrate Subtype Consistency among Primary and Recurrent or Metastatic Breast Cancer. Transl. Oncol. 2019, 12, 493–501. [Google Scholar] [CrossRef]
- Gasparini, P.; Cascione, L.; Fassan, M.; Lovat, F.; Guler, G.; Balci, S.; Irkkan, C.; Morrison, C.; Croce, C.M.; Shapiro, C.L.; et al. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget 2014, 5, 1174–1184. [Google Scholar] [CrossRef]
- Hon, J.D.; Singh, B.; Sahin, A.; Du, G.; Wang, J.; Wang, V.Y.; Deng, F.M.; Zhang, D.Y.; Monaco, M.E.; Lee, P. Breast cancer molecular subtypes: From TNBC to QNBC. Am. J. Cancer Res. 2016, 6, 1864–1872. [Google Scholar] [PubMed]
- Jovanović, B.; Sheng, Q.; Seitz, R.S.; Lawrence, K.D.; Morris, S.W.; Thomas, L.R.; Hout, D.R.; Schweitzer, B.L.; Guo, Y.; Pietenpol, J.A.; et al. Comparison of triple-negative breast cancer molecular subtyping using RNA from matched fresh-frozen versus formalin-fixed paraffin-embedded tissue. BMC Cancer 2017, 17, 241. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.; Tripathi, S.; Hughley, R.; He, Q.; Bae, S.; Karanam, B.; Martini, R.; Newman, L.; Colomb, W.; Grizzle, W.; et al. AR negative triple negative or “quadruple negative” breast cancers in African American women have an enriched basal and immune signature. PLoS ONE 2018, 13, e0196909. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wu, J.; Ling, R.; Li, N. Quadruple negative breast cancer. Breast Cancer 2020, 27, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, S.; Sugita, B.M.; Bortoletto, S.M.; Fonseca, A.S.; Cavalli, L.R.; Aneja, R. QNBC Is Associated with High Genomic Instability Characterized by Copy Number Alterations and miRNA Deregulation. Int. J. Mol. Sci. 2021, 22, 11548. [Google Scholar] [CrossRef]
- Bignold, L.P.; Coghlan, B.L.; Jersmann, H.P. Hansemann, Boveri, chromosomes and the gametogenesis-related theories of tumours. Cell Biol. Int. 2006, 30, 640–644. [Google Scholar] [CrossRef]
- Nigg, E.A. Centrosome duplication: Of rules and licenses. Trends Cell Biol. 2007, 17, 215–221. [Google Scholar] [CrossRef]
- Ogden, A.; Rida, P.C.; Aneja, R. Prognostic value of CA20, a score based on centrosome amplification-associated genes, in breast tumors. Sci. Rep. 2017, 21, 262. [Google Scholar] [CrossRef]
- Ogden, A.; Rida, P.C.; Aneja, R. Let’s huddle to prevent a muddle: Centrosome declustering as an attractive anticancer strategy. Cell Death Differ. 2012, 19, 1255–1267. [Google Scholar] [CrossRef]
- Kwon, M.; Godinho, S.A.; Chandhok, N.S.; Ganem, N.J.; Azioune, A.; Thery, M.; Pellman, D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008, 22, 2189–2203. [Google Scholar] [CrossRef]
- Ganem, N.J.; Godinho, S.A.; Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009, 460, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Silkworth, W.T.; Nardi, I.K.; Scholl, L.M.; Cimini, D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE 2009, 4, e6564. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, A.A.; Martinez, A.C.; van Wely, K.H. Merotelic attachments and non-homologous end joining are the basis of chromosomal instability. Cell Div. 2010, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Leber, B.; Maier, B.; Fuchs, F.; Chi, J.; Riffel, P.; Anderhub, S.; Wagner, L.; Ho, A.D.; Salisbury, J.L.; Boutros, M.; et al. Proteins required for centrosome clustering in cancer cells. Sci. Transl. Med. 2010, 2, 33ra38. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Maier, B.; Bartek, J. Centrosome clustering and chromosomal (in)stability: A matter of life and death. Mol. Oncol. 2011, 5, 324–335. [Google Scholar] [CrossRef]
- D’Assoro, A.B.; Lingle, W.L.; Salisbury, J.L. Centrosome amplification and the development of cancer. Oncogene 2002, 21, 6146–6153. [Google Scholar] [CrossRef]
- Saunders, W. Centrosomal amplification and spindle multipolarity in cancer cells. Semin. Cancer Biol. 2005, 15, 25–32. [Google Scholar] [CrossRef]
- Maresca, T.J.; Salmon, E.D. Welcome to a new kind of tension: Translating kinetochore mechanics into a wait-anaphase signal. J. Cell Sci. 2010, 123, 825–835. [Google Scholar] [CrossRef]
- Mountain, V.; Simerly, C.; Howard, L.; Ando, A.; Schatten, G.; Compton, D.A. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 1999, 147, 351–366. [Google Scholar] [CrossRef]
- Cai, S.; Weaver, L.N.; Ems-McClung, S.C.; Walczak, C.E. Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol. Biol. Cell 2009, 20, 1348–1359. [Google Scholar] [CrossRef]
- Pannu, V.; Mittal, K.; Cantuaria, G.; Reid, M.D.; Li, X.; Donthamsetty, S.; McBride, M.; Klimov, S.; Osan, R.; Gupta, M.V.; et al. Rampant centrosome amplification underlies more aggressive disease course of triple negative breast cancers. Oncotarget 2015, 6, 10487–10497. [Google Scholar] [CrossRef] [PubMed]
- Pannu, V.; Rida, P.C.; Ogden, A.; Turaga, R.C.; Donthamsetty, S.; Bowen, N.J.; Rudd, K.; Gupta, M.V.; Reid, M.D.; Cantuaria, G.; et al. HSET overexpression fuels tumor progression via centrosome clustering-independent mechanisms in breast cancer patients. Oncotarget 2015, 6, 6076–6091. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Jinna, N.; Yuan, Y.C.; Rida, P. Kinesin Family Member C1 (KIFC1/HSET) Underlies Aggressive Disease in Androgen Receptor-Low and Basal-Like Triple-Negative Breast Cancers. Int. J. Mol. Sci. 2023, 24, 16072. [Google Scholar] [CrossRef]
- Jeyaprakash, A.A.; Klein, U.R.; Lindner, D.; Ebert, J.; Nigg, E.A.; Conti, E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 2007, 131, 271–285. [Google Scholar] [CrossRef]
- Trivedi, P.; Palomba, F.; Niedzialkowska, E.; Digman, M.A.; Gratton, E.; Stukenberg, P.T. The inner centromere is a biomolecular condensate scaffolded by the chromosomal passenger complex. Nat. Cell Biol. 2019, 21, 1127–1137. [Google Scholar] [CrossRef]
- Hindriksen, S.; Meppelink, A.; Lens, S.M. Functionality of the chromosomalpassenger complex in cancer. Biochem. Soc. Trans. 2015, 43, 23–32. [Google Scholar]
- Thiru, P.; Kern, D.M.; McKinley, K.L.; Monda, J.K.; Rago, F.; Su, K.C.; Tsinman, T.; Yarar, D.; Bell, G.W.; Cheeseman, I.M. Kinetochore genes are coordinately up-regulated in human tumors as part of a FoxM1-related cell division program. Mol. Biol. Cell 2014, 25, 1983–1994. [Google Scholar] [CrossRef]
- Glover, D.M.; Leibowitz, M.H.; McLean, D.A.; Parry, H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 1995, 81, 95–105. [Google Scholar] [CrossRef]
- Kettenbach, A.N.; Schweppe, D.K.; Faherty, B.K.; Pechenick, D.; Pletnev, A.A.; Gerber, S.A. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal 2011, 4, rs5. [Google Scholar] [CrossRef]
- Koch, A.; Krug, K.; Pengelley, S.; Macek, B.; Hauf, S. Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci. Signal 2011, 4, rs6. [Google Scholar] [CrossRef] [PubMed]
- Hengeveld, R.C.; Hertz, N.T.; Vromans, M.J. Development of a chemical genetic approach for human aurora B kinase identifies novel substrates of the chromosomal passenger complex. Mol. Cell Proteom. 2012, 11, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Yoo, T.Y.; Choi, J.M.; Conway, W.; Yu, C.H.; Pappu, R.V.; Needleman, D.J. Measuring NDC80 binding reveals the molecular basis of tension-dependent kinetochore-microtubule attachments. ELife 2018, 25, e36392. [Google Scholar] [CrossRef] [PubMed]
- Welburn, J.P.; Vleugel, M.; Liu, D.; Yates, J.R.; Lampson, M.A.; Fukagawa, T.; Cheeseman, I.M. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 2010, 38, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Zhang, Z.; Chen, Q.; Yan, H.; Zhang, M.; Zhou, L.; Xu, J.; Lu, W.; Wang, F. Centromere-localized Aurora B kinase is required for the fidelity of chromosome segregation. J. Cell Biol. 2020, 219, e201907092. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef]
- Gregan, J.; Polakova, S.; Zhang, L.; Tolić-Nørrelykke, I.M.; Cimini, D. Merotelic kinetochore attachment: Causes and effects. Trends Cell Biol. 2011, 21, 374–381. [Google Scholar] [CrossRef]
- Ricke, R.M.; Jeganathan, K.B.; van Deursen, J.M. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J. Cell Biol. 2011, 193, 1049–1064. [Google Scholar] [CrossRef]
- Tatsuka, M.; Katayama, H.; Ota, T.; Tanaka, T.; Odashima, S.; Suzuki, F.; Terada, Y. Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 1998, 58, 4811–4816. [Google Scholar]
- Ota, T.; Suto, S.; Katayama, H. Increased mitotic phosphorylation of histone H3 attributable to AIM-1/Aurora-B overexpression contributes to chromosome number instability. Cancer Res. 2002, 62, 5168–5177. [Google Scholar]
- Nguyen, H.G.; Chinnappan, D.; Urano, T.; Ravid, K. Mechanism of Aurora-B degradation and its dependency on intact KEN and A-boxes: Identification of an aneuploidy-promoting property. Mol. Cell Biol. 2005, 25, 4977–4992. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.L.; Eklund, A.C.; Kohane, I.S.; Harris, L.N.; Szallasi, Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006, 38, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Uren, A.G.; Beilharz, T.; O’Connell, M.J.; Bugg, S.J.; van Driel, R.; Vaux, D.L.; Lithgow, T. Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc. Natl. Acad. Sci. USA 1999, 96, 10170–10175. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ambrosini, G.; Chu, E.Y.; Plescia, J.; Tognin, S.; Marchisio, P.C.; Altieri, D.C. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 1998, 396, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, S.P.; Altieri, D.C. Survivin at a glance. J. Cell Sci. 2019, 132, jcs223826. [Google Scholar] [CrossRef]
- Beardmore, V.A.; Ahonen, L.J.; Gorbsky, G.J.; Kallio, M.J. Survivin dynamics increases at centromeres during G2/M phase transition and is regulated by microtubule-attachment and Aurora B kinase activity. J. Cell Sci. 2004, 117, 4033–4042. [Google Scholar] [CrossRef]
- Rosa, J.; Canovas, P.; Islam, A.; Altieri, D.C.; Doxsey, S.J. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol. Biol. Cell 2006, 17, 1483–1493. [Google Scholar] [CrossRef]
- Babkoff, A.; Cohen-Kfir, E.; Aharon, H.; Ronen, D.; Rosenberg, M.; Wiener, R.; Ravid, S. A direct interaction between survivin and myosin II is required for cytokinesis. J. Cell Sci. 2019, 132, jcs233130. [Google Scholar] [CrossRef]
- Gassmann, R.; Carvalho, A.; Henzing, A.J.; Ruchaud, S.; Hudson, D.F.; Honda, R.; Nigg, E.A.; Gerloff, D.L.; Earnshaw, W.C. Borealin: A novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 2004, 166, 179–191. [Google Scholar] [CrossRef]
- Bohnert, K.A.; Chen, J.S.; Clifford, D.M.; Vander Kooi, C.W.; Gould, K.L. A link between aurora kinase and Clp1/Cdc14 regulation uncovered by the identification of a fission yeast borealin-like protein. Mol. Biol. Cell 2009, 20, 3646–3659. [Google Scholar] [CrossRef]
- Van der Waal, M.S.; Hengeveld, R.C.; van der Horst, A.; Lens, S.M. Cell division control by the Chromosomal Passenger Complex. Exp. Cell Res. 2012, 318, 1407–1420. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, X.; Liu, T.; Meng, X.; Yin, X.; Fang, C.; Huang, D.; Cao, Y.; Weng, H.; Zeng, X.; et al. Identification of Biomarkers Correlated with the TNM Staging and Overall Survival of Patients with Bladder Cancer. Front. Physiol. 2017, 8, 947. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.L.; Chen, T.H.; Wang, C.F.; Chiang, Y.H.; Huang, Y.L.; Wong, F.H.; Chou, C.K.; Chen, C.M. Borealin/Dasra B is a cell cycle-regulated chromosomal passenger protein and its nuclear accumulation is linked to poor prognosis for human gastric cancer. Exp. Cell Res. 2006, 312, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Hayama, S.; Daigo, Y.; Yamabuki, T.; Hirata, D.; Kato, T.; Miyamoto, M.; Ito, T.; Tsuchiya, E.; Kondo, S.; Nakamura, Y. Phosphorylation and activation of cell division cycle associated 8 by aurora kinase B plays a significant role in human lung carcinogenesis. Cancer Res. 2007, 67, 4113–4122. [Google Scholar] [CrossRef] [PubMed]
- Narayan, G.; Bourdon, V.; Chaganti, S.; Arias-Pulido, H.; Nandula, S.V.; Rao, P.H.; Gissmann, L.; Dürst, M.; Schneider, A.; Pothuri, B.; et al. Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: Identification of candidate amplified and overexpressed genes. Genes Chromosomes Cancer 2007, 46, 373–384. [Google Scholar] [CrossRef]
- Klein, U.R.; Nigg, E.A.; Gruneberg, U. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol. Biol. Cell 2006, 17, 2547–2558. [Google Scholar] [CrossRef]
- Nguyen, H.G.; Ravid, K. Tetraploidy/aneuploidy and stem cells in cancer promotion: The role of chromosome passenger proteins. J. Cell Physioly 2006, 208, 12–22. [Google Scholar] [CrossRef]
- Tsukahara, T.; Tanno, Y.; Watanabe, Y. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature 2010, 467, 719–723. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2018, 45, 98–102. [Google Scholar] [CrossRef]
- De Almeida, B.P.; Vieira, A.F.; Paredes, J.; Bettencourt-Dias, M.; Barbosa-Morais, N.L. Pan-cancer association of a centrosome amplification gene expression signature with genomic alterations and clinical outcome. PLoS Comput. Biol. 2019, 15, e1006832. [Google Scholar] [CrossRef]
- Győrffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021, 19, 4101–4109. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Rodriguez, I.P.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, A.; Khoury, T.; Stein, L.; Shrikant, P.; Sood, A.K. Prostate derived Ets transcription factor and Carcinoembryonic antigen related cell adhesion molecule 6 constitute a highly active oncogenic axis in breast cancer. Oncotarget 2013, 4, 610–621. [Google Scholar] [CrossRef]
- Tarapore, P.; Fukasawa, K. Loss of p53 and centrosome hyperamplification. Oncogene 2002, 21, 6234–6240. [Google Scholar] [CrossRef]
- Blons, H.; Laccourreye, O.; Houllier, A.M.; Carnot, F.; Brasnu, D.; Beaune, P.; Zucman-Rossi, J.; Laurent-Puig, P. Delineation and candidate gene mutation screening of the 18q22 minimal region of deletion in head and neck squamous cell carcinoma. Oncogene 2002, 21, 5016–5023. [Google Scholar] [CrossRef]
- Wang, I.C.; Chen, Y.J.; Hughes, D.; Petrovic, V.; Major, M.L.; Park, H.J.; Tan, Y.; Ackerson, T.; Costa, R.H. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 2005, 25, 10875–10894. [Google Scholar] [CrossRef]
- Gartel, A.L. FOXM1 in Cancer: Interactions and Vulnerabilities. Cancer Resh. 2017, 77, 3135–3139. [Google Scholar] [CrossRef]
- Li, L.; Wu, D.; Yu, Q.; Li, L.; Wu, P. Prognostic value of FOXM1 in solid tumors: A systematic review and meta-analysis. Oncotarget 2017, 8, 32298–32308. [Google Scholar] [CrossRef]
- Millour, J.; de Olano, N.; Horimoto, Y.; Monteiro, L.J.; Langer, J.K.; Aligue, R.; Hajji, N.; Lam, E.W. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol. Cancer Ther. 2011, 10, 1046–1058. [Google Scholar] [CrossRef]
- Hollern, D.P.; Swiatnicki, M.R.; Rennhack, J.P.; Misek, S.A.; Matson, B.C.; McAuliff, A.; Gallo, K.A.; Caron, K.M.; Andrechek, E.R. E2F1 Drives Breast Cancer Metastasis by Regulating the Target Gene FGF13 and Altering Cell Migration. Sci. Rep. 2019, 9, 10718. [Google Scholar] [CrossRef] [PubMed]
- Logotheti, S.; Marquardt, S.; Gupta, S.K.; Richter, C.; Edelhäuser, B.A.H.; Engelmann, D.; Brenmoehl, J.; Söhnchen, C.; Murr, N.; Alpers, M.; et al. LncRNA-SLC16A1-AS1 induces metabolic reprogramming during Bladder Cancer progression as target and co-activator of E2F1. Theranostics 2020, 10, 9620–9643. [Google Scholar] [CrossRef] [PubMed]
- Ciró, M.; Prosperini, E.; Quarto, M.; Grazini, U.; Walfridsson, J.; McBlane, F.; Nucifero, P.; Pacchiana, G.; Capra, M.; Christensen, J.; et al. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009, 69, 8491–8498. [Google Scholar] [CrossRef] [PubMed]
- Revenko, A.S.; Kalashnikova, E.V.; Gemo, A.T.; Zou, J.X.; Chen, H.W. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol. Cell. Biol. 2010, 30, 5260–5272. [Google Scholar] [CrossRef]
- Liu, H.; Wen, Q.; Yan, S.; Zeng, W.; Zou, Y.; Liu, Q.; Zhang, G.; Zou, J.; Zou, X. Tumor-Promoting ATAD2 and Its Preclinical Challenges. Biomolecules 2022, 12, 1040. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Győrffy, B. muTarget: A platform linking gene expression changes and mutation status in solid tumors. Int. J. Cancer 2021, 148, 502–511. [Google Scholar] [CrossRef]
- Venet, D.; Dumont, J.E.; Detours, V. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput. Biol. 2011, 7, e1002240. [Google Scholar] [CrossRef]
- Gully, C.P.; Velazquez-Torres, G.; Shin, J.H. Aurora B kinase phosphorylates and instigates degradation of p53. Proc. Natl. Acad. Sci. USA 2012, 109, 1513–1522. [Google Scholar] [CrossRef]
- Marxer, M.; Ma, H.T.; Man, W.Y.; Poon, R.Y. p53 deficiency enhances mitotic arrest and slippage induced by pharmacological inhibition of Aurora kinases. Oncogene 2014, 33, 3550–3560. [Google Scholar] [CrossRef]
- Wiedemuth, R.; Klink, B.; Töpfer, K.; Schröck, E.; Schackert, G.; Tatsuka, M.; Temme, A. Survivin safeguards chromosome numbers and protects from aneuploidy independently from p53. Mol. Cancer 2014, 13, 107. [Google Scholar] [CrossRef]
- Mirza, A.; McGuirk, M.; Hockenberry, T.N.; Wu, Q.; Ashar, H.; Black, S.; Wen, S.F.; Wang, L.; Kirschmeier, P.; Bishop, W.R.; et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene 2002, 21, 2613–2622. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, W.H.; Biade, S.; Zilfou, J.T.; Chen, J.; Murphy, M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem. 2002, 277, 3247–3257. [Google Scholar] [CrossRef] [PubMed]
- Raj, D.; Liu, T.; Samadashwily, G.; Li, F.; Grossman, D. Survivin repression by p53, Rb and E2F2 in normal human melanocytes. Carcinogenesis 2008, 29, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qin, Y.; Chen, X.; Wang, X. Cell division cycle associated 8 promotes the growth and inhibits the apoptosis of endometrial cancer cells by regulating cell cycle and P53/Rb signaling pathway. Am. J. Transl. Res. 2023, 15, 3864–3881. [Google Scholar] [PubMed]
- Date, D.A.; Jacob, C.J.; Bekier, M.E.; Stiff, A.C.; Jackson, M.W.; Taylor, W.R. Borealin is repressed in response to p53/Rb signaling. Cell Biol. Int. 2007, 31, 1470–1481. [Google Scholar] [CrossRef]
- Wingender, E.; Schoeps, T.; Haubrock, M.; Krull, M.; Dönitz, J. TFClass: Expanding the classification of human transcription factors to their mammalian orthologs. Nucleic Acids Res. 2018, 46, 343–347. [Google Scholar] [CrossRef]
- Messeguer, X.; Escudero, R.; Farré, D.; Núñez, O.; Martínez, J.; Albà, M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef]
- Farré, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Roselló, L.; Albà, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Leroy, B.; Anderson, M.; Soussi, T. TP53 mutations in human cancer: Database reassessment and prospects for the next decade. Hum. Mutat. 2014, 35, 672–688. [Google Scholar] [CrossRef]
- Joerger, A.C.; Fersht, A.R. The tumor suppressor p53: From structures to drug discovery. Cold Spring Harb. Perspect. Biol. 2010, 2, a000919. [Google Scholar]
- Meek, D.W.; Anderson, C.W. Posttranslational modification of p53: Cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 2009, 1, a000950. [Google Scholar]
- Braithwaite, A.W.; Del Sal, G.; Lu, X. Some p53-binding proteins that can function as arbiters of life and death. Cell Death Differ. 2006, 13, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Brandt, T.; Townsley, F.M.; Teufel, D.P.; Freund, S.M.; Veprintsev, D.B. Molecular basis for modulation of the p53 target selectivity by KLF4. PLoS ONE 2012, 7, e48252. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jézéquel, P.; Frénel, J.S.; Campion, L.; Guérin-Charbonnel, C.; Gouraud, W.; Ricolleau, G.; Campone, M. bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database 2013, 2013, bas060. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, A.; Forcados, G.E.; Katsayal, B.S.; Bako, R.S.; Aminu, S.; Sadiq, I.Z.; Abubakar, M.B.; Yusuf, A.P.; Malami, I.; Faruk, M.; et al. Potential epigenetic modifications implicated in triple- to quadruple-negative breast cancer transition: A review. Epigenomics 2022, 14, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Korver, W.; Roose, J.; Clevers, H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997, 25, 1715–1719. [Google Scholar] [CrossRef]
- Ye, H.; Kelly, T.F.; Samadani, U.; Lim, L.; Rubio, S.; Overdier, D.G.; Roebuck, K.A.; Costa, R.H. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol. Cell. Biol. 1997, 17, 1626–1641. [Google Scholar] [CrossRef]
- Myatt, S.S.; Lam, E.W. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer 2007, 7, 847–859. [Google Scholar] [CrossRef]
- Wierstra, I. FOXM1 (Forkhead box M1) in tumorigenesis: Overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv. Cancer Res. 2013, 119, 191–419. [Google Scholar]
- Saba, R.; Alsayed, A.; Zacny, J.P.; Dudek, A.Z. The Role of Forkhead Box Protein M1 in Breast Cancer Progression and Resistance to Therapy. Int. J. Breast Cancer 2016, 2016, 9768183. [Google Scholar] [CrossRef]
- Hong, H.; Zhu, H.; Zhao, S.; Wang, K.; Zhang, N.; Tian, Y.; Li, Y.; Wang, Y.; Lv, X.; Wei, T.; et al. The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis. 2019, 10, 950. [Google Scholar] [CrossRef] [PubMed]
- Nestal de Moraes, G.; Delbue, D.; Silva, K.L.; Robaina, M.C.; Khongkow, P.; Gomes, A.R.; Zona, S.; Crocamo, S.; Mencalha, A.L.; Magalhães, L.M.; et al. FOXM1 targets XIAP and Survivin to modulate breast cancer survival and chemoresistance. Cell. Signal. 2015, 27, 2496–2505. [Google Scholar] [CrossRef] [PubMed]
- Katzenellenbogen, B.S.; Guillen, V.S.; Katzenellenbogen, J.A. Targeting the oncogenic transcription factor FOXM1 to improve outcomes in all subtypes of breast cancer. Breast Cancer Res. 2023, 25, 76. [Google Scholar] [CrossRef] [PubMed]
- Barger, C.J.; Branick, C.; Chee, L.; Karpf, A.R. Pan-Cancer Analyses Reveal Genomic Features of FOXM1 Overexpression in Cancer. Cancers 2019, 11, 251. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Quaas, M.; Steiner, L.; Engeland, K. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res. 2016, 44, 164–174. [Google Scholar] [CrossRef]
- Sadasivam, S.; DeCaprio, J.A. The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer 2013, 13, 585–595. [Google Scholar] [CrossRef]
- Litovchick, L.; Sadasivam, S.; Florens, L.; Zhu, X.; Swanson, S.K.; Velmurugan, S.; Chen, R.; Washburn, M.P.; Liu, X.S.; DeCaprio, J.A. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 2007, 26, 539–551. [Google Scholar] [CrossRef]
- Tedesco, D.; Lukas, J.; Reed, S.I. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 2002, 16, 2946–2957. [Google Scholar] [CrossRef]
- Park, H.J.; Costa, R.H.; Lau, L.F.; Tyner, A.L.; Raychaudhuri, P. Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol. Cell. Biol. 2008, 28, 5162–5171. [Google Scholar] [CrossRef]
- Okumura, F.; Joo-Okumura, A.; Nakatsukasa, K.; Kamura, T. Hypoxia-inducible factor-2α stabilizes the von Hippel-Lindau (VHL) disease suppressor, Myb-related protein 2. PLoS ONE 2017, 12, e0175593. [Google Scholar] [CrossRef]
- Schade, A.E.; Oser, M.G.; Nicholson, H.E.; DeCaprio, J.A. Cyclin D-CDK4 relieves cooperative repression of proliferation and cell cycle gene expression by DREAM and RB. Oncogene 2019, 38, 4962–4976. [Google Scholar] [CrossRef] [PubMed]
- Kalashnikova, E.V.; Revenko, A.S.; Gemo, A.T.; Andrews, N.P.; Tepper, C.G.; Zou, J.X.; Cardiff, R.D.; Borowsky, A.D.; Chen, H.W. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 2010, 70, 9402–9412. [Google Scholar] [CrossRef] [PubMed]
- Pattschull, G.; Walz, S.; Gründl, M.; Schwab, M.; Rühl, E.; Baluapuri, A.; Cindric-Vranesic, A.; Kneitz, S.; Wolf, E.; Ade, C.P.; et al. The Myb-MuvB Complex Is Required for YAP-Dependent Transcription of Mitotic Genes. Cell Rep. 2019, 27, 3533–3546.e7. [Google Scholar] [CrossRef] [PubMed]
- Eisinger-Mathason, T.S.; Mucaj, V.; Biju, K.M.; Nakazawa, M.S.; Gohil, M.; Cash, T.P.; Yoon, S.S.; Skuli, N.; Park, K.M.; Gerecht, S.; et al. Deregulation of the Hippo pathway in soft-tissue sarcoma promotes FOXM1 expression and tumorigenesis. Proc. Natl. Acad. Sci. USA 2015, 112, E3402-11. [Google Scholar] [CrossRef]
- Nilsson, M.B.; Sun, H.; Robichaux, J.; Pfeifer, M.; McDermott, U.; Travers, J.; Diao, L.; Xi, Y.; Tong, P.; Shen, L.; et al. A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components. Sci. Transl. Med. 2020, 12, eaaz4589. [Google Scholar] [CrossRef]
- Kohler, R.; Engeland, K. A-MYB substitutes for B-MYB in activating cell cycle genes and in stimulating proliferation. Nucleic Acids Res. 2024, 52, 6830–6849. [Google Scholar] [CrossRef]
- Saldivar, J.C.; Hamperl, S.; Bocek, M.J.; Chung, M.; Bass, T.E.; Cisneros-Soberanis, F.; Samejima, K.; Xie, L.; Paulson, J.R.; Earnshaw, W.C.; et al. An intrinsic S/G2 checkpoint enforced by ATR. Science 2018, 361, 806–810. [Google Scholar] [CrossRef]
- Kalathil, D.; John, S.; Nair, A.S. FOXM1 and Cancer: Faulty Cellular Signaling Derails Homeostasis. Front. Oncol. 2021, 10, 626836. [Google Scholar] [CrossRef]
- Cheng, X.H.; Black, M.; Ustiyan, V.; Le, T.; Fulford, L.; Sridharan, A.; Medvedovic, M.; Kalinichenko, V.V.; Whitsett, J.A.; Kalin, T.V. SPDEF inhibits prostate carcinogenesis by disrupting a positive feedback loop in regulation of the Foxm1 oncogene. PLoS Genet. 2014, 10, e1004656. [Google Scholar] [CrossRef]
- Jiao, D.C.; Lu, Z.D.; Qiao, J.H.; Yan, M.; Cui, S.D.; Liu, Z.Z. Expression of CDCA8 correlates closely with FOXM1 in breast cancer: Public microarray data analysis and immunohistochemical study. Neoplasma 2015, 62, 464–469. [Google Scholar] [CrossRef]
- El-Deiry, W.S.; Tokino, T.; Velculescu, V.E.; Levy, D.B.; Parsons, R.; Trent, J.M.; Lin, D.; Mercer, W.E.; Kinzler, K.W.; Vogelstein, B. WAF1, a potential mediator of p53 tumor suppression. Cell 1993, 75, 817–825. [Google Scholar] [CrossRef]
- Pfister, K.; Pipka, J.L.; Chiang, C.; Liu, Y.; Clark, R.A.; Keller, R.; Skoglund, P.; Guertin, M.J.; Hall, I.M.; Stukenberg, P.T. Identification of Drivers of Aneuploidy in Breast Tumors. Cell Rep. 2018, 23, 2758–2769. [Google Scholar] [CrossRef] [PubMed]
- Oettgen, P.; Finger, E.; Sun, Z.; Akbarali, Y.; Thamrongsak, U.; Boltax, J.; Grall, F.; Dube, A.; Weiss, A.; Brown, L.; et al. PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J. Biol. Chem. 2000, 275, 1216–1225. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.P.; Roth, A.; Goya, R.; Oloumi, A.; Ha, G.; Zhao, Y.; Turashvili, G.; Ding, J.; Tse, K.; Haffari, G.; et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012, 486, 395–399. [Google Scholar] [CrossRef]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Merjaneh, N.; Hajjar, M.; Lan, Y.W.; Kalinichenko, V.V.; Kalin, T.V. The Promise of Combination Therapies with FOXM1 Inhibitors for Cancer Treatment. Cancers 2024, 16, 756. [Google Scholar] [CrossRef]
- Luo, G.; Lin, X.; Vega-Medina, A.; Xiao, M.; Li, G.; Wei, H.; Velázquez-Martínez, C.A.; Xiang, H. Targeting of the FOXM1 Oncoprotein by E3 Ligase-Assisted Degradation. J. Med. Chem. 2021, 64, 17098–17114. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rida, P.; Baker, S.; Saidykhan, A.; Bown, I.; Jinna, N. FOXM1 Transcriptionally Co-Upregulates Centrosome Amplification and Clustering Genes and Is a Biomarker for Poor Prognosis in Androgen Receptor-Low Triple-Negative Breast Cancer. Cancers 2024, 16, 3191. https://doi.org/10.3390/cancers16183191
Rida P, Baker S, Saidykhan A, Bown I, Jinna N. FOXM1 Transcriptionally Co-Upregulates Centrosome Amplification and Clustering Genes and Is a Biomarker for Poor Prognosis in Androgen Receptor-Low Triple-Negative Breast Cancer. Cancers. 2024; 16(18):3191. https://doi.org/10.3390/cancers16183191
Chicago/Turabian StyleRida, Padmashree, Sophia Baker, Adam Saidykhan, Isabelle Bown, and Nikita Jinna. 2024. "FOXM1 Transcriptionally Co-Upregulates Centrosome Amplification and Clustering Genes and Is a Biomarker for Poor Prognosis in Androgen Receptor-Low Triple-Negative Breast Cancer" Cancers 16, no. 18: 3191. https://doi.org/10.3390/cancers16183191
APA StyleRida, P., Baker, S., Saidykhan, A., Bown, I., & Jinna, N. (2024). FOXM1 Transcriptionally Co-Upregulates Centrosome Amplification and Clustering Genes and Is a Biomarker for Poor Prognosis in Androgen Receptor-Low Triple-Negative Breast Cancer. Cancers, 16(18), 3191. https://doi.org/10.3390/cancers16183191






