Cancer-Associated Fibroblast Proteins as Potential Targets against Colorectal Cancers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. Immunohistochemistry
2.3. Sequencing of Mutations
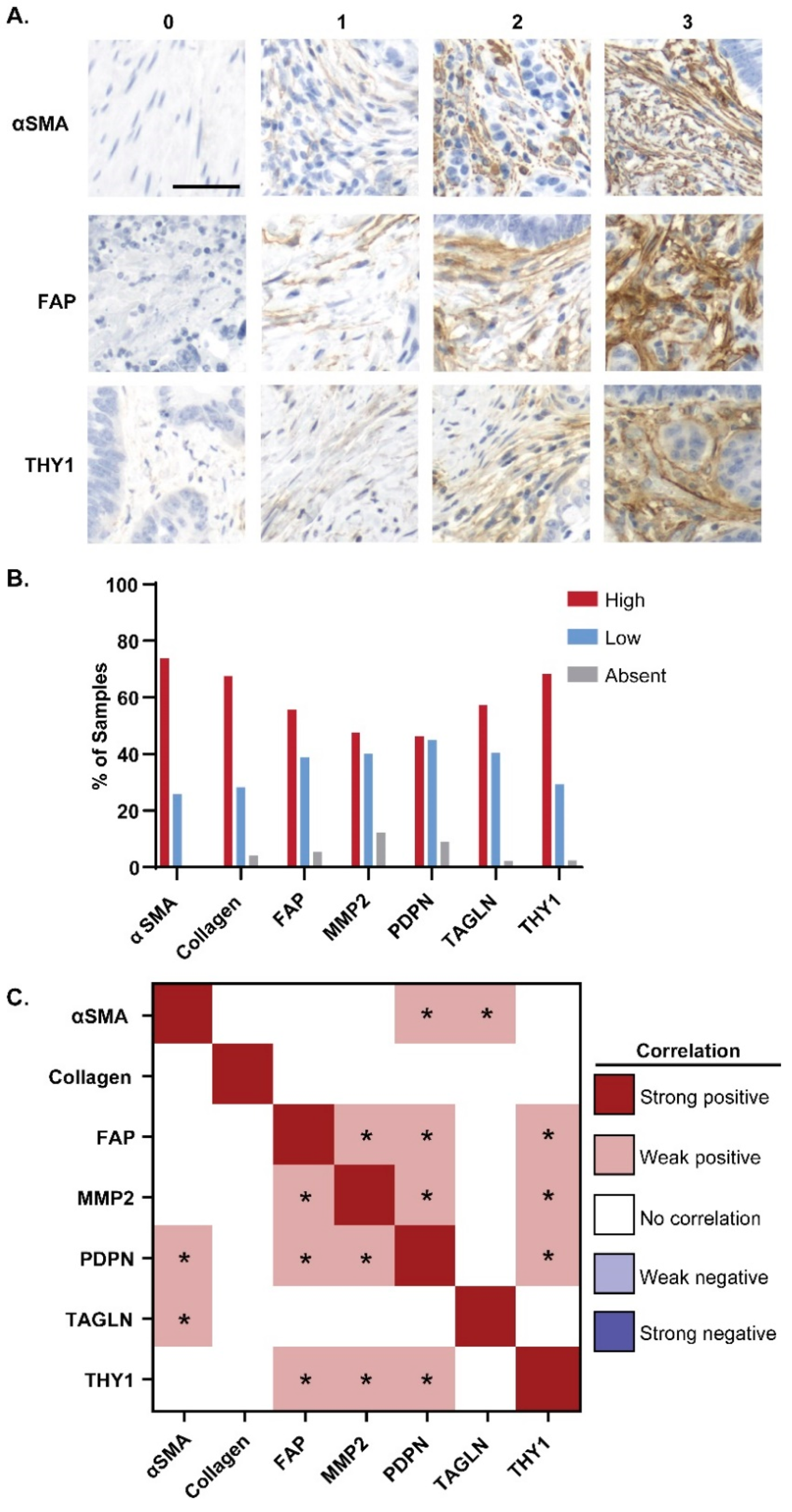
2.4. Scoring Tissue Sections
2.5. TCGA Data Analysis
2.6. Gene Set Enrichment Analysis
2.7. Statistical Methods
3. Results
3.1. Expression of CAF Markers in Cancerous Tissue
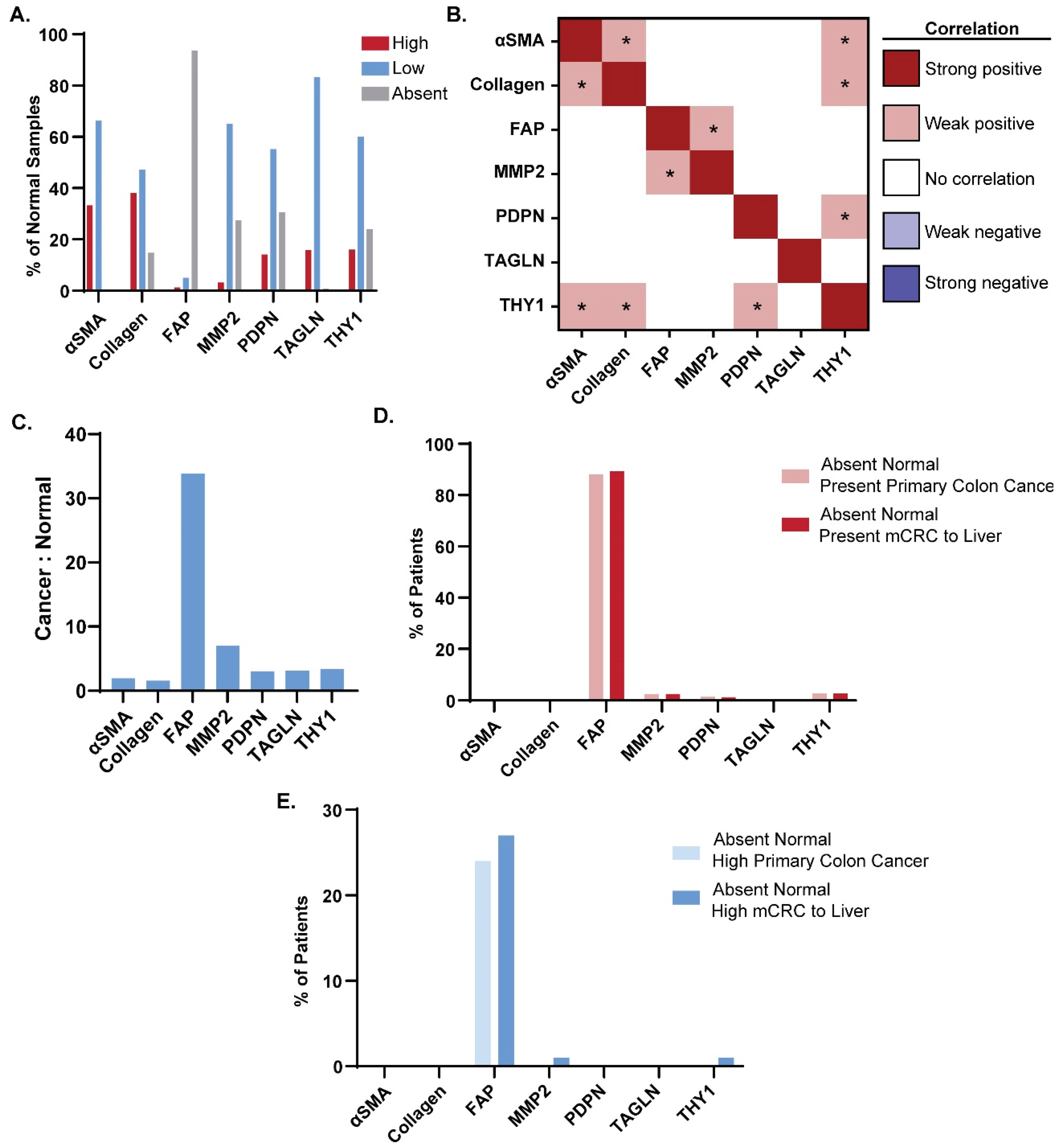
3.2. Expression of CAF Markers in Normal Tissue
3.3. Cancer-Specific Marker Expression
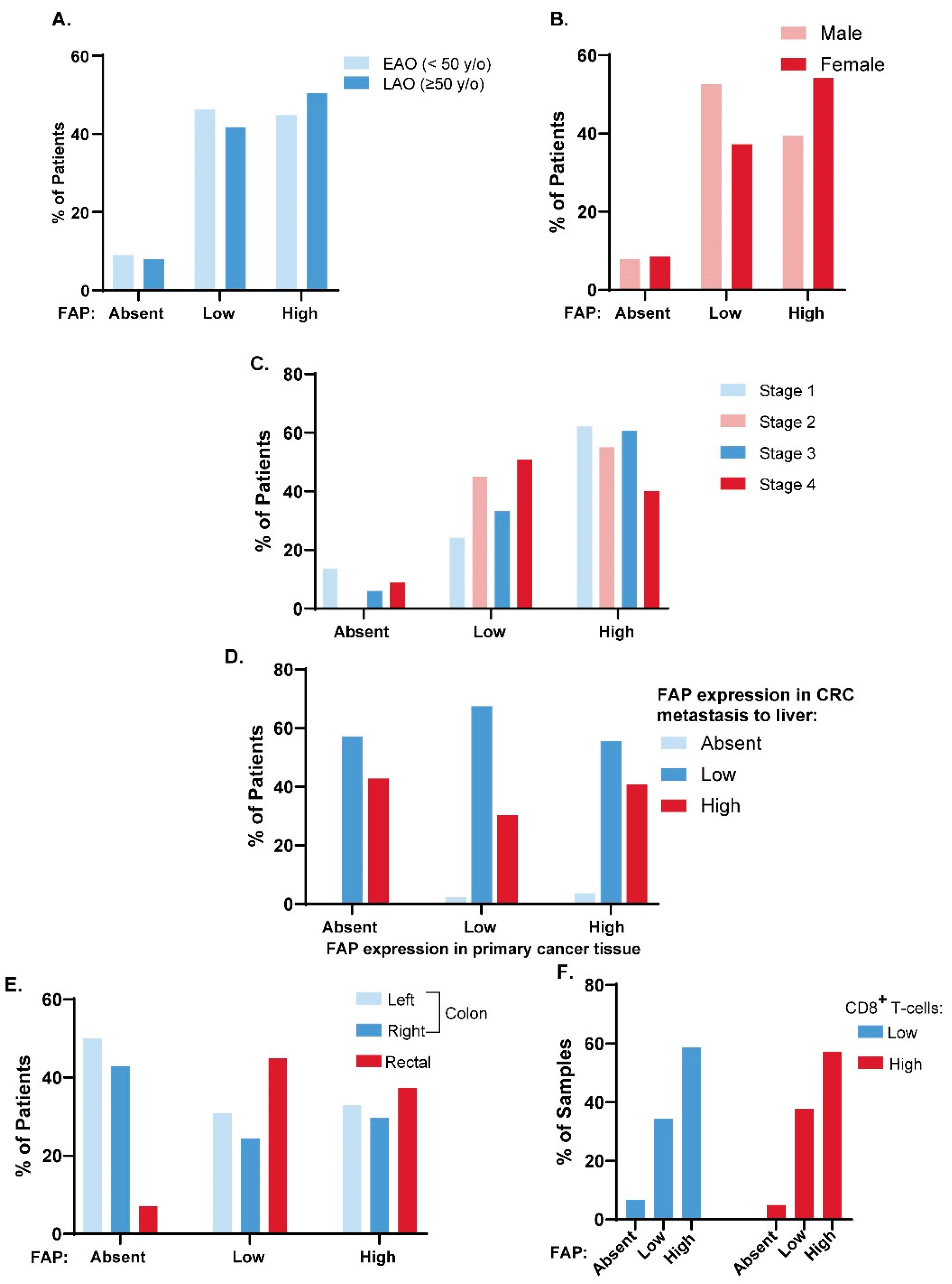
3.4. FAP Expression across Clinical Disease Subtypes
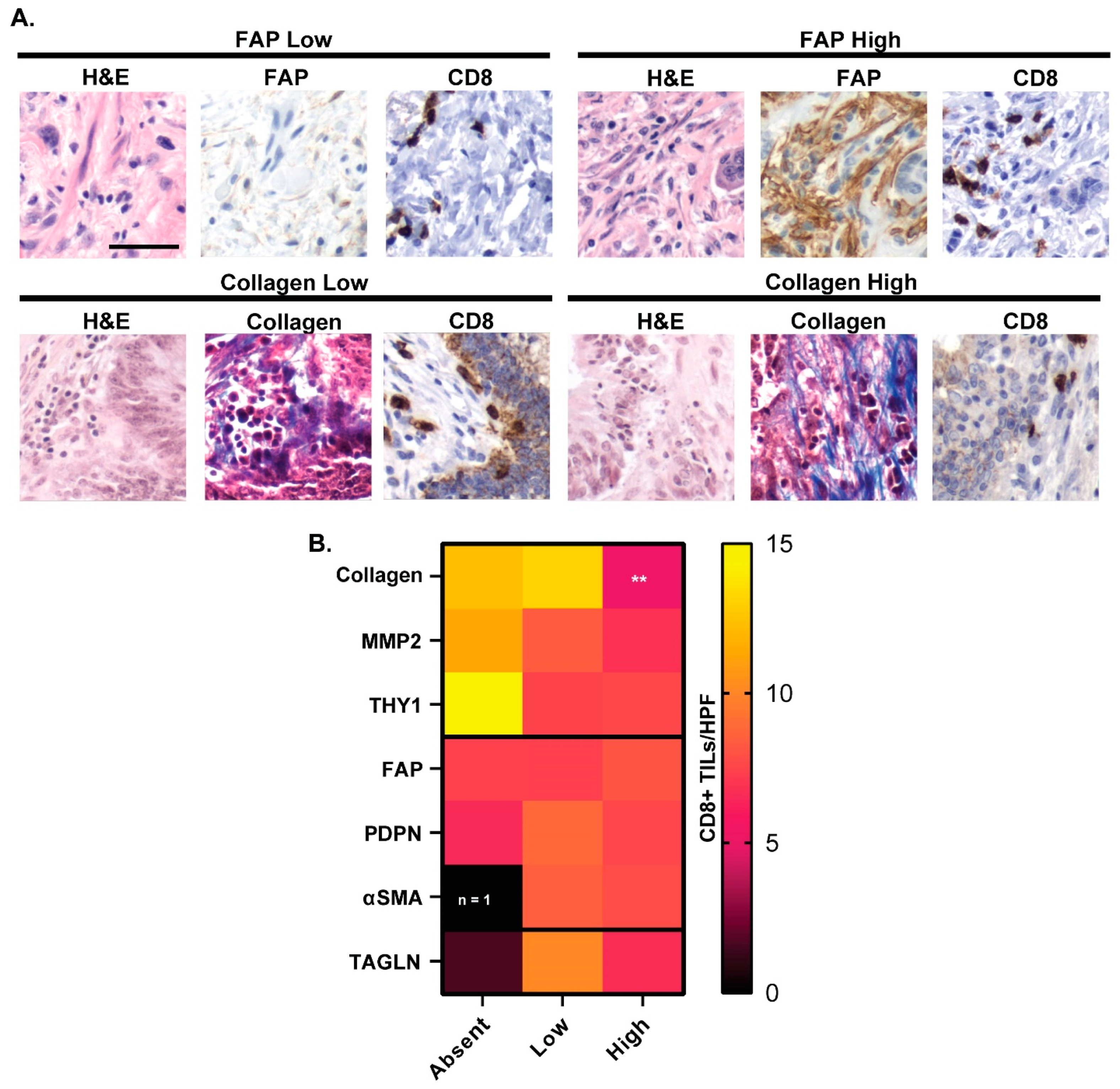
3.5. FAP Expression and CD8+ T Cell Tumor Infiltration
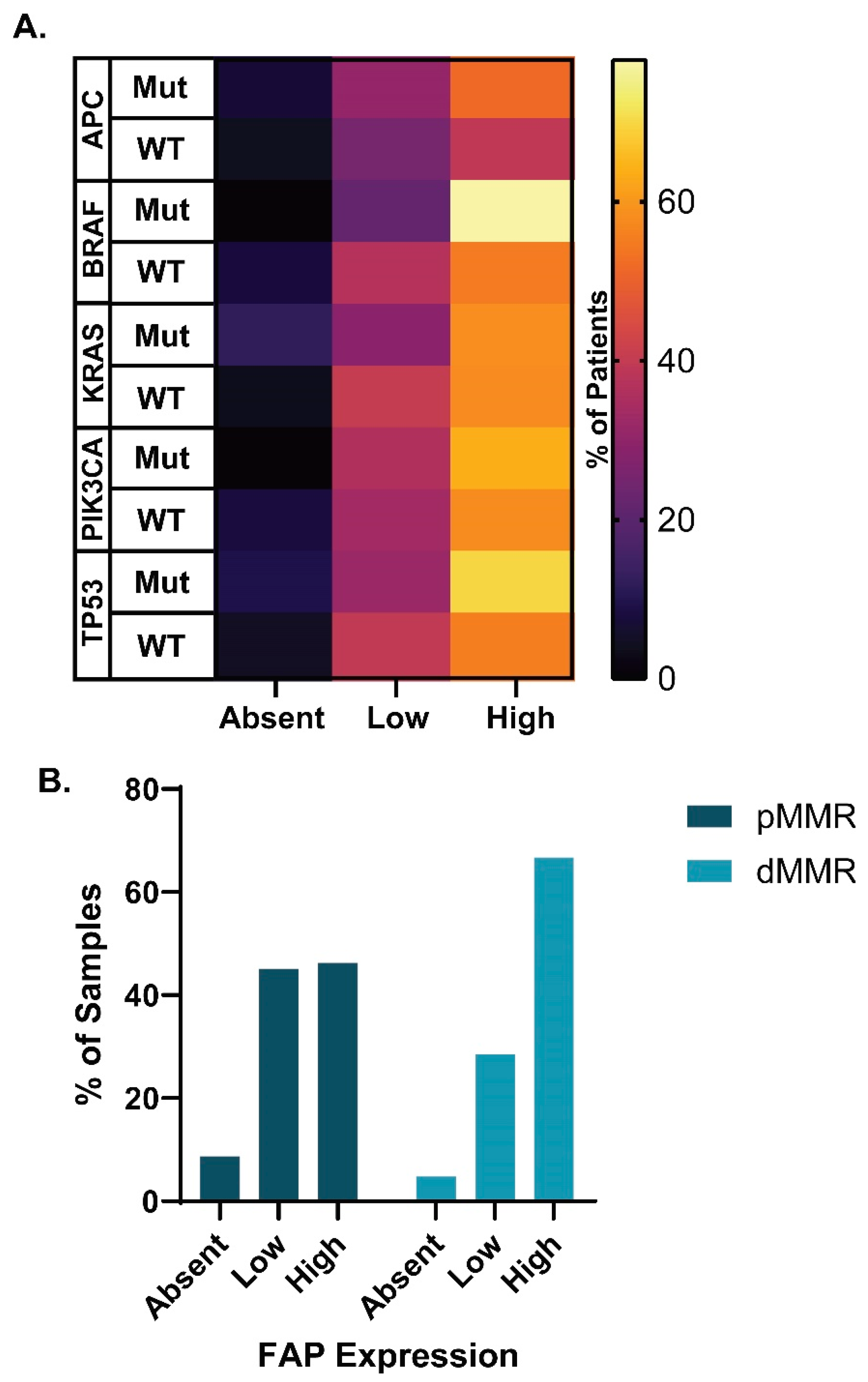
3.6. FAP Expression across Molecular Disease Subtypes
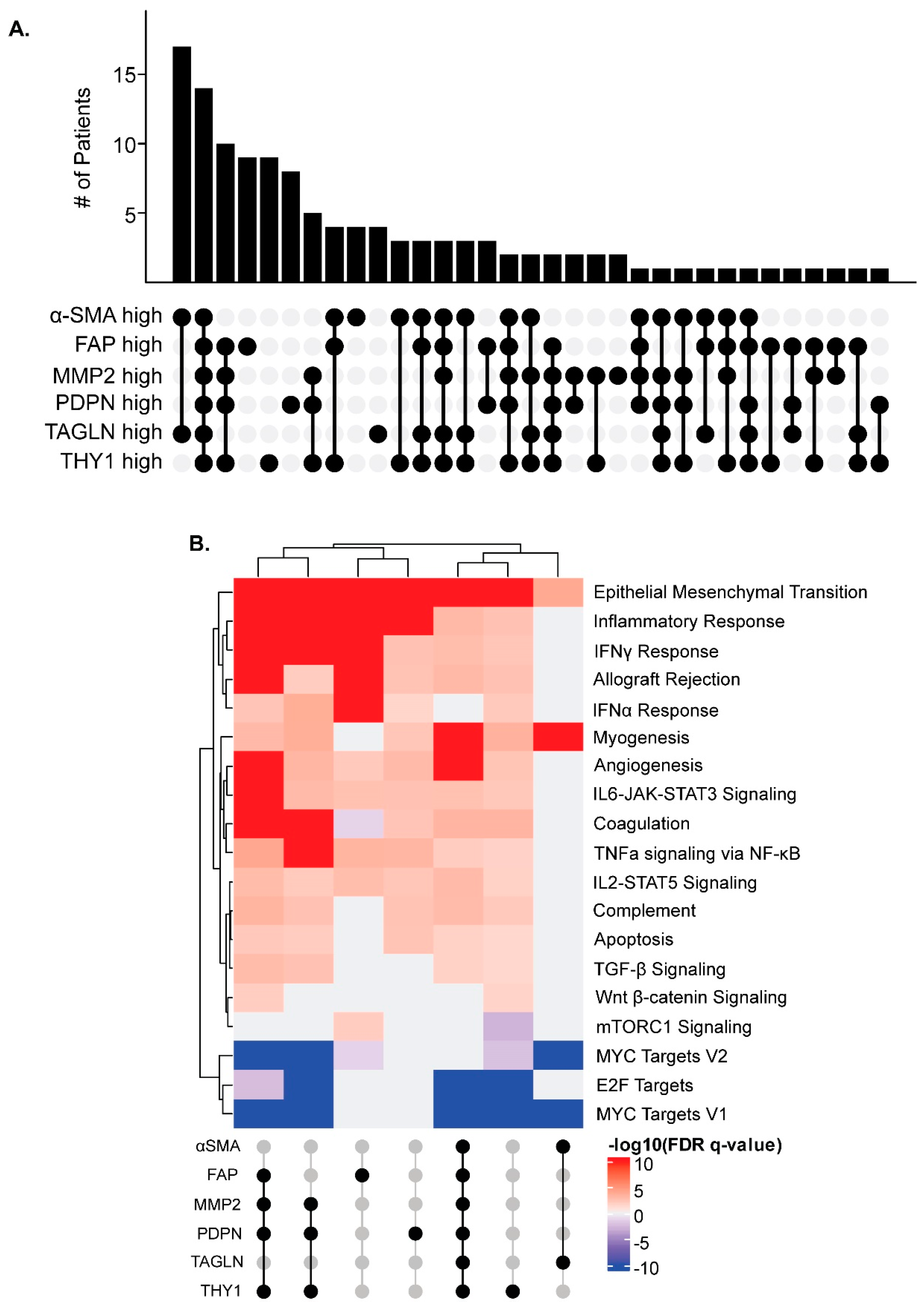
3.7. CAF Marker Expression and CD8+ T Cell Infiltration
3.8. CAF Marker Expression and Upregulated Pathways in Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The American Cancer Society Key Statistics for Colorectal Cancer. Available online: https://www.cancer.org/cancer/types/colon-rectal-cancer/about/key-statistics.html (accessed on 18 April 2024).
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Arain, M.A.; Chen, Y.-J.; Ciombor, K.K.; Cohen, S.; Cooper, H.S.; Deming, D.; Farkas, L.; et al. Colon Cancer, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 329–359. [Google Scholar] [CrossRef] [PubMed]
- Abbasian, M.; Mousavi, E.; Arab-Bafrani, Z.; Sahebkar, A. The Most Reliable Surface Marker for the Identification of Colorectal Cancer Stem-like Cells: A Systematic Review and Meta-Analysis. J. Cell. Physiol. 2019, 234, 8192–8202. [Google Scholar] [CrossRef] [PubMed]
- Belov, L.; Zhou, J.; Christopherson, R.I. Cell Surface Markers in Colorectal Cancer Prognosis. Int. J. Mol. Sci. 2011, 12, 78–113. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A Framework for Advancing Our Understanding of Cancer-Associated Fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Nurmik, M.; Ullmann, P.; Rodriguez, F.; Haan, S.; Letellier, E. In Search of Definitions: Cancer-Associated Fibroblasts and Their Markers. Int. J. Cancer 2020, 146, 895–905. [Google Scholar] [CrossRef]
- Lee, H.O.; Hong, Y.; Etlioglu, H.E.; Cho, Y.B.; Pomella, V.; Van den Bosch, B.; Vanhecke, J.; Verbandt, S.; Hong, H.; Min, J.W.; et al. Lineage-Dependent Gene Expression Programs Influence the Immune Landscape of Colorectal Cancer. Nat. Genet. 2020, 52, 594–603. [Google Scholar] [CrossRef]
- Tlsty, T.D.; Hein, P.W. Know Thy Neighbor: Stromal Cells Can Contribute Oncogenic Signals. Curr. Opin. Genet. Dev. 2001, 11, 54–59. [Google Scholar] [CrossRef]
- LeBeau, A.M.; Brennen, W.N.; Aggarwal, S.; Denmeade, S.R. Targeting the Cancer Stroma with a Fibroblast Activation Protein-Activated Promelittin Protoxin. Mol. Cancer Ther. 2009, 8, 1378–1386. [Google Scholar] [CrossRef]
- Garin-Chesa, P.; Old, L.J.; Rettig, W.J. Cell Surface Glycoprotein of Reactive Stromal Fibroblasts as a Potential Antibody Target in Human Epithelial Cancers. Proc. Natl. Acad. Sci. USA 1990, 87, 7235–7239. [Google Scholar] [CrossRef]
- Yuan, Z.; Hu, H.; Zhu, Y.; Zhang, W.; Fang, Q.; Qiao, T.; Ma, T.; Wang, M.; Huang, R.; Tang, Q.; et al. Colorectal Cancer Cell Intrinsic Fibroblast Activation Protein Alpha Binds to Enolase1 and Activates NF-ΚB Pathway to Promote Metastasis. Cell Death Dis. 2021, 12, 543. [Google Scholar] [CrossRef]
- Rolin, C.; Zimmer, J.; Seguin-Devaux, C. Bridging the Gap with Multispecific Immune Cell Engagers in Cancer and Infectious Diseases. Cell Mol. Immunol. 2024, 21, 643–661. [Google Scholar] [CrossRef] [PubMed]
- Tapia-Galisteo, A.; Sánchez Rodríguez, Í.; Aguilar-Sopeña, O.; Harwood, S.L.; Narbona, J.; Ferreras Gutierrez, M.; Navarro, R.; Martín-García, L.; Corbacho, C.; Compte, M.; et al. Trispecific T-Cell Engagers for Dual Tumor-Targeting of Colorectal Cancer. Oncoimmunology 2022, 11, 2034355. [Google Scholar] [CrossRef] [PubMed]
- Fenis, A.; Demaria, O.; Gauthier, L.; Vivier, E.; Narni-Mancinelli, E. New Immune Cell Engagers for Cancer Immunotherapy. Nat. Rev. Immunol. 2024, 24, 471–486. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Lei, R.; Ding, S.W.; Zhu, S. Skewer: A Fast and Accurate Adapter Trimmer for next-Generation Sequencing Paired-End Reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Girardot, C.; Scholtalbers, J.; Sauer, S.; Su, S.Y.; Furlong, E.E.M. Je, a Versatile Suite to Handle Multiplexed NGS Libraries with Unique Molecular Identifiers. BMC Bioinform. 2016, 17, 419. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Källberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and Accurate Calling of Germline and Somatic Variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain W1118; Iso-2; Iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; Van Den Beek, M.; Bouvier, D.; Ech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Sander, C.; Stuart, J.M.; Chang, K.; Creighton, C.J.; et al. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: http://www.R-project.org (accessed on 18 July 2024).
- Gu, Z. Complex Heatmap Visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Fuchs, T.L.; Sioson, L.; Sheen, A.; Jafari-Nejad, K.; Renaud, C.J.; Andrici, J.; Ahadi, M.; Chou, A.; Gill, A.J. Assessment of Tumor-Infiltrating Lymphocytes Using International TILs Working Group (ITWG) System Is a Strong Predictor of Overall Survival in Colorectal Carcinoma: A Study of 1034 Patients. Am. J. Surg. Pathol. 2020, 44, 536–544. [Google Scholar] [CrossRef]
- Overman, M.J.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.J.; Gelsomino, F.; Aglietta, M.; Morse, M.A.; Van Cutsem, E.; McDermott, R.; Hill, A.; et al. Durable Clinical Benefit with Nivolumab plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J. Clin. Oncol. 2018, 36, 773–779. [Google Scholar] [CrossRef]
- Le, D.T.; Kim, T.W.; van Cutsem, E.; Geva, R.; Jäger, D.; Hara, H.; Burge, M.; O’Neil, B.; Kavan, P.; Yoshino, T.; et al. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability–High/Mismatch Repair–Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J. Clin. Oncol. 2020, 38, 11–19. [Google Scholar] [CrossRef]
- Henry, L.R.; Lee, H.O.; Lee, J.S.; Klein-Szanto, A.; Watts, P.; Ross, E.A.; Chen, W.T.; Cheng, J.D. Clinical Implications of Fibroblast Activation Protein in Patients with Colon Cancer. Clin. Cancer Res. 2007, 13, 1736–1741. [Google Scholar] [CrossRef]
- Liu, R.; Li, H.; Liu, L.; Yu, J.; Ren, X. Fibroblast Activation Protein: A Potential Therapeutic Target in Cancer. Cancer Biol. Ther. 2012, 13, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Greimelmaier, K.; Klopp, N.; Mairinger, E.; Wessolly, M.; Borchert, S.; Steinborn, J.; Schmid, K.W.; Wohlschlaeger, J.; Mairinger, F.D. Fibroblast Activation Protein-α Expression in Fibroblasts Is Common in the Tumor Microenvironment of Colorectal Cancer and May Serve as a Therapeutic Target. Pathol. Oncol. Res. 2023, 29, 1611163. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, B.C.; Pentcheva-Hoang, T.; Carstens, J.L.; Zheng, X.; Wu, C.C.; Simpson, T.R.; Laklai, H.; Sugimoto, H.; Kahlert, C.; Novitskiy, S.V.; et al. Depletion of Carcinoma-Associated Fibroblasts and Fibrosis Induces Immunosuppression and Accelerates Pancreas Cancer with Reduced Survival. Cancer Cell 2014, 25, 719–734. [Google Scholar] [CrossRef] [PubMed]
- Rhim, A.D.; Oberstein, P.E.; Thomas, D.H.; Mirek, E.T.; Palermo, C.F.; Sastra, S.A.; Dekleva, E.N.; Saunders, T.; Becerra, C.P.; Tattersall, I.W.; et al. Stromal Elements Act to Restrain, Rather than Support, Pancreatic Ductal Adenocarcinoma. Cancer Cell 2014, 25, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Hofheinz, R.D.; Al-Batran, S.E.; Hartmann, F.; Hartung, G.; Jäger, D.; Renner, C.; Tanswell, P.; Kunz, U.; Amelsberg, A.; Kuthan, H.; et al. Stromal Antigen Targeting by a Humanised Monoclonal Antibody: An Early Phase II Trial of Sibrotuzumab in Patients with Metastatic Colorectal Cancer. Onkologie 2003, 26, 44–48. [Google Scholar] [CrossRef]
- Scott, A.M.; Wiseman, G.; Welt, S.; Adjei, A.; Lee, F.T.; Hopkins, W.; Divgi, C.R.; Hanson, L.H.; Mitchell, P.; Gansen, D.N.; et al. A Phase I Dose-Escalation Study of Sibrotuzumab in Patients with Advanced or Metastatic Fibroblast Activation Protein-Positive Cancer. Clin. Cancer Res. 2003, 9, 1639–1647. [Google Scholar]
- Fabre, M.; Ferrer, C.; Domínguez-Hormaetxe, S.; Bockorny, B.; Murias, L.; Seifert, O.; Eisler, S.A.; Kontermann, R.E.; Pfizenmaier, K.; Lee, S.Y.; et al. OMTX705, a Novel FAP-Targeting ADC Demonstrates Activity in Chemotherapy and Pembrolizumab-Resistant Solid Tumor Models. Clin. Cancer Res. 2020, 26, 3420–3430. [Google Scholar] [CrossRef]
- Zana, A.; Puig-Moreno, C.; Bocci, M.; Gilardoni, E.; Di Nitto, C.; Principi, L.; Ravazza, D.; Rotta, G.; Prodi, E.; De Luca, R.; et al. A Comparative Analysis of Fibroblast Activation Protein-Targeted Small Molecule-Drug, Antibody-Drug, and Peptide-Drug Conjugates. Bioconjug. Chem. 2023, 34, 1205–1211. [Google Scholar] [CrossRef]
- Lindner, T.; Giesel, F.L.; Kratochwil, C.; Serfling, S.E. Radioligands Targeting Fibroblast Activation Protein (FAP). Cancers 2021, 13, 5744. [Google Scholar] [CrossRef]
- Xu, M.; Chen, J.; Zhang, P.; Cai, J.; Song, H.; Li, Z.; Liu, Z. An Antibody-Radionuclide Conjugate Targets Fibroblast Activation Protein for Cancer Therapy. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 3214–3224. [Google Scholar] [CrossRef]
- Kiani, M.; Jokar, S.; Hassanzadeh, L.; Behnammanesh, H.; Bavi, O.; Beiki, D.; Assadi, M. Recent Clinical Implications of FAPI: Imaging and Therapy. Clin. Nucl. Med. 2024; online ahead of print. [Google Scholar]
- Coto-Llerena, M.; Ercan, C.; Kancherla, V.; Taha-Mehlitz, S.; Eppenberger-Castori, S.; Soysal, S.D.; Ng, C.K.Y.; Bolli, M.; von Flüe, M.; Nicolas, G.P.; et al. High Expression of FAP in Colorectal Cancer Is Associated with Angiogenesis and Immunoregulation Processes. Front. Oncol. 2020, 10, 979. [Google Scholar] [CrossRef] [PubMed]
- Hintz, H.M.; Cowan, A.E.; Shapovalova, M.; LeBeau, A.M. Development of a Cross-Reactive Monoclonal Antibody for Detecting the Tumor Stroma. Bioconjug. Chem. 2019, 30, 1466–1476. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, R.; Johnson, K.A.; Lippert, A.E.L.; Kraus, S.G.; Emmerich, P.B.; Pasch, C.A.; Zhang, W.; Matkowskyj, K.A.; LeBeau, A.M.; Deming, D.A. Cancer-Associated Fibroblast Proteins as Potential Targets against Colorectal Cancers. Cancers 2024, 16, 3158. https://doi.org/10.3390/cancers16183158
Shah R, Johnson KA, Lippert AEL, Kraus SG, Emmerich PB, Pasch CA, Zhang W, Matkowskyj KA, LeBeau AM, Deming DA. Cancer-Associated Fibroblast Proteins as Potential Targets against Colorectal Cancers. Cancers. 2024; 16(18):3158. https://doi.org/10.3390/cancers16183158
Chicago/Turabian StyleShah, Ruchi, Katherine A. Johnson, Anna E. L. Lippert, Sean G. Kraus, Philip B. Emmerich, Cheri A. Pasch, Wei Zhang, Kristina A. Matkowskyj, Aaron M. LeBeau, and Dustin A. Deming. 2024. "Cancer-Associated Fibroblast Proteins as Potential Targets against Colorectal Cancers" Cancers 16, no. 18: 3158. https://doi.org/10.3390/cancers16183158
APA StyleShah, R., Johnson, K. A., Lippert, A. E. L., Kraus, S. G., Emmerich, P. B., Pasch, C. A., Zhang, W., Matkowskyj, K. A., LeBeau, A. M., & Deming, D. A. (2024). Cancer-Associated Fibroblast Proteins as Potential Targets against Colorectal Cancers. Cancers, 16(18), 3158. https://doi.org/10.3390/cancers16183158




