The Role of ctDNA and Liquid Biopsy in the Diagnosis and Monitoring of Head and Neck Cancer: Towards Precision Medicine
Abstract
Simple Summary
Abstract
1. Introduction
2. Identification of Studies Included in the Present Review
3. Liquid Biopsy
4. Head and Neck Squamous Cell Carcinoma
5. EBV+ Nasopharyngeal Carcinoma
6. Other Types of Head and Neck Cancers
7. Challenges and Limitations
8. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Mody, M.D.; Rocco, J.W.; Yom, S.S.; Haddad, R.I.; Saba, N.F. Head and neck cancer. Lancet 2021, 398, 2289–2299. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B. AJCC Cancer Staging Manual, 8th Up to date with Vulva Version 9 protocol ed.; Mahul, B., Amin, S.B.E., Eds.; American College of Surgeons: Essex, CT, USA, 2017. [Google Scholar]
- Gormley, M.; Creaney, G.; Schache, A.; Ingarfield, K.; Conway, D.I. Reviewing the epidemiology of head and neck cancer: Definitions, trends and risk factors. Br. Dent. J. 2022, 233, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Mandel, P.; Metais, P. Nucleic acids of human blood plasma. Comptes Rendus Seances Soc. Biol. Ses Fil. 1948, 142, 241–243. [Google Scholar]
- Pös, O.; Biró, O.; Szemes, T.; Nagy, B. Circulating cell-free nucleic acids: Characteristics and applications. Eur. J. Hum. Genet. 2018, 26, 937–945. [Google Scholar] [CrossRef]
- Grabuschnig, S.; Bronkhorst, A.J.; Holdenrieder, S.; Rosales Rodriguez, I.; Schliep, K.P.; Schwendenwein, D.; Ungerer, V.; Sensen, C.W. Putative origins of cell-free DNA in humans: A review of active and passive nucleic acid release mechanisms. Int. J. Mol. Sci. 2020, 21, 8062. [Google Scholar] [CrossRef]
- Spector, B.L.; Harrell, L.; Sante, D.; Wyckoff, G.J.; Willig, L. The methylome and cell-free DNA: Current applications in medicine and pediatric disease. Pediatr. Res. 2023, 94, 89–95. [Google Scholar] [CrossRef]
- Celec, P.; Vlková, B.; Lauková, L.; Bábíčková, J.; Boor, P. Cell-free DNA: The role in pathophysiology and as a biomarker in kidney diseases. Expert Rev. Mol. Med. 2018, 20, e1. [Google Scholar] [CrossRef]
- Polina, I.A.; Ilatovskaya, D.V.; DeLeon-Pennell, K.Y. Cell free DNA as a diagnostic and prognostic marker for cardiovascular diseases. Clin. Chim. Acta 2020, 503, 145–150. [Google Scholar] [CrossRef]
- Xu, Y.; Song, Y.; Chang, J.; Zhou, X.; Qi, Q.; Tian, X.; Li, M.; Zeng, X.; Xu, M.; Zhang, W. High levels of circulating cell-free DNA are a biomarker of active SLE. Eur. J. Clin. Investig. 2018, 48, e13015. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.; Shapiro, B.; Sklaroff, D.; Yaros, M. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar] [PubMed]
- Dao, J.; Conway, P.J.; Subramani, B.; Meyyappan, D.; Russell, S.; Mahadevan, D. Using cfDNA and ctDNA as oncologic markers: A path to clinical validation. Int. J. Mol. Sci. 2023, 24, 13219. [Google Scholar] [CrossRef] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.-D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Alix-Panabières, C.; Pantel, K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef]
- Bellairs, J.A.; Hasina, R.; Agrawal, N. Tumor DNA: An emerging biomarker in head and neck cancer. Cancer Metastasis Rev. 2017, 36, 515–523. [Google Scholar] [CrossRef]
- Pessoa, L.S.; Heringer, M.; Ferrer, V.P. ctDNA as a cancer biomarker: A broad overview. Crit. Rev. Oncol. Hematol. 2020, 155, 103109. [Google Scholar] [CrossRef]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef]
- Mastoraki, S.; Strati, A.; Tzanikou, E.; Chimonidou, M.; Politaki, E.; Voutsina, A.; Psyrri, A.; Georgoulias, V.; Lianidou, E. ESR1 methylation: A liquid biopsy–based epigenetic assay for the follow-up of patients with metastatic breast cancer receiving endocrine treatment. Clin. Cancer Res. 2018, 24, 1500–1510. [Google Scholar] [CrossRef]
- Méhes, G. Liquid biopsy for predictive mutational profiling of solid cancer: The pathologist’s perspective. J. Biotechnol. 2019, 297, 66–70. [Google Scholar] [CrossRef]
- Huang, X.; Leo, P.; Jones, L.; Duijf, P.H.; Hartel, G.; Kenny, L.; Vasani, S.; Punyadeera, C. A comparison between mutational profiles in tumour tissue DNA and circulating tumour DNA in head and neck squamous cell carcinoma—A systematic review. Mutat. Res. Rev. Mutat. Res. 2023, 793, 108477. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.L.; D’Agostino, R.B., Jr.; Meegalla, N.; Petro, R.; Commander, S.; Topaloglu, U.; Zhang, W.; Porosnicu, M. The prognostic and therapeutic value of the mutational profile of blood and tumor tissue in head and neck squamous cell carcinoma. Oncologist 2021, 26, e279–e289. [Google Scholar] [CrossRef]
- Kampel, L.; Feldstein, S.; Tsuriel, S.; Hannes, V.; Carmel Neiderman, N.N.; Horowitz, G.; Warshavsky, A.; Leider-Trejo, L.; Hershkovitz, D.; Muhanna, N. Mutated TP53 in Circulating Tumor DNA as a Risk Level Biomarker in Head and Neck Squamous Cell Carcinoma Patients. Biomolecules 2023, 13, 1418. [Google Scholar] [CrossRef]
- Misawa, K.; Imai, A.; Matsui, H.; Kanai, A.; Misawa, Y.; Mochizuki, D.; Mima, M.; Yamada, S.; Kurokawa, T.; Nakagawa, T. Identification of novel methylation markers in HPV-associated oropharyngeal cancer: Genome-wide discovery, tissue verification and validation testing in ctDNA. Oncogene 2020, 39, 4741–4755. [Google Scholar] [CrossRef] [PubMed]
- Mydlarz, W.K.; Hennessey, P.T.; Wang, H.; Carvalho, A.L.; Califano, J.A. Serum biomarkers for detection of head and neck squamous cell carcinoma. Head Neck 2016, 38, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Wan, Y.; Vagenas, D.; Ovchinnikov, D.A.; Perry, C.F.; Davis, M.J.; Punyadeera, C. Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer 2016, 16, 749. [Google Scholar] [CrossRef]
- Pall, A.H.; Jakobsen, K.K.; Grønhøj, C.; von Buchwald, C. Circulating tumour DNA alterations as biomarkers for head and neck cancer: A systematic review. Acta Oncol. 2020, 59, 845–850. [Google Scholar] [CrossRef]
- Tian, F.; Yip, S.P.; Kwong, D.L.W.; Lin, Z.; Yang, Z.; Wu, V.W.C. Promoter hypermethylation of tumor suppressor genes in serum as potential biomarker for the diagnosis of nasopharyngeal carcinoma. Cancer Epidemiol. 2013, 37, 708–713. [Google Scholar] [CrossRef]
- Yang, X.; Dai, W.; Kwong, D.L.w.; Szeto, C.Y.; Wong, E.H.w.; Ng, W.T.; Lee, A.W.; Ngan, R.K.; Yau, C.C.; Tung, S.Y. Epigenetic markers for noninvasive early detection of nasopharyngeal carcinoma by methylation-sensitive high resolution melting. Int. J. Cancer 2015, 136, E127–E135. [Google Scholar] [CrossRef]
- Wong, T.-S.; Kwong, D.L.-W.; Sham, J.S.-T.; Wei, W.I.; Kwong, Y.-L.; Yuen, A.P.-W. Quantitative plasma hypermethylated DNA markers of undifferentiated nasopharyngeal carcinoma. Clin. Cancer Res. 2004, 10, 2401–2406. [Google Scholar] [CrossRef]
- Tan, R.; Phua, S.K.A.; Soong, Y.L.; Oon, L.L.E.; Chan, K.S.; Lucky, S.S.; Mong, J.; Tan, M.H.; Lim, C.M. Clinical utility of Epstein-Barr virus DNA and other liquid biopsy markers in nasopharyngeal carcinoma. Cancer Commun. 2020, 40, 564–585. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, K.; Mal, S.; Ghosh, M.; Chattopadhyay, N.R.; Roy, S.D.; Chakraborty, K.; Mukherjee, S.; Aier, M.; Choudhuri, T. Blood-based DNA methylation in advanced Nasopharyngeal Carcinoma exhibited distinct CpG methylation signature. Sci. Rep. 2023, 13, 22086. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.S.; Kerrigan, K.C.; Yang, D. Circulating tumor DNA profiling and serial analysis in salivary gland carcinomas reveal unique mutational subsets and actionable alterations. J. Clin. Oncol. 2022, 40, 6097. [Google Scholar] [CrossRef]
- Metcalf, R.; Mohan, S.; Hilton, S.; Pierce, J.; Hudson, J.; Betts, G.; Chaturvedi, A.; Homer, J.; Leong, H.; Schofield, P. The application of liquid biopsies in metastatic salivary gland cancer to identify candidate therapeutic targets. Ann. Oncol. 2017, 28, vii8. [Google Scholar] [CrossRef]
- Cabezas-Camarero, S.; de la Orden García, V.; García-Barberán, V.; Mediero-Valeros, B.; Subhi-Issa, A.I.; Llovet García, P.; Bando-Polaino, I.; Merino Menéndez, S.; Pérez-Segura, P.; Díaz-Rubio, E. Nasoethmoidal intestinal-type adenocarcinoma treated with cetuximab: Role of liquid biopsy and BEAMing in predicting response to anti-epidermal growth factor receptor therapy. Oncologist 2019, 24, 293–300. [Google Scholar] [CrossRef]
- Freiberger, S.N.; Turko, P.; Hüllner, M.; Dummer, R.; Morand, G.B.; Levesque, M.P.; Holzmann, D.; Rupp, N.J. Who’s driving? Switch of drivers in immunotherapy-treated progressing sinonasal melanoma. Cancers 2021, 13, 2725. [Google Scholar] [CrossRef]
- Tarasova, V.D.; Tsai, J.; Masannat, J.; Hernandez Prera, J.C.; Hallanger Johnson, J.; Veloski, C.; Agosto Salgado, S.; McIver, B.; Drusbosky, L.M.; Chung, C.H. Characterization of the Thyroid Cancer Genomic Landscape by Plasma-Based Circulating Tumor DNA Next-Generation Sequencing. Thyroid 2024, 34, 197–205. [Google Scholar] [CrossRef]
- Wang, W.; Zheng, Z.; Lei, J. CTC, ctDNA, and exosome in thyroid cancers: A review. Int. J. Mol. Sci. 2023, 24, 13767. [Google Scholar] [CrossRef]
- Sandulache, V.C.; Williams, M.D.; Lai, S.Y.; Lu, C.; William, W.N.; Busaidy, N.L.; Cote, G.J.; Singh, R.R.; Luthra, R.; Cabanillas, M.E. Real-time genomic characterization utilizing circulating cell-free DNA in patients with anaplastic thyroid carcinoma. Thyroid 2017, 27, 81–87. [Google Scholar] [CrossRef]
- Lee, T.H.; Jeon, H.J.; Choi, J.H.; Kim, Y.J.; Hwangbo, P.-N.; Park, H.S.; Son, C.Y.; Choi, H.-G.; Kim, H.N.; Chang, J.W. A high-sensitivity cfDNA capture enables to detect the BRAF V600E mutation in papillary thyroid carcinoma. Korean J. Chem. Eng. 2023, 40, 429–435. [Google Scholar] [CrossRef]
- Almubarak, H.; Qassem, E.; Alghofaili, L.; Alzahrani, A.S.; Karakas, B. Non-invasive molecular detection of minimal residual disease in papillary thyroid cancer patients. Front. Oncol. 2020, 9, 1510. [Google Scholar] [CrossRef] [PubMed]
- Khatami, F.; Larijani, B.; Heshmat, R.; Nasiri, S.; Haddadi-Aghdam, M.; Teimoori-Toolabi, L.; Tavangar, S.M. Hypermethylated RASSF1 and SLC5A8 promoters alongside BRAFV600E mutation as biomarkers for papillary thyroid carcinoma. J. Cell. Physiol. 2020, 235, 6954–6968. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Wang, Y.; Gao, J.; Li, Z.; Pang, R.; Zhai, T.; Ma, Y.; Wang, Z.; Meng, X. Detection of BRAFV600E mutation of thyroid cancer in circulating tumor DNA by an electrochemical-enrichment assisted ARMS-qPCR assay. Microchem. J. 2022, 179, 107452. [Google Scholar] [CrossRef]
- Ciampi, R.; Romei, C.; Ramone, T.; Matrone, A.; Prete, A.; Gambale, C.; Materazzi, G.; De Napoli, L.; Torregrossa, L.; Basolo, F. Pre-and post-operative circulating tumoral DNA in patients with medullary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2022, 107, e3420–e3427. [Google Scholar] [CrossRef] [PubMed]
- Santos, V.; Freitas, C.; Fernandes, M.G.; Sousa, C.; Reboredo, C.; Cruz-Martins, N.; Mosquera, J.; Hespanhol, V.; Campelo, R. Liquid biopsy: The value of different bodily fluids. Biomark. Med. 2022, 16, 127–145. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Circulating tumour cells in cancer patients: Challenges and perspectives. Trends Mol. Med. 2010, 16, 398–406. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Liquid biopsy: From discovery to clinical application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef]
- Schmidt, H.; Kulasinghe, A.; Perry, C.; Nelson, C.; Punyadeera, C. A liquid biopsy for head and neck cancers. Expert Rev. Mol. Diagn. 2016, 16, 165–172. [Google Scholar] [CrossRef]
- Schmidt, H.; Kulasinghe, A.; Kenny, L.; Punyadeera, C. The development of a liquid biopsy for head and neck cancers. Oral Oncol. 2016, 61, 8–11. [Google Scholar] [CrossRef]
- Spector, M.E.; Farlow, J.L.; Haring, C.T.; Brenner, J.C.; Birkeland, A.C. The potential for liquid biopsies in head and neck cancer. Discov. Med. 2018, 25, 251. [Google Scholar] [PubMed]
- Hsu, C.-L.; Chang, Y.-S.; Li, H.-P. Molecular Diagnosis of Nasopharyngeal Carcinoma: Past and Future. Biomed. J. 2024, 23, 100748. [Google Scholar] [CrossRef] [PubMed]
- Jakimovska, F.; Stojkovski, I.; Kjosevska, E. Nasal Cavity and Paranasal Sinus Cancer: Diagnosis and Treatment. Curr. Oncol. Rep. 2024, 2024, 1–13. [Google Scholar] [CrossRef]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 131. [Google Scholar] [CrossRef]
- Kong, L.; Birkeland, A.C. Liquid biopsies in head and neck cancer: Current state and future challenges. Cancers 2021, 13, 1874. [Google Scholar] [CrossRef]
- Patel, A.; Patel, S.; Patel, P.; Tanavde, V. Saliva based liquid biopsies in head and neck cancer: How far are we from the clinic? Front. Oncol. 2022, 12, 828434. [Google Scholar] [CrossRef]
- Mishra, V.; Singh, A.; Chen, X.; Rosenberg, A.J.; Pearson, A.T.; Zhavoronkov, A.; Savage, P.A.; Lingen, M.W.; Agrawal, N.; Izumchenko, E. Application of liquid biopsy as multi-functional biomarkers in head and neck cancer. Br. J. Cancer 2022, 126, 361–370. [Google Scholar] [CrossRef]
- Cabezas-Camarero, S.; Pérez-Segura, P. Liquid biopsy in head and neck cancer: Current evidence and future perspective on squamous cell, salivary gland, paranasal sinus and nasopharyngeal cancers. Cancers 2022, 14, 2858. [Google Scholar] [CrossRef]
- Rutkowski, T.W.; Mazurek, A.M.; Śnietura, M.; Hejduk, B.; Jędrzejewska, M.; Bobek-Billewicz, B.; d’Amico, A.; Pigłowski, W.; Wygoda, A.; Składowski, K.; et al. Circulating HPV16 DNA may complement imaging assessment of early treatment efficacy in patients with HPV-positive oropharyngeal cancer. J. Transl. Med. 2020, 18, 167. [Google Scholar] [CrossRef]
- Lele, S.J.; Adilbay, D.; Lewis, E.; Pang, J.; Asarkar, A.A.; Nathan, C.A.O. ctDNA as an Adjunct to Posttreatment PET for Head and Neck Cancer Recurrence Risk. JAMA Otolaryngol.-Head Neck Surg. 2024, 171, 439–444. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Ferrandino, R.M.; Chen, S.; Kappauf, C.; Barlow, J.; Gold, B.S.; Berger, M.H.; Westra, W.H.; Teng, M.S.; Khan, M.N.; Posner, M.R.; et al. Performance of Liquid Biopsy for Diagnosis and Surveillance of Human Papillomavirus-Associated Oropharyngeal Cancer. JAMA Otolaryngol.-Head Neck Surg. 2023, 149, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Mattox, A.K.; D’Souza, G.; Khan, Z.; Allen, H.; Henson, S.; Seiwert, T.Y.; Koch, W.; Pardoll, D.M.; Fakhry, C. Comparison of next generation sequencing, droplet digital PCR, and quantitative real-time PCR for the earlier detection and quantification of HPV in HPV-positive oropharyngeal cancer. Oral Oncol. 2022, 128, 105805. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; O’Boyle, C.J.; Varmeh, S.; Queenan, N.; Michel, A.; Stein, J.; Thierauf, J.; Sadow, P.M.; Faquin, W.C.; Perry, S.K. Cell-free HPV DNA provides an accurate and rapid diagnosis of HPV-associated head and neck cancer. Clin. Cancer Res. 2022, 28, 719–727. [Google Scholar] [CrossRef]
- Chera, B.S.; Kumar, S.; Beaty, B.T.; Marron, D.; Jefferys, S.; Green, R.; Goldman, E.C.; Amdur, R.; Sheets, N.; Dagan, R. Rapid clearance profile of plasma circulating tumor HPV type 16 DNA during chemoradiotherapy correlates with disease control in HPV-associated oropharyngeal cancer. Clin. Cancer Res. 2019, 25, 4682–4690. [Google Scholar] [CrossRef]
- Damerla, R.R.; Lee, N.Y.; You, D.; Soni, R.; Shah, R.; Reyngold, M.; Katabi, N.; Wu, V.; McBride, S.M.; Tsai, C.J. Detection of early human papillomavirus–associated cancers by liquid biopsy. JCO Precis. Oncol. 2019, 3, 1–17. [Google Scholar] [CrossRef]
- Chan, K.A.; Woo, J.K.; King, A.; Zee, B.C.; Lam, W.J.; Chan, S.L.; Chu, S.W.; Mak, C.; Tse, I.O.; Leung, S.Y. Analysis of plasma Epstein–Barr virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med. 2017, 377, 513–522. [Google Scholar] [CrossRef]
- Ahn, S.M.; Chan, J.Y.; Zhang, Z.; Wang, H.; Khan, Z.; Bishop, J.A.; Westra, W.; Koch, W.M.; Califano, J.A. Saliva and plasma quantitative polymerase chain reaction–based detection and surveillance of human papillomavirus–related head and neck cancer. JAMA Otolaryngol.-Head Neck Surg. 2014, 140, 846–854. [Google Scholar] [CrossRef]
- Rapado-González, Ó.; Rodríguez-Ces, A.M.; López-López, R.; Suárez-Cunqueiro, M.M. Liquid biopsies based on cell-free DNA as a potential biomarker in head and neck cancer. Jpn. Dent. Sci. Rev. 2023, 59, 289–302. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Prim. 2020, 6, 92. [Google Scholar] [CrossRef]
- Marur, S.; Forastiere, A.A. Head and neck squamous cell carcinoma: Update on epidemiology, diagnosis, and treatment. Mayo Clin. Proc. 2016, 91, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Pannone, G.; Santoro, A.; Papagerakis, S.; Lo Muzio, L.; De Rosa, G.; Bufo, P. The role of human papillomavirus in the pathogenesis of head & neck squamous cell carcinoma: An overview. Infect. Agents Cancer 2011, 6, 4. [Google Scholar]
- Economopoulou, P.; Kotsantis, I.; Kyrodimos, E.; Lianidou, E.; Psyrri, A. Liquid biopsy: An emerging prognostic and predictive tool in head and neck squamous cell carcinoma (HNSCC). Focus on circulating tumor cells (CTCs). Oral Oncol. 2017, 74, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Payne, K.; Spruce, R.; Beggs, A.; Sharma, N.; Kong, A.; Martin, T.; Parmar, S.; Praveen, P.; Nankivell, P.; Mehanna, H. Circulating tumor DNA as a biomarker and liquid biopsy in head and neck squamous cell carcinoma. Head Neck 2018, 40, 1598–1604. [Google Scholar] [CrossRef]
- Payne, K.F.; Brotherwood, P.; Suriyanarayanan, H.; Brooks, J.M.; Batis, N.; Beggs, A.D.; Gendoo, D.M.; Mehanna, H.; Nankivell, P. Circulating tumour DNA detects somatic variants contributing to spatial and temporal intra-tumoural heterogeneity in head and neck squamous cell carcinoma. Front. Oncol. 2024, 14, 1374816. [Google Scholar] [CrossRef]
- Hudečková, M.; Koucký, V.; Rottenberg, J.; Gál, B. Gene mutations in circulating tumour DNA as a diagnostic and prognostic marker in head and neck cancer—A systematic review. Biomedicines 2021, 9, 1548. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Sougnez, C.; Lichtenstein, L.; Cibulskis, K.; Lander, E.; Gabriel, S.B.; Getz, G.; Ally, A.; Balasundaram, M.; Birol, I.; et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.-X.; Zhang, J.; Wang, J. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011, 333, 1154–1157. [Google Scholar] [CrossRef]
- Nwachuku, K.; Johnson, D.E.; Grandis, J.R. The mutational landscape of head and neck squamous cell carcinoma: Opportunities for detection and monitoring via analysis of circulating tumor DNA. In Early Detection and Treatment of Head & Neck Cancers: Theoretical Background and Newly Emerging Research; Springer: Berlin/Heidelberg, Germany, 2021; pp. 107–122. [Google Scholar]
- Brauswetter, D.; Dános, K.; Gurbi, B.; Félegyházi, É.F.; Birtalan, E.; Meggyesházi, N.; Krenács, T.; Tamás, L.; Peták, I. Copy number gain of PIK3CA and MET is associated with poor prognosis in head and neck squamous cell carcinoma. Virchows Arch. 2016, 468, 579–587. [Google Scholar] [CrossRef]
- Kim, K.T.; Kim, B.S.; Kim, J.H. Association between FAT1 mutation and overall survival in patients with human papillomavirus–negative head and neck squamous cell carcinoma. Head Neck 2016, 38, E2021–E2029. [Google Scholar] [CrossRef]
- Cochicho, D.; Esteves, S.; Rito, M.; Silva, F.; Martins, L.; Montalvão, P.; Cunha, M.; Magalhães, M.; Gil da Costa, R.M.; Felix, A. PIK3CA gene mutations in HNSCC: Systematic review and correlations with HPV status and patient survival. Cancers 2022, 14, 1286. [Google Scholar] [CrossRef]
- Hanna, G.; Supplee, J.; Kuang, Y.; Mahmood, U.; Lau, C.; Haddad, R.; Jänne, P.; Paweletz, C. Plasma HPV cell-free DNA monitoring in advanced HPV-associated oropharyngeal cancer. Ann. Oncol. 2018, 29, 1980–1986. [Google Scholar] [CrossRef] [PubMed]
- Ferrier, S.T.; Tsering, T.; Sadeghi, N.; Zeitouni, A.; Burnier, J.V. Blood and saliva-derived ctDNA is a marker of residual disease after treatment and correlates with recurrence in human papillomavirus-associated head and neck cancer. Cancer Med. 2023, 12, 15777–15787. [Google Scholar] [CrossRef] [PubMed]
- Bhambhani, C.; Kang, Q.; Hovelson, D.H.; Sandford, E.; Olesnavich, M.; Dermody, S.M.; Wolfgang, J.; Tuck, K.L.; Brummel, C.; Bhangale, A.D. ctDNA transiting into urine is ultrashort and facilitates noninvasive liquid biopsy of HPV+ oropharyngeal cancer. JCI Insight 2024, 9, e177759. [Google Scholar] [CrossRef]
- Naegele, S.; Efthymiou, V.; Das, D.; Sadow, P.M.; Richmon, J.D.; Iafrate, A.J.; Faden, D.L. Detection and monitoring of circulating tumor HPV DNA in HPV-associated sinonasal and nasopharyngeal cancers. JAMA Otolaryngol.-Head Neck Surg. 2023, 149, 179–181. [Google Scholar] [CrossRef]
- Chantre-Justino, M.; Alves, G.; Delmonico, L. Clinical applications of liquid biopsy in HPV-negative and HPV-positive head and neck squamous cell carcinoma: Advances and challenges. Explor. Target. Anti-Tumor Ther. 2022, 3, 533. [Google Scholar] [CrossRef]
- Chen, Y.-P.; Chan, A.T.; Le, Q.-T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef]
- Tsao, S.W.; Yip, Y.L.; Tsang, C.M.; Pang, P.S.; Lau, V.M.Y.; Zhang, G.; Lo, K.W. Etiological factors of nasopharyngeal carcinoma. Oral Oncol. 2014, 50, 330–338. [Google Scholar] [CrossRef]
- Lee, H.M.; Okuda, K.S.; González, F.E.; Patel, V. Current perspectives on nasopharyngeal carcinoma. In Human Cell Transformation—Advances in Cell Models for the Study of Cancer and Aging; Springer: Berlin/Heidelberg, Germany, 2019; pp. 11–34. [Google Scholar]
- Su, Z.Y.; Siak, P.Y.; Lwin, Y.Y.; Cheah, S.-C. Epidemiology of nasopharyngeal carcinoma: Current insights and future outlook. Cancer Metastasis Rev. 2024, 43, 919–939. [Google Scholar] [CrossRef]
- Lo, Y.D.; Chan, L.Y.; Lo, K.-W.; Leung, S.-F.; Zhang, J.; Chan, A.T.; Lee, J.C.; Hjelm, N.M.; Johnson, P.J.; Huang, D.P. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999, 59, 1188–1191. [Google Scholar]
- Peng, L.; Yang, Y.; Guo, R.; Mao, Y.P.; Xu, C.; Chen, Y.P.; Sun, Y.; Ma, J.; Tang, L.L. Relationship between pretreatment concentration of plasma Epstein-Barr virus DNA and tumor burden in nasopharyngeal carcinoma: An updated interpretation. Cancer Med. 2018, 7, 5988–5998. [Google Scholar] [CrossRef]
- Lv, J.; Wu, C.; Li, J.; Chen, F.; He, S.; He, Q.; Zhou, G.; Ma, J.; Sun, Y.; Wei, D. Improving on-treatment risk stratification of cancer patients with refined response classification and integration of circulating tumor DNA kinetics. BMC Med. 2022, 20, 268. [Google Scholar] [CrossRef]
- Lam, W.J.; Jiang, P.; Chan, K.A.; Cheng, S.H.; Zhang, H.; Peng, W.; Tse, O.O.; Tong, Y.K.; Gai, W.; Zee, B.C. Sequencing-based counting and size profiling of plasma Epstein–Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. USA 2018, 115, E5115–E5124. [Google Scholar] [CrossRef]
- Nicholls, J.M.; Lee, V.H.-F.; Chan, S.-K.; Tsang, K.-C.; Choi, C.-W.; Kwong, D.L.-W.; Lam, K.-O.; Chan, S.-Y.; Tong, C.-C.; So, T.-H. Negative plasma Epstein-Barr virus DNA nasopharyngeal carcinoma in an endemic region and its influence on liquid biopsy screening programmes. Br. J. Cancer 2019, 121, 690–698. [Google Scholar] [CrossRef]
- Zheng, X.H.; Deng, C.M.; Zhou, T.; Li, X.Z.; Tang, C.L.; Jiang, C.T.; Liao, Y.; Wang, T.M.; He, Y.Q.; Jia, W.H. Saliva biopsy: Detecting the difference of EBV DNA methylation in the diagnosis of nasopharyngeal carcinoma. Int. J. Cancer 2023, 153, 882–892. [Google Scholar] [CrossRef]
- Liu, T.; Liu, J.; Wang, G.; Chen, C.; He, L.; Wang, R.; Ouyang, C. Circulating tumor cells: A valuable indicator for locally advanced nasopharyngeal carcinoma. Eur. Arch. Oto-Rhino-Laryngol. 2024, 2024, 1–10. [Google Scholar] [CrossRef]
- Wu, L.; Wang, J.; Zhu, D.; Zhang, S.; Zhou, X.; Zhu, W.; Zhu, J.; He, X. Circulating Epstein-Barr virus microRNA profile reveals novel biomarker for nasopharyngeal carcinoma diagnosis. Cancer Biomark. 2020, 27, 365–375. [Google Scholar] [CrossRef]
- Wei, J.; Meng, X.; Wei, X.; Zhu, K.; Du, L.; Wang, H. Down-regulated lncRNA ROR in tumor-educated platelets as a liquid-biopsy biomarker for nasopharyngeal carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 4403–4409. [Google Scholar] [CrossRef]
- Britze, T.E.; Jakobsen, K.K.; Grønhøj, C.; von Buchwald, C. A systematic review on the role of biomarkers in liquid biopsies and saliva samples in the monitoring of salivary gland cancer. Acta Oto-Laryngol. 2023, 143, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Cappelletti, V.; Miodini, P.; Reduzzi, C.; Alfieri, S.; Daidone, M.; Licitra, L.; Locati, L. Tailoring treatment of salivary duct carcinoma (SDC) by liquid biopsy: ARv7 expression in circulating tumor cells. Ann. Oncol. 2018, 29, 1599–1601. [Google Scholar] [CrossRef]
- Fisher, B.M.; Tang, K.; Warkiani, M.; Punyadeera, C.; Batstone, M. A pilot study for presence of circulating tumour cells in adenoid cystic carcinoma. Int. J. Oral Maxillofac. Surg. 2021, 50, 994–998. [Google Scholar] [CrossRef]
- Bigagli, E.; Maggiore, G.; Cinci, L.; D’Ambrosio, M.; Locatello, L.G.; Nardi, C.; Palomba, A.; Leopardi, G.; Orlando, P.; Licci, G. Low levels of miR-34c in nasal washings as a candidate marker of aggressive disease in wood and leather exposed workers with sinonasal intestinal-type adenocarcinomas (ITACs). Transl. Oncol. 2022, 25, 101507. [Google Scholar] [CrossRef]
- Buglione, M.; Grisanti, S.; Almici, C.; Mangoni, M.; Polli, C.; Consoli, F.; Verardi, R.; Costa, L.; Paiar, F.; Pasinetti, N. Circulating tumour cells in locally advanced head and neck cancer: Preliminary report about their possible role in predicting response to non-surgical treatment and survival. Eur. J. Cancer 2012, 48, 3019–3026. [Google Scholar] [CrossRef]
- Zeyghami, W.; Hansen, M.-L.U.; Jakobsen, K.K.; Groenhøj, C.; Feldt-Rasmussen, U.; von Buchwald, C.; Hahn, C.H. Liquid biopsies in thyroid cancers: A systematic review and meta-analysis. Endocr. Relat. Cancer 2023, 30, e230002. [Google Scholar] [CrossRef]
- Porter, A.; Natsuhara, M.; Daniels, G.A.; Patel, S.P.; Sacco, A.G.; Bykowski, J.; Banks, K.C.; Cohen, E.E. Next generation sequencing of cell free circulating tumor DNA in blood samples of recurrent and metastatic head and neck cancer patients. Transl. Cancer Res. 2020, 9, 203. [Google Scholar] [CrossRef]
- Lanman, R.B.; Mortimer, S.A.; Zill, O.A.; Sebisanovic, D.; Lopez, R.; Blau, S.; Collisson, E.A.; Divers, S.G.; Hoon, D.S.; Kopetz, E.S. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS ONE 2015, 10, e0140712. [Google Scholar] [CrossRef]
- Nonaka, T.; Wong, D. Liquid biopsy in head and neck cancer: Promises and challenges. J. Dent. Res. 2018, 97, 701–708. [Google Scholar] [CrossRef]
- Araujo, A.L.D.; Santos-Silva, A.R.; Kowalski, L.P. Diagnostic accuracy of liquid biopsy for Oral potentially malignant disorders and head and neck cancer: An overview of systematic reviews. Curr. Oncol. Rep. 2023, 25, 279–292. [Google Scholar] [CrossRef]
- Whale, A.S.; Huggett, J.F.; Tzonev, S. Fundamentals of multiplexing with digital PCR. Biomol. Detect. Quantif. 2016, 10, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Nekrutenko, A.; Taylor, J. Next-generation sequencing data interpretation: Enhancing reproducibility and accessibility. Nat. Rev. Genet. 2012, 13, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Iacoangeli, A.; Al Khleifat, A.; Sproviero, W.; Shatunov, A.; Jones, A.; Morgan, S.; Pittman, A.; Dobson, R.; Newhouse, S.; Al-Chalabi, A. DNAscan: Personal computer compatible NGS analysis, annotation and visualisation. BMC Bioinform. 2019, 20, 213. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.A.; Beck, J.; Leu, M.; Oellerich, M.; Rave-Fränk, M.; Walson, P.D.; Schütz, E.; Canis, M. Cell-free plasma DNA for disease stratification and prognosis in head and neck cancer. Clin. Chem. 2018, 64, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Rosing, F.; Meier, M.; Schroeder, L.; Laban, S.; Hoffmann, T.; Kaufmann, A.; Siefer, O.; Wuerdemann, N.; Klußmann, J.P.; Rieckmann, T. Quantification of human papillomavirus cell-free DNA from low-volume blood plasma samples by digital PCR. Microbiol. Spectr. 2024, 12, e00024. [Google Scholar] [CrossRef] [PubMed]
- Zwirner, K.; Hilke, F.J.; Demidov, G.; Ossowski, S.; Gani, C.; Rieß, O.; Zips, D.; Welz, S.; Schroeder, C. Circulating cell-free DNA: A potential biomarker to differentiate inflammation and infection during radiochemotherapy. Radiother. Oncol. 2018, 129, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.O. Circulating cell-free DNA differentiates severity of inflammation. Biol. Res. Nurs. 2016, 18, 477–488. [Google Scholar] [CrossRef]
- Ma, M.; Zhu, H.; Zhang, C.; Sun, X.; Gao, X.; Chen, G. “Liquid biopsy”-ctDNA detection with great potential and challenges. Ann. Transl. Med. 2015, 3, 235. [Google Scholar] [CrossRef]
- Di Capua, D.; Bracken-Clarke, D.; Ronan, K.; Baird, A.-M.; Finn, S. The liquid biopsy for lung cancer: State of the art, limitations and future developments. Cancers 2021, 13, 3923. [Google Scholar] [CrossRef]
- Allen, T.A. The Role of Circulating Tumor Cells as a Liquid Biopsy for Cancer: Advances, Biology, Technical Challenges, and Clinical Relevance. Cancers 2024, 16, 1377. [Google Scholar] [CrossRef]
- Rossi, G.; Ignatiadis, M. Promises and pitfalls of using liquid biopsy for precision medicine. Cancer Res. 2019, 79, 2798–2804. [Google Scholar] [CrossRef]
- Boukovala, M.; Westphalen, C.B.; Probst, V. Liquid biopsy into the clinics: Current evidence and future perspectives. J. Liq. Biopsy 2024, 4, 100146. [Google Scholar] [CrossRef]
- Heidrich, I.; Ačkar, L.; Mossahebi Mohammadi, P.; Pantel, K. Liquid biopsies: Potential and challenges. Int. J. Cancer 2021, 148, 528–545. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-C. Circulating tumor DNA in liquid biopsy: Current diagnostic limitation. World J. Gastroenterol. 2024, 30, 2175. [Google Scholar] [CrossRef] [PubMed]
- Haring, C.T.; Kana, L.A.; Dermody, S.M.; Brummel, C.; McHugh, J.B.; Casper, K.A.; Chinn, S.B.; Malloy, K.M.; Mierzwa, M.; Prince, M.E. Patterns of recurrence in head and neck squamous cell carcinoma to inform personalized surveillance protocols. Cancer 2023, 129, 2817–2827. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.X.; Kut, C.; Quon, H.; Seiwert, T.Y.; D’souza, G.; Fakhry, C. Clinical uncertainties of circulating tumor DNA in human papillomavirus–related oropharyngeal squamous cell carcinoma in the absence of National Comprehensive Cancer Network Guidelines. J. Clin. Oncol. 2023, 41, 2483. [Google Scholar] [CrossRef]
- Hanna, G.J.; Patel, N.; Tedla, S.G.; Baugnon, K.L.; Aiken, A.; Agrawal, N. Personalizing surveillance in head and neck cancer. Am. Soc. Clin. Oncol. Educ. Book 2023, 43, e389718. [Google Scholar] [CrossRef]
- Mannelli, C. Tissue vs. liquid biopsies for cancer detection: Ethical issues. J. Bioethical Inq. 2019, 16, 551–557. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Febbo, P.G.; Allo, M.; Alme, E.B.; Cuyun Carter, G.; Dumanois, R.; Essig, A.; Kiernan, E.; Kubler, C.B.; Martin, N.; Popescu, M.C. Recommendations for the equitable and widespread implementation of liquid biopsy for cancer care. JCO Precis. Oncol. 2024, 8, e2300382. [Google Scholar] [CrossRef]
- Guan, J. A Bidirectional Study in Exploring the Dynamic Changes of Plasma and Urine Metabolites During the Occurrence and Development of Head and Neck Cancer in Southern China. ClinicalTrials.gov identifier: NCT05969262. 2023. Available online: https://clinicaltrials.gov/study/NCT05969262?cond=Head%20and%20Neck%20Cancer&intr=ctDNA&term=NCT05969262%20&rank=1 (accessed on 3 September 2024).
- Ofo, E. Liquid Biopsy for Early DiagNosis of Squamous Cell Carcinoma of the Head and Neck Region (ENHANCE Study). ClinicalTrials.gov Identifier: NCT05645783. 2023. Available online: https://clinicaltrials.gov/study/NCT05645783?term=NCT05645783%20&rank=1 (accessed on 3 September 2024).
- Pilka, R. Liquid Biopsies—A Possible Tool for Treatment Monitoring and Early Recurrence Detection in HPV-associated Diseases. ClinicalTrials.gov Identifier: NCT05774561. 2024. Available online: https://clinicaltrials.gov/study/NCT05774561?term=NCT05774561%20&rank=1 (accessed on 3 September 2024).
- Guan, J. A Bidirectional Study in Exploring the Dynamic Changes of Plasma and Urine Metabolites During the Occurrence and Development of Nasopharyngeal Carcinoma in Southern China. ClinicalTrials.gov Identifier: NCT05682703. 2023. Available online: https://clinicaltrials.gov/study/NCT05682703?term=NCT05682703%20&rank=1 (accessed on 3 September 2024).
- Jonsson Comprehensive Cancer Centre. Isolation and Characterization of Extracellular Vesicles in Patients with Thyroid Nodules and Thyroid Cancer. ClinicalTrials.gov Identifier: NCT04742608. 2024. Available online: https://clinicaltrials.gov/study/NCT04742608?term=NCT04742608%20&rank=1 (accessed on 3 September 2024).
- Princess Margaret Cancer Center. Real-Time Detection of ctDNA and/or HPV DNA in High-Risk Locally-Advanced Head and Neck Squamous Cell Carcinoma (LA-HNSCC): The Pre-MERIDIAN (Molecular Residual Disease Interception in High-Risk LA-HNSCC) Study. ClinicalTrials.gov Identifier: NCT04599309. 2024. Available online: https://clinicaltrials.gov/study/NCT04599309?term=NCT04599309%20&rank=1 (accessed on 3 September 2024).
- Princess Margaret Cancer Center. Study of Circulating Tumor DNA (ctDNA) Kinetics in Immuno-oncology: Intense Dynamic Monitoring of ctDNA in Advanced/Metastatic Head and Neck Squamous Cell Carcinoma (HNSCC) Patients Treated with Immune Checkpoint Inhibitors. ClinicalTrials.gov Identifier: NCT04606940. 2022. Available online: https://clinicaltrials.gov/study/NCT04606940?term=NCT04606940%20&rank=1 (accessed on 3 September 2024).
- Pharmissist Ltd. A Clinical Performance Study to Validate the Use of Novel Molecular Diagnostic Assays for the Detection of Cancer Biomarkers in Peripheral Blood and Primary Tumor Tissue of Patients With Recurrent/Metastatic HNSCC, NSCLC or Melanoma. ClinicalTrials.gov Identifier: NCT04490564. 2023. Available online: https://clinicaltrials.gov/study/NCT04490564?term=NCT04490564%20&rank=1 (accessed on 3 September 2024).
- Minn, H. Genetic Profiling by Liquid Biopsy for Initial Characterization and Response Monitoring of Head and Neck Squamous Cell Carcinoma (HNSCC). ClinicalTrials.gov Identifier: NCT03926468. 2019. Available online: https://clinicaltrials.gov/study/NCT03926468?term=NCT03926468%20&rank=1 (accessed on 3 September 2024).
- Princess Margaret Cancer Centre. Multi-Omic Assessment of Squamous Cell Cancers Receiving Systemic Therapy. ClinicalTrials.gov Identifier: NCT03712566. 2024. Available online: https://clinicaltrials.gov/study/NCT03712566?term=NCT03712566%20&rank=1 (accessed on 3 September 2024).
- Grønhøj, C. Cell-Free Tumor DNAand HPV-DNA in Blood Samples From Newly Diagnosed Patients with Head and Neck Cancer. ClinicalTrial.gov Identifier: NCT03942380. 2021. Available online: https://clinicaltrials.gov/study/NCT03942380?term=NCT03942380%20&rank=1 (accessed on 3 September 2024).
- Wirth, M. Liquid Biopsy of Head and Neck Cancer Patients in Blood and Saliva. ClinicalTrial.gov Identifier: NCT05122507. 2023. Available online: https://clinicaltrials.gov/study/NCT05122507?term=NCT05122507%20&rank=1 (accessed on 3 September 2024).

| Type of Malignancy | Identified Altered ctDNA Genes |
|---|---|
| Head and neck squamous cell carcinomas [22,23,24,25,26,27,28] |
|
| Nasopharyngeal carcinoma [29,30,31,32,33] |
|
| Salivary gland carcinomas [34,35] |
|
| Sinonasal carcinomas [36,37] |
|
| Thyroid carcinomas [38,39,40,41,42,43,44,45] |
|
| Type of Malignancy | Diagnostic Standard |
|---|---|
| Head and neck squamous cell carcinomas [51] |
|
| Nasopharyngeal carcinoma [53] |
|
| Salivary gland carcinomas |
|
| Sinonasal carcinomas [54] |
|
| Thyroid carcinomas [39] |
|
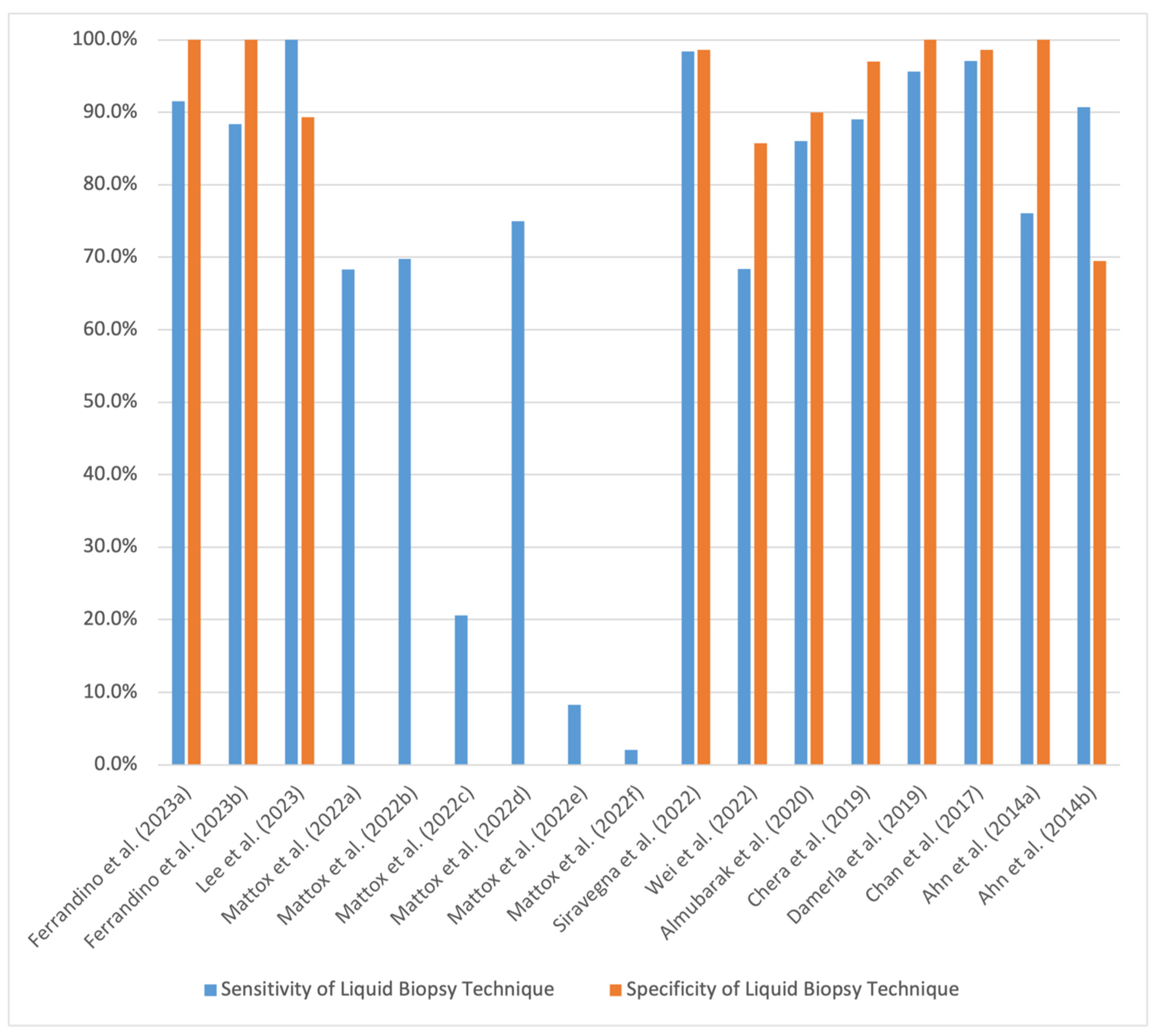
| Study | Aim | Type of Cancer | Sample Size | Method and Sample Used | Sensitivity of Liquid Biopsy Technique | Specificity of Liquid Biopsy Technique | Method Compared to | Conclusions | Limitations |
|---|---|---|---|---|---|---|---|---|---|
| Ferrandino et al. (2023) [63] | Determining performance metrics of ctHPVDNA in the diagnosis of oropharyngeal SCC | Oropharyngeal SCC | 163; 152 with HPV-positive SCC and 11 with HPV-negative SCC | TTMV-HPV DNA testing; plasma | 91.5% | 100% | Using tumor samples, p16 staining confirmed diagnosis in 98.7% of patients, HPV PCR in 75%, and in situ hybridization assays in 10.5% | Approximately 1 in 10 false negatives will result; ctHPVDNA assays should be used in conjunction with other tests | Ascertainment bias as patients were known to have HPV-positive oropharyngeal SCC |
| Determining performance metrics of ctHPVDNA in the detection of recurrence of HPV-positive oropharyngeal SCC at 3 months post treatment completion | HPV-positive oropharyngeal SCC | 290 | TTMV-HPV DNA testing; plasma | 88.4% | 100% | Using tumor samples, p16 staining confirmed diagnosis in 95.5% of patients, HPV PCR in 84.8%, and in situ hybridization assays in 4.8% | Prospective study where most patients did not have pretreatment ctHPVDNA measurements available; conservative definition of false-negative recurrence at 3-month follow-up potentially lowered the calculated sensitivity of the assay | ||
| Lee et al. (2023) [41] | Detecting multifocality of papillary thyroid carcinoma (PTC) | PTC | 37 | PDA/ SiO2-coated bead cfDNA detection assay; serum | 100% | 89.3% | Free T4 had a sensitivity of 33.3% and specificity of 82.1%; TSH had a sensitivity of 66.7% and specificity of 60.7%; Tg had a sensitivity of 11.1% and specificity of 64.3%; TgAb had a sensitivity of 33.3% and specificity of 82.1% | PDA/SiO2-coated bead liquid biopsy assays effectively capture ctDNA that can permit analysis of multiple mutations associated with PTC | Small sample size, no control subjects |
| Mattox et al. (2022) [64] | Comparing sensitivity of NGS, ddPCR, and qPCR assays in the detection of ctHPVDNA in plasma and oral rinse samples | HPV-16-positive oropharyngeal SCC | 66 | NGS; plasma | 68.3% | Not reported | Compared qPCR, ddPCR, and NGS sensitivity values for analysis of plasma and oral rinse samples | NGS and ddPCR have significantly higher sensitivity values for the detection of ctHPVDNA in plasma samples compared to qPCR, while NGS has a significantly higher sensitivity value for the detection of ctHPVDNA in oral rinse samples compared to ddPCR and qPCR. Levels of ctHPVDNA detected by NGS in plasma samples may reflect the clinical course of patients with HPV-positive oropharyngeal SCC | Small sample size, no control subjects |
| ddPCR; plasma | 69.8% | ||||||||
| qPCR; plasma | 20.6% | ||||||||
| NGS; oral rinse | 75% | ||||||||
| ddPCR; oral rinse | 8.3% | ||||||||
| qPCR; oral rinse | 2.1% | ||||||||
| Siravegna et al. (2022) [65] | Comparing effectiveness of a non-invasive diagnostic approach using ctHPVDNA liquid biopsy and imaging/physical exam to a standard diagnostic clinical workup with tumor biopsy | HPV-positive HNSCC (oropharyngeal, nasopharyngeal, and sinonasal SCCs) | 131, 61 patients with HPV-positive HNSCC, 45 controls with HPV-negative HNSCC, and 25 healthy controls | ddPCR; plasma | 98.4% | 98.6% | Standard clinical workup with tumor biopsy | Non-invasive techniques with liquid biopsy were significantly more effective in diagnosing HPV-positive HNSCC compared to the current standard of care with tumor biopsy (Youden index of 0.937 vs. 0.070) | Selection and information biases due to observational study design, lack of details present in referenced outside medical records |
| Wei et al. (2022) [44] | Testing the effectiveness of EC-ARMS-qPCR assay in the detection of BRAFV600E mutation in ctDNA from plasma samples of patients with PTC | PTC | 74; 54 patients with PTC and 20 patients with benign thyroid nodules | EC-ARMS-qPCR assay; plasma | 68.42% | 85.71% | EC-ARMS-qPCR assay using FNA samples (concordance of 73.08%) | EC-ARMS-qPCR assay can detect BRAFV600E ctDNA mutations in plasma samples and is in good concordance with test results of EC-ARMS-qPCR assay performed using FNA tissue samples | Small sample size, case–control design, only 26 patients (22 with PTC, 4 with benign thyroid nodules) underwent FNA for comparison |
| Almubarak et al. (2020) [42] | Assessing the use of liquid biopsy to detect BRAFV600E mutations in plasma ctDNA of patients with PTC for monitoring of minimal residual tumor presence | PTC | 38 | 3D digital PCR; plasma | 86% | 90% | Serum Tg (sensitivity of 78%, sensitivity of 65%) | The 3D digital PCR plasma assay using ctDNA had greater sensitivity and specificity for detecting minimal residual PTC tumors than serum Tg levels. The use of both techniques in conjunction could further increase sensitivity and specificity values | Small sample size |
| Chera et al. (2019) [66] | Determining performance metrics of ddPCR in diagnosing non-metastatic HPV-positive oropharyngeal SCC and testing for disease control in patients post chemoradiotherapy using blood plasma ctDNA | HPV-positive oropharyngeal SCC | 103 | ddPCR; plasma | 89% | 97% | Did not have a method to compare to; patients were eligible if they had their diagnoses confirmed by tumor biopsy | ctHPVDNA is detectable in patients with newly diagnosed HPV-positive oropharyngeal SCC and liquid biopsy assays quantifying plasma ctHPVDNA levels can stratify patients by risk in the post-treatment period | Low power due to low rates of disease persistence, disease recurrence, and patient follow-up |
| Damerla et al. (2019) [67] | Assess effectiveness of ddPCR using plasma ctHPVDNA in the detection of early-stage HPV-associated SCC | HPV-positive oropharyngeal SCC | 132; 97 patients with HPV-positive oropharyngeal SCC, 8 patients with HPV-positive anal SCC, 7 controls with HPV-negative oropharyngeal SCC, and 20 healthy controls without cancer | ddPCR; plasma | 95.6% (only accounting for oropharyngeal SCC patients) | 100% (only accounting for oropharyngeal SCC patients) | p16 immunohistochemistry assays and HPV DNA or RNA in situ hybridization assays using tumor tissue (sensitivity and specificity values not reported) | ctHPVDNA has high sensitivity and specificity for the detection of intact HPV-positive tumors, even in patients with low tumor burden. This implies clinical utility in screening and treatment response monitoring | Did not genotype all pathological specimens to identify specific HPV subtype, patients with low tumor burden had locoregional disease with potential micrometastatic lesions and thus may not be representative of all patients with early subclinical disease |
| Chan et al. (2017) [68] | Determining utility of screening for nasopharyngeal carcinoma (NPC) using EBV DNA in the plasma of asymptomatic patients | NPC | 20,174 | qPCR; plasma | 97.1% | 98.6% | No screening; diagnosis per standard of care using endoscopy and MRI. Screening cohort had a significantly higher proportion of stage I and II disease (71% vs. 20%) and significantly greater rates of 3-year progression-free survival (97% vs. 70%) | Screening asymptomatic individuals for NPC using EBV DNA plasma levels is associated with earlier diagnosis and better outcomes compared to individuals not undergoing screening | Sampling bias as participants were ethnically Chinese men aged 40 to 62 in Hong Kong, where NPC is endemic |
| Ahn et al. (2014) [69] | Determining effectiveness of liquid biopsy assays using ctHPVDNA from plasma and oral rinses in detecting oropharyngeal SCC prior to beginning treatment | Oropharyngeal SCC | 93; 81 patients with HPV-16 positive SCC and 12 with HPV-16 negative SCC | qPCR; plasma and oral rinses | 76.1% (combined plasma and oral rinse sample results) | 100% (combined plasma and oral rinse sample results) | Compared use of oral rinse samples and plasma samples to one another in effectiveness of corroborating HPV status of tumor biopsy | Using combination of findings from assays analyzing plasma and oral rinse samples increases the sensitivity of HPV-16 liquid biopsy assays in the screening for HPV-positive oropharyngeal SCC compared to use of either sample type alone. Liquid biopsy assays using these samples provide valuable prognostic information on recurrence free survival and overall survival | Small sample size, retrospective design |
| Determining effectiveness of liquid biopsy assays using ctHPVDNA from plasma and oral rinses in predicting 3-year recurrence of oropharyngeal SCC | 90.7% (combined plasma and oral rinse sample results) | 69.5% (combined plasma and oral rinse sample results) |
| ClinicalTrials.gov Study ID | Aim | Study Design | Type of HNC | Sample Size | Samples Used | Enrollment Status | Year of Study Start Date | Estimated Year of Study Completion |
|---|---|---|---|---|---|---|---|---|
| NCT05969262 [132] | Development of early intervention, detection, and treatment strategies for HNCs using combined proteomic and liquid biopsy techniques for analysis of patient plasma and urine samples | Mixed methods study utilizing a retrospective cohort and prospective cohort | HNCs; histopathological and anatomical subtypes not specified | 500; 250 in the retrospective cohort (125 HNC patients, 125 healthy controls) and 250 in the prospective cohort (125 HNC patients, 125 healthy controls) | Plasma and urine | Recruiting | 2023 | 2025 |
| NCT05645783 [133] | Determining the sensitivity and specificity of a NGS liquid biopsy assay in detecting HNSCC in high-risk patients with head and neck lesions | Prospective observational study | HNSCC | 170 | Blood | Recruiting | 2023 | 2024 |
| NCT05774561 [134] | Evaluate use of liquid biopsy in risk stratification of patients with HPV-positive HNC and cervical cancer according to disease recurrence | Mixed methods study utilizing retrospective and prospective designs; prospective portion includes newly diagnosed patients and retrospective portion includes patients post-treatment follow-up | HPV-positive oropharyngeal carcinoma | 480; 200 patients with oropharyngeal cancer and 280 patients with cervical cancer or high-grade cervical intraepithelial lesions | Oropharyngeal swabs, oral rinses, exhaled breath condensate, and blood | Recruiting | 2022 | 2026 |
| NCT05682703 [135] | Examining changes in plasma and urine metabolites of patients with NPC at different points in the disease and treatment course | Observational cohort study | NPC | 2000 | Plasma and urine | Recruiting | 2022 | 2025 |
| NCT04742608 [136] | Determining the sensitivity and specificity of extracellular vesicles liquid biopsy techniques in the diagnosis of thyroid cancer | Prospective cohort study | Thyroid cancer; histopathological subtype not specified | 250 | Blood | Suspended | 2020 | 2026 |
| NCT04599309 [137] | Comparing ctDNA and/or ctHPVDNA levels in blood samples of patients with locally advanced HNSCC before and after undergoing standard treatment | Prospective cohort study | Locally advanced stage III/IV HNSCC | 35 | Blood | Active, not recruiting | 2020 | 2024 |
| NCT04606940 [138] | Characterizing the levels of ctDNA in blood samples of patients with recurrent or metastatic HNSCC following the first dose of anti-PD1 antibody immune checkpoint inhibitor therapy | Prospective cohort study | HNSCC | 18 | Blood | Completed | 2020 | 2021 |
| NCT04490564 [139] | Establishing performance metrics for liquid biopsy assay that detects PD-L1 expression by CTCs in peripheral blood samples of patients with metastatic/recurrent HNSCC, Non-Small Cell Lung Cancer, or metastatic melanoma | Prospective cohort study | HNSCC | 155; 25 patients with metastatic/recurrent HNSCC, 120 patients with Non-Small Cell Lung Cancer, 10 patients with metastatic melanoma, and 30 healthy controls | Plasma | Active, not recruiting | 2019 | 2023 |
| NCT03926468 [140] | Determining diagnostic performance of ddPCR assay measuring ctDNA in peripheral blood samples of patients with stage III/IV HNSCC at baseline and 3 months after completing treatment | Prospective cohort study | Stage III/IV HNSCC | 30 | Blood | Unknown status | 2019 | 2022 |
| NCT03712566 [141] | Serially characterizing the changes in genomic, epigenetic, and immune profiling attributes of peripheral blood samples obtained from patients with recurrent or metastatic SCC of the head and neck, esophagus, or anus who are undergoing treatment with platinum-based chemotherapy or immunotherapy | Prospective cohort study | Recurrent or metastatic HNSCC | 39 | Blood | Active, not recruiting | 2018 | 2024 |
| NCT03942380 [142] | Testing if liquid biopsy assays using ctDNA, ctHPVDNA, or ctRNA in blood samples of patients can detect newly diagnosed or recurrent HNSCC | Interventional, non-randomized study | HNSCC | 500 | Blood | Recruiting | 2017 | 2025 |
| NCT05122507 [143] | Evaluating the use of NGS, ELISA, and PCR assays in monitoring tumor-associated nucleic acids and protein biomarkers to assess patient response to treatment, early detection of recurrence, and overall prognosis | Prospective cohort study | HNSCC | 200 | Plasma, serum, and saliva | Recruiting | 2017 | 2023 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nassar, S.I.; Suk, A.; Nguyen, S.A.; Adilbay, D.; Pang, J.; Nathan, C.-A.O. The Role of ctDNA and Liquid Biopsy in the Diagnosis and Monitoring of Head and Neck Cancer: Towards Precision Medicine. Cancers 2024, 16, 3129. https://doi.org/10.3390/cancers16183129
Nassar SI, Suk A, Nguyen SA, Adilbay D, Pang J, Nathan C-AO. The Role of ctDNA and Liquid Biopsy in the Diagnosis and Monitoring of Head and Neck Cancer: Towards Precision Medicine. Cancers. 2024; 16(18):3129. https://doi.org/10.3390/cancers16183129
Chicago/Turabian StyleNassar, Sami I., Amber Suk, Shaun A. Nguyen, Dauren Adilbay, John Pang, and Cherie-Ann O. Nathan. 2024. "The Role of ctDNA and Liquid Biopsy in the Diagnosis and Monitoring of Head and Neck Cancer: Towards Precision Medicine" Cancers 16, no. 18: 3129. https://doi.org/10.3390/cancers16183129
APA StyleNassar, S. I., Suk, A., Nguyen, S. A., Adilbay, D., Pang, J., & Nathan, C.-A. O. (2024). The Role of ctDNA and Liquid Biopsy in the Diagnosis and Monitoring of Head and Neck Cancer: Towards Precision Medicine. Cancers, 16(18), 3129. https://doi.org/10.3390/cancers16183129







