Exploring TSGA10 Function: A Crosstalk or Controlling Mechanism in the Signaling Pathway of Carcinogenesis?
Abstract
Simple Summary
Abstract
1. Introduction
2. A State-of-the-Art Literature Review of TSGA10 Role in the Physiologic Development
3. A State-of-the-Art Literature Review of TSGA10 Role in Human Malignancies
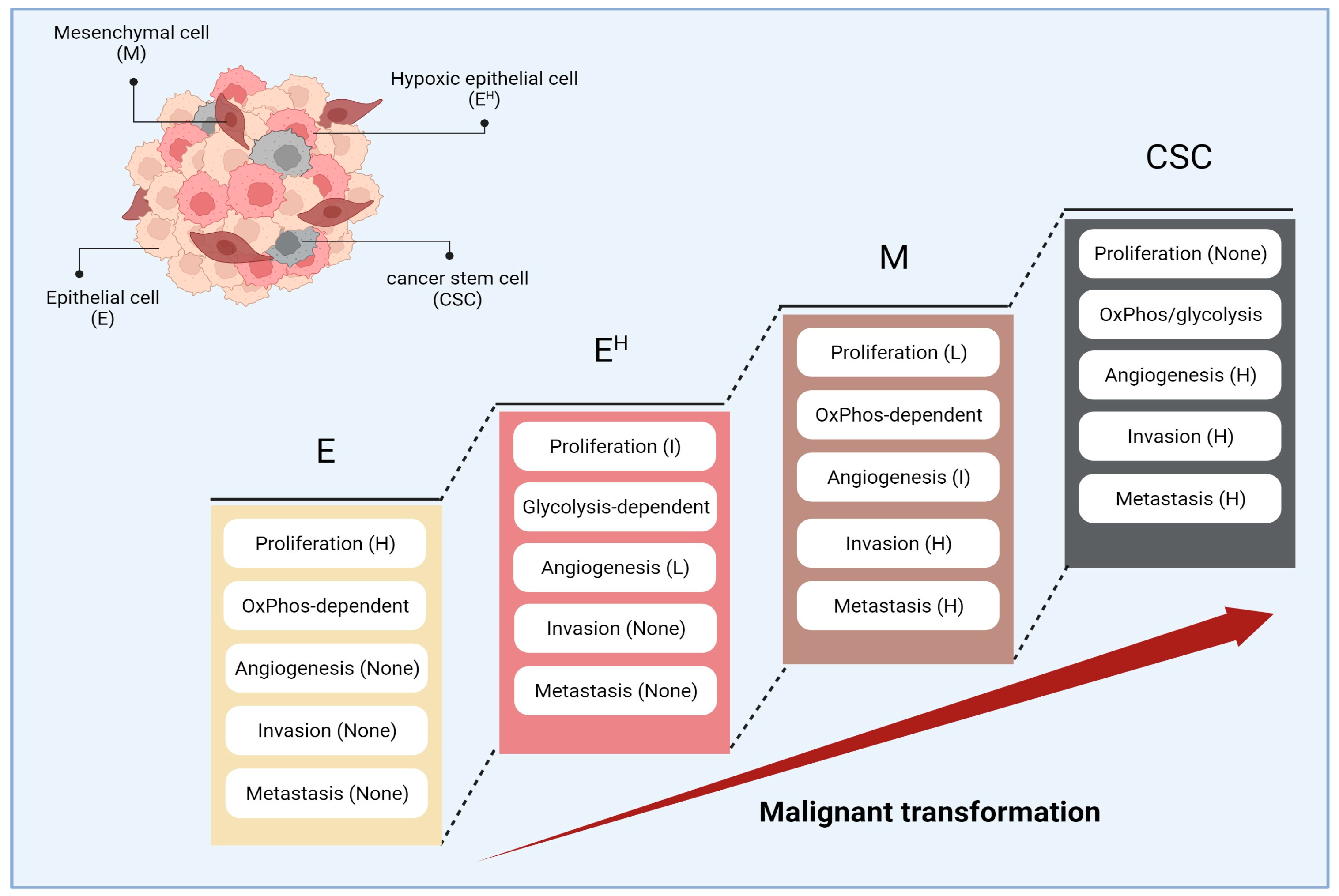
4. Stepwise Cancer Progression and Cancer Hallmarks
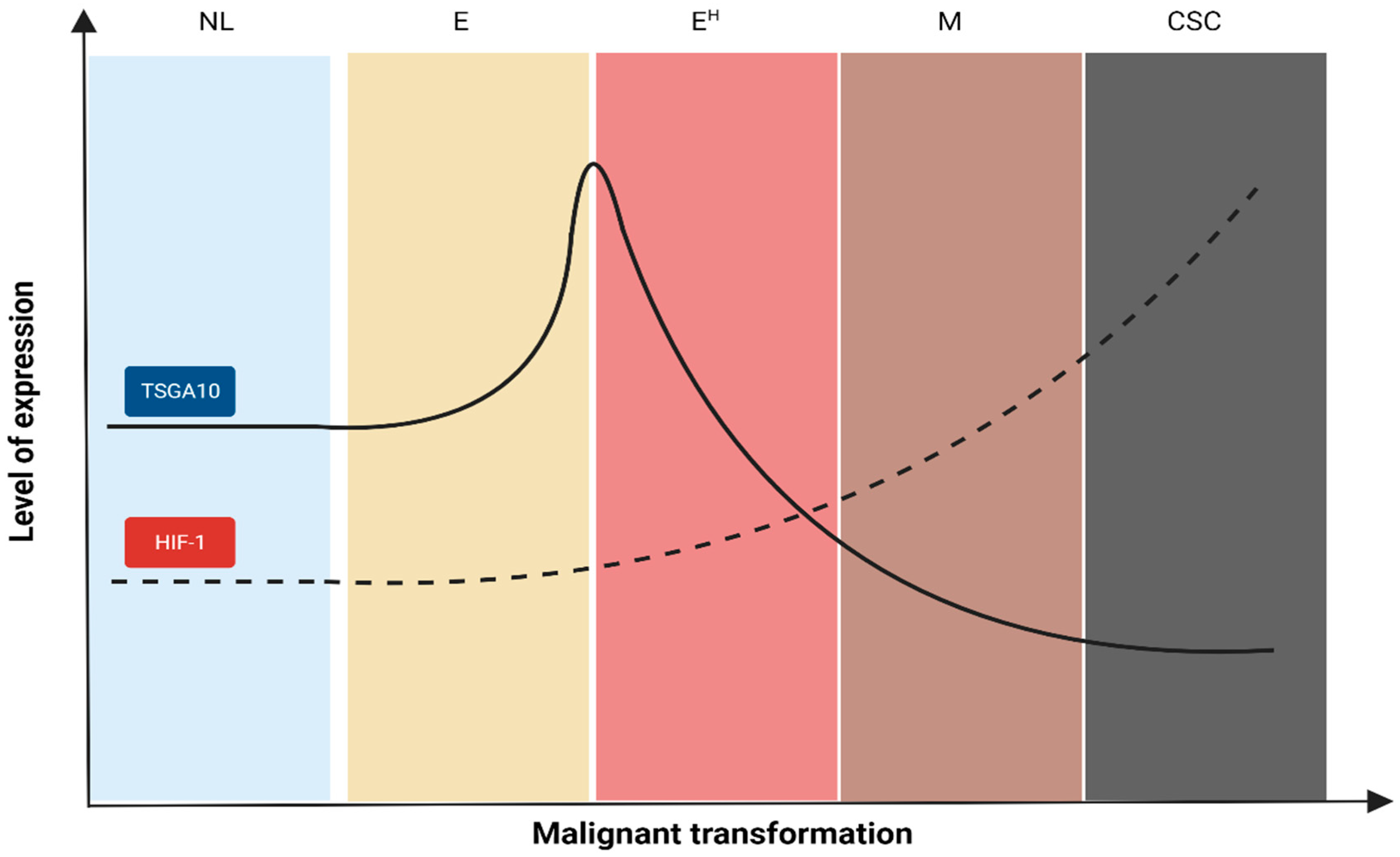
5. Interpretation of TSGA10 Studies Based on the Malignant Transformation
6. Interpretation of TSGA10 Studies Based on the Cancer Hallmarks
- (a)
- Mitochondria trafficking is an essential component in malignant transformation. It has been demonstrated that cancer cells with high affinity to metastasis had fragmented mitochondria in their periphery, likely to provide enough energy for invasion. However, mitochondria in cancer cells with less metastatic affinity are mainly located in the perinuclear region in the fused form [61];
- (b)
7. The Potential Clinical Implications of TSGA10 Upregulation in Cancer Cells
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Li, X.F.; Ren, P.; Shen, W.Z.; Jin, X.; Zhang, J. The Expression, Modulation and Use of Cancer-Testis Antigens as Potential Biomarkers for Cancer Immunotherapy. Am. J. Transl. Res. 2020, 12, 7002–7019. [Google Scholar] [PubMed]
- Tanaka, R.; Ono, T.; Sato, S.; Nakada, T.; Koizumi, F.; Hasegawa, K.; Nakagawa, K.; Okumura, H.; Yamashita, T.; Ohtsuka, M.; et al. Over-Expression of the Testis-Specific Gene TSGA10 in Cancers and Its Immunogenicity. Microbiol. Immunol. 2004, 48, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Salehipour, P.; Nematzadeh, M.; Mobasheri, M.B.; Afsharpad, M.; Mansouri, K.; Modarressi, M.H. Identification of New TSGA10 Transcript Variants in Human Testis with Conserved Regulatory RNA Elements in 5’untranslated Region and Distinct Expression in Breast Cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Modarressi, M.H.; Cameron, J.; Taylor, K.E.; Wolfe, J. Identification and Characterisation of a Novel Gene, TSGA10, Expressed in Testis. Gene 2001, 262, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Roghanian, A.; Jones, D.C.; Pattisapu, J.V.; Wolfe, J.; Young, N.T.; Behnam, B. Filament-Associated TSGA10 Protein Is Expressed in Professional Antigen Presenting Cells and Interacts with Vimentin. Cell. Immunol. 2010, 265, 120–126. [Google Scholar] [CrossRef]
- Asgari, R.; Bakhtiari, M.; Rezazadeh, D.; Yarani, R.; Esmaeili, F.; Mansouri, K. TSGA10 as a Potential Key Factor in the Process of Spermatid Differentiation/Maturation: Deciphering Its Association with Autophagy Pathway. Reprod. Sci. 2021, 28, 3228–3240. [Google Scholar] [CrossRef] [PubMed]
- Behnam, B.; Modarressi, M.H.; Conti, V.; Taylor, K.E.; Puliti, A.; Wolfe, J. Expression of Tsga10 Sperm Tail Protein in Embryogenesis and Neural Development: From Cilium to Cell Division. Biochem. Biophys. Res. Commun. 2006, 344, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Reimand, K.; Perheentupa, J.; Link, M.; Krohn, K.; Peterson, P.; Uibo, R. Testis-expressed protein TSGA10 an auto-antigen in autoimmune polyendocrine syndrome type I. Int. Immunol. 2008, 20, 39–44. [Google Scholar] [CrossRef][Green Version]
- Smith, C.J.A.; Oscarson, M.; Rönnblom, L.; Alimohammadi, M.; Perheentupa, J.; Husebye, E.S.; Gustafsson, J.; Nordmark, G.; Meloni, A.; Crock, P.A.; et al. TSGA10—A Target for Autoantibodies in Autoimmune Polyendocrine Syndrome Type 1 and Systemic Lupus Erythematosus. Scand. J. Immunol. 2011, 73, 147–153. [Google Scholar] [CrossRef]
- Mobasheri, M.B.; Modarressi, M.H.; Shabani, M.; Asgarian, H.; Sharifian, R.A.; Vossough, P.; Shokri, F. Expression of the Testis-Specific Gene, TSGA10, in Iranian Patients with Acute Lymphoblastic Leukemia (ALL). Leuk. Res. 2006, 30, 883–889. [Google Scholar] [CrossRef]
- Mobasheri, M.B.; Jahanzad, I.; Mohagheghi, M.A.; Aarabi, M.; Farzan, S.; Modarressi, M.H. Expression of Two Testis-Specific Genes, TSGA10 and SYCP3, in Different Cancers Regarding to Their Pathological Features. Cancer Detect. Prev. 2007, 31, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Behnam, B.; Chahlavi, A.; Pattisapu, J.; Wolfe, J. TSGA10 Is Specifically Expressed in Astrocyte and Over-Expressed in Brain Tumors. Avicenna J. Med. Biotechnol. 2009, 1, 161–166. [Google Scholar]
- Mansouri, K.; Mostafie, A.; Rezazadeh, D.; Shahlaei, M.; Modarressi, M.H. New Function of TSGA10 Gene in Angiogenesis and Tumor Metastasis: A Response to a Challengeable Paradox. Hum. Mol. Genet. 2016, 25, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Jahani, M.; Shahlaei, M.; Norooznezhad, F.; Miraghaee, S.S.; Hosseinzadeh, L.; Moasefi, N.; Khodarahmi, R.; Farokhi, A.; Mahnam, A.; Mansouri, K. TSGA10 Over Expression Decreases Metastasic and Metabolic Activity by Inhibiting HIF-1 in Breast Cancer Cells. Arch. Med. Res. 2020, 51, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Valipour, E.; Nooshabadi, V.T.; Mahdipour, S.; Shabani, S.; Farhady-Tooli, L.; Majidian, S.; Noroozi, Z.; Mansouri, K.; Motevaseli, E.; Modarressi, M.H. Anti-Angiogenic Effects of Testis-Specific Gene Antigen 10 on Primary Endothelial Cells. Gene 2020, 754, 144856. [Google Scholar] [CrossRef]
- Taghizadeh-Hesary, F. “Reinforcement” by Tumor Microenvironment: The Seventh “R” of Radiobiology. Int. J. Radiat. Oncol. Biol. Phys. 2023, 119, 727–733. [Google Scholar] [CrossRef]
- Aarabi, M.; Soltanghoraee, H.; Amirjannati, N.; Ghaffari, M.; Sadeghi, M.R.; Akhondi, M.M.; Modarresi, M.H. Testis Specific Gene 10 Expression in the Testes of Patients with Non-Obstructive Azoospermia. J. Reprod. Infertil. 2006, 7, 179–186. [Google Scholar]
- Hägele, S.; Behnam, B.; Borter, E.; Wolfe, J.; Paasch, U.; Lukashev, D.; Sitkovsky, M.; Wenger, R.H.; Katschinski, D.M. TSGA10 Prevents Nuclear Localization of the Hypoxia-Inducible Factor (HIF)-1α. FEBS Lett. 2006, 580, 3731–3738. [Google Scholar] [CrossRef]
- Hoseinkhani, Z.; Rastegari-Pouyani, M.; Oubari, F.; Mozafari, H.; Rahimzadeh, A.B.; Maleki, A.; Amini, S.; Mansouri, K. Contribution and Prognostic Value of TSGA10 Gene Expression in Patients with Acute Myeloid Leukemia (AML). Pathol. Res. Pract. 2019, 215, 506–511. [Google Scholar] [CrossRef]
- Yuan, X.; He, J.; Sun, F.; Gu, J. Effects and Interactions of MiR-577 and TSGA10 in Regulating Esophageal Squamous Cell Carcinoma. Int. J. Clin. Exp. Pathol. 2013, 6, 2651–2667. [Google Scholar]
- Asgharzadeh, M.R.; Pourseif, M.M.; Barar, J.; Eskandani, M.; Jafari Niya, M.; Mashayekhi, M.R.; Omidi, Y. Functional Expression and Impact of Testis-Specific Gene Antigen 10 in Breast Cancer: A Combined in Vitro and in Silico Analysis. Bioimpacts 2019, 9, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Kazerani, R.; Salehipour, P.; Shah Mohammadi, M.; Amanzadeh Jajin, E.; Modarressi, M.H. Identification of TSGA10 and GGNBP2 Splicing Variants in 5′ Untranslated Region with Distinct Expression Profiles in Brain Tumor Samples. Front. Oncol. 2023, 13, 1075638. [Google Scholar] [CrossRef]
- Bao, L.; You, B.; Shi, S.; Shan, Y.; Zhang, Q.; Yue, H.; Zhang, J.; Zhang, W.; Shi, Y.; Liu, Y.; et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene 2018, 37, 2873–2889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, J.; Fu, Z.; Dong, L.; Tang, Y.; Xu, C.; Wang, H.; Zhang, T.; Wu, Y.; Dong, C.; et al. Hypoxia-Induced microRNA-10b-3p Promotes Esophageal Squamous Cell Carcinoma Growth and Metastasis by Targeting TSGA10. Aging 2019, 11, 10374–10384. [Google Scholar] [CrossRef] [PubMed]
- Theinert, S.M.; Pronest, M.M.; Peris, K.; Sterry, W.; Walden, P. Identification of the Testis-Specific Protein 10 (TSGA10) as Serologically Defined Tumour-Associated Antigen in Primary Cutaneous T-Cell Lymphoma: CTCL-Associated Antigen TSGA10. Br. J. Dermatol. 2005, 153, 639–641. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Kao, C.; Yang, F.; Wang, F.; Yin, G.; Wang, Y.; He, Y.; Ji, J.; Liu, L. Integrated Multi-Omics Analysis Model to Identify Biomarkers Associated With Prognosis of Breast Cancer. Front. Oncol. 2022, 12, 899900. [Google Scholar] [CrossRef]
- Dianatpour, M.; Mehdipour, P.; Nayernia, K.; Mobasheri, M.-B.; Ghafouri-Fard, S.; Savad, S.; Modarressi, M.H. Expression of Testis Specific Genes TSGA10, TEX101 and ODF3 in Breast Cancer. Iran. Red Crescent Med. J. 2012, 14, 730–734. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, Z.; Chen, W.; Luo, C.; Zhang, W. Exploring a Four-Gene Risk Model Based on Doxorubicin Resistance-Associated lncRNAs in Hepatocellular Carcinoma. Front. Pharmacol. 2022, 13, 1015842. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Taghizadeh-Hesary, F.; Houshyari, M.; Farhadi, M. Mitochondrial Metabolism: A Predictive Biomarker of Radiotherapy Efficacy and Toxicity. J. Cancer Res. Clin. Oncol. 2023, 149, 6719–6741. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Gregorio, J.; Petricca, S.; Iorio, R.; Toniato, E.; Flati, V. Mitochondrial and Metabolic Alterations in Cancer Cells. Eur. J. Cell Biol. 2022, 101, 151225. [Google Scholar] [CrossRef]
- Shi, R.; Liao, C.; Zhang, Q. Hypoxia-Driven Effects in Cancer: Characterization, Mechanisms, and Therapeutic Implications. Cells 2021, 10, 678. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.C.; Rathore, A.; Younas, H.; Gilkes, D.; Polotsky, V.Y. Hypoxia-Inducible Factors and Cancer. Curr. Sleep. Med. Rep. 2017, 3, 1–10. [Google Scholar] [CrossRef]
- Harrison, H.; Pegg, H.J.; Thompson, J.; Bates, C.; Shore, P. HIF1-Alpha Expressing Cells Induce a Hypoxic-like Response in Neighbouring Cancer Cells. BMC Cancer 2018, 18, 674. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, Z.; Zhou, C.; Liu, L.; Huang, C. Epithelial-Mesenchymal Transition: The History, Regulatory Mechanism, and Cancer Therapeutic Opportunities. MedComm 2020, 3, 144. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.V.; Conroy, S.; Pavlov, K.; Sontakke, P.; Tomar, T.; Eggens-Meijer, E.; Balasubramaniyan, V.; Wagemakers, M.; Dunnen, W.F.A.; Kruyt, F.A.E. Hypoxia Enhances Migration and Invasion in Glioblastoma by Promoting a Mesenchymal Shift Mediated by the HIF1α–ZEB1 Axis. Cancer Lett. 2015, 359, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.H.; Wu, M.Z.; Chiou, S.H.; Chen, P.M.; Chang, S.Y.; Liu, C.J.; Teng, S.C.; Wu, K.J. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat. Cell Biol. 2008, 10, 295–305. [Google Scholar] [CrossRef]
- Lv, L.; Yuan, J.; Huang, T.; Zhang, C.; Zhu, Z.; Wang, L.; Jiang, G.; Zeng, F. Stabilization of Snail by HIF-1α and TNF-α Is Required for Hypoxia-Induced Invasion in Prostate Cancer PC3 Cells. Mol. Biol. Rep. 2014, 41, 4573–4582. [Google Scholar] [CrossRef] [PubMed]
- Chaffer, C.L.; Weinberg, R.A. How Does Multistep Tumorigenesis Really Proceed? Cancer Discov. 2015, 5, 22–24. [Google Scholar] [CrossRef]
- Bisht, S.; Nigam, M.; Kunjwal, S.S.; Sergey, P.; Mishra, A.P.; Sharifi-Rad, J. Cancer Stem Cells: From an Insight into the Basics to Recent Advances and Therapeutic Targeting. Stem Cells Int. 2022, 2022, 9653244. [Google Scholar] [CrossRef]
- Wang, H.; Unternaehrer, J.J. Epithelial-Mesenchymal Transition and Cancer Stem Cells: At the Crossroads of Differentiation and Dedifferentiation. Dev. Dyn. 2019, 248, 10–20. [Google Scholar] [CrossRef]
- Yasuda, T.; Ishimoto, T.; Baba, H. Conflicting Metabolic Alterations in Cancer Stem Cells and Regulation by the Stromal Niche. Regen. Ther. 2021, 17, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Taghizadeh-Hesary, F.; Heike, Y.; Bahadori, M. Cell Energy: A New Hypothesis in Decoding Cancer Evolution. Arch. Iran. Med. 2019, 22, 733–735. [Google Scholar] [PubMed]
- Behnam, B.; Mobahat, M.; Fazilaty, H.; Wolfe, J.; Omran, H. TSGA10 Is a Centrosomal Protein, Interacts with ODF2 and Localizes to Basal Body. J. Cell Sci. Ther. 2015, 6, 217. [Google Scholar]
- Ma, Z.; Xiang, X.; Li, S.; Xie, P.; Gong, Q.; Goh, B.-C.; Wang, L. Targeting Hypoxia-Inducible Factor-1, for Cancer Treatment: Recent Advances in Developing Small-Molecule Inhibitors from Natural Compounds. Semin. Cancer Biol. 2022, 80, 379–390. [Google Scholar] [CrossRef]
- Zaffryar-Eilot, S.; Marshall, D.; Voloshin, T.; Bar-Zion, A.; Spangler, R.; Kessler, O.; Ghermazien, H.; Brekhman, V.; Suss-Toby, E.; Adam, D.; et al. Lysyl Oxidase-like-2 Promotes Tumour Angiogenesis and Is a Potential Therapeutic Target in Angiogenic Tumours. Carcinogenesis 2013, 34, 2370–2379. [Google Scholar] [CrossRef][Green Version]
- Amoorahim, M.; Valipour, E.; Hoseinkhani, Z.; Mahnam, A.; Rezazadeh, D.; Ansari, M.; Shahlaei, M.; Gamizgy, Y.H.; Moradi, S.; Mansouri, K. TSGA10 Overexpression Inhibits Angiogenesis of HUVECs: A HIF-2α Biased Perspective. Microvasc. Res. 2020, 128, 103952. [Google Scholar] [CrossRef]
- Robinson, N.J.; Schiemann, W.P. Telomerase in Cancer: Function, Regulation, and Clinical Translation. Cancers 2022, 14, 808. [Google Scholar] [CrossRef] [PubMed]
- Yatabe, N.; Kyo, S.; Maida, Y.; Nishi, H.; Nakamura, M.; Kanaya, T.; Tanaka, M.; Isaka, K.; Ogawa, S.; Inoue, M. HIF-1-mediated activation of telomerase in cervical cancer cells. Oncogene 2004, 23, 3708–3715. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Lyu, Y.; Tran, L.; Lan, J.; Xie, Y.; Yang, Y.; Murugan, N.L.; Wang, Y.J.; Semenza, G.L. HIF-1 Recruits NANOG as a Coactivator for TERT Gene Transcription in Hypoxic Breast Cancer Stem Cells. Cell Rep. 2021, 36, 109757. [Google Scholar] [CrossRef]
- Mittal, K.; Kaur, J.; Jaczko, M.; Wei, G.; Toss, M.S.; Rakha, E.A.; Janssen, E.A.M.; Søiland, H.; Kucuk, O.; Reid, M.D.; et al. Centrosome Amplification: A Quantifiable Cancer Cell Trait with Prognostic Value in Solid Malignancies. Cancer Metastasis Rev. 2021, 40, 319–339. [Google Scholar] [CrossRef]
- Lin, Y.C.; Chang, C.W.; Hsu, W.B.; Tang, C.J.; Lin, Y.N.; Chou, E.J.; Wu, C.T.; Tang, T.K. Human Microcephaly Protein CEP135 Binds to hSAS-6 and CPAP, and Is Required for Centriole Assembly. EMBO J. 2013, 32, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Ganapathi Sankaran, D.; Stemm-Wolf, A.J.; Pearson, C.G. CEP135 Isoform Dysregulation Promotes Centrosome Amplification in Breast Cancer Cells. Mol. Biol. Cell 2019, 30, 1230–1244. [Google Scholar] [CrossRef] [PubMed]
- Carvalho-Santos, Z.; Machado, P.; Branco, P.; Tavares-Cadete, F.; Rodrigues-Martins, A.; Pereira-Leal, J.B.; Bettencourt-Dias, M. Stepwise Evolution of the Centriole-Assembly Pathway. J. Cell Sci. 2010, 123, 1414–1426. [Google Scholar] [CrossRef]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N.R. Hypoxia-Modified Cancer Cell Metabolism. Front. Cell Dev. Biol. 2019, 7, 4. [Google Scholar] [CrossRef]
- Taghizadeh-Hesary, F.; Akbari, H.; Bahadori, M.; Behnam, B. Targeted Anti-Mitochondrial Therapy: The Future of Oncology. Genes 2022, 13, 1728. [Google Scholar] [CrossRef]
- Luo, G.; Hou, M.; Wang, B.; Liu, Z.; Liu, W.; Han, T.; Zhang, D.; Zhou, X.; Jia, W.; Tan, Y.; et al. Tsga10 Is Essential for Arrangement of Mitochondrial Sheath and Male Fertility in Mice. Andrology 2021, 9, 368–375. [Google Scholar] [CrossRef]
- Boulton, D.P.; Caino, M.C. Mitochondrial Fission and Fusion in Tumor Progression to Metastasis. Front. Cell Dev. Biol. 2022, 10, 849962. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento-Salinas, F.L.; Delgado-Magallón, A.; Montes-Alvarado, J.B.; Ramírez-Ramírez, D.; Flores-Alonso, J.C.; Cortés-Hernández, P.; Reyes-Leyva, J.; Herrera-Camacho, I.; Anaya-Ruiz, M.; Pelayo, R.; et al. Breast Cancer Subtypes Present a Differential Production of Reactive Oxygen Species (ROS) and Susceptibility to Antioxidant Treatment. Front. Oncol. 2019, 9, 480. [Google Scholar] [CrossRef]
- Castaneda, M.; Hollander, P.; Kuburich, N.A.; Rosen, J.M.; Mani, S.A. Mechanisms of cancer metastasis. Semin. Cancer Biol. 2022, 87, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Rao, L.; Giannico, D.; Leone, P.; Solimando, A.G.; Maiorano, E.; Caporusso, C.; Duda, L.; Tamma, R.; Mallamaci, R.; Susca, N.; et al. HB-EGF–EGFR Signaling in Bone Marrow Endothelial Cells Mediates Angiogenesis Associated with Multiple Myeloma. Cancers 2020, 12, 173. [Google Scholar] [CrossRef] [PubMed]
- Tam, S.Y.; Wu, V.W.C.; Law, H.K.W. Hypoxia-Induced Epithelial-Mesenchymal Transition in Cancers: HIF-1α and Beyond. Front. Oncol. 2020, 10, 486. [Google Scholar] [CrossRef] [PubMed]
- Kortleve, D.; Coelho, R.M.L.; Hammerl, D.; Debets, R. Cancer Germline Antigens and Tumor-Agnostic CD8(+) T Cell Evasion. Trends Immunol. 2022, 43, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Nocquet, L.; Juin, P.P.; Souazé, F. Mitochondria at Center of Exchanges between Cancer Cells and Cancer-Associated Fibroblasts during Tumor Progression. Cancers 2020, 12, 3017. [Google Scholar] [CrossRef]
- Ippolito, L.; Morandi, A.; Taddei, M.L.; Parri, M.; Comito, G.; Iscaro, A.; Raspollini, M.R.; Magherini, F.; Rapizzi, E.; Masquelier, J.; et al. Cancer-Associated Fibroblasts Promote Prostate Cancer Malignancy via Metabolic Rewiring and Mitochondrial Transfer. Oncogene 2019, 38, 5339–5355. [Google Scholar] [CrossRef]
- Wang, Y.; Gan, G.; Wang, B.; Wu, J.; Cao, Y.; Zhu, D.; Xu, Y.; Wang, X.; Han, H.; Li, X.; et al. Cancer-Associated Fibroblasts Promote Irradiated Cancer Cell Recovery Through Autophagy. EBioMedicine 2017, 17, 45–56. [Google Scholar] [CrossRef]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between Cancer-Associated Fibroblasts and Immune Cells in the Tumor Microenvironment: New Findings and Future Perspectives. Mol. Cancer 2021, 20, 131. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Franco, P.I.; Rodrigues, A.P.; Menezes, L.B.; Pacheco Miguel, M. Tumor Microenvironment Components: Allies of Cancer Progression. Pathol. Res. Pract. 2020, 216, 152729. [Google Scholar] [CrossRef]
- Xiao, S.; Liu, L.; Lu, X.; Long, J.; Zhou, X.; Fang, M. The Prognostic Significance of Bromodomain PHD-Finger Transcription Factor in Colorectal Carcinoma and Association with Vimentin and E-Cadherin. J. Cancer Res. Clin. Oncol. 2015, 141, 1465–1474. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Marshall, W.F. Centrosome positioning in vertebrate development. J. Cell Sci. 2012, 125, 4951–4961. [Google Scholar] [CrossRef] [PubMed]
- Das, L. Epigenetic Alterations Impede Epithelial-Mesenchymal Transition by Modulating Centrosome Amplification and Myc/RAS Axis in Triple Negative Breast Cancer Cells. Sci. Rep. 2023, 13, 2458. [Google Scholar] [CrossRef]
- Olivares-Urbano, M.A.; Griñán-Lisón, C.; Marchal, J.A.; Núñez, M.I. CSC Radioresistance: A Therapeutic Challenge to Improve Radiotherapy Effectiveness in Cancer. Cells 2020, 9, 1651. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and Their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.; Merhej, G.; Saravanan, S.; Chen, H. Cancer Resistance to Immunotherapy: What Is the Role of Cancer Stem Cells? Cancer Drug Resist. 2022, 5, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key Players in Cancer and Potential Therapeutic Strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Fadavi, P.; Nafissi, N.; Mahdavi, S.R.; Jafarnejadi, B.; Javadinia, S.A. Outcome of Hypofractionated Breast Irradiation and Intraoperative Electron Boost in Early Breast Cancer: A Randomized Non-Inferiority Clinical Trial. Cancer Rep. 2021, 4, 1376. [Google Scholar] [CrossRef]
- Ameri, A.; Norouzi, S.; Sourati, A.; Azghandi, S.; Novin, K.; Taghizadeh-Hesary, F. Randomized Trial on Acute Toxicities of Weekly vs Three-weekly cisplatin-based Chemoradiation in Head and Neck Cancer. Cancer Rep. 2022, 5, e1425. [Google Scholar] [CrossRef]
- Borrego-Soto, G.; Ortiz-López, R.; Rojas-Martínez, A. Ionizing Radiation-Induced DNA Injury and Damage Detection in Patients with Breast Cancer. Genet. Mol. Biol. 2015, 38, 420–432. [Google Scholar] [CrossRef]
- Syljuåsen, R.G. Cell Cycle Effects in Radiation Oncology. In Radiation Oncology; Wenz, F., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–8. [Google Scholar]
- Suwa, T.; Kobayashi, M.; Nam, J.-M.; Harada, H. Tumor microenvironment and radioresistance. Exp. Mol. Med. 2021, 53, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lin, Q.; Yun, Z. Cellular and molecular mechanisms underlying oxygen-dependent radiosensitivity. Radiat. Res. 2015, 183, 487–496. [Google Scholar] [CrossRef]
- You, L.; Wu, W.; Wang, X.; Fang, L.; Adam, V.; Nepovimova, E.; Wu, Q.; Kuca, K. The Role of Hypoxia-Inducible Factor 1 in Tumor Immune Evasion. Med. Res. Rev. 2021, 41, 1622–1643. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Multhoff, G. Hypoxia-/HIF-1α-Driven Factors of the Tumor Microenvironment Impeding Antitumor Immune Responses and Promoting Malignant Progression. Adv. Exp. Med. Biol. 2018, 1072, 171–175. [Google Scholar] [CrossRef]
- Shurin, M.R.; Umansky, V. Cross-Talk between HIF and PD-1/PD-L1 Pathways in Carcinogenesis and Therapy. J. Clin. Investig. 2022, 132, e159473. [Google Scholar] [CrossRef] [PubMed]
- Sethumadhavan, S.; Silva, M.; Philbrook, P.; Nguyen, T.; Hatfield, S.M.; Ohta, A.; Sitkovsky, M.V. Hypoxia and Hypoxia-Inducible Factor (HIF) Downregulate Antigen-Presenting MHC Class I Molecules Limiting Tumor Cell Recognition by T Cells. PLoS ONE 2017, 12, 0187314. [Google Scholar] [CrossRef]
- Chiu, D.K.; Tse, A.P.; Xu, I.M.; Cui, J.; Lai, R.K.; Li, L.L.; Koh, H.Y.; Tsang, F.H.; Wei, L.L.; Wong, C.M. Hypoxia Inducible Factor HIF-1 Promotes Myeloid-Derived Suppressor Cells Accumulation through ENTPD2/CD39L1 in Hepatocellular Carcinoma. Nat. Commun. 2017, 8, 517. [Google Scholar] [CrossRef] [PubMed]
- Groth, C.; Hu, X.; Weber, R.; Fleming, V.; Altevogt, P.; Utikal, J.; Umansky, V. Immunosuppression Mediated by Myeloid-Derived Suppressor Cells (MDSCs) during Tumour Progression. Br. J. Cancer 2019, 120, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Behnam, B.; Taghizadeh-Hesary, F. Mitochondrial Metabolism: A New Dimension of Personalized Oncology. Cancers 2023, 15, 4058. [Google Scholar] [CrossRef] [PubMed]

| Cancer Types | Discussed Mechanisms | TSGA10 over Expression | TSGA10 Downregulation |
|---|---|---|---|
| Esophageal Squamous Cell Carcinoma [20,24] | TSGA10 acts as a tumor suppressor as it inhibits tumor growth by regulating the cell cycle and inducing apoptosis. Typically, downregulated in more advanced stages, larger and poorly differentiated ESCC, which leads to increased cell proliferation and malignancy. | Can it help regulate tumorigenesis? | MiR-577 functions as an oncomir as it promotes cancer progression by targeting and downregulating TSGA10. Under hypoxic conditions, the expression of miR-10b-3p would be enhanced, therefore targeting TSGA10 and reducing its expression. |
| Primary cutaneous T-cell lymphoma (CTCL) [25] | TSGA10 acts as a tumor-associated antigen and a candidate for targeted immunotherapy in primary CTCL and suggests a role in the immune response against tumor cells. | TSGA10 is overexpressed as a potential tumor-associated antigen in primary CTCL. | Likely reduce the immune system’s ability to recognize and target the cancer cells; hence, less effective immune surveillance and, potentially, cancer progression. |
| Breast Cancer [3,14,21,26,27] | A paradoxical relationship is observed between TSGA10 expression and cellular migration. The high-affinity interaction of TSGA10 C-terminal domain with HIF-1α affects 8 key proteins (VEGFA, HSP90AA1, AKT1, ARNT, TP53, VHL, JUN, and EGFR) in cancer progression. | TSGA10 overexpression is associated with reduced metastasis. TSGA10 overexpression decreases metastatic and metabolic activities, thereby reducing cell proliferation and metastasis. | TSGA10 is typically downregulated in breast cancer, which leads to cancer progression and metastasis. |
| Brain Tumor [12,22] | Unknown. TSGA10 is specifically expressed in astrocytes. | TSGA10 is overexpressed in brain tumors. | TSGA10 Downregulation may disrupt normal cell cycle control, which could lead to decreased cell proliferation. |
| Nasopharyngeal Carcinoma [23] | miR-23a regulated angiogenesis by directly targeting TSGA10. Metastasis-associated miR-23a from NPC-derived exosomes plays an important role in mediating angiogenesis by targeting TSGA10. | Overexpression of TSGA10 can counteract the effects of miR-23a and result in inhibiting proliferation, angiogenesis, and cell migration and invasion. | Suppression of TSGA10 is associated with tumorigenesis via enhancing the migration of endothelial cells, suggesting that angiogenesis is regulated by miR-23a as it directly targets TSGA10 and represses its antiangiogenic functions. |
| Hepatocellular Carcinoma (HCC) [28] | TSGA10 acts as an immunogenic protein that can elicit an immune response; hence, TSGA10 plays a significant role in the progression and prognosis of hepatocellular carcinoma. | TSGA10’s overexpression is linked to tumor aggressiveness, poor patient outcomes, and serves as a potential immunogenic target. | Downregulation of TSGA10 is associated with increased cell proliferation and reduced apoptosis. |
| AML/ALL [10,19] | TSGA10 acts as a tumor suppressor gene in AML, as it negatively regulates the expression of VEGF by interacting with HIF-1α. TSGA10 may be involved in the proliferation of leukemic cells. | TSGA10 Overexpression leads to VEGF and HIF-1α downregulation, consequently inhibiting tumor growth and angiogenesis. TSGA10 is overexpressed in ALL, leading to proliferation of leukemic cells. | Decreased expression of TSGA10 in AML leads to increased VEGF and HIF-1α levels, promoting tumor growth and angiogenesis. |
Pan-cancer studies
| Irregular expression of TSGA10 in various cancers can affect the proliferation of cancer cells, suggesting its role in tumorigenesis. | TSGA10 is overexpressed in a subset of melanoma (5%), colon cancer (5%), HCC (20%), ovarian cancer (35%), and prostate cancer (15%), leading to increased cell division and growth, altered apoptosis, enhanced cell migration and invasion, and activation of oncogenic pathways. | Downregulation of TSGA10 can lead to a potential tumor suppression. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taghizadeh-Hesary, F.; Ghadyani, M.; Kashanchi, F.; Behnam, B. Exploring TSGA10 Function: A Crosstalk or Controlling Mechanism in the Signaling Pathway of Carcinogenesis? Cancers 2024, 16, 3044. https://doi.org/10.3390/cancers16173044
Taghizadeh-Hesary F, Ghadyani M, Kashanchi F, Behnam B. Exploring TSGA10 Function: A Crosstalk or Controlling Mechanism in the Signaling Pathway of Carcinogenesis? Cancers. 2024; 16(17):3044. https://doi.org/10.3390/cancers16173044
Chicago/Turabian StyleTaghizadeh-Hesary, Farzad, Mobina Ghadyani, Fatah Kashanchi, and Babak Behnam. 2024. "Exploring TSGA10 Function: A Crosstalk or Controlling Mechanism in the Signaling Pathway of Carcinogenesis?" Cancers 16, no. 17: 3044. https://doi.org/10.3390/cancers16173044
APA StyleTaghizadeh-Hesary, F., Ghadyani, M., Kashanchi, F., & Behnam, B. (2024). Exploring TSGA10 Function: A Crosstalk or Controlling Mechanism in the Signaling Pathway of Carcinogenesis? Cancers, 16(17), 3044. https://doi.org/10.3390/cancers16173044








