Rare Germline Variants in DNA Repair Genes Detected in BRCA-Negative Finnish Patients with Early-Onset Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Turku Whole-Exome Sequencing Set
2.1.1. Study Subjects
2.1.2. Sample Preparation and Whole-Exome Sequencing
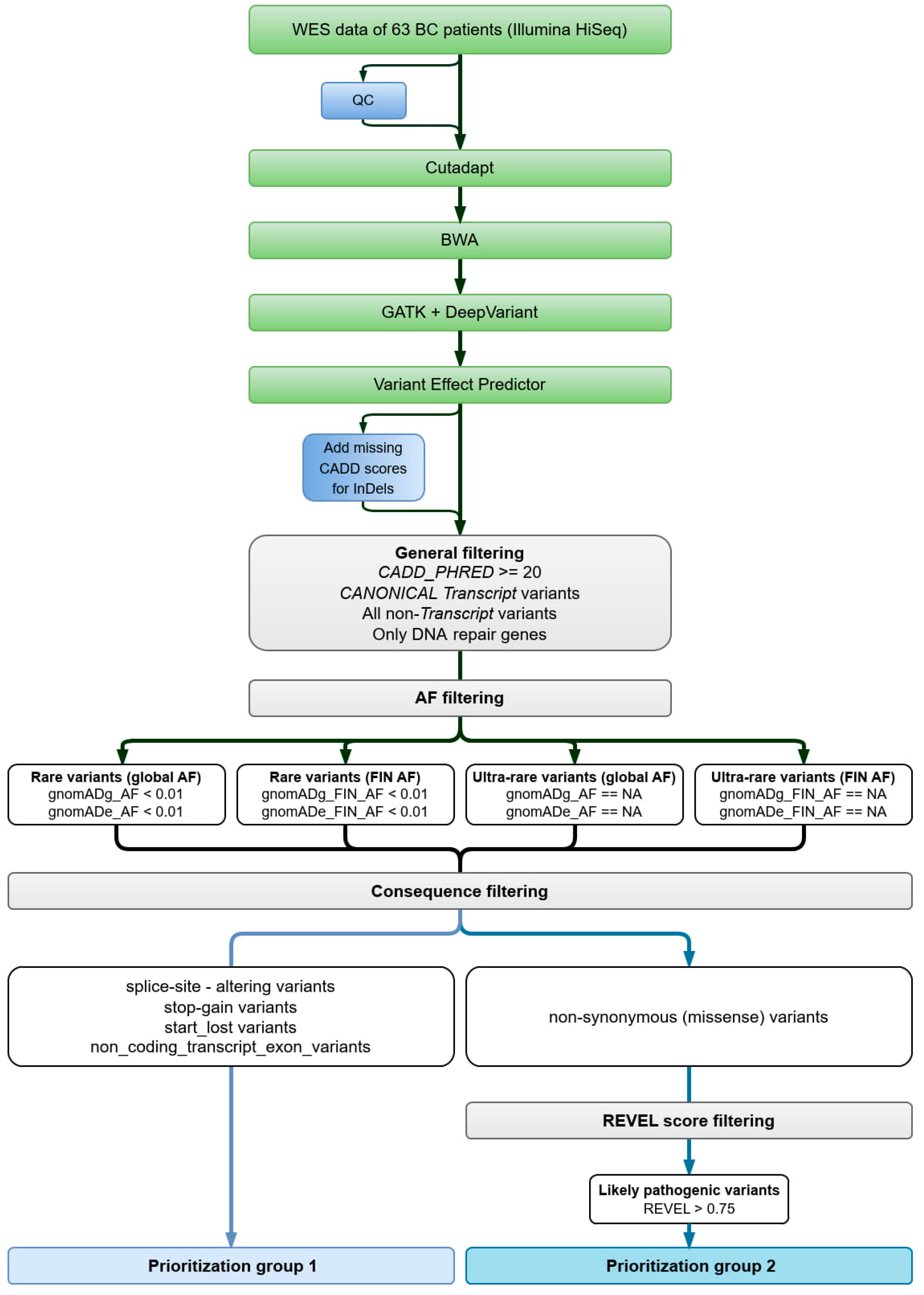
2.1.3. Data Analysis
2.1.4. Variant Filtering
2.2. Validation Set Helsinki
2.3. Protein Structure Modeling
3. Results
3.1. Turku Whole-Exome Sequencing Set Results
3.2. Validation Set Helsinki Results
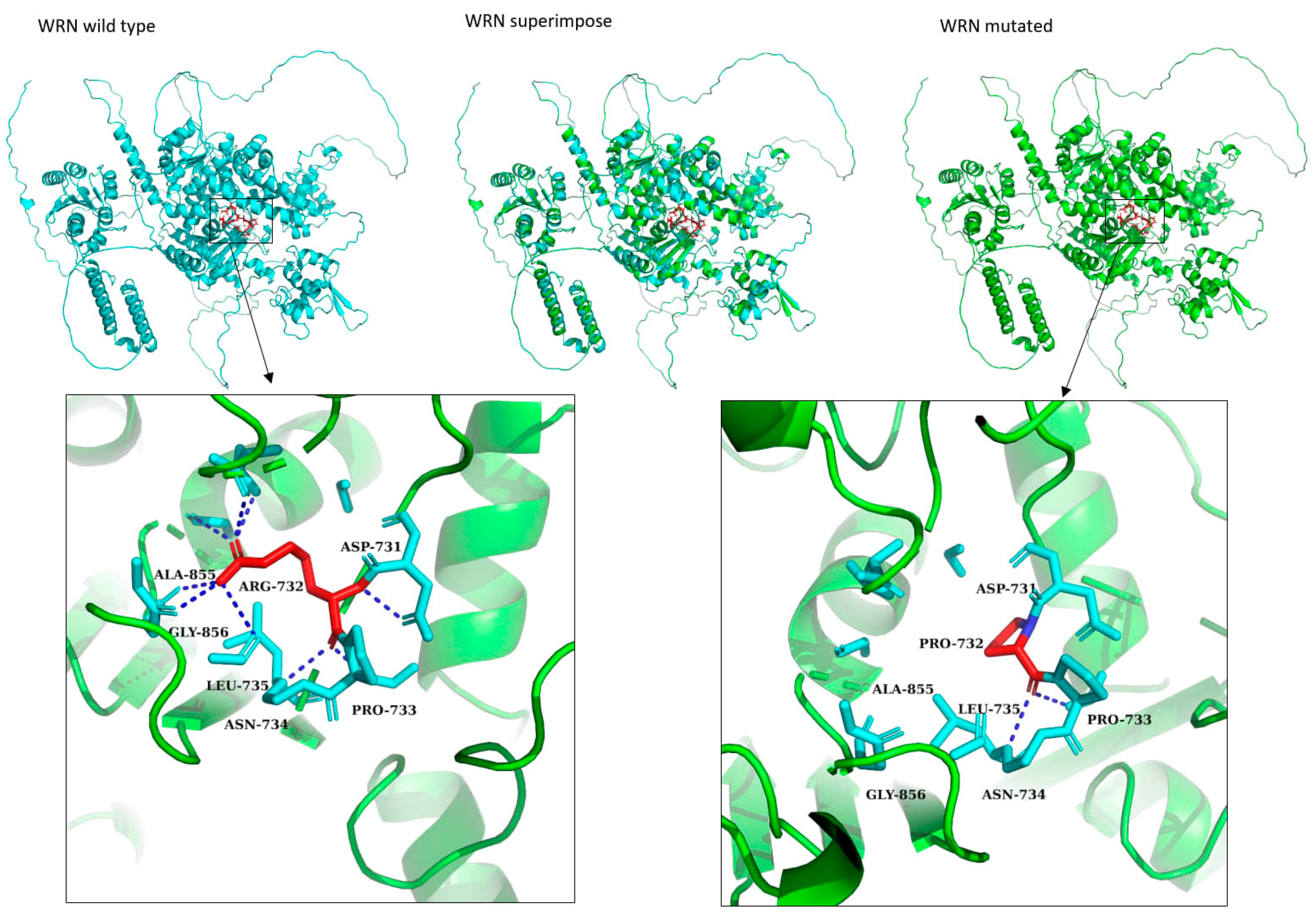
3.3. Protein Structure Modeling of Novel Variants
4. Discussion
4.1. Novel Variants Detected
4.2. Previously Known Cancer Variants Detected
4.3. Polygenic Variants Detected
4.4. The Homogenic Group of the Study’s EOBC Patients
4.5. Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | Breast cancer |
| DCIS | Ductal carcinoma in situ |
| DSB | Double-strand break |
| EOBC | Early-onset breast cancer |
| ER | Estrogen receptor |
| HER2 | Human epidermal growth factor receptor 2 |
| LCIS | Lobular carcinoma in situ |
| PR | Progesterone receptor |
| PRS | Polygenic risk score |
| PV | pathogenic variant |
| TNBC | Triple-negative breast cancer |
| VUS | Variant of unknown significance |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Narod, S.A. Breast cancer in young women. Nat. Rev. Clin. Oncol. 2012, 9, 460–470. [Google Scholar] [CrossRef]
- Assi, H.A.; Khoury, K.E.; Dbouk, H.; Khalil, L.E.; Mouhieddine, T.H.; el Saghir, S.N. Epidemiology and prognosis of breast cancer in young women. J. Thorac. Dis. 2013, 5 (Suppl. 1), S2–S8. [Google Scholar] [PubMed]
- Chelmow, D.; Pearlman, M.D.; Young, A.; Bozzuto, L.; Dayaratna, S.; Jeudy, M.; Kremer, M.E.; Scott, D.M.; O’Hara, J.S. Executive Summary of the Early-Onset Breast Cancer Evidence Review Conference. Obstet. Gynecol. 2020, 135, 1457–1478. [Google Scholar] [CrossRef] [PubMed]
- Tinterri, C.; Grimaldi, S.D.M.; Sagona, A.; Barbieri, E.; Darwish, S.; Bottini, A.; Canavese, G.; Gentile, D. Comparison of Long-Term Oncological Results in Young Women with Breast Cancer between BRCA-Mutation Carriers Versus Non-Carriers: How Tumor and Genetic Risk Factors Influence the Clinical Prognosis. Cancers 2023, 15, 4177. [Google Scholar] [CrossRef]
- Harbeck, N.; Gnant, M. Breast cancer. The lancet (North American edition). N. Am. 2017, 389, 1134–1150. [Google Scholar]
- Alluri, P.; Newman, L.A. Basal-Like and Triple-Negative Breast Cancers. Surg. Oncol. Clin. N. Am. 2014, 23, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Azim, H.A.; Partridge, A.H. Biology of breast cancer in young women. Breast Cancer Res. 2014, 16, 427. [Google Scholar] [CrossRef]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef] [PubMed]
- Määttä, K.; Rantapero, T.; Lindström, A.; Nykter, M.; Kankuri-Tammilehto, M.; Laasanen, S.-L.; Schleutker, J. Whole-exome sequencing of Finnish hereditary breast cancer families. Eur. J. Hum. Genet. 2016, 25, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Vahteristo, P.; Eerola, H.; Tamminen, A.; Blomqvist, C.; Nevanlinna, H. A probability model for predicting BRCA1 and BRCA2 mutations in breast and breast-ovarian cancer families. Br. J. Cancer 2001, 84, 704. [Google Scholar] [CrossRef] [PubMed]
- Pallonen, T.A.S.; Lempiäinen, S.M.M.; Joutsiniemi, T.K.; Aaltonen, R.I.; Pohjola, P.E.; Kankuri-Tammilehto, M.K. Genetic, clinic and histopathologic characterization of BRCA-associated hereditary breast and ovarian cancer in southwestern Finland. Sci. Rep. 2022, 12, 6704. [Google Scholar] [CrossRef]
- Nurmi, A.K.; Suvanto, M.; Dennis, J.; Aittomäki, K.; Blomqvist, C.; Nevanlinna, H. Pathogenic Variant Spectrum in Breast Cancer Risk Genes in Finnish Patients. Cancers 2022, 14, 6158. [Google Scholar] [CrossRef] [PubMed]
- Mars, N.; Widén, E.; Kerminen, S.; Meretoja, T.; Pirinen, M.; Parolo, P.d.B.; Palta, P.; Palotie, A.; Kaprio, J.; Joensuu, H.; et al. The role of polygenic risk and susceptibility genes in breast cancer over the course of life. Nat. Commun. 2020, 11, 6383. [Google Scholar] [CrossRef] [PubMed]
- Muranen, T.A.; Mavaddat, N.; Khan, S.; Fagerholm, R.; Pelttari, L.; Lee, A.; Aittomäki, K.; Blomqvist, C.; Easton, D.F.; Nevanlinna, H. Polygenic risk score is associated with increased disease risk in 52 Finnish breast cancer families. Breast Cancer Res. Treat. 2016, 158, 463–469. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Momozawa, Y.; Mizukami, K. Unique roles of rare variants in the genetics of complex diseases in humans. J. Hum. Genet. 2021, 66, 11–23. [Google Scholar] [CrossRef]
- Marmolejo, D.H.; Wong, M.Y.Z.; Bajalica-Lagercrantz, S.; Tischkowitz, M.; Balmaña, J.; Patócs, A.B.; Chappuis, P.; Colas, C.; Genuardi, M.; Haanpää, M.; et al. Overview of hereditary breast and ovarian cancer (HBOC) guidelines across Europe. Eur. J. Med. Genet. 2021, 64, 104350. [Google Scholar] [CrossRef] [PubMed]
- Welsh, A.W.; Moeder, C.B.; Kumar, S.; Gershkovich, P.; Alarid, E.T.; Harigopal, M.; Haffty, B.G.; Rimm, D.L. Standardization of Estrogen Receptor Measurement in Breast Cancer Suggests False-Negative Results Are a Function of Threshold Intensity Rather Than Percentage of Positive Cells. J. Clin. Oncol. 2011, 29, 2978–2984. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Poplin, R.; Chang, P.-C.; Alexander, D.; Schwartz, S.; Colthurst, T.; Ku, A.; Newburger, D.; Dijamco, J.; Nguyen, N.; Afshar, P.T.; et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018, 36, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Kircher, M.; Witten, D.M.; Jain, P.; O’roak, B.J.; Cooper, G.M.; Shendure, J. A General Framework for Estimating the Relative Pathogenicity of Human Genetic Variants; Nature Publishing Group: New York, NY, USA, 2014. [Google Scholar]
- Wood, R.D.; Mitchell, M.; Sgouros, J.; Lindahl, T. Human DNA repair genes. Science 2001, 291, 1284–1289. [Google Scholar] [CrossRef] [PubMed]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef]
- Chen, S.; Francioli, L.C.; Goodrich, J.K.; Collins, R.L.; Kanai, M.; Wang, Q.; Alföldi, J.; Watts, N.A.; Vittal, C.; Gauthier, L.D.; et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature 2024, 625, 92–100. [Google Scholar] [CrossRef]
- Ioannidis, N.M.; Rothstein, J.H.; Pejaver, V.; Middha, S.; McDonnell, S.K.; Baheti, S.; Musolf, A.; Li, Q.; Holzinger, E.; Karyadi, D.; et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016, 99, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Syrjakoski, K.; Vahteristo, P.; Eerola, H.; Tamminen, A.; Kivinummi, K.; Sarantaus, L.; Holli, K.; Blomqvist, C.; Kallioniemi, O.-P.; Kainu, T.; et al. Population-based study of BRCA1 and BRCA2 mutations in 1035 unselected Finnish breast cancer patients. J. Natl. Cancer Inst. JNCI 2000, 92, 1529–1531. [Google Scholar] [CrossRef] [PubMed]
- Kilpivaara, O.; Bartkova, J.; Eerola, H.; Syrjäkoski, K.; Vahteristo, P.; Lukas, J.; Blomqvist, C.; Holli, K.; Heikkilä, P.; Sauter, G.; et al. Correlation of CHEK2 protein expression and c.1100delC mutation status with tumor characteristics among unselected breast cancer patients. Int. J. Cancer 2005, 113, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Fagerholm, R.; Hofstetter, B.; Tommiska, J.; Aaltonen, K.; Vrtel, R.; Syrjäkoski, K.; Kallioniemi, A.; Kilpivaara, O.; Mannermaa, A.; Kosma, V.-M.; et al. NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat. Genet. 2008, 40, 844–853. [Google Scholar] [CrossRef]
- Eerola, H.; Blomqvist, C.; Pukkala, E.; Pyrhönen, S.; Nevanlinna, H. Familial breast cancer in southern Finland: How prevalent are breast cancer families and can we trust the family history reported by patients? Eur. J. Cancer 2000, 36, 1143–1148. [Google Scholar] [CrossRef]
- Vahteristo, P.; Bartkova, J.; Eerola, H.; Syrjäkoski, K.; Ojala, S.; Kilpivaara, O.; Tamminen, A.; Kononen, J.; Aittomäki, K.; Heikkilä, P.; et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am. J. Hum. Genet. 2002, 71, 432–438. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Chen, L.; Oshima, J. Werner Syndrome. J. Biomed. Biotechnol. 2002, 2, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, G.; Sun, F.; Dong, N.; Sun, Z.; Jiang, D. Association Between WRN Cys1367Arg (T>C) and Cancer Risk: A Meta-analysis. Technol. Cancer Res. Treat. 2016, 15, 20–27. [Google Scholar] [CrossRef]
- Zins, K.; Frech, B.; Taubenschuss, E.; Schneeberger, C.; Abraham, D.; Schreiber, M. Association of the rs1346044 Polymorphism of the Werner Syndrome Gene RECQL2 with Increased Risk and Premature Onset of Breast Cancer. Int. J. Mol. Sci. 2015, 16, 29643–29653. [Google Scholar] [CrossRef]
- Sokolenko, A.P.; Preobrazhenskaya, E.V.; Aleksakhina, S.N.; Iyevleva, A.G.; Mitiushkina, N.V.; Zaitseva, O.A.; Yatsuk, O.S.; Tiurin, V.I.; Strelkova, T.N.; Togo, A.V.; et al. Candidate gene analysis of BRCA1/2 mutation-negative high-risk Russian breast cancer patients. Cancer Lett. 2015, 359, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Lim, B.W.X.; Thompson, E.R.; McInerny, S.; Zethoven, M.; Cheasley, D.; Rowley, S.M.; Wong-Brown, M.W.; Devereux, L.; Gorringe, K.L.; et al. Investigation of monogenic causes of familial breast cancer: Data from the BEACCON case-control study. NPJ Breast Cancer 2021, 7, 76. [Google Scholar] [CrossRef]
- Oshima, J.; Sidorova, J.M.; Monnat, R.J. Werner syndrome: Clinical features, pathogenesis and potential therapeutic interventions. Ageing Res. Rev. 2017, 33, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.-H.; Kusumoto, R.; Opresko, P.L.; Sui, X.; Huang, S.; Nicolette, M.L.; Paull, T.T.; Campisi, J.; Seidman, M.; Bohr, V.A. Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res. 2006, 34, 2751–2760. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.C.; Chen, Y.R.; Kensicki, E.; Li, A.Y.J.; Kong, M.; Li, Y.; Mohney, R.P.; Shen, H.M.; Stiles, B.; Mizushima, N.; et al. Autophagy: Resetting glutamine-dependent metabolism and oxygen consumption. Autophagy 2012, 8, 1477–1493. [Google Scholar] [CrossRef]
- Zhou, T.; Yi, F.; Wang, Z.; Guo, Q.; Liu, J.; Bai, N.; Li, X.; Dong, X.; Ren, L.; Cao, L.; et al. The Functions of DNA Damage Factor RNF8 in the Pathogenesis and Progression of Cancer. Int. J. Biol. Sci. 2019, 15, 909–918. [Google Scholar] [CrossRef]
- Li, L.; Halaby, M.J.; Hakem, A.; Cardoso, R.; El Ghamrasni, S.; Harding, S.; Chan, N.; Bristow, R.; Sanchez, O.; Durocher, D.; et al. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J. Exp. Med. 2010, 207, 983–997. [Google Scholar] [CrossRef]
- Cao, Z.; Qin, X.; Liu, F.; Zhou, L. Tryptophan-induced pathogenesis of breast cancer. Afr. Health Sci. 2015, 15, 982–985. [Google Scholar] [CrossRef]
- COSMIC, RNF4. Available online: https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=RNF4 (accessed on 12 October 2021).
- Galanty, Y.; Belotserkovskaya, R.; Coates, J.; Jackson, S.P. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012, 26, 1179–1195. [Google Scholar] [CrossRef]
- Shimada, T.; Kudoh, Y.; Noguchi, T.; Kagi, T.; Suzuki, M.; Tsuchida, M.; Komatsu, H.; Takahashi, M.; Hirata, Y.; Matsuzawa, A. The E3 Ubiquitin-Protein Ligase RNF4 Promotes TNF-α-Induced Cell Death Triggered by RIPK1. Int. J. Mol. Sci. 2021, 22, 5796. [Google Scholar] [CrossRef]
- Rajagopal, T.; Seshachalam, A.; Rathnam, K.K.; Jothi, A.; Viswanathan, S.; Talluri, S.; Dunna, N.R. DNA repair genes hOGG1, XRCC1 and ERCC2 polymorphisms and their molecular mapping in breast cancer patients from India. Mol. Biol. Rep. 2020, 47, 5081–5090. [Google Scholar] [CrossRef]
- Zhao, R.; Ying, M.F. Association between ERCC1 and ERCC2 polymorphisms and breast cancer risk in a Chinese population. Genet. Mol. Res. 2016, 15, 15017263. [Google Scholar] [CrossRef] [PubMed]
- Hardi, H.; Melki, R.; Boughaleb, Z.; el Harroudi, T.; Aissaoui, S.; Boukhatem, N. Significant association between ERCC2 and MTHR polymorphisms and breast cancer susceptibility in Moroccan population: Genotype and haplotype analysis in a case-control study. BMC Cancer 2018, 18, 292. [Google Scholar] [CrossRef]
- Lindor, N.M.; Guidugli, L.; Wang, X.; Vallée, M.P.; Monteiro, A.N.; Tavtigian, S.; Goldgar, D.E.; Couch, F.J. A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS). Hum. Mutat. 2012, 33, 8–21. [Google Scholar] [CrossRef]
- Meeks, H.D.; Song, H.; Michailidou, K.; Bolla, M.K.; Dennis, J.; Wang, Q.; Barrowdale, D.; Frost, D.; Embrace; McGuffog, L.; et al. BRCA2 Polymorphic Stop Codon K3326X and the Risk of Breast, Prostate, and Ovarian Cancers. J. Natl. Cancer Inst. 2016, 108, djv315. [Google Scholar] [CrossRef] [PubMed]
- ClinVar. RCV000077387.24. 2024. ClinVar Database: ID rs55712212. Available online: https://www.ncbi.nlm.nih.gov/clinvar/RCV000077387/ (accessed on 18 July 2023).
- Koivuluoma, S.; Winqvist, R.; Keski-Filppula, R.; Kuismin, O.; Moilanen, J.; Pylkäs, K. Evaluating the role of MLH3 p.Ser1188Ter variant in inherited breast cancer predisposition. Genet. Med. 2020, 22, 663–664. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.; Silva, S.N.; Azevedo, A.P.; Teixeira, V.; Pina, J.E.; Rueff, J.; Gaspar, J.F. Association of common variants in mismatch repair genes and breast cancer susceptibility: A multigene study. BMC Cancer 2009, 9, 344. [Google Scholar] [CrossRef]
- Miao, H.K.; Chen, L.P.; Cai, D.P.; Kong, W.J.; Xiao, L.; Lin, J. MSH3 rs26279 polymorphism increases cancer risk: A meta-analysis. Int. J. Clin. Exp. Pathol. 2015, 8, 11060–11067. [Google Scholar]
- ClinVar. RCV000131434.23. 2024. ClinVar Database ID: rs587782401. Available online: https://www.ncbi.nlm.nih.gov/clinvar/RCV000131434/ (accessed on 5 May 2023).
- Boonen, R.A.C.M.; Vreeswijk, M.P.G.; van Attikum, H. CHEK2 variants: Linking functional impact to cancer risk. Trends Cancer 2022, 8, 759–770. [Google Scholar] [CrossRef]
- Sanoguera-Miralles, L.; Valenzuela-Palomo, A.; Bueno-Martínez, E.; Esteban-Sánchez, A.; Lorca, V.; Llinares-Burguet, I.; García-Álvarez, A.; Pérez-Segura, P.; Infante, M.; Easton, D.F.; et al. Systematic Minigene-Based Splicing Analysis and Tentative Clinical Classification of 52 CHEK2 Splice-Site Variants. Clin. Chem. 2023, 70, 319–338. [Google Scholar] [CrossRef]
- Yadav, S.; Boddicker, N.J.; Na, J.; Polley, E.C.; Hu, C.; Hart, S.N.; Gnanaolivu, R.D.; Larson, N.; Dunn, C.; Holtegaard, S.; et al. Contralateral Breast Cancer Risk Among Carriers of Germline Pathogenic Variants in ATM, BRCA1, BRCA2, CHEK2, and PALB2. J. Clin. Oncol. 2023, 41, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Girard, E.; Eon-Marchais, S.; Olaso, R.; Renault, A.; Damiola, F.; Dondon, M.; Barjhoux, L.; Goidin, D.; Meyer, V.; Le Gal, D.; et al. Familial breast cancer and DNA repair genes: Insights into known and novel susceptibility genes from the GENESIS study, and implications for multigene panel testing. Int. J. Cancer 2019, 144, 1962–1974. [Google Scholar] [CrossRef]
- Bonache, S.; Esteban, I.; Moles-Fernández, A.; Tenés, A.; Duran-Lozano, L.; Montalban, G.; Bach, V.; Carrasco, E.; Gadea, N.; López-Fernández, A.; et al. Multigene panel testing beyond BRCA1/2 in breast/ovarian cancer Spanish families and clinical actionability of findings. J. Cancer Res. Clin. Oncol. 2018, 144, 2495–2513. [Google Scholar] [CrossRef] [PubMed]
- ClinVar. VCV000210988.58. 2024. ClinVar Database ID: rs759217526. Available online: https://www.ncbi.nlm.nih.gov/clinvar/RCV000192919/ (accessed on 16 February 2024).
- Wang, H.C.; Chiu, C.F.; Tsai, R.Y.; Kuo, Y.S.; Chen, H.S.; Wang, R.F.; Tsai, C.W.; Chang, C.H.; Lin, C.C.; Bau, D.T. Association of Genetic Polymorphisms of EXO1 Gene with Risk of Breast Cancer in Taiwan. Anticancer Res. 2009, 29, 3897. Available online: http://ar.iiarjournals.org/content/29/10/3897.abstract (accessed on 21 August 2024).
- Liu, J.; Zhang, J. Elevated EXO1 expression is associated with breast carcinogenesis and poor prognosis. Ann. Transl. Med. 2021, 9, 135. [Google Scholar] [CrossRef] [PubMed]
- Higgins, G.S.; Harris, A.L.; Prevo, R.; Helleday, T.; McKenna, W.G.; Buffa, F.M. Overexpression of POLQ Confers a Poor Prognosis in Early Breast Cancer Patients. Oncotarget 2010, 1, 175. [Google Scholar] [CrossRef]
- Zhou, J.; Gelot, C.; Pantelidou, C.; Li, A.; Yücel, H.; Davis, R.E.; Färkkilä, A.; Kochupurakkal, B.; Syed, A.; Shapiro, G.I.; et al. A first-in-class polymerase theta inhibitor selectively targets homologous-recombination-deficient tumors. Nat. Cancer 2021, 2, 598–610. [Google Scholar] [CrossRef]
- Lee, A.; Mavaddat, N.; Wilcox, A.N.; Cunningham, A.P.; Carver, T.; Hartley, S.; Babb de Villiers, C.; Izquierdo, A.; Simard, J.; Schmidt, M.K.; et al. BOADICEA: A comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet. Med. 2019, 21, 1708–1718. [Google Scholar] [CrossRef]
- Yanes, T.; Young, M.A.; Meiser, B.; James, P.A. Clinical applications of polygenic breast cancer risk: A critical review and perspectives of an emerging field. Breast Cancer Res. 2020, 22, 21. [Google Scholar] [CrossRef]
| Variant | Gene | Location | Nucleotide Change | Effect on Protein | Consequence | Subcategory | gnomADe FIN AF | gnomADg FIN AF | SIFT | PolyPhen | REVEL | CADD PHRED |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs193219754 | MLH3 | chr14:75039918 | c.3563C>G | p.Ser1188Ter | stop gained | 0.002491 | 0.003159 | NA | NA | NA | 42 | |
| rs180177100 | PALB2 | chr16:23635306 | c.1240C>T | p.Arg414Ter | stop gained | 0 | 0 | NA | NA | NA | 37 | |
| rs11574410 | WRN | chr8:31173019 | c.4216C>T | p.Arg1406Ter | stop gained | 0.0006469 | 0.0002832 | NA | NA | NA | 36 | |
| rs81002862 | BRCA2 | chr13:32380005 | c.9118-2A>G | NA | splice acceptor | 0.0003697 | 0.0001885 | NA | NA | NA | 35 | |
| rs11571833 | BRCA2 | chr13:32398489 | c.9976A>T | p.Lys3326Ter | stop gained | 0.01086 | 0.01045 | NA | NA | NA | 35 | |
| rs5745908 | NEIL1 | chr15:75349341 | c.434+2T>C | NA | splice donor | 0.0018 | 0.001508 | NA | NA | NA | 34 | |
| rs587782401 | CHEK2 | chr22:28734401 | c.319+2T>A | NA | splice donor | 0.0005548 | 0.0003764 | NA | NA | NA | 34 | |
| rs766240074 | RAD54L | chr1:46273748 | c.1610+1G>A | NA | splice donor | 4.968 × 10−5 | 0 | NA | NA | NA | 33 | |
| chr4:2490336 G/T | RNF4 | chr4:2490336 | c.-157-1G>T | NA | splice acceptor | NA | NA | NA | NA | NA | 33 | |
| rs756188698 | RAD18 | chr3:8899048 | c.1169-1G>C | NA | splice acceptor | 0.0002398 | 9.457 × 10−5 | NA | NA | NA | 28.9 | |
| rs11572913 | GTF2H3 | chr12:123633862 | c.3G>A | p.Met1? | start lost | 0.0007112 | 0.000565 | 0.02lc | 0.273 | NA | 24.8 | |
| rs187418762 | SPIDR | chr8:47713484 | c.2189-5G>A | NA | splice region, splice polypyrimidine tract, intron | 0.01286 | 0.01198 | NA | NA | NA | 20.5 | |
| rs149243307 | FANCI | chr15:89260841 | c.286G>A | p.Glu96Lys | missense, splice region | 2 | 0.001848 | 0.001507 | 0 | 1 | 0.448 | 33 |
| rs41540016 | POLQ | chr3:121436275 | c.7390G>A | p.Ala2464Thr | missense, splice region | 2 | 0.01381 | 0.01208 | 0 | 1 | 0.84 | 33 |
| chr1:241861448 -/T | EXO1 | chr1:241861447-241861448 | c.987dup | p.Lys330Ter | frameshift | 1 | NA | NA | NA | NA | NA | 33 |
| rs773570504 | ATM | chr11:108326152-108326153 | c.6908dup | p.Glu2304GlyfsTer69 | frameshift | 1 | 4.625 × 10−5 | 9.56 × 10−5 | NA | NA | NA | 33 |
| rs762390984 | FANCI | chr15:89301393-89301405 | c.2957_2969del | p.Val986AlafsTer39 | frameshift | 1 | 0.002957 | 0.002454 | NA | NA | NA | 33 |
| rs759217526 | FANCL | chr2:58159793-58159794 | c.1096_1099dup | p.Thr367AsnfsTer13 | frameshift | 1 | 0.001814 | 0.002649 | NA | NA | NA | 33 |
| rs199791286 | MSH3 | chr5:80778737 | c.2336G>A | p.Arg779His | missense | 1 | 0.001294 | 0.001415 | 0 | 1 | 0.884 | 32 |
| chr8:31111721 G/C | WRN | chr8:31111721 | c.2195G>C | p.Arg732Pro | missense | 2 | NA | NA | 0 | 1 | 0.499 | 31 |
| chr12:132677395 -/T | POLE | chr12:132677394-132677395 | c.769dup | p.Ile257AsnfsTer6 | frameshift | 1 | NA | NA | NA | NA | NA | 31 |
| rs28363284 | RAD51D | chr17:35103294 | c.698A>G | p.Glu233Gly | missense | 1 | 0.001185 | 0.001797 | 0.04 | 0.973 | NA | 29.9 |
| rs746243211 | MSH3 | chr5:80672754 | c.923A>T | p.Lys308Met | missense | 1 | 0 | 0 | 0 | 1 | 0.823 | 29.7 |
| rs137923123 | PER1 | chr17:8147713 | c.1349G>A | p.Arg450His | missense | 1 | 0.003681 | 0.002823 | 0 | 1 | NA | 29.2 |
| rs373080718 | TOP3A | chr17:18277728 | c.2774C>T | p.Pro925Leu | missense | 1 | 4.623 × 10−5 | 0 | 0.01 | 0.999 | NA | 29 |
| rs2306211 | POLQ | chr3:121432937 | c.7640C>T | p.Ala2547Val | missense | 2 | 0.007347 | 0.006223 | 0.03 | 1 | 0.571 | 28.8 |
| rs200535477 | RECQL5 | chr17:75627670 | c.2828G>A | p.Arg943His | missense | 1 | 0.009798 | 0.008201 | 0 | 1 | NA | 28.7 |
| rs202068855 | RPA1 | chr17:1880615 | c.1165C>T | p.Arg389Trp | missense | 2 | 0.01502 | 0.0152 | 0 | 1 | NA | 28.1 |
| rs144564120 | ERCC2 | chr19:45352249 | c.2150C>G | p.Ala717Gly | missense | 1 | 4.624 × 10−5 | 0 | 0.01 | 0.015 | NA | 28.1 |
| rs28897758 | BRCA2 | chr13:32394734 | c.1267T>G | p.Cys423Gly | missense | 1 | 0.0004619 | 0.000659 | 0 | 1 | 0.788 | 28 |
| rs34001746 | TOP3A | chr17:18285268 | c.1751T>G | p.Leu584Arg | missense | 1 | 0.0004619 | 0.000659 | 0.02 | 0.803 | NA | 27.3 |
| rs121913016 | ERCC2 | chr19:45357368 | c.1381C>G | p.Leu461Val | missense | 1 | 4.633e-05 | 0 | 0 | 0.982 | NA | 27.3 |
| rs1805378 | NTHL1 | chr16:2044652 | c.503T>C | p.Ile168Thr | missense | 1 | 0.0004352 | 0.0003766 | 0 | 1 | 0.876 | 27.1 |
| rs34642881 | RECQL4 | chr8:144517415 | c.212A>G | p.Glu71Gly | missense, splice region | 1 | 0.004511 | 0.005941 | 0.02 | 0.803 | NA | 26.6 |
| rs17879961 | CHEK2 | chr22:28725099 | c.470T>C | p.Ile157Thr | missense | 1 | 0.004511 | 0.005941 | 0.07 | 0.514 | NA | 26.5 |
| rs142213781 | NEIL1 | chr15:75353853 | c.833C>T | p.Thr278Ile | missense | 2 | 0.002086 | 0.002352 | 0 | 1 | 0.563 | 26.4 |
| rs562132292 | ERCC2 | chr19:45357290 | c.1459C>T | p.Arg487Trp | missense | 1 | 0.0003272 | 0.0006591 | 0 | 0.998 | NA | 26.4 |
| chr6:37360499 C/G | RNF8 | chr6:37360499 | c.165C>G | p.Cys55Trp | missense | 2 | NA | NA | 0 | 1 | 0.747 | 26.3 |
| rs149253459 | FAAP100 | chr17:81547649 | c.1433A>G | p.Gln478Arg | missense | 1 | 0.002894 | 0.0016 | 0.01 | 0.997 | NA | 26.2 |
| rs750771205 | ATM | chr11:108289000 | c.4133C>T | p.Pro1378Leu | missense | 2 | 0 | 0 | 0 | 0.969 | 0.546 | 26.1 |
| rs1801673 | ATM | chr11:108304736 | c.5558A>T | p.Asp1853Val | missense | 2 | 0.003699 | 0.00292 | 0 | 0.987 | 0.589 | 26.1 |
| rs1799802 | ERCC4 | chr16:13934224 | c.1135C>T | p.Pro379Ser | missense | 2 | 0.007207 | 0.009714 | 0 | 1 | 0.526 | 25.4 |
| rs11212587 | ATM | chr11:108315883 | c.6067G>A | p.Gly2023Arg | missense | 2 | 0.0008318 | 0.0007553 | 0 | 0.364 | 0.511 | 25.3 |
| rs61752784 | POLG | chr15:89330133 | c.803G>C | p.Gly268Ala | missense | 1 | 0.004158 | 0.00546 | 0 | 0.999 | 0.967 | 25.3 |
| rs200981995 | LIG3 | chr17:34999827 | c.2302T>C | p.Tyr768His | missense | 2 | 0.01173 | 0.009134 | 0.11 | 0.994 | NA | 25.1 |
| chr17:18292743 T/A | TOP3A | chr17:18292743 | c.1183A>T | p.Ser395Cys | missense | 1 | NA | NA | 0.01 | 0.976 | NA | 25 |
| rs201920810 | SPIDR | chr8:47712713 | c.2029G>A | p.Asp677Asn | missense | 2 | 0 | 0 | 0 | 1 | 0.561 | 24.9 |
| rs140566004 | POLE | chr12:132673646 | c.1288G>A | p.Ala430Thr | missense | 2 | 0.0001395 | 0 | 0 | 0.997 | 0.487 | 24.7 |
| rs78488552 | WRN | chr8:31154721 | c.3785C>G | p.Thr1262Arg | missense | 2 | 0.000231 | 9.432 × 10−5 | 0 | 0.999 | 0.413 | 24.5 |
| rs546221341 | POLQ | chr3:121488663-121488669 | c.4262_4268del | p.Ile1421ArgfsTer8 | frameshift | 1 | 0.006209 | 0.005384 | NA | NA | NA | 24.3 |
| rs145289229 | POLG | chr15:89328532 | c.1174C>G | p.Leu392Val | missense | 1 | 0.007823 | 0.01085 | 0.06 | 0.999 | 0.796 | 24.2 |
| rs55748151 | POLQ | chr3:121533021 | c.929T>G | p.Val310Gly | missense | 2 | 0.001063 | 0.001413 | 0 | 0.552 | 0.498 | 24.1 |
| rs150018949 | EXO5 | chr1:40515573-40515574 | c.1029_1030insG | p.Arg344AlafsTer10 | frameshift | 1 | 0.004352 | 0.003488 | NA | NA | NA | 24.1 |
| rs771308001 | ERCC1 | chr19:45407145-45407146 | NA | NA | downstream gene | 1 | 0.003376 | 0.00311 | NA | NA | NA | 23.7 |
| rs565251228 | RECQL5 | chr17:75624891 | NA | NA | downstream gene | 1 | 0.0001461 | 0.0002827 | NA | NA | NA | 23.4 |
| rs55801750 | ATM | chr11:108330296 | c.7390T>C | p.Cys2464Arg | missense | 2 | 4.619 × 10−5 | 0 | 0.1 | 0.005 | 0.668 | 22.9 |
| rs201503405 | PNKP | chr19:49862573 | c.901C>T | p.Arg301Trp | missense | 1 | 0.006464 | 0.006775 | 0 | 0.911 | NA | 22.9 |
| chr19:45352306 T/C | ERCC2 | chr19:45352306 | c.2093A>G | p.Gln698Arg | missense | 1 | NA | NA | 0.54 | 0.218 | NA | 22.8 |
| chrX:153444976 C/G | TREX2 | chrX:153444976 | c.455G>C | p.Arg152Pro | missense | 1 | NA | NA | 0 | 0.999 | NA | 22.7 |
| rs775001669 | MLH3 | chr14:75048767 | c.889C>T | p.Arg297Trp | missense | 2 | 0.00134 | 0.001132 | 0 | 1 | 0.655 | 22.6 |
| rs41549716 | POLG | chr15:89321842 | c.2492A>G | p.Tyr831Cys | missense | 2 | 0.01803 | 0.0161 | 0.02 | 0.995 | 0.732 | 22.6 |
| rs201414369 | EME1 | chr17:50380824 | c.1598G>A | p.Arg533His | missense | 1 | 0.0001386 | 0.0001884 | 0.22 | 0.994 | NA | 22.6 |
| rs144340710 | TP53 | chr17:7674259 | c.704A>G | p.Asn235Ser | missense | 1 | 0.0002772 | 0.0004745 | 0.22 | 0.385 | NA | 22.5 |
| rs4987202 | RAD23A | chr19:12948812 | c.599C>T | p.Thr200Met | missense, splice region | 1 | 0.004359 | 0.003299 | 0.43 | 0.02 | NA | 22.5 |
| rs146309259 | LIG1 | chr19:48121298 | c.2257G>A | p.Val753Met | missense | 1 | 0.002451 | 0.001788 | 0.06 | 0.407 | NA | 22.5 |
| rs28897689 | BRCA1 | chr17:43091492 | c.4039A>G | p.Arg1347Gly | missense | 1 | 0.001944 | 0.001788 | 0.09 | 0.255 | NA | 22.2 |
| rs55712212 | BRCA2 | chr13:32341176 | c.6821G>T | p.Gly2274Val | missense | 2 | 0.01303 | 0.01349 | 0.37 | 0.966 | 0.481 | 21.8 |
| rs144276604 | XAB2 | chr19:7625912 | c.790G>A | p.Asp264Asn | missense | 1 | 0.0002318 | 0 | 0.09 | 0.846 | NA | 21.7 |
| rs3730947 | LIG1 | chr19:48140013 | c.1045G>A | p.Val349Met | missense | 2 | 0.01564 | 0.01648 | 0 | 0.997 | NA | 21.5 |
| rs776329282 | ERCC4 | chr16:13926750-13926755 | c.580_584+1del | NA | in-frame deletion, splice region | 1 | 9.256 × 10−5 | 0.0001884 | NA | NA | NA | 20.5 |
| rs763165669 | NTHL1 | chr16:2038404-2038416 | NA | NA | downstream gene | 2 | 0.01363 | 0.007775 | NA | NA | NA | NA |
| rs41547220 | POLQ | chr3:121489857-121489859 | c.3072_3074del | p.Lys1025del | in-frame deletion | 2 | 0.01831 | 0.01925 | NA | NA | NA | NA |
| ID* | Found var | Age | Histology, Grade, ER, PR, Her2 | ||||
|---|---|---|---|---|---|---|---|
| 424 | rs145289229 | rs11571833 | rs2306211 | rs546221341 | 25 | Ductal, G3, ER+, PR-, Her2+ | |
| POLG | BRCA2 | POLQ | POLQ | ||||
| 425 | 27 | Ductal, G3, ER+, PR+, Her2- | |||||
| 426 | 27 | Ductal, G3, ER-, PR-, NA | |||||
| 427 | 28 | Ductal, G3, ER+, PR+, Her2+ | |||||
| 428 | rs11571833 | rs3730947 | 28 | Ductal, G3, ER-, PR-, Her2- | |||
| BRCA2 | LIG1 | ||||||
| 429 | 28 | Ductal, G3, ER+, PR+, Her2+ | |||||
| 431 | rs140566004 | rs4987202 | 28 | Ductal, G3, ER-, PR+, Her2+ | |||
| POLE | RAD23A | ||||||
| 432- | rs201920810 | rs773570504 | 30 | Ductal, G3, ER+, PR+, Her2- | |||
| SPIDR | ATM | ||||||
| 433 | 31, 35 | Ductal, G3, ER-, PR-, Her2-; 2 Ductal, ER+, Her2- ^ | |||||
| 434- | rs200535477 | rs2306211 | 31 | Ductal, G3, ER-, PR-, Her2- | |||
| RECQL5 | POLQ | ||||||
| 435- | Novel RNF8 | rs78488552 | rs565251228 | 32 | Ductal, G3, ER-, PR-, Her2+ | ||
| WRN | RECQL5 | ||||||
| 436- | rs1801673 | rs762390984 | rs763165669 | 32 | Micropapillar, ER+, PR+, Her2- | ||
| ATM | FANCI | NTHL1 | |||||
| 437 | rs373080718 | rs202068855 | 33 | DCIS, G3 | |||
| TOP3A | RPA1 | ||||||
| 438- | rs187418762 | 33 | Ductal, G3, ER-, PR-, Her2- | ||||
| SPIDR | |||||||
| 440 | rs55712212 | rs137923123 | novel ERCC2 | 34, 37 | Ductal, G3, ER-, PR-; 2 LCIS | ||
| BRCA2 | PER1 | ||||||
| 441- | 34 | Ductal, G3, ER+, PR+, Her2- | |||||
| 442 | rs41547220 | rs149253459 | 34 | Ductal, G3, ER+, PR+, Her2- | |||
| POLQ | FAAP100 | ||||||
| 443- | rs766240074 | rs562132292 | 34 | Ductal, G3, ER-, PR-, Her2- | |||
| RAD54L | ERCC2 | ||||||
| 444- | 35 | Ductal, G3, ER-, PR-, Her2+ | |||||
| 445 | rs587782401 | 35,43 | Ductal, G3, ER-, PR-, Her2+; 2 Ductal, G3, ER-, PR-, Her2+ | ||||
| CHEK2 | |||||||
| 446- | rs771308001 | 34 | Ductal, G2, ER+, PR+, Her2- | ||||
| ERCC1 | |||||||
| 610 | rs28897758 | rs1805378 | 40, 40 | Ductal, G1, ER+, PR+, Her2-; 2 G3, ER+, PR+, Her2- | |||
| BRCA2 | NTHL1 | ||||||
| 611 | 40, 54 | Ductal, G2, ER+, PR-, Her2-; 2 ductal, G2, ER+, PR+, Her2- | |||||
| 612 | rs81002862 | rs11212587 | 40 | DCIS | |||
| BRCA2 | ATM | ||||||
| 613 | 35 | ||||||
| 614 | 26 | DCIS, G3 | |||||
| 615 | rs55748151 | 30 | Ductal, G3, ER-, PR-, Her2- | ||||
| POLQ | |||||||
| 616 | rs55801750 | rs34001746 | rs144276604 | 38 | Ductal, G3, ER-, PR-, Her2+ | ||
| ATM | TOP3A | XAB2 | |||||
| 617 | rs746243211 | Novel Trex2 | 40, 67 | 2 Ductal G3, ER+, PR+, Her2- | |||
| MSH3 | |||||||
| 618 | rs776329282 | 27 | Ductal, G2, ER+, PR+, Her2- | ||||
| ERCC4 | |||||||
| 619 | rs5745908 | rs41549716 | Novel Top3A | rs41540016 | 27 | Ductal, G3, ER-, PR-, Her2- | |
| NEIL1 | POLG | POLQ | |||||
| 61 | rs121913016 | rs180177100 | rs144564120 | 36 | Ductal, ER+, PR+, Her2- | ||
| ERCC2 | PALB2 | ERCC2 | |||||
| 620 | 35 | Ductal, G2, ER+, PR+, Her2- | |||||
| 621 | rs55712212 | 39 | Lobular | ||||
| BRCA2 | |||||||
| 622 | 38 | Ductal, G2, ER+, PR+, Her2+ | |||||
| 623 | Novel RNF4 | 31 | Ductal, G2, ER+, PR+, Her2+ | ||||
| 624 | rs149243307 | rs61752784 | rs17879961 | Novel EXO1 | 23 | Ductal, G3, ER+, PR+, Her2+ | |
| FANCI | POLG | CHEK2 | |||||
| 625 | rs3730947 | 28 | Lobular, G2, ER+, PR+, Her2- | ||||
| LIG1 | |||||||
| 626 | rs750771205 | rs145289229 | rs146309259 | rs3730947 | rs587782401 | 34, 34 | Ductal, ER+, PR+, Her2+; 2 DCIS |
| ATM | POLG | LIG1 | LIG1 | CHEK2 | |||
| 627 | 33 | ||||||
| 628 | rs28363284 | rs41540016 | 28 | Ductal, G3, ER-, PR-, Her2- | |||
| RAD51D | POLQ | ||||||
| 629- | rs150018949 | 40 | |||||
| EXO5 | |||||||
| 62 | 36 | DCIS | |||||
| 630 | rs1799802 | 26 | Ductal, G3, ER-, PR-, Her2- | ||||
| ERCC4 | |||||||
| 631- | rs193219754 | 31 | Ductal, G2, ER+, PR-, Her2- | ||||
| MLH3 | |||||||
| 632 | rs187418762 | rs4987202 | rs201503405 | 38, 67 | Ductal; 2 ductal, G3, ER+, PR+, Her2- | ||
| SPIDR | RAD23A | PNKP | |||||
| 633- | rs41540016 | 32 | Lobular, G2, ER+, PR+, Her2- | ||||
| POLQ | |||||||
| 634- | Novel POLE | rs28897689 | Novel WRN | rs11574410 | 34 | Ductal, G3, ER-, PR-, Her2- | |
| BRCA1 | WRN | ||||||
| 635- | rs144340710 | rs3730947 | 30 | Ductal, G3, ER+, PR-, Her2- | |||
| TP53 | LIG1 | ||||||
| 636 | rs4987202 | 24 | |||||
| RAD23A | |||||||
| 637 | rs150018949 | 31 | Ductal, G3, ER+, PR+, Her2+ | ||||
| EXO5 | |||||||
| 639- | rs759217526 | 24 | Ductal, G3, ER+, PR+, Her2- | ||||
| FANCL | |||||||
| 63 | rs11572913 | 37, 46 | Ductal, G2; 2 Ductal, G2, ER+, PR+, Her2- | ||||
| GTF2H3 | |||||||
| 64 | 37, 57 | Ductal, G3, ER+, PR+, Her2-; 2 | |||||
| 65 | rs34642881 | 38 | |||||
| RECQL4 | |||||||
| 66 | rs202068855 | rs756188698 | 39 | Ductal, G3, ER+, PR+, Her2- | |||
| RPA1 | RAD18 | ||||||
| 67 | rs142213781 | 39 | Ductal, G1, ER+, PR+, Her2- | ||||
| NEIL1 | |||||||
| 68 | 39, 47 | Lobular, G2, ER+, PR+, Her2-; 2 DCIS, G3 | |||||
| 69 | rs200981995 | rs17879961 | rs201503405 | 40 | Ductal, G3, ER-, PR-,Her2- | ||
| LIG3 | CHEK2 | PNKP | |||||
| 71 | 29 | Ductal, G3, ER-, PR-, Her2- | |||||
| 72- | rs775001669 | rs41540016 | rs199791286 | 39 | Ductal, G3, ER+, PR+, Her2+ | ||
| MLH3 | POLQ | MSH3 | |||||
| 73- | 33 | DCIS, G3 | |||||
| 74- | rs201414369 | rs150018949 | 36, 36 | Ductal, G3, ER-, PR-, Her2-; 2 ductal, G3, ER-, PR-, Her2- | |||
| EME1 | EXO5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurkilahti, V.; Rathinakannan, V.S.; Nynäs, E.; Goel, N.; Aittomäki, K.; Nevanlinna, H.; Fey, V.; Kankuri-Tammilehto, M.; Schleutker, J. Rare Germline Variants in DNA Repair Genes Detected in BRCA-Negative Finnish Patients with Early-Onset Breast Cancer. Cancers 2024, 16, 2955. https://doi.org/10.3390/cancers16172955
Kurkilahti V, Rathinakannan VS, Nynäs E, Goel N, Aittomäki K, Nevanlinna H, Fey V, Kankuri-Tammilehto M, Schleutker J. Rare Germline Variants in DNA Repair Genes Detected in BRCA-Negative Finnish Patients with Early-Onset Breast Cancer. Cancers. 2024; 16(17):2955. https://doi.org/10.3390/cancers16172955
Chicago/Turabian StyleKurkilahti, Viivi, Venkat Subramaniam Rathinakannan, Erja Nynäs, Neha Goel, Kristiina Aittomäki, Heli Nevanlinna, Vidal Fey, Minna Kankuri-Tammilehto, and Johanna Schleutker. 2024. "Rare Germline Variants in DNA Repair Genes Detected in BRCA-Negative Finnish Patients with Early-Onset Breast Cancer" Cancers 16, no. 17: 2955. https://doi.org/10.3390/cancers16172955
APA StyleKurkilahti, V., Rathinakannan, V. S., Nynäs, E., Goel, N., Aittomäki, K., Nevanlinna, H., Fey, V., Kankuri-Tammilehto, M., & Schleutker, J. (2024). Rare Germline Variants in DNA Repair Genes Detected in BRCA-Negative Finnish Patients with Early-Onset Breast Cancer. Cancers, 16(17), 2955. https://doi.org/10.3390/cancers16172955






