DM–AHR: A Self-Supervised Conditional Diffusion Model for AI-Generated Hairless Imaging for Enhanced Skin Diagnosis Applications
Abstract
Simple Summary
Abstract
1. Introduction
- We introduce DM–AHR, a customized self-supervised diffusion model for automatic hair removal in dermoscopic images. This model enhances the clarity and diagnostic quality of skin images by effectively differentiating hair from skin features.
- We demonstrate significant improvement in skin lesion detection accuracy after applying DM–AHR as a preprocessing step before the classification model.
- We introduce the novel DERMAHAIR dataset, advancing research in the task of automatic hair removal from skin images.
- We introduce a new self-supervised technique specifically tailored for the task of automatic hair removal from skin images.
- We demonstrate DM–AHR’s potential in broader skin diagnosis applications and automated dermatological systems.
2. Related Works
3. Proposed Method
3.1. Dataset and Problem Formulation
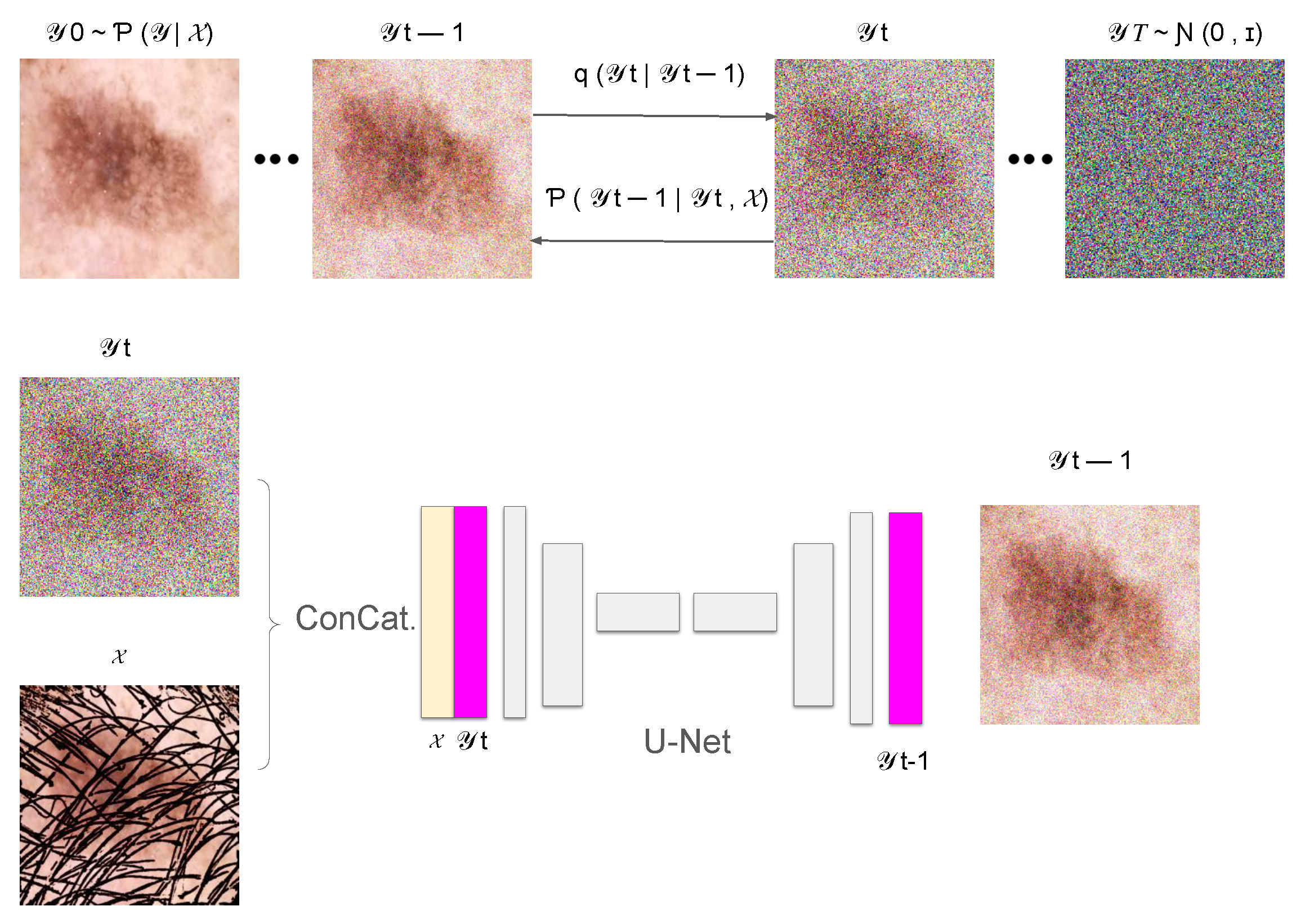
3.2. Conditional Denoising Diffusion Process
3.2.1. Forward Diffusion Process
3.2.2. Reverse Diffusion Process
3.3. Training of the Denoising Model
3.3.1. Loss Function
3.3.2. Training Algorithm
| Algorithm 1: Training algorithm for the DM–AHR Model |
while not converged do for each batch in the training dataset do Sample a pair from the batch for to T do Sample noise level from a predefined noise schedule Generate noise Create a noisy image using the formula: Compute the loss function for backpropagation: Update by applying the gradient descent step: end for end for end while |
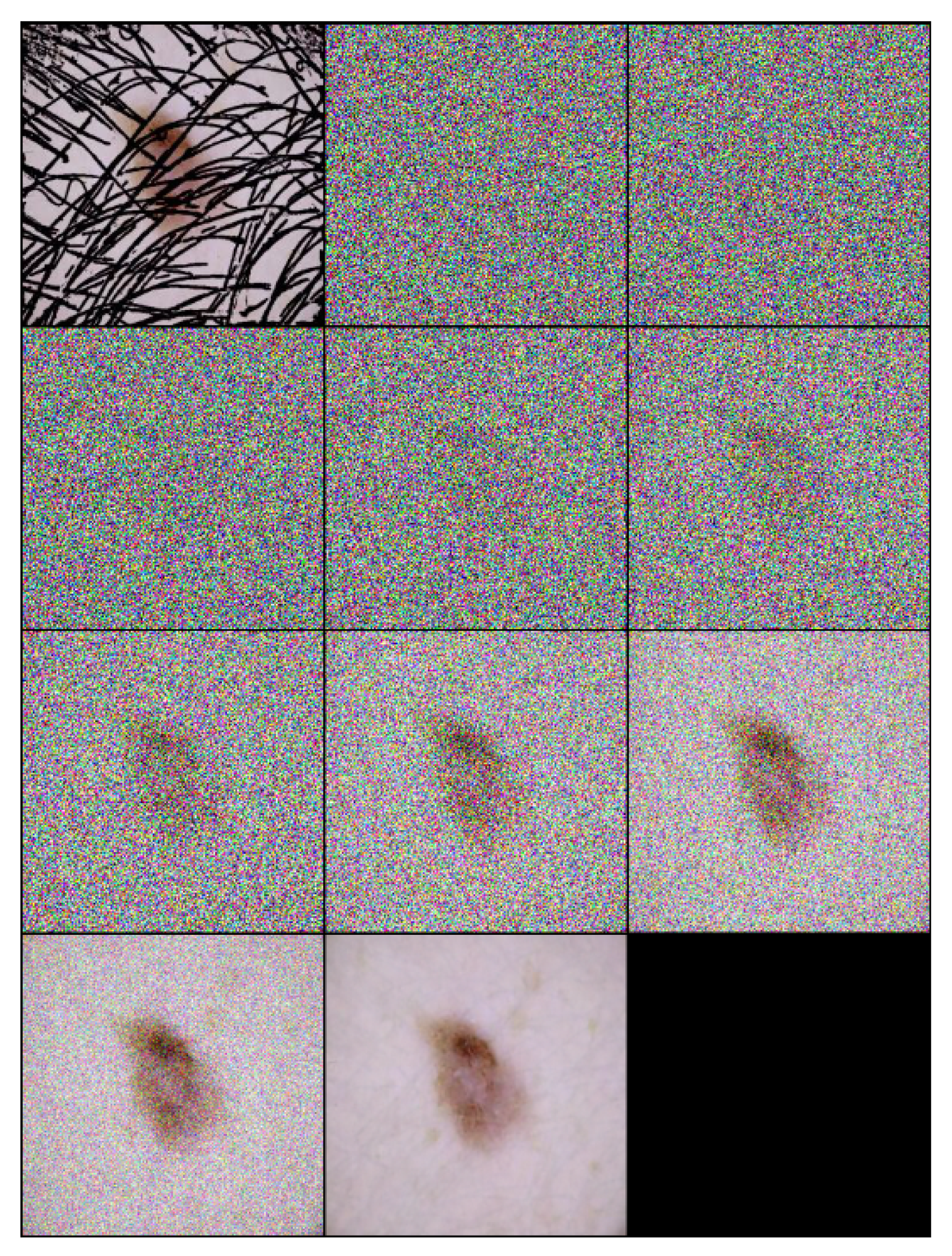
3.4. Inference Process of the DM–AHR Model
| Algorithm 2: Inference process for hair removal in DM–AHR |
Initialize for down to 1 do Sample noise Apply the denoising model: end for return , the final hair-free image |
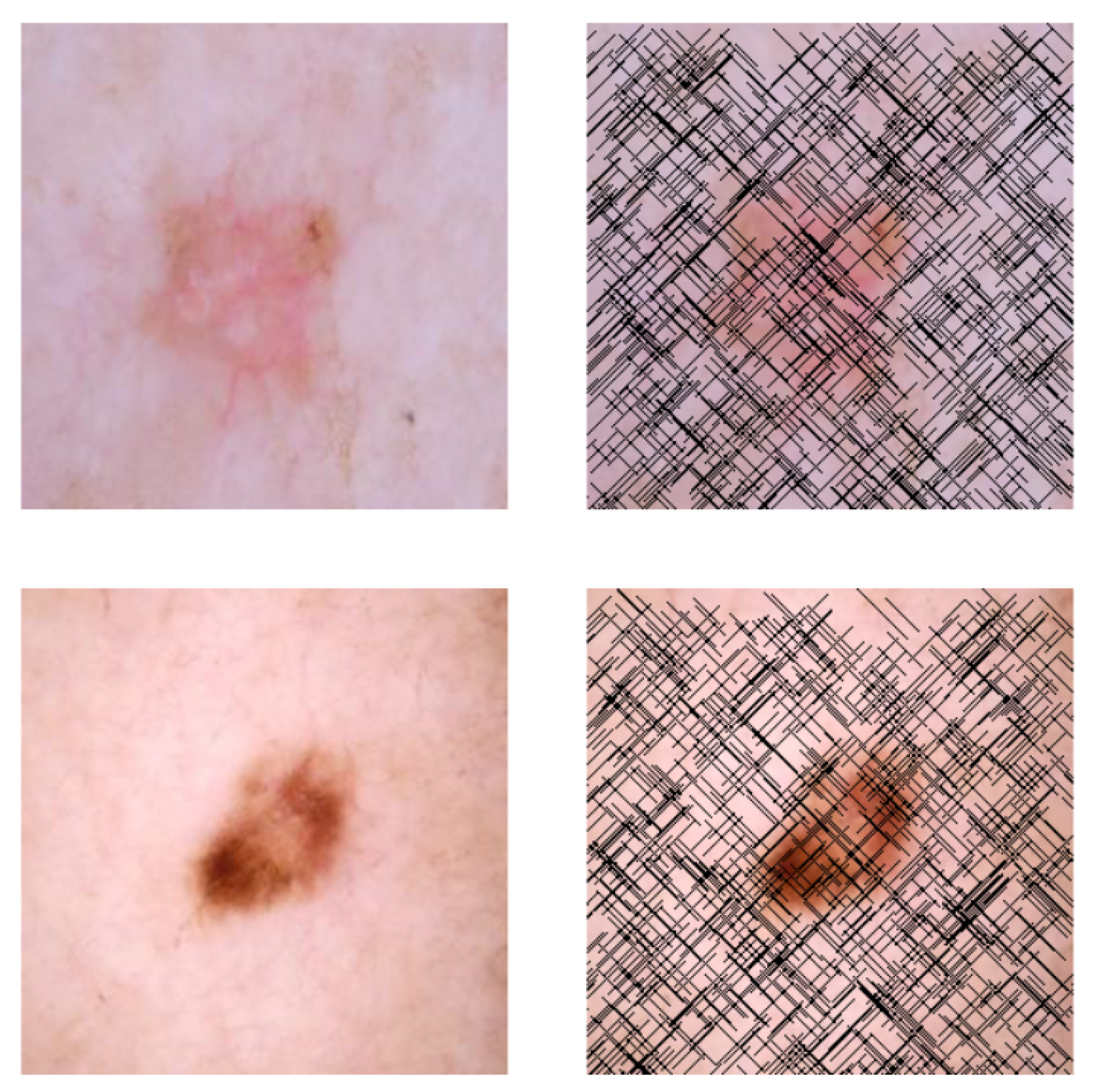
3.5. Self-Supervised Technique
| Algorithm 3: Self-supervised technique customized for training skin hair removal models |
Input: {the original hair-free dermoscopic image} Output: {Image with the small artificial lines simulating hair-like patterns} {Fix the number of lines to draw} Size of {Get the input image dimensions} for to do {pick a random integer in } {pick a random integer in } {pick a random integer in } {pick a random direction for diagonal orientation} {End x-coordinate} {End y-coordinate} Draw line on I from to on end for return |
4. Experiments
4.1. Experimental Settings
4.2. Dataset Description
4.2.1. Description of the HAM10000 Dataset
4.2.2. Description of the Artificial Hair Addition Process
4.3. Evaluation Metrics
4.3.1. Evaluation Metrics of the DM–AHR Performance
4.3.2. Evaluation Metrics of the Skin Lesion Classification
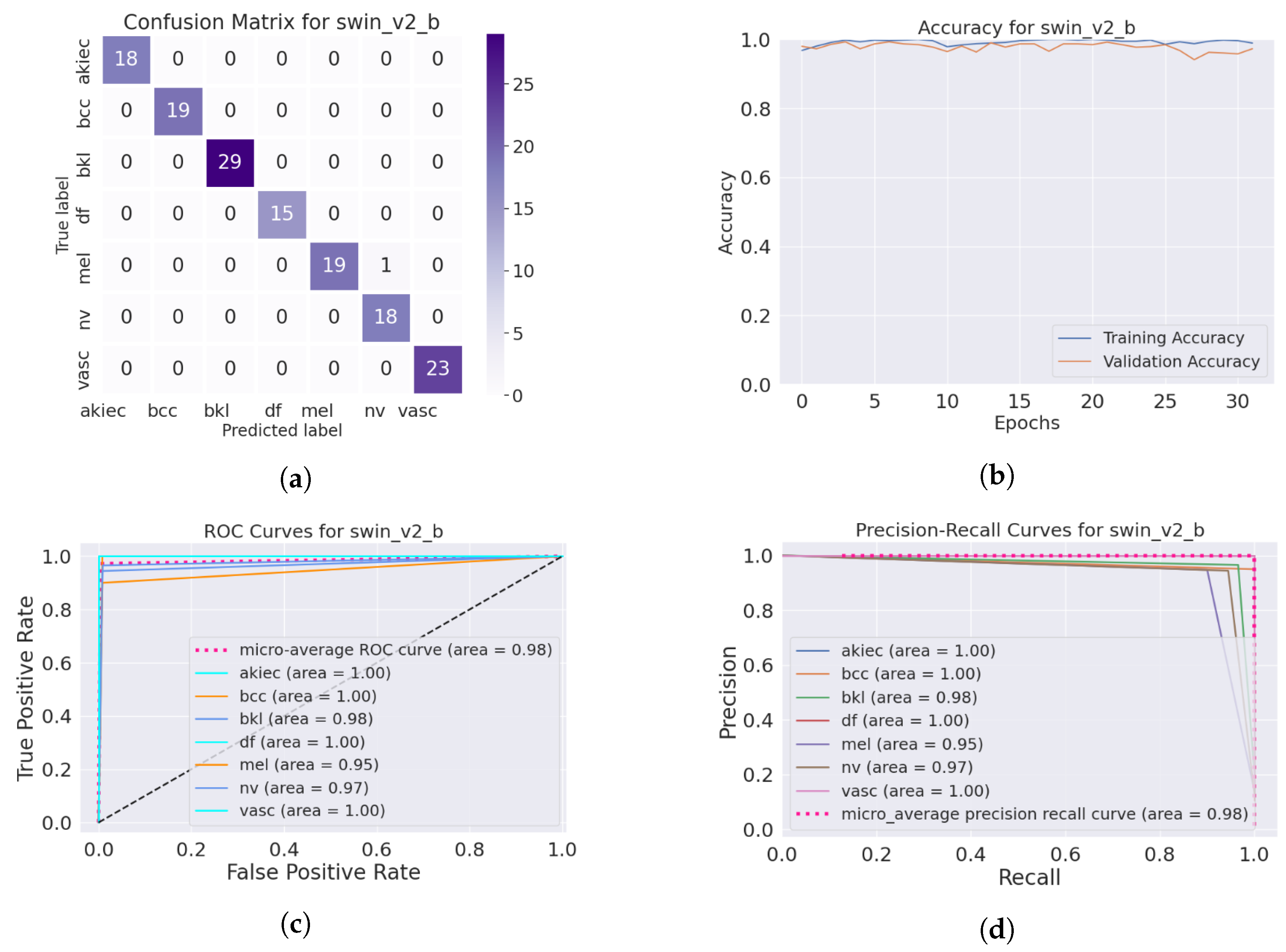
4.4. Performance of the Skin Image Enhancement Using the DM–AHR Model
4.5. Improvement of the Medical Skin Diagnosis after Using DM–AHR
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Ultraviolet (UV) Radiation and Skin Cancer. 2023. Available online: https://www.who.int/news-room/questions-and-answers/item/radiation-ultraviolet-(uv)-radiation-and-skin-cancer (accessed on 25 December 2023).
- Mahmood, T.; Saba, T.; Rehman, A.; Alamri, F.S. Harnessing the power of radiomics and deep learning for improved breast cancer diagnosis with multiparametric breast mammography. Expert Syst. Appl. 2024, 249, 123747. [Google Scholar] [CrossRef]
- Omar Bappi, J.; Rony, M.A.T.; Shariful Islam, M.; Alshathri, S.; El-Shafai, W. A Novel Deep Learning Approach for Accurate Cancer Type and Subtype Identification. IEEE Access 2024, 12, 94116–94134. [Google Scholar] [CrossRef]
- Abu Tareq Rony, M.; Shariful Islam, M.; Sultan, T.; Alshathri, S.; El-Shafai, W. MediGPT: Exploring Potentials of Conventional and Large Language Models on Medical Data. IEEE Access 2024, 12, 103473–103487. [Google Scholar] [CrossRef]
- Soleimani, M.; Harooni, A.; Erfani, N.; Khan, A.R.; Saba, T.; Bahaj, S.A. Classification of cancer types based on microRNA expression using a hybrid radial basis function and particle swarm optimization algorithm. Microsc. Res. Tech. 2024, 87, 1052–1062. [Google Scholar] [CrossRef]
- Emara, H.M.; El-Shafai, W.; Algarni, A.D.; Soliman, N.F.; El-Samie, F.E.A. A Hybrid Compressive Sensing and Classification Approach for Dynamic Storage Management of Vital Biomedical Signals. IEEE Access 2023, 11, 108126–108151. [Google Scholar] [CrossRef]
- Vestergaard, M.; Macaskill, P.; Holt, P.; Menzies, S. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: A meta-analysis of studies performed in a clinical setting. Br. J. Dermatol. 2008, 159, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Hammad, M.; Pławiak, P.; ElAffendi, M.; El-Latif, A.A.A.; Latif, A.A.A. Enhanced deep learning approach for accurate eczema and psoriasis skin detection. Sensors 2023, 23, 7295. [Google Scholar] [CrossRef] [PubMed]
- Alyami, J.; Rehman, A.; Sadad, T.; Alruwaythi, M.; Saba, T.; Bahaj, S.A. Automatic skin lesions detection from images through microscopic hybrid features set and machine learning classifiers. Microsc. Res. Tech. 2022, 85, 3600–3607. [Google Scholar] [CrossRef]
- Nawaz, M.; Mehmood, Z.; Nazir, T.; Naqvi, R.A.; Rehman, A.; Iqbal, M.; Saba, T. Skin cancer detection from dermoscopic images using deep learning and fuzzy k-means clustering. Microsc. Res. Tech. 2022, 85, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Jain, A.; Abbeel, P. Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 2020, 33, 6840–6851. [Google Scholar]
- Sohl-Dickstein, J.; Weiss, E.; Maheswaranathan, N.; Ganguli, S. Deep unsupervised learning using nonequilibrium thermodynamics. In Proceedings of the International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 2256–2265. [Google Scholar]
- Song, Y.; Sohl-Dickstein, J.; Kingma, D.P.; Kumar, A.; Ermon, S.; Poole, B. Score-based generative modeling through stochastic differential equations. arXiv 2020, arXiv:2011.13456. [Google Scholar]
- Farooq, M.A.; Yao, W.; Schukat, M.; Little, M.A.; Corcoran, P. Derm-t2im: Harnessing synthetic skin lesion data via stable diffusion models for enhanced skin disease classification using vit and cnn. arXiv 2024, arXiv:2401.05159. [Google Scholar]
- Abuzaghleh, O.; Barkana, B.D.; Faezipour, M. Automated skin lesion analysis based on color and shape geometry feature set for melanoma early detection and prevention. In Proceedings of the IEEE Long Island Systems, Applications and Technology (LISAT) Conference 2014, Farmingdale, NY, USA, 2 May 2014; pp. 1–6. [Google Scholar]
- Maglogiannis, I.; Doukas, C.N. Overview of advanced computer vision systems for skin lesions characterization. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Wang, S.; Zhang, W.; Li, C. Reconstruction of Neural Radiance Fields With Vivid Scenes in the Metaverse. IEEE Trans. Consum. Electron. 2023, 70, 3222–3231. [Google Scholar] [CrossRef]
- Bao, Q.; Liu, Y.; Gang, B.; Yang, W.; Liao, Q. S2Net: Shadow Mask-Based Semantic-Aware Network for Single-Image Shadow Removal. IEEE Trans. Consum. Electron. 2022, 68, 209–220. [Google Scholar] [CrossRef]
- Ji, Z.; Zheng, H.; Zhang, Z.; Ye, Q.; Zhao, Y.; Xu, M. Multi-Scale Interaction Network for Low-Light Stereo Image Enhancement. IEEE Trans. Consum. Electron. 2023, 70, 3626–3634. [Google Scholar] [CrossRef]
- Anand, V.; Gupta, S.; Nayak, S.R.; Koundal, D.; Prakash, D.; Verma, K. An automated deep learning models for classification of skin disease using Dermoscopy images: A comprehensive study. Multimed. Tools Appl. 2022, 81, 37379–37401. [Google Scholar] [CrossRef]
- Li, W.; Raj, A.N.J.; Tjahjadi, T.; Zhuang, Z. Digital hair removal by deep learning for skin lesion segmentation. Pattern Recognit. 2021, 117, 107994. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, J.; He, T.; Wang, W.; Abbas, H.; Lv, Z. DS-CNN: Dual-Stream Convolutional Neural Networks-Based Heart Sound Classification for Wearable Devices. IEEE Trans. Consum. Electron. 2023, 69, 1186–1194. [Google Scholar] [CrossRef]
- Kim, D.; Hong, B.W. Unsupervised feature elimination via generative adversarial networks: Application to hair removal in melanoma classification. IEEE Access 2021, 9, 42610–42620. [Google Scholar] [CrossRef]
- Dong, L.; Zhou, Y.; Jiang, J. Dual-Clustered Conditioning towards GAN-based Diverse Image Generation. IEEE Trans. Consum. Electron. 2024, 70, 2817–2825. [Google Scholar] [CrossRef]
- He, Y.; Jin, X.; Jiang, Q.; Cheng, Z.; Wang, P.; Zhou, W. LKAT-GAN: A GAN for Thermal Infrared Image Colorization Based on Large Kernel and AttentionUNet-Transformer. IEEE Trans. Consum. Electron. 2023, 69, 478–489. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Delibasis, K.; Moutselos, K.; Vorgiazidou, E.; Maglogiannis, I. Automated hair removal in dermoscopy images using shallow and deep learning neural architectures. Comput. Methods Programs Biomed. Update 2023, 4, 100109. [Google Scholar] [CrossRef]
- Abbas, Q.; Celebi, M.E.; García, I.F. Hair removal methods: A comparative study for dermoscopy images. Biomed. Signal Process. Control 2011, 6, 395–404. [Google Scholar] [CrossRef]
- Saharia, C.; Ho, J.; Chan, W.; Salimans, T.; Fleet, D.J.; Norouzi, M. Image super-resolution via iterative refinement. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 4713–4726. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015, Proceedings, Part III 18; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Digital Hair Dataset. Available online: https://www.kaggle.com/datasets/weilizai/digital-hair-dataset (accessed on 17 December 2023).
- Zhang, R.; Isola, P.; Efros, A.A.; Shechtman, E.; Wang, O. The unreasonable effectiveness of deep features as a perceptual metric. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 586–595. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014, 27, 2672–2680. [Google Scholar]
- Liang, J.; Cao, J.; Sun, G.; Zhang, K.; Van Gool, L.; Timofte, R. Swinir: Image restoration using swin transformer. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 1833–1844. [Google Scholar]
- Rombach, R.; Blattmann, A.; Lorenz, D.; Esser, P.; Ommer, B. High-resolution image synthesis with latent diffusion models. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 10684–10695. [Google Scholar]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1492–1500. [Google Scholar]
- Tu, Z.; Talebi, H.; Zhang, H.; Yang, F.; Milanfar, P.; Bovik, A.; Li, Y. Maxvit: Multi-axis vision transformer. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2022; pp. 459–479. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
| Skin Cancer Type | SRGAN | SwinIR | SDM | DM–AHR | DM–AHR(SS) |
|---|---|---|---|---|---|
| Actinic keratosis | 32.401835 | 39.401370 | 33.731562 | 39.095776 | 41.280412 |
| Basal cell carcinoma | 31.700830 | 39.625874 | 34.317885 | 38.633978 | 40.806729 |
| Benign Keratosis | 31.644829 | 39.894135 | 34.109435 | 39.228128 | 41.026910 |
| Dermatofibroma | 31.807330 | 39.207628 | 34.196231 | 39.148305 | 40.910219 |
| Melanoma | 30.917425 | 38.493721 | 33.536934 | 38.728289 | 39.883010 |
| Melanocytic nevi | 32.242417 | 39.974818 | 34.960045 | 39.091819 | 41.212679 |
| Vascular lesions | 32.633316 | 41.737362 | 35.790086 | 39.963068 | 42.170609 |
| Average | 31.906854 | 39.762129 | 34.377454 | 39.12705 | 41.041512 |
| Skin Cancer Type | SRGAN | SwinIR | SDM | DM–AHR | DM–AHR(SS) |
|---|---|---|---|---|---|
| Actinic keratosis | 0.881940 | 0.959481 | 0.865522 | 0.960036 | 0.967910 |
| Basal cell carcinoma | 0.882850 | 0.957559 | 0.872703 | 0.955464 | 0.964175 |
| Benign Keratosis | 0.879369 | 0.959514 | 0.871176 | 0.956925 | 0.965370 |
| Dermatofibroma | 0.869542 | 0.957726 | 0.865824 | 0.956125 | 0.964412 |
| Melanoma | 0.868869 | 0.953418 | 0.860095 | 0.951718 | 0.961027 |
| Melanocytic nevi | 0.879389 | 0.955402 | 0.892593 | 0.954957 | 0.962382 |
| Vascular lesions | 0.899865 | 0.968740 | 0.917110 | 0.967328 | 0.973995 |
| Average | 0.880261 | 0.958834 | 0.87786 | 0.957508 | 0.965613 |
| Skin Cancer Type | SRGAN | SwinIR | SDM | DM–AHR | DM–AHR(SS) |
|---|---|---|---|---|---|
| Actinic keratosis | 0.171810 | 0.042776 | 0.050473 | 0.018420 | 0.012620 |
| Basal cell carcinoma | 0.175114 | 0.038973 | 0.046981 | 0.020546 | 0.013838 |
| Benign Keratosis | 0.185645 | 0.036158 | 0.047129 | 0.020156 | 0.013638 |
| Dermatofibroma | 0.182939 | 0.042536 | 0.045433 | 0.020481 | 0.013376 |
| Melanoma | 0.192164 | 0.045766 | 0.049873 | 0.023361 | 0.015010 |
| Melanocytic nevi | 0.164768 | 0.040105 | 0.036954 | 0.019475 | 0.012884 |
| Vascular lesions | 0.131372 | 0.025131 | 0.027423 | 0.013758 | 0.008439 |
| Average | 0.171973 | 0.038777 | 0.043467 | 0.019457 | 0.012829 |
| Original Skin Image | Skin Image with Hair | Skin Image after DM–AHR | |
|---|---|---|---|
| Result | 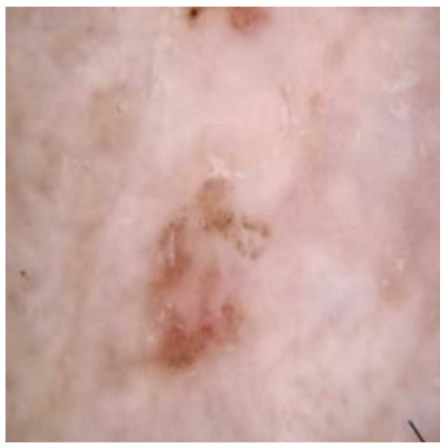 |  | 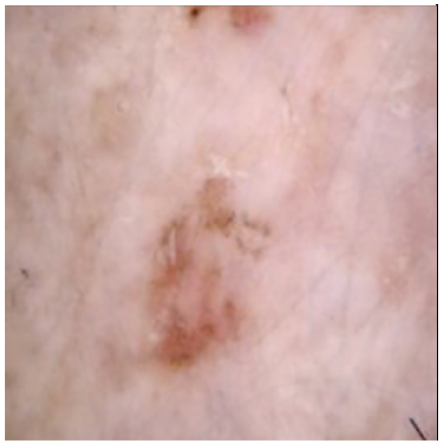 |
| Histogram | 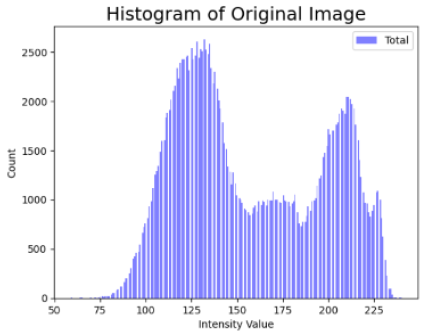 | 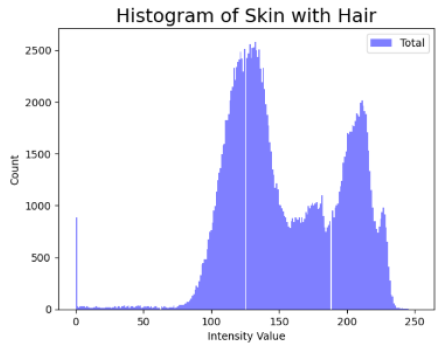 | 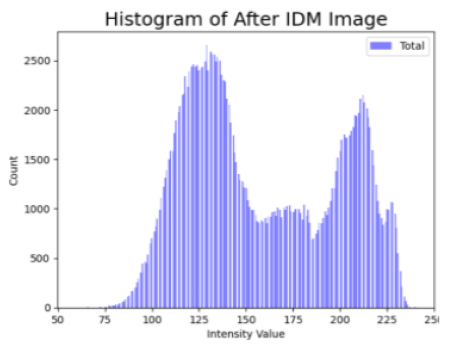 |
| Cancer Type | Original | After Hair | After DM–AHR |
|---|---|---|---|
| Actinic keratosis | 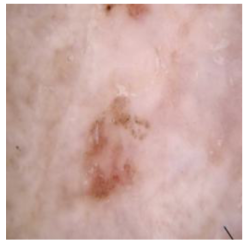 |  | 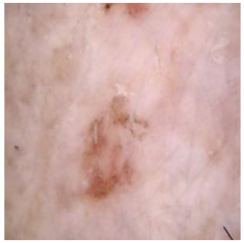 |
| Basal cell carcinoma | 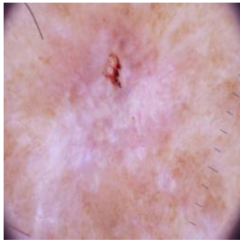 |  | 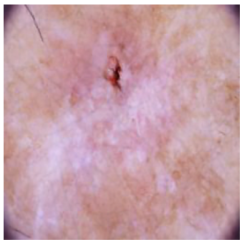 |
| Benign keratosis | 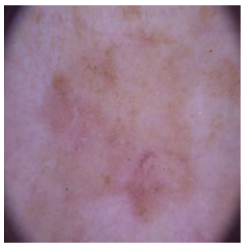 |  | 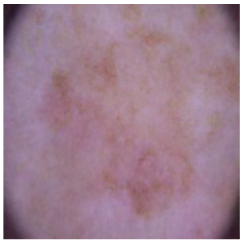 |
| Dermatofibroma | 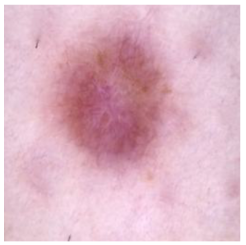 | 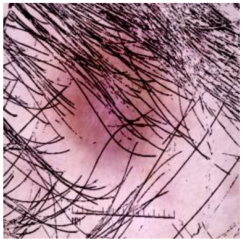 | 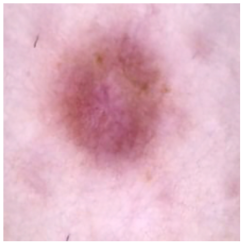 |
| Melanoma | 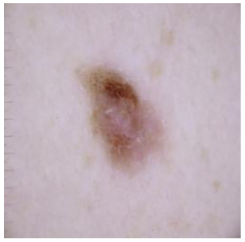 |  | 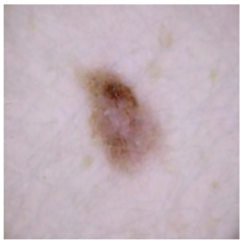 |
| Melanocytic nevi | 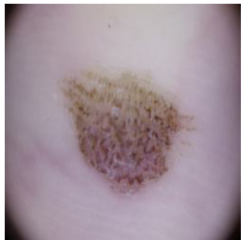 |  | 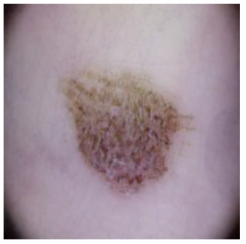 |
| Vascular lesions | 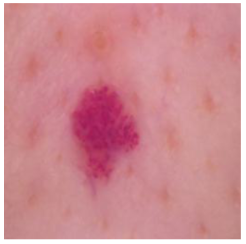 |  | 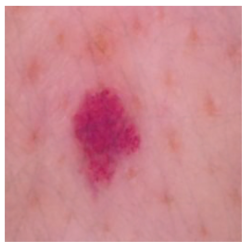 |
| Metric | Original | After Adding Hair | After DM–AHR Hair Removal |
|---|---|---|---|
| Accuracy | 0.985242 | 0.879514 | 0.972731 |
| Loss | 0.050478 | 0.565113 | 0.099951 |
| Precision | 0.9856 | 0.8624 | 0.9825 |
| Recall | 0.9852 | 0.8795 | 0.9727 |
| Specificity | 0.9973 | 0.9778 | 0.9964 |
| F1-score | 0.9853 | 0.8660 | 0.9766 |
| Metric | Original | After Adding Hair | After DM–AHR Hair Removal |
|---|---|---|---|
| Accuracy | 0.988786 | 0.950656 | 0.992857 |
| Loss | 0.042649 | 0.168300 | 0.024280 |
| Precision | 0.9889 | 0.9436 | 0.9940 |
| Recall | 0.9888 | 0.9507 | 0.9929 |
| Specificity | 0.9980 | 0.9906 | 0.9988 |
| F1-score | 0.9888 | 0.9462 | 0.9933 |
| Metric | Original | After Adding Hair | After IDM Hair Removal |
|---|---|---|---|
| Accuracy | 0.983420 | 0.967687 | 0.992957 |
| Loss | 0.066955 | 0.114440 | 0.027340 |
| Precision | 0.9837 | 0.9689 | 0.9925 |
| Recall | 0.9834 | 0.9677 | 0.9929 |
| Specificity | 0.9970 | 0.9941 | 0.9988 |
| F1-score | 0.9834 | 0.9665 | 0.9925 |
| Model | Model Size (MB) | Training Time |
|---|---|---|
| resnext101_32x8d | 1340.72 | 403 m 32 s |
| maxvit_t | 834.43 | 196 m 23 s |
| SwinTransformer | 911.96 | 418 m 49 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benjdira, B.; M. Ali, A.; Koubaa, A.; Ammar, A.; Boulila, W. DM–AHR: A Self-Supervised Conditional Diffusion Model for AI-Generated Hairless Imaging for Enhanced Skin Diagnosis Applications. Cancers 2024, 16, 2947. https://doi.org/10.3390/cancers16172947
Benjdira B, M. Ali A, Koubaa A, Ammar A, Boulila W. DM–AHR: A Self-Supervised Conditional Diffusion Model for AI-Generated Hairless Imaging for Enhanced Skin Diagnosis Applications. Cancers. 2024; 16(17):2947. https://doi.org/10.3390/cancers16172947
Chicago/Turabian StyleBenjdira, Bilel, Anas M. Ali, Anis Koubaa, Adel Ammar, and Wadii Boulila. 2024. "DM–AHR: A Self-Supervised Conditional Diffusion Model for AI-Generated Hairless Imaging for Enhanced Skin Diagnosis Applications" Cancers 16, no. 17: 2947. https://doi.org/10.3390/cancers16172947
APA StyleBenjdira, B., M. Ali, A., Koubaa, A., Ammar, A., & Boulila, W. (2024). DM–AHR: A Self-Supervised Conditional Diffusion Model for AI-Generated Hairless Imaging for Enhanced Skin Diagnosis Applications. Cancers, 16(17), 2947. https://doi.org/10.3390/cancers16172947








