Prognostic Significance of Elevated UCHL1, SNRNP200, and PAK4 Expression in High-Grade Clear Cell Renal Cell Carcinoma: Insights from LC-MS/MS Analysis and Immunohistochemical Validation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Cohorts
2.2. Immunohistochemistry and Pathology Evaluation
+ (% of cells stained at intensity 3 × 3)
2.3. Digital Image Acquisition and Storage
2.4. Statistical Analysis
2.5. Reactome Pathway Enrichment Analysis
3. Results
3.1. Protein Expression Values in CPTAC CCRCC
3.2. Immunohistochemical Analysis of UCHL1, SNRNP200, and PAK4 Reveals Protein Candidates for Further Correlation with Patients’ Clinical Outcomes
3.3. PAK4, SNRNP200, and UCHL1 Protein Expression as Measured by IHC
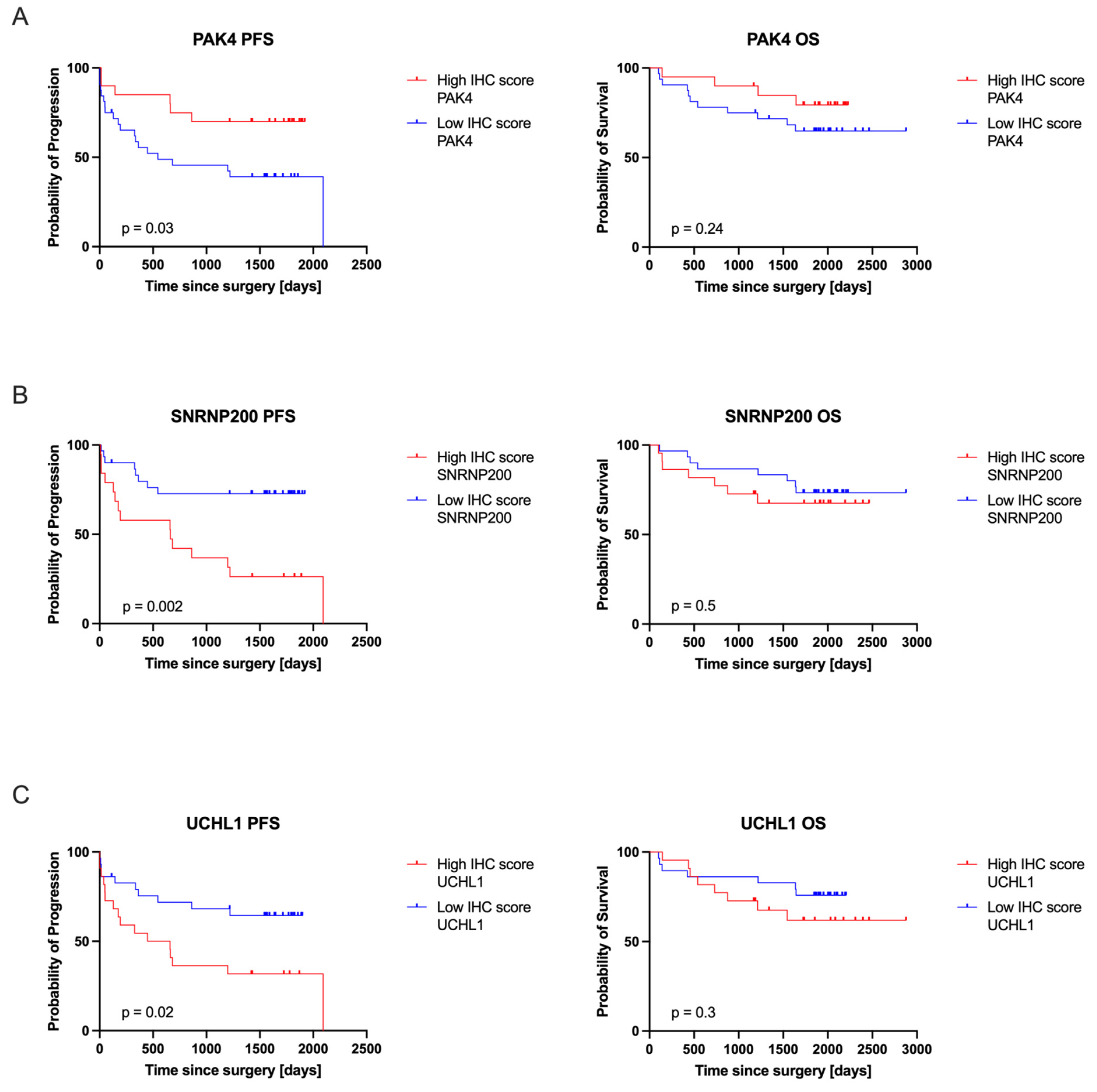
3.4. PAK4, SNRNP200, and UCHL1 Protein Expression Correlated with Patients’ Clinical Outcomes
3.5. Identification of the Top Significantly Enriched Reactome Pathways Associated with PAK4, SNRNP200, and UCHL1
4. Discussion
5. Conclusions
6. Study Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer Incidence and Mortality Patterns in Europe: Estimates for 40 Countries and 25 Major Cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef] [PubMed]
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of Renal Cell Carcinoma. World J. Oncol. 2020, 11, 79–87. [Google Scholar] [CrossRef]
- Bukavina, L.; Bensalah, K.; Bray, F.; Carlo, M.; Challacombe, B.; Karam, J.A.; Kassouf, W.; Mitchell, T.; Montironi, R.; O’Brien, T.; et al. Epidemiology of Renal Cell Carcinoma: 2022 Update. Eur. Urol. 2022, 82, 529–542. [Google Scholar] [CrossRef]
- DeCastro, G.J.; McKiernan, J.M. Epidemiology, Clinical Staging, and Presentation of Renal Cell Carcinoma. Urol. Clin. N. Am. 2008, 35, 581–592. [Google Scholar] [CrossRef]
- Creighton, C.J.; Hernandez-Herrera, A.; Jacobsen, A.; Levine, D.A.; Mankoo, P.; Schultz, N.; Du, Y.; Zhang, Y.; Larsson, E.; Sheridan, R.; et al. Integrated Analyses of MicroRNAs Demonstrate Their Widespread Influence on Gene Expression in High-Grade Serous Ovarian Carcinoma. PLoS ONE 2012, 7, e34546. [Google Scholar] [CrossRef] [PubMed]
- Scelo, G.; Larose, T.L. Epidemiology and Risk Factors for Kidney Cancer. J. Clin. Oncol. 2018, 36, 3574–3581. [Google Scholar] [CrossRef]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review, 1975–2016; National Cancer Institute: Bethesda, MA, USA, 2019. [Google Scholar]
- van de Pol, J.A.A.; George, L.; van den Brandt, P.A.; Baldewijns, M.M.L.L.; Schouten, L.J. Etiologic Heterogeneity of Clear-Cell and Papillary Renal Cell Carcinoma in the Netherlands Cohort Study. Int. J. Cancer 2021, 148, 67–76. [Google Scholar] [CrossRef]
- Kase, A.M.; George, D.J.; Ramalingam, S. Clear Cell Renal Cell Carcinoma: From Biology to Treatment. Cancers 2023, 15, 665. [Google Scholar] [CrossRef]
- Morgan, T.M.; Seeley, E.H.; Fadare, O.; Caprioli, R.M.; Clark, P.E. Imaging the Clear Cell Renal Cell Carcinoma Proteome. J. Urol. 2013, 189, 1097–1103. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chinello, C.; L’Imperio, V.; Stella, M.; Smith, A.J.; Bovo, G.; Grasso, A.; Grasso, M.; Raimondo, F.; Pitto, M.; Pagni, F.; et al. The Proteomic Landscape of Renal Tumors. Expert Rev. Proteom. 2016, 13, 1103–1120. [Google Scholar] [CrossRef]
- Song, Y.; Zhong, L.; Zhou, J.; Lu, M.; Xing, T.; Ma, L.; Shen, J. Data-Independent Acquisition-Based Quantitative Proteomic Analysis Reveals Potential Biomarkers of Kidney Cancer. Proteom. Clin. Appl. 2017, 11, 1700066. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics Connects Somatic Mutations to Signalling in Breast Cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, J.; Wang, X.; Zhu, J.; Liu, Q.; Shi, Z.; Chambers, M.C.; Zimmerman, L.J.; Shaddox, K.F.; Kim, S.; et al. Proteogenomic Characterization of Human Colon and Rectal Cancer. Nature 2014, 513, 382–387. [Google Scholar] [CrossRef]
- Clark, D.J.; Dhanasekaran, S.M.; Petralia, F.; Pan, J.; Song, X.; Hu, Y.; da Veiga Leprevost, F.; Reva, B.; Lih, T.S.M.; Chang, H.Y.; et al. Integrated Proteogenomic Characterization of Clear Cell Renal Cell Carcinoma. Cell 2019, 179, 964–983.e31. [Google Scholar] [CrossRef]
- Jara, J.H.; Frank, D.D.; Özdinler, P.H. Could Dysregulation of UPS Be a Common Underlying Mechanism for Cancer and Neurodegeneration? Lessons from UCHL1. Cell Biochem. Biophys. 2013, 67, 45–53. [Google Scholar] [CrossRef]
- Matuszczak, E.; Tylicka, M.; Dębek, W.; Tokarzewicz, A.; Gorodkiewicz, E.; Hermanowicz, A. Concentration of UHCL1 in the Serum of Children with Acute Appendicitis, Before and After Surgery, and Its Correlation with CRP and Prealbumin. J. Investig. Surg. 2018, 31, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Kastner, B.; Will, C.L.; Stark, H.; Lührmann, R. Structural Insights into Nuclear Pre-MRNA Splicing in Higher Eukaryotes. Cold Spring Harb. Perspect. Biol. 2019, 11, a032417. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Vacas, J.M.; Herrero-Aguayo, V.; Montero-Hidalgo, A.J.; Gómez-Gómez, E.; Fuentes-Fayos, A.C.; León-González, A.J.; Sáez-Martínez, P.; Alors-Pérez, E.; Pedraza-Arévalo, S.; González-Serrano, T.; et al. Dysregulation of the Splicing Machinery Is Directly Associated to Aggressiveness of Prostate Cancer. eBioMedicine 2020, 51, 102547. [Google Scholar] [CrossRef] [PubMed]
- Jaffer, Z.M.; Chernoff, J. P21-Activated Kinases: Three More Join the Pak. Int. J. Biochem. Cell Biol. 2002, 34, 713–717. [Google Scholar] [CrossRef]
- Dummler, B.; Ohshiro, K.; Kumar, R.; Field, J. Pak Protein Kinases and Their Role in Cancer. Cancer Metastasis Rev. 2009, 28, 51–63. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Varella-Garcia, M.; Bunn, P.A.; Di Maria, M.V.; Veve, R.; Bremnes, R.M.; Barón, A.E.; Zeng, C.; Franklin, W.A. Epidermal Growth Factor Receptor in Non-Small-Cell Lung Carcinomas: Correlation between Gene Copy Number and Protein Expression and Impact on Prognosis. J. Clin. Oncol. 2003, 21, 3798–3807. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, N.; Li, M.; Hong, T.; Meng, W.; Ouyang, T. Ubiquitin C-terminal Hydrolase-L1: A New Cancer Marker and Therapeutic Target with Dual Effects (Review). Oncol. Lett. 2023, 25, 123. [Google Scholar] [CrossRef]
- Li, L.; Tao, Q.; Jin, H.; Van Hasselt, A.; Poon, F.F.; Wang, X.; Zeng, M.S.; Jia, W.H.; Zeng, Y.X.; Chan, A.T.C.; et al. The Tumor Suppressor UCHL1 Forms a Complex with P53/MDM2/ARF to Promote P53 Signaling and Is Frequently Silenced in Nasopharyngeal Carcinoma. Clin. Cancer Res. 2010, 16, 2949–2958. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, W.; Zhou, B.; Jin, C.; Wang, Z.; Yang, Y.; Wang, Z.; Chen, Y.; Feng, X. The Diagnosis Value of Promoter Methylation of UCHL1 in the Serum for Progression of Gastric Cancer. BioMed Res. Int. 2015, 2015, 741030. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jin, C.; Yu, W.; Lou, X.; Zhou, F.; Han, X.; Zhao, N.; Lin, B. UCHL1 Is a Putative Tumor Suppressor in Ovarian Cancer Cells and Contributes to Cisplatin Resistance. J. Cancer 2013, 4, 662–670. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 26 June 2024).
- Takano, T.; Miyauchi, A.; Matsuzuka, F.; Yoshida, H.; Nakata, Y.; Kuma, K.; Amino, N. PGP9.5 MRNA Could Contribute to the Molecular-Based Diagnosis of Medullary Thyroid Carcinoma. Eur. J. Cancer 2004, 40, 614–618. [Google Scholar] [CrossRef]
- Kagara, I.; Enokida, H.; Kawakami, K.; Matsuda, R.; Toki, K.; Nishimura, H.; Chiyomaru, T.; Tatarano, S.; Itesako, T.; Kawamoto, K.; et al. CpG Hypermethylation of the UCHL1 Gene Promoter Is Associated with Pathogenesis and Poor Prognosis in Renal Cell Carcinoma. J. Urol. 2008, 180, 343–351. [Google Scholar] [CrossRef]
- Seliger, B.; Handke, D.; Schabel, E.; Bukur, J.; Lichtenfels, R.; Dammann, R. Epigenetic Control of the Ubiquitin Carboxyl Terminal Hydrolase 1 in Renal Cell Carcinoma. J. Transl. Med. 2009, 7, 90. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, Y.M.; Lim, S.; Nam, Y.K.; Jeong, J.; Kim, H.J.; Lee, K.J. Ubiquitin C-Terminal Hydrolase-L1 Is a Key Regulator of Tumor Cell Invasion and Metastasis. Oncogene 2009, 28, 117–127. [Google Scholar] [CrossRef]
- Hussain, S.; Foreman, O.; Perkins, S.L.; Witzig, T.E.; Miles, R.R.; Van Deursen, J.; Galardy, P.J. The De-Ubiquitinase UCH-L1 Is an Oncogene That Drives the Development of Lymphoma in Vivo by Deregulating PHLPP1 and Akt Signaling. Leukemia 2010, 24, 1641–1655. [Google Scholar] [CrossRef]
- Hennessy, B.T.; Smith, D.L.; Ram, P.T.; Lu, Y.; Mills, G.B. Exploiting the PI3K/AKT Pathway for Cancer Drug Discovery. Nat. Rev. Drug Discov. 2005, 4, 988–1004. [Google Scholar] [CrossRef] [PubMed]
- Yoeli-Lerner, M.; Toker, A. Akt/PKB Signaling in Cancer: A Function in Cell Motility and Invasion. Cell Cycle 2006, 5, 603–605. [Google Scholar] [CrossRef]
- Harada, T.; Harada, C.; Wang, Y.L.; Osaka, H.; Amanai, K.; Tanaka, K.; Takizawa, S.; Setsuie, R.; Sakurai, M.; Sato, Y.; et al. Role of Ubiquitin Carboxy Terminal Hydrolase-L1 in Neural Cell Apoptosis Induced by Ischemic Retinal Injury in Vivo. Am. J. Pathol. 2004, 164, 59–64. [Google Scholar] [CrossRef][Green Version]
- Mani, A.; Gelmann, E.P. The Ubiquitin-Proteasome Pathway and Its Role in Cancer. J. Clin. Oncol. 2005, 23, 4776–4789. [Google Scholar] [CrossRef]
- Ummanni, R.; Jost, E.; Braig, M.; Lohmann, F.; Mundt, F.; Barett, C.; Schlomm, T.; Sauter, G.; Senff, T.; Bokemeyer, C.; et al. Ubiquitin Carboxyl-Terminal Hydrolase 1 (UCHL1) Is a Potential Tumour Suppressor in Prostate Cancer and Is Frequently Silenced by Promoter Methylation. Mol. Cancer 2011, 10, 129. [Google Scholar] [CrossRef]
- Cheng, J.; Liu, H.; Shen, Y.; Ding, J.; He, H.; Mao, S.; Chen, L.; Zhang, C.; Zhou, J. Deubiquitinase UCHL1 Stabilizes KDM4B to Augment VEGF Signaling and Confer Bevacizumab Resistance in Clear Cell Renal Cell Carcinoma. Transl. Oncol. 2024, 45, 101987. [Google Scholar] [CrossRef]
- Gottschalk, A.; Kastner, B.; Lührmann, R.; Fabrizio, P. The Yeast U5 SnRNP Coisolated with the U1 SnRNP Has an Unexpected Protein Composition and Includes the Splicing Factor Aar2p. RNA 2001, 7, 1554–1565. [Google Scholar]
- Ye, Z.; Bing, A.; Zhao, S.; Yi, S.; Zhan, X. Comprehensive Analysis of Spliceosome Genes and Their Mutants across 27 Cancer Types in 9070 Patients: Clinically Relevant Outcomes in the Context of 3P Medicine. EPMA J. 2022, 13, 335–350. [Google Scholar] [CrossRef]
- Zhan, Y.T.; Li, L.; Zeng, T.T.; Zhou, N.N.; Guan, X.Y.; Li, Y. SNRPB-Mediated RNA Splicing Drives Tumor Cell Proliferation and Stemness in Hepatocellular Carcinoma. Aging 2021, 13, 537–554. [Google Scholar] [CrossRef]
- Li, X.; Turanli, B.; Juszczak, K.; Kim, W.; Arif, M.; Sato, Y.; Ogawa, S.; Turkez, H.; Nielsen, J.; Boren, J.; et al. Classification of Clear Cell Renal Cell Carcinoma Based on PKM Alternative Splicing. Heliyon 2020, 6, e03440. [Google Scholar] [CrossRef]
- Zhang, T.; Bai, J.; Zhang, X.; Zheng, X.; Lu, N.; Liang, Z.; Lin, L.; Chen, Y. SNRNP200 Mutations Cause Autosomal Dominant Retinitis Pigmentosa. Front. Med. 2021, 7, 588991. [Google Scholar] [CrossRef] [PubMed]
- Knorr, K.; Rahman, J.; Erickson, C.; Wang, E.; Monetti, M.; Li, Z.; Ortiz-Pacheco, J.; Jones, A.; Lu, S.X.; Stanley, R.F.; et al. Systematic Evaluation of AML-Associated Antigens Identifies Anti-U5 SNRNP200 Therapeutic Antibodies for the Treatment of Acute Myeloid Leukemia. Nat. Cancer 2023, 4, 1675–1692. [Google Scholar] [CrossRef]
- Bradley, R.K.; Anczuków, O. RNA Splicing Dysregulation and the Hallmarks of Cancer. Nat. Rev. Cancer 2023, 23, 135–155. [Google Scholar] [CrossRef]
- Ivanova, O.M.; Anufrieva, K.S.; Kazakova, A.N.; Malyants, I.K.; Shnaider, P.V.; Lukina, M.M.; Shender, V.O. Non-Canonical Functions of Spliceosome Components in Cancer Progression. Cell Death Dis. 2023, 14, 77. [Google Scholar] [CrossRef]
- Kichina, J.V.; Goc, A.; Al-Husein, B.; Somanath, P.R.; Kandel, E.S. PAK1 as a Therapeutic Target. Expert Opin. Ther. Targets 2010, 14, 703–725. [Google Scholar] [CrossRef]
- Ye, D.Z.; Field, J. PAK Signaling in Cancer. Cell. Logist. 2012, 2, 105–116. [Google Scholar] [CrossRef]
- Yeo, D.; He, H.; Baldwin, G.S.; Nikfarjam, M. The Role of P21-Activated Kinases in Pancreatic Cancer. Pancreas 2015, 44, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, K.; Dong, Z. The Role of P21-Activated Kinases in Cancer and Beyond: Where Are We Heading? Front. Cell Dev. Biol. 2021, 9, 641381. [Google Scholar] [CrossRef]
- Kumar, R.; Sanawar, R.; Li, X.; Li, F. Structure, Biochemistry, and Biology of PAK Kinases. Gene 2017, 605, 20–31. [Google Scholar] [CrossRef]
- O’Sullivan, G.C.; Tangney, M.; Casey, G.; Ambrose, M.; Houston, A.; Barry, O.P. Modulation of P21-Activated Kinase 1 Alters the Behavior of Renal Cell Carcinoma. Int. J. Cancer 2007, 121, 1930–1940. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yang, Y.; Liu, Y.; Liu, H.; Zhang, W.; Xu, L.; Zhu, Y.; Xu, J. P21-Activated Kinase 4 Predicts Early Recurrence and Poor Survival in Patients with Nonmetastatic Clear Cell Renal Cell Carcinoma. Urol. Oncol. Semin. Orig. Investig. 2015, 33, 205.e13–205.e21. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.W.; Piao, X.M.; Lee, H.Y.; Kim, K.; Seo, S.P.; Ha, Y.S.; Kim, Y.U.; Kim, W.T.; Kim, Y.J.; Lee, S.C.; et al. Expression of Phosphorylated P21-Activated Kinase 4 Is Associated with Aggressive Histologic Characteristics and Poor Prognosis in Patients with Surgically Treated Renal Cell Carcinoma. Investig. Clin. Urol. 2021, 62, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Linehan, W.M.; Ricketts, C.J. The Cancer Genome Atlas of Renal Cell Carcinoma: Findings and Clinical Implications. Nat. Rev. Urol. 2019, 16, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Park, J.J.; Park, M.H.; Oh, E.H.; Soung, N.K.; Lee, S.J.; Jung, J.K.; Lee, O.J.; Yun, S.J.; Kim, W.J.; Shin, E.Y.; et al. The P21-Activated Kinase 4-Slug Transcription Factor Axis Promotes Epithelial−mesenchymal Transition and Worsens Prognosis in Prostate Cancer. Oncogene 2018, 37, 5147–5159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Guo, Q.; Wang, Y.; Zhou, Y.; Peng, H.; Cheng, M.; Zhao, D.; Li, F. LCH-7749944, a Novel and Potent P21-Activated Kinase 4 Inhibitor, Suppresses Proliferation and Invasion in Human Gastric Cancer Cells. Cancer Lett. 2012, 317, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Ryu, B.J.; Lee, H.; Kim, S.H.; Heo, J.N.; Choi, S.W.; Yeon, J.T.; Lee, J.; Lee, J.; Cho, J.Y.; Kim, S.H.; et al. PF-3758309, P21-Activated Kinase 4 Inhibitor, Suppresses Migration and Invasion of A549 Human Lung Cancer Cells via Regulation of CREB, NF-ΚB, and β-Catenin Signalings. Mol. Cell. Biochem. 2014, 389, 69–77. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Xu, L.; Tao, Y.; Yang, C.; Chen, X.; Fang, F.; Wu, Y.; Ding, X.; Zhao, H.; et al. Inhibition of Neuroblastoma Proliferation by PF-3758309, a Small-Molecule Inhibitor That Targets P21-Activated Kinase 4. Oncol. Rep. 2017, 38, 2705–2716. [Google Scholar] [CrossRef]




| Antibody | Source | Identifier | Clone | Dilution |
|---|---|---|---|---|
| Rabbit polyclonal anti-UCHL1 | Atlas Antibodies, Stockholm, Sweden | Cat# HPA005993, RRID:AB_1858560 | Polyclonal | 1:3000 |
| Rabbit polyclonal anti-SNRNP200 | Atlas Antibodies, Stockholm, Sweden | Cat# HPA029321, RRID:AB_10604096 | Polyclonal | 1:225 |
| Mouse monoclonal anti-PAK4 | ThermoFisher Scientific, Waltham, MA, USA | Cat# MA5-26859, RRID:AB_2723443 | OTI1C7 | 1:150 |
| Protein | H-Score Range—High IHC Score Group | H-Score Range—Low IHC Score Group | Reaction |
|---|---|---|---|
| PAK4 | 50–210 | 0–40 | Cytoplasmic, membranous |
| SNRNP200 | 50–150 | 0–40 | Nuclear |
| UCHL1 | 65–300 | 0–60 | Cytoplasmic, membranous, nuclear |
| Progression-Free Survival (PFS) | Overall Survival (OS) | |||
|---|---|---|---|---|
| Hazard Ratio (HR) | 95% CI of HR | Hazard Ratio (HR) | 95% CI of HR | |
| PAK4 | 2.57 | 1.181–5.569 | 1.96 | 0.7027–5.476 |
| SNRNP200 | 3.35 | 1.427–7.842 | 1.41 | 0.498–3.995 |
| UCHL1 | 2.39 | 1.092–5.240 | 1.69 | 0.602–4.738 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasperczak, M.; Bromiński, G.; Kołodziejczak-Guglas, I.; Antczak, A.; Wiznerowicz, M. Prognostic Significance of Elevated UCHL1, SNRNP200, and PAK4 Expression in High-Grade Clear Cell Renal Cell Carcinoma: Insights from LC-MS/MS Analysis and Immunohistochemical Validation. Cancers 2024, 16, 2844. https://doi.org/10.3390/cancers16162844
Kasperczak M, Bromiński G, Kołodziejczak-Guglas I, Antczak A, Wiznerowicz M. Prognostic Significance of Elevated UCHL1, SNRNP200, and PAK4 Expression in High-Grade Clear Cell Renal Cell Carcinoma: Insights from LC-MS/MS Analysis and Immunohistochemical Validation. Cancers. 2024; 16(16):2844. https://doi.org/10.3390/cancers16162844
Chicago/Turabian StyleKasperczak, Michał, Gabriel Bromiński, Iga Kołodziejczak-Guglas, Andrzej Antczak, and Maciej Wiznerowicz. 2024. "Prognostic Significance of Elevated UCHL1, SNRNP200, and PAK4 Expression in High-Grade Clear Cell Renal Cell Carcinoma: Insights from LC-MS/MS Analysis and Immunohistochemical Validation" Cancers 16, no. 16: 2844. https://doi.org/10.3390/cancers16162844
APA StyleKasperczak, M., Bromiński, G., Kołodziejczak-Guglas, I., Antczak, A., & Wiznerowicz, M. (2024). Prognostic Significance of Elevated UCHL1, SNRNP200, and PAK4 Expression in High-Grade Clear Cell Renal Cell Carcinoma: Insights from LC-MS/MS Analysis and Immunohistochemical Validation. Cancers, 16(16), 2844. https://doi.org/10.3390/cancers16162844






