CCL2/CCR2 Expression in Locally Advanced Prostate Cancer and Patient Long-Term Outcome: 10-Year Results from the TROG 03.04 RADAR Trial
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Prostate Cancer Patient Cohort
2.2. Immunohistochemistry
2.3. Digital Quantification of Immunohistochemistry
2.4. Enzyme-Linked Immunosorbent Assay (ELISA)
2.5. Statistical Analyses
3. Results
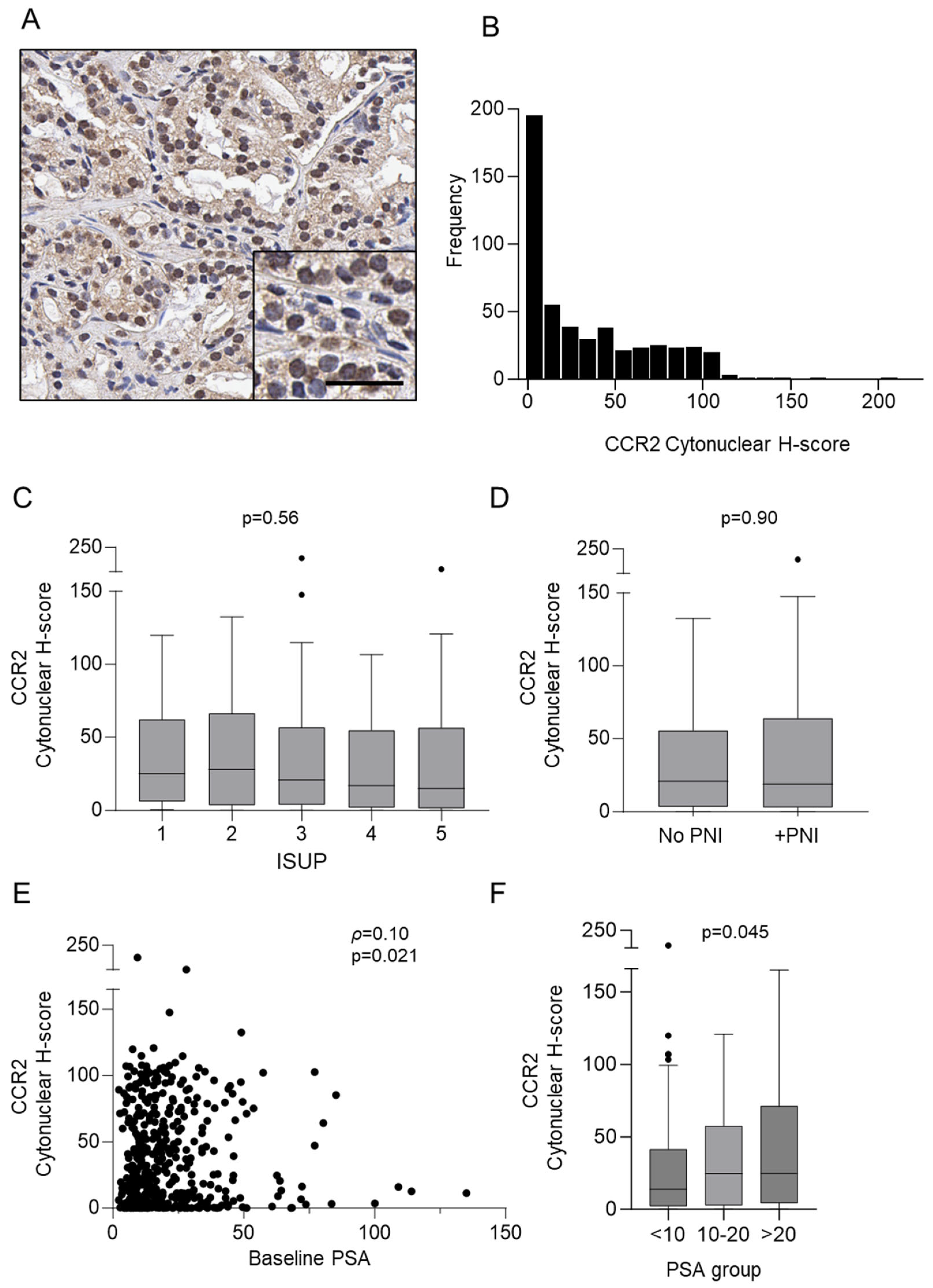
3.1. Detection of CCR2 Protein in Prostate Cancer Biopsies
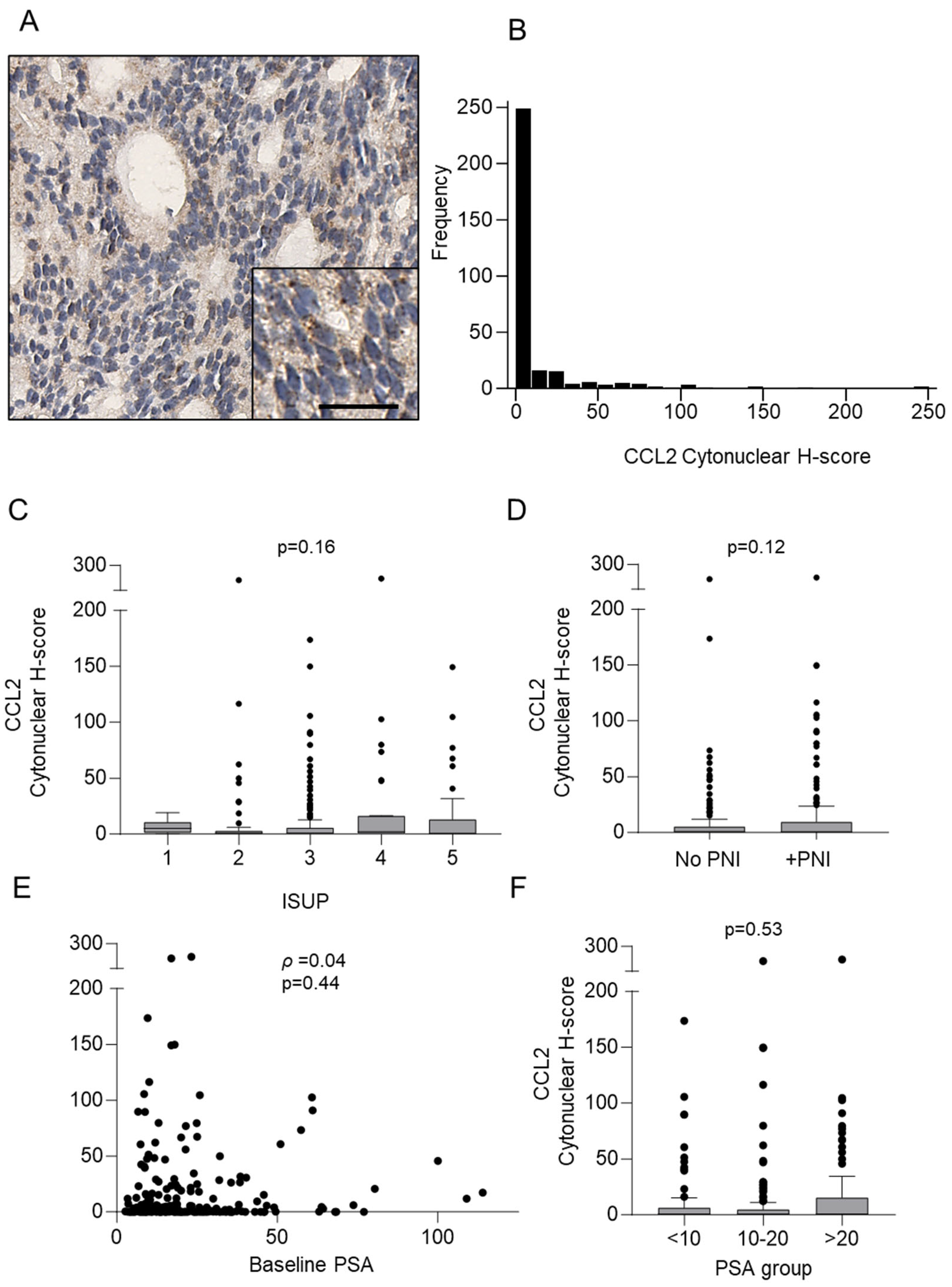
3.2. Detection of CCL2 Protein in Prostate Cancer Core Biopsies
3.3. CCL2 Serum Concentration in Patients with Prostate Cancer Compared to Normal and Benign Prostatic Hyperplasia
3.4. Correlation of CCL2/CCR2 Expression in Prostate Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van der Kwast, T.; Bristow, R.G. Prostate cancer. Nat. Rev. Dis. Primers 2021, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, S.; Moore, C.M.; Chiong, E.; Beltran, H.; Bristow, R.G.; Williams, S.G. Prostate cancer. Lancet 2021, 398, 1075–1090. [Google Scholar] [CrossRef] [PubMed]
- Australian Institute of Health and Welfare. Prostate Cancer in Australia; AIHW: Canberra, Australia, 2013. [Google Scholar]
- Knipper, S.; Ott, S.; Schlemmer, H.P.; Grimm, M.O.; Graefen, M.; Wiegel, T. Options for Curative Treatment of Localized Prostate Cancer. Dtsch. Arztebl. Int. 2021, 118, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Paller, C.J.; Antonarakis, E.S. Management of biochemically recurrent prostate cancer after local therapy: Evolving standards of care and new directions. Clin. Adv. Hematol. Oncol. 2013, 11, 14–23. [Google Scholar] [PubMed]
- Benjamin, D.J.; Rezazadeh Kalebasty, A. Characterization and Survival Benefit of Drug Approvals for Metastatic Prostate Cancer, 2004 to 2022. Clin. Med. Insights Oncol. 2024, 18, 11795549241227413. [Google Scholar] [CrossRef] [PubMed]
- Saad, F.; Bögemann, M.; Suzuki, K.; Shore, N. Treatment of nonmetastatic castration-resistant prostate cancer: Focus on second-generation androgen receptor inhibitors. Prostate Cancer Prostatic Dis. 2021, 24, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Förster, S.; Muders, M. The Role of Perineural Invasion in Prostate Cancer and Its Prognostic Significance. Cancers 2022, 14, 4065. [Google Scholar] [CrossRef] [PubMed]
- Delahunt, B.; Murray, J.D.; Steigler, A.; Atkinson, C.; Christie, D.; Duchesne, G.; Egevad, L.; Joseph, D.; Matthews, J.; Oldmeadow, C.; et al. Perineural invasion by prostate adenocarcinoma in needle biopsies predicts bone metastasis: Ten year data from the TROG 03.04 RADAR Trial. Histopathology 2020, 77, 284–292. [Google Scholar] [CrossRef]
- Iwamoto, H.; Izumi, K.; Nakagawa, R.; Toriumi, R.; Aoyama, S.; Shimada, T.; Kano, H.; Makino, T.; Kadomoto, S.; Yaegashi, H.; et al. Usefulness of serum CCL2 as prognostic biomarker in prostate cancer: A long-term follow-up study. Jpn. J. Clin. Oncol. 2022, 52, 1337–1344. [Google Scholar] [CrossRef]
- Yang, Y.I.; Wang, Y.Y.; Ahn, J.H.; Kim, B.H.; Choi, J.H. CCL2 overexpression is associated with paclitaxel resistance in ovarian cancer cells via autocrine signaling and macrophage recruitment. Biomed. Pharmacother. 2022, 153, 113474. [Google Scholar] [CrossRef]
- Feng, H.; Liu, K.; Shen, X.; Liang, J.; Wang, C.; Qiu, W.; Cheng, X.; Zhao, R. Targeting tumor cell-derived CCL2 as a strategy to overcome Bevacizumab resistance in ETV5(+) colorectal cancer. Cell Death Dis. 2020, 11, 916. [Google Scholar] [CrossRef] [PubMed]
- Gschwandtner, M.; Derler, R.; Midwood, K.S. More Than Just Attractive: How CCL2 Influences Myeloid Cell Behavior Beyond Chemotaxis. Front. Immunol. 2019, 10, 2759. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Chen, W.Y.; Abou-Kheir, W.; Zeng, T.; Yin, J.J.; Bahmad, H.; Lee, Y.C.; Liu, Y.N. Androgen deprivation therapy-induced epithelial-mesenchymal transition of prostate cancer through downregulating SPDEF and activating CCL2. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.H.; Izumi, K.; Lee, S.O.; Lin, W.J.; Yeh, S.; Chang, C. Anti-androgen receptor ASC-J9 versus anti-androgens MDV3100 (Enzalutamide) or Casodex (Bicalutamide) leads to opposite effects on prostate cancer metastasis via differential modulation of macrophage infiltration and STAT3-CCL2 signaling. Cell Death Dis. 2013, 4, e764. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, H.; Izumi, K.; Nakagawa, R.; Toriumi, R.; Aoyama, S.; Kamijima, T.; Shimada, T.; Kano, H.; Makino, T.; Naito, R.; et al. Serum CCL2 Is a Prognostic Biomarker for Non-Metastatic Castration-Sensitive Prostate Cancer. Biomedicines 2022, 10, 2369. [Google Scholar] [CrossRef]
- Hao, Q.; Vadgama, J.V.; Wang, P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun. Signal 2020, 18, 82. [Google Scholar] [CrossRef] [PubMed]
- Kadomoto, S.; Izumi, K.; Mizokami, A. Roles of CCL2-CCR2 Axis in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 8530. [Google Scholar] [CrossRef] [PubMed]
- Denham, J.W.; Joseph, D.; Lamb, D.S.; Spry, N.A.; Duchesne, G.; Matthews, J.; Atkinson, C.; Tai, K.H.; Christie, D.; Kenny, L.; et al. Short-term androgen suppression and radiotherapy versus intermediate-term androgen suppression and radiotherapy, with or without zoledronic acid, in men with locally advanced prostate cancer (TROG 03.04 RADAR): An open-label, randomised, phase 3 factorial trial. Lancet Oncol. 2014, 15, 1076–1089. [Google Scholar] [CrossRef] [PubMed]
- Denham, J.W.; Joseph, D.; Lamb, D.S.; Spry, N.A.; Duchesne, G.; Matthews, J.; Atkinson, C.; Tai, K.H.; Christie, D.; Kenny, L.; et al. Short-term androgen suppression and radiotherapy versus intermediate-term androgen suppression and radiotherapy, with or without zoledronic acid, in men with locally advanced prostate cancer (TROG 03.04 RADAR): 10-year results from a randomised, phase 3, factorial trial. Lancet Oncol. 2019, 20, 267–281. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, M.; Smith, W.; D’Este, C.; Duke, J.; Peel, R.; Schofield, P.; Scott, R.; Byles, J.; Henry, D.; Ewald, B.; et al. Cohort profile: The Hunter Community Study. Int. J. Epidemiol. 2010, 39, 1452–1463. [Google Scholar] [CrossRef]
- Marsland, M.; Dowdell, A.; Faulkner, S.; Jobling, P.; Rush, R.A.; Gedye, C.; Lynam, J.; Griffin, C.P.; Baker, M.; Marsland, J.; et al. ProNGF Expression and Targeting in Glioblastoma Multiforme. Int. J. Mol. Sci. 2023, 24, 1616. [Google Scholar] [CrossRef] [PubMed]
- Marsland, M.; Dowdell, A.; Jiang, C.C.; Wilmott, J.S.; Scolyer, R.A.; Zhang, X.D.; Hondermarck, H.; Faulkner, S. Expression of NGF/proNGF and Their Receptors TrkA, p75(NTR) and Sortilin in Melanoma. Int. J. Mol. Sci. 2022, 23, 4260. [Google Scholar] [CrossRef] [PubMed]
- Horai, Y.; Mizukawa, M.; Nishina, H.; Nishikawa, S.; Ono, Y.; Takemoto, K.; Baba, N. Quantification of histopathological findings using a novel image analysis platform. J. Toxicol. Pathol. 2019, 32, 319–327. [Google Scholar] [CrossRef]
- March, B.; Lockhart, K.R.; Faulkner, S.; Smolny, M.; Rush, R.; Hondermarck, H. ELISA-based quantification of neurotrophic growth factors in urine from prostate cancer patients. FASEB Bioadv. 2021, 3, 888–896. [Google Scholar] [CrossRef]
- Lu, Y.; Cai, Z.; Xiao, G.; Liu, Y.; Keller, E.T.; Yao, Z.; Zhang, J. CCR2 expression correlates with prostate cancer progression. J. Cell Biochem. 2007, 101, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Cai, Z.; Galson, D.L.; Xiao, G.; Liu, Y.; George, D.E.; Melhem, M.F.; Yao, Z.; Zhang, J. Monocyte chemotactic protein-1 (MCP-1) acts as a paracrine and autocrine factor for prostate cancer growth and invasion. Prostate 2006, 66, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Izumi, K.; Fang, L.Y.; Mizokami, A.; Namiki, M.; Li, L.; Lin, W.J.; Chang, C. Targeting the androgen receptor with siRNA promotes prostate cancer metastasis through enhanced macrophage recruitment via CCL2/CCR2-induced STAT3 activation. EMBO Mol. Med. 2013, 5, 1383–1401. [Google Scholar] [CrossRef] [PubMed]
- Zareba, P.; Flavin, R.; Isikbay, M.; Rider, J.R.; Gerke, T.A.; Finn, S.; Pettersson, A.; Giunchi, F.; Unger, R.H.; Tinianow, A.M.; et al. Perineural Invasion and Risk of Lethal Prostate Cancer. Cancer Epidemiol. Biomark. Prev. 2017, 26, 719–726. [Google Scholar] [CrossRef] [PubMed]
- He, S.; He, S.; Chen, C.H.; Deborde, S.; Bakst, R.L.; Chernichenko, N.; McNamara, W.F.; Lee, S.Y.; Barajas, F.; Yu, Z.; et al. The chemokine (CCL2-CCR2) signaling axis mediates perineural invasion. Mol. Cancer Res. 2015, 13, 380–390. [Google Scholar] [CrossRef]
- Huang, T.; Fan, Q.; Wang, Y.; Cui, Y.; Wang, Z.; Yang, L.; Sun, X.; Wang, Y. Schwann Cell-Derived CCL2 Promotes the Perineural Invasion of Cervical Cancer. Front. Oncol. 2020, 10, 19. [Google Scholar] [CrossRef]

| Characteristic | ||
|---|---|---|
| Age, median (IQR), years | 68.7 | (63.9–73.1) |
| PSA, median (IQR), ng/mL | 15.2 | (9.6–25.6) |
| <10 | 136 | (27.2) |
| 10–20 | 197 | (39.3) |
| >20 | 168 | (33.5) |
| ISUP Grade | ||
| 1 | 17 | (3.4) |
| 2 | 112 | (22.4) |
| 3 | 201 | (40.1) |
| 4 | 96 | (19.2) |
| 5 | 75 | (15.0) |
| Clinical T-stage | ||
| T2 | 335 | (66.9) |
| T3,4 | 166 | (33.1) |
| NCCN risk group | ||
| Intermediate | 164 | (32.7) |
| High | 337 | (67.3) |
| Perineural invasion | ||
| Absent | 260 | (51.9) |
| Present | 236 | (47.1) |
| Inevaluable | 5 | (1.0) |
| Univariable (N = 501) | Multivariable (N = 492) † | |||||
|---|---|---|---|---|---|---|
| Endpoints | n | HR (95% CI) | p | n | HR (95% CI) | p |
| PSA progression | 222 | 1.00 (0.97–1.04) | 0.87 | 220 | 1.00 (0.96–1.04) | 0.96 |
| Bone progression | 107 | 0.99 (0.94–1.04) | 0.67 | 106 | 1.00 (0.95–1.06) | 0.94 |
| Distant progression | 140 | 1.00 (0.95–1.04) | 0.89 | 139 | 1.01 (0.97–1.05) | 0.68 |
| PC-specific mortality | 64 | 0.98 (0.92–1.05) | 0.58 | 64 | 0.99 (0.93–1.06) | 0.86 |
| All-cause mortality | 174 | 0.98 (0.94–1.03) | 0.44 | 172 | 0.98 (0.94–1.02) | 0.38 |
| Characteristic | ||
|---|---|---|
| Age, median (IQR), years | 68.7 | (63.7–72.5) |
| PSA, median (IQR), ng/mL | 15.5 | (9.9–24.4) |
| <10 | 79 | (25.2) |
| 10–20 | 133 | (42.4) |
| >20 | 102 | (32.5) |
| ISUP Grade | ||
| 1 | 8 | (2.6) |
| 2 | 78 | (24.8) |
| 3 | 141 | (44.9) |
| 4 | 29 | (9.2) |
| 5 | 58 | (18.5) |
| Clinical T-stage | ||
| T2 | 206 | (65.6) |
| T3,4 | 108 | (34.4) |
| NCCN risk group | ||
| Intermediate | 110 | (35.0) |
| High | 204 | (65.0) |
| Perineural invasion | ||
| Absent | 145 | (46.2) |
| Present | 166 | (52.9) |
| Inevaluable | 3 | (1.0) |
| Univariable (N = 314) | Multivariable (N = 307) † | |||||
|---|---|---|---|---|---|---|
| Endpoints | n | HR (95% CI) | p | n | HR (95% CI) | p |
| PSA progression | 134 | 1.03 (0.99–1.07) | 0.09 | 132 | 1.03 (0.97–1.08) | 0.36 |
| Bone progression | 71 | 1.03 (0.98–1.09) | 0.26 | 70 | 1.03 (0.96–1.11) | 0.36 |
| Distant progression | 92 | 1.03 (0.98–1.08) | 0.25 | 91 | 1.03 (0.96–1.10) | 0.41 |
| PC-specific mortality | 44 | 1.00 (0.90–1.11) | 0.97 | 43 | 1.00 (0.88–1.13) | 0.99 |
| All-cause mortality | 111 | 0.99 (0.93–1.06) | 0.81 | 109 | 0.99 (0.93–1.06) | 0.83 |
| Characteristic | ||
|---|---|---|
| Age, median (IQR), years | 68.2 | (62.9–73.0) |
| PSA, median (IQR), ng/mL | 14.0 | (8.8–23.0) |
| <10 | 67 | (30.5) |
| 10–20 | 89 | (40.5) |
| >20 | 64 | (29.1) |
| ISUP Grade | ||
| 1 | 10 | (4.6) |
| 2 | 60 | (27.3) |
| 3 | 59 | (26.8) |
| 4 | 48 | (21.8) |
| 5 | 43 | (19.6) |
| Clinical T-stage | ||
| T2 | 105 | (47.7) |
| T3,4 | 115 | (52.3) |
| NCCN risk group | ||
| Intermediate | 60 | (27.3) |
| High | 160 | (72.7) |
| Perineural invasion | ||
| Absent | 140 | (63.6) |
| Present | 78 | (35.5) |
| Inevaluable | 2 | (0.9) |
| Univariable (N = 220) | Multivariable (N = 218) † | |||||
|---|---|---|---|---|---|---|
| Endpoints | n | HR (95% CI) | p | n | HR (95% CI) | p |
| PSA progression | 90 | 1.50 (1.07–2.09) | 0.017 | 90 | 1.19 (0.83–1.69) | 0.35 |
| Bone progression | 49 | 1.20 (0.63–2.27) | 0.58 | 49 | 0.91 (0.48–1.75) | 0.78 |
| Distant progression | 69 | 1.44 (0.91–2.30) | 0.12 | 69 | 1.10 (0.65–1.86) | 0.72 |
| PC-specific mortality | 30 | 1.17 (0.53–2.60) | 0.70 | 30 | 0.72 (0.31–1.63) | 0.43 |
| All-cause mortality | 86 | 0.86 (0.48–1.54) | 0.61 | 85 | 0.76 (0.40–1.44) | 0.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marsland, M.; Jiang, C.C.; Faulkner, S.; Steigler, A.; McEwan, K.; Jobling, P.; Oldmeadow, C.; Delahunt, B.; Denham, J.W.; Hondermarck, H. CCL2/CCR2 Expression in Locally Advanced Prostate Cancer and Patient Long-Term Outcome: 10-Year Results from the TROG 03.04 RADAR Trial. Cancers 2024, 16, 2794. https://doi.org/10.3390/cancers16162794
Marsland M, Jiang CC, Faulkner S, Steigler A, McEwan K, Jobling P, Oldmeadow C, Delahunt B, Denham JW, Hondermarck H. CCL2/CCR2 Expression in Locally Advanced Prostate Cancer and Patient Long-Term Outcome: 10-Year Results from the TROG 03.04 RADAR Trial. Cancers. 2024; 16(16):2794. https://doi.org/10.3390/cancers16162794
Chicago/Turabian StyleMarsland, Mark, Chen Chen Jiang, Sam Faulkner, Allison Steigler, Kristen McEwan, Phillip Jobling, Christopher Oldmeadow, Brett Delahunt, James W. Denham, and Hubert Hondermarck. 2024. "CCL2/CCR2 Expression in Locally Advanced Prostate Cancer and Patient Long-Term Outcome: 10-Year Results from the TROG 03.04 RADAR Trial" Cancers 16, no. 16: 2794. https://doi.org/10.3390/cancers16162794
APA StyleMarsland, M., Jiang, C. C., Faulkner, S., Steigler, A., McEwan, K., Jobling, P., Oldmeadow, C., Delahunt, B., Denham, J. W., & Hondermarck, H. (2024). CCL2/CCR2 Expression in Locally Advanced Prostate Cancer and Patient Long-Term Outcome: 10-Year Results from the TROG 03.04 RADAR Trial. Cancers, 16(16), 2794. https://doi.org/10.3390/cancers16162794








