Simple Summary
The expansion of mutant cells that outgrow and displace normal blood cells is called clonal hematopoiesis (CH). CH starts with the acquisition of mutations in blood stem cells that change their normal behavior and improve their fitness. Low numbers of mutant blood cells are present in many middle-aged and elderly people without noticeable effects. However, CH can progress to leukemia and contribute to diseases in other tissues, including the heart, liver, and pancreas. As these diseases progress and affect health when the number of mutant blood cells increases, preventing and/or reducing the expansion of mutant cells might delay this process. Identifying the factors driving mutant blood cell expansion is thus critical and has garnered heightened interest in the subject. Here, we review and discuss recent progress in the field that suggests that mutant blood stem cells are more resilient than normal cells and thrive in inflammatory conditions.
Abstract
Clonal hematopoiesis (CH), the relative expansion of mutant clones, is derived from hematopoietic stem cells (HSCs) with acquired somatic or cytogenetic alterations that improve cellular fitness. Individuals with CH have a higher risk for hematological and non-hematological diseases, such as cardiovascular disease, and have an overall higher mortality rate. Originally thought to be restricted to a small fraction of elderly people, recent advances in single-cell sequencing and bioinformatics have revealed that CH with multiple expanded mutant clones is universal in the elderly population. Just a few years ago, phylogenetic reconstruction across the human lifespan and novel sensitive sequencing techniques showed that CH can start earlier in life, decades before it was thought possible. These studies also suggest that environmental factors acting through aberrant inflammation might be a common theme promoting clonal expansion and disease progression. However, numerous aspects of this phenomenon remain to be elucidated and the precise mechanisms, context-specific drivers, and pathways of clonal expansion remain to be established. Here, we review our current understanding of the cellular mechanisms driving CH and specifically focus on how pro-inflammatory factors affect normal and mutant HSC fates to promote clonal selection.
1. Introduction
The formation of blood relies on a small number of hematopoietic stem cells (HSCs) located in the bone marrow. HSCs can self-renew to create new stem cells and differentiate to produce all mature blood cell types for the entirety of the lifespan [1]. The ability of HSCs to regenerate the hematopoietic system throughout life is extensive, but it is not endless and the production of new blood cells changes and declines over time [2]. Contrary to that, mutant HSCs can self-renew indefinitely and are thought to drive hematological malignancies and therapy resistance [3]. These cells are sometimes called leukemic stem cells and their ability to self-renew is acquired by mutations in numerous genes affecting cell behavior and fates such as survival, proliferation, self-renewal, and differentiation. While these efforts are ongoing, and novel rare mutations are still being discovered, next-generation sequencing (NGS) studies with large cohorts of cancer patients identified the mutations that are most frequently associated with hematological diseases [4]. Surprisingly, these studies found that many of the somatic mutations associated with malignancies were already present in healthy individuals. Despite having no symptoms, healthy individuals had clonally expanded mutant hematopoietic cells [5,6]. As these mutations seemed to drive clonal hematopoiesis (CH) but did not lead to cytopenias and dysplastic hematopoiesis characteristic of other pre-malignant disorders, the benign clonal expansion was termed clonal hematopoiesis of indeterminate potential (CHIP) [5]. CHIP was originally defined as a subset of CH with a variant allele frequency ≥2% of somatic mutations in genes associated with hematological malignancies [7]. However, as we will discuss in more detail later, the 2% threshold is somewhat arbitrary and underestimates the frequency of CH, as smaller clones can be detected reliably. Importantly, although individuals with CH lack overt clinical symptoms, clonally restricted hematopoiesis increases the risk of myeloid [8] and lymphoid malignancies [9] as well as non-hematological diseases, including atherosclerotic cardiovascular disease, pulmonary disease, type 2 diabetes, acute kidney disease, chronic liver disease, and osteoporosis (Figure 1) [5,10,11,12].
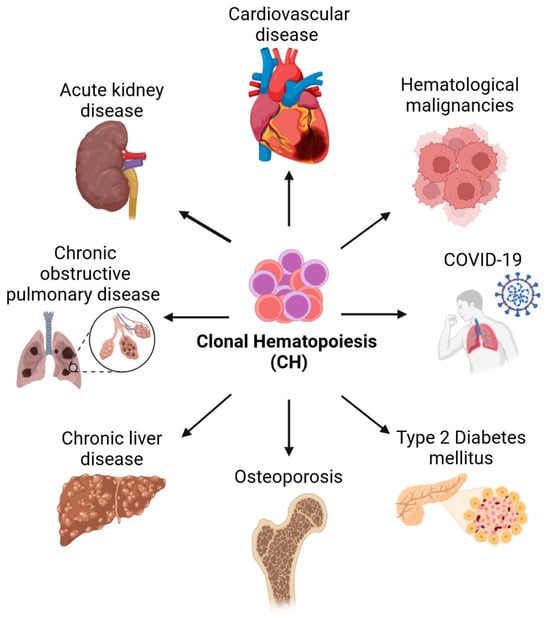
Figure 1.
Diseases associated with clonal hematopoiesis. Individuals with clonal hematopoiesis have a higher risk of hematologic and non-hematologic diseases and an overall higher rate of mortality. Hematologic diseases associated with CH include myelodysplastic syndrome and myeloid and lymphoid malignancies. Non-hematologic diseases associated with CH include, among others, cardiovascular disease, acute kidney disease, chronic obstructive pulmonary disease, chronic liver disease, osteoporosis, type 2 diabetes, and COVID-19. Created with https://biorender.com.
Individuals with CH also have a higher risk for severe infection with COVID-19 and bacteria and show an increased overall mortality [13]. Due to its association with multiple hematological and non-hematological diseases, CH has gained considerable attention from the scientific community in recent years. Despite these efforts, many questions remain and the mechanisms regulating clonal expansion and progression from a benign preleukemic to a malignant leukemic state remain poorly understood. As the field moves forward quickly, we attempt to summarize what we have learned thus far, highlight key findings, and discuss how recent technological advances provide new insights and challenge old views. We also discuss the critical next steps and how new tools can help to improve our understanding of CH.
2. Early Evidence for Clonal Expansion
In 1962, Mary Lyon proposed that one of the two mammalian X-chromosomes present in females is randomly inactivated during ontogeny [14]. This hypothesis, known today as X-chromosome inactivation (XCI), explained why female mice heterozygous for sex-linked genes have mosaic coat colors by suggesting that genetically identical cells (=clones) are present in patches with the same fur color. In the same year, Beutler et al. experimentally validated this theory in women heterozygous for the X-linked gene glucose-6-phosphate dehydrogenase (G6PD). Based on earlier observations, Beutler reasoned that if erythrocyte precursors of heterozygous women were a mixture of cells with normal and deficient G6PD enzyme activity, these mosaics should have intermediate G6PD activity in their blood. Using the stability of glutathione as a readout for G6PD activity, Beutler verified his hypothesis by comparing the glutathione stability of blood samples from heterozygous, hemizygous, and normal individuals with known family pedigrees for G6PD heterozygosity [15]. A few years later, in 1965, Linder and Gartler studied leiomyoma and myometrium tumor samples isolated from women heterozygous for two G6PD variants. Using starch gel electrophoresis, they found that the majority of tumors expressed the same G6PD variants, suggesting these tumors were derived from a single cell and of clonal origin [16]. Using the same system, Fialkow and colleagues studied blood samples of women who were heterozygous for G6PD with chronic myeloid leukemia (CML). Similar to Linder and Gartler, they found only one G6PD variant in the granulocytes and erythrocytes, while the skin fibroblasts expressed both G6PD forms, suggesting a clonal origin of CML [17]. In the 1990s, the development of novel assays, based on the high rate of polymorphism of the androgen-receptor gene made clonal studies based on XCI feasible in the majority of women by performing a simple PCR [18]. Using these assays on different tissues of the same women [19] and in women of different ages [20,21] revealed that clonal outgrowth (assessed by the strength of skewed XCI) is higher in the hematopoietic system and occurs more frequently in older individuals. In Champion et al., the XCI skewing of purified blood cells in normal elderly women showed that 63.4% of female patients with myeloproliferative disorder exhibit a clonal outgrowth in granulocytes and T cells. Based on these observations, he proposed clonal expansion might precede the development of myelodysplastic or myeloproliferative diseases [20].
3. Identifying the Driver of Clonal Expansion: The Genomic Revolution
While these early studies provided evidence for the expansion of hematopoietic clones, the analysis of G6PD variants and/or AR polymorphisms was limited, as they relied on low levels of heterozygosity in the general population and were of limited throughput, precluding the analysis of rare disorders and large cohorts [22]. Hence, the mutations and mechanisms driving CH remained largely unknown. This changed with the advent of affordable high-throughput sequencing, as mutations in blood cells of large cohorts of healthy individuals could be identified at scale with relatively little effort. Using these tools, in 2014, three independent groups discovered the most commonly mutated genes in benign and malignant hematopoietic clones [23,24,25], among which the loss-of-function mutations in the epigenetic modifiers DNMT3A, TET2, and ASXL1 were most frequently observed (Figure 2A). Importantly, many CH mutations were shown to persist in AML patients after treatment and were found after relapse, supporting the idea that mutant clones precede malignancies [26,27].
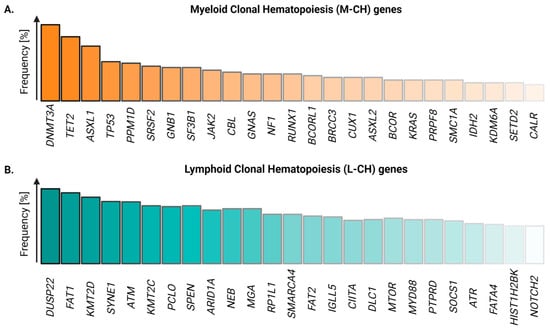
Figure 2.
Frequencies of the most common somatic variants associated with myeloid and lymphoid clonal hematopoiesis. (A) The top 25 driver genes mutated in myeloid CH (M-CH) and (B) lymphoid CH (L-CH), respectively (adapted from [28]). Created with https://biorender.com.
Although some of the identified mutations in these studies were associated with lymphoid malignancies [24], their frequency was low. The majority of the field thus focused on the more abundant mutations that increase the risk for myeloid malignancies. However, in their recently published seminal work, Niroula et al. investigated 46,706 samples of healthy individuals and identified 234 mutated genes associated with an increased risk of chronic lymphocytic leukemia [28]. As the majority of these mutations did not give rise to myeloid malignancies, the authors classified the mutant variants into genes associated with myeloid clonal hematopoiesis (M-CH) and lymphoid clonal hematopoiesis (L-CH). Similar to M-CH, L-CH occurs more often in the elderly population. However, the overall frequency is lower and the most frequently found mutant variants in L-CH are more evenly distributed compared to M-CH [9] (Figure 2B).
The size of these mutant clones is determined using the variant allele frequency (VAF), which represents the fraction of variant sequencing reads found in a genetic locus. In other words, VAF indicates how often a mutation is found in the sequenced DNA and provides an estimate of the relative number of mutant cells present in the analyzed sample. Based on a VAF cutoff of ≥2%, these early NGS studies identified a strong correlation between CH and age [23,24,29] and reported CH in 10–20% of individuals older than 70. At the same time, using the 2% threshold, benign clones were rarely found in younger individuals below 60 years of age at the time [30]. While a VAF of 2% rarely progresses to a malignant state, a growing body of evidence suggests that individuals with a VAF ≥10% (which corresponds to ≥20% of the nucleated blood cells with the mutation) exhibit a high risk of developing malignancies [31,32].
However, the sensitivity of these early sequencing studies was limited and clones with lower VAFs could not be detected reliably. More recent studies with improved sensitivity using ultra-sensitive sequencing [33], error-corrected sequencing [30], or single molecule molecular inversion probe sequencing [34], showed that DNMT3A and TET2 mutant clones are detectable in 95% of studied cases in middle-aged individuals, suggesting that the acquisition of mutant clones is almost universal [30]. It is also increasingly becoming clear that mutant hematopoietic clones can be found decades before CH can be detected in the peripheral blood and that numerous independently expanded clones are present per individual [4].
Importantly, longitudinal studies following individual clones over 20 years showed that the originally proposed VAF > 2% threshold for CH can miss fast-growing, potentially harmful variants and that the number of mutant clones is highly variable over time and can grow, shrink, or remain steady (Figure 3) [34,35]. The phylogenetic reconstruction and analysis of mutant clones in monozygotic and dizygotic twins further showed that mutant clones can arise even in utero or during childhood. Interestingly, these studies did not find evidence of a genetic predisposition for CH [12], as suggested in earlier studies [36]. Another study that tracked individuals for a median time of 13 years and used phylogenetic reconstruction found that clones with different mutations expand at vastly different rates and at different times in life [12]. While the growth of mutant clones was overall exponential, clones with DNMT3A and TP53 mutations were found to grow 5% per year, while clones with mutations in the splicing factor SRSF2P95H grew by over 50% annually. Interestingly, and for unclear reasons, clones with mutations in splicing factors appeared only later in life in aged individuals, while TET2 clones emerged in young, middle-aged, and old age and expanded at a constant rate of 6.8% per year. DNMT3A mutant clones, on the other hand, grew faster early in life but progressively slowed down during aging [12].
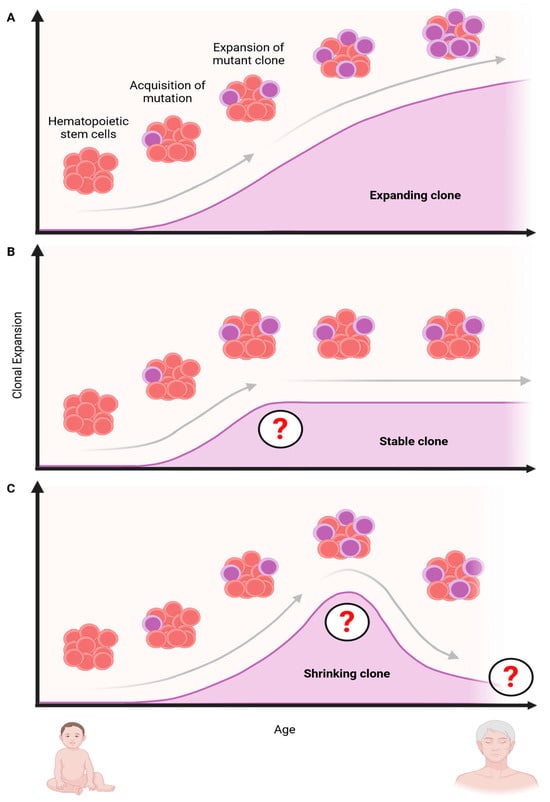
Figure 3.
The number of mutant clones throughout an individual’s lifetime can change dynamically and grow, shrink, or remain stable for extended periods. Clonal hematopoiesis is initiated when HSCs acquire mutations that confer a competitive advantage over wild-type HSCs. (A) Over time, this mutation allows the mutant clone (blue) to expand relative to normal HSCs (red). (B) Alternatively, mutant clones can remain stable for extended periods and/or the entire lifespan. (C) Mutant clones that started to expand can also shrink again. This can happen if the environmental conditions become less favorable and/or if another clone with increased evolutionary fitness outcompetes the original clone. The red question mark indicates changes in clonal growth rates. The reasons for these changes are currently unclear. Created with https://biorender.com.
These studies provide novel insights and question the still widely spread assumption that CH starts in elderly men and women. Instead, recent evidence suggests that, at least in some individuals, CH can start during development and/or early in life and that the presence of small mutant clones is universal in middle-aged individuals. These studies are also starting to reveal how different mutations affect clonal evolution and provide insights into the context-dependent role of environmental factors. These observations raise important questions, such as the following: Does CH always start during development? And if so, at what stage of blood formation do HSCs acquire these mutations? What is the cell of origin and why do these mutations occur in the first place?
4. Cell of Origin
Because mutant clones persist for many decades and contribute to myeloid and lymphoid cells, it is thought that CH begins with mutations in HSCs [37]. However, the precise timing when these mutations occur remains poorly understood. While all blood cells are derived from a common ancestor preceding gastrulation [38], human HSCs form at the aorta–gonad–mesonephros region of the embryo at approximately 4–6 weeks after fertilization [39]. The HSCs then migrate to the placenta, fetal liver, and finally, the bone marrow, where they acquire the quiescent state typical for adult HSCs. While CH is still often thought to start with mutant HSCs in the adult bone marrow, recent phylogenetic analyses suggest that CH can start as early as in utero. Williams et al. estimated that the mutations in individuals with mutant JAKV617F and DNMT3A could have been acquired as early as 33 and 8 weeks of gestation, respectively, while a PPM1D mutation was acquired in an individual by 5.8 years of age [40]. Studies of mutant clones in monozygotic twins found rare somatic mutations can be shared in both [41], suggesting these mutations can occur as early as the morula or blastocyst stage, when the embryo consists only of a few dozen to a few hundred cells. These observations raise an important question: how representative are reports of clonal growth in the embryo and how often are mutations acquired early in life? As the phylogenetic reconstruction of genomic sequencing is a rather new approach [42] and relatively few individuals have been studied in great detail, the chances that extremely rare cases of embryonic clonal acquisition were discovered by chance seem unlikely. These findings thus suggest that the acquisition of mutations during embryogenesis might be far more common than expected. Indeed, estimates that cells acquire between 0.069 and 0.858 exonic mutations every year [43] and approximately 1–2 mutations per division during embryogenesis [38] suggest that the acquisition of mutations is the rule rather than the exception. Over time and many cell divisions, our bodies thus become complex genetic mosaics of clones with coding and non-coding mutations—a phenomenon known as somatic mosaicism—that was recently analyzed in the context of a DNMT3A-mutant individual with germline mosaicism [44]. For further reading on the causes and consequences of somatic mosaicism and its relevance to CH, we refer the reader to an excellent recently published review [45].
While somatic mosaicism and the acquisition of mutations in HSCs during development and adult life can create clones of varying fitness, another source of heterogeneity is HSC subtypes with different stable properties. While the exact number of HSC subtypes remains unknown, forms of myeloid-biased, lymphoid-biased, and HSCs with balanced lineage output have been reported by several laboratories in mice [46,47] and humans [48]. However, the relationship between HSC subtypes, somatic mosaicism, and clonal outgrowth is unclear, and whether mutations in different HSC subtypes affect clonal outgrowth, fitness, and risk for transformation has not been studied. As distinct HSC subpopulations react differently to TGF-β [49], context-specific effects might promote the clonal expansion of some mutant HSC subtypes but act inhibitory on others—a poorly understood phenomenon that requires further investigation.
5. Mechanisms of Clonal Expansion
After a decade of research, and identifying the major mutations associated with CH, it is now well established that mutant HSCs possess a selective advantage over their normal counterparts [50,51,52]. Despite their fitness advantage, how CH is initiated and how mutant HSCs and their clones expand remain poorly understood [53]. Physiological stress, including repeated infections, autoimmune dysregulation, systemic disease, and metabolic stress promoting inflammation, have been suggested to provide the positive selection pressure required for the expansion of mutant clones [54]. Understanding these mechanisms and identifying the contributing molecular pathways is critical to developing therapeutic interventions that prevent clonal growth, disease progression, and malignant transformation.
As numerous mutations have been associated with CH, patterns of deregulated cellular pathways have emerged and mutant genes can broadly be categorized into regulators of (1) epigenetic modifications, (2) RNA splicing, (3) the DNA damage response, (4) transcription, (5) chromatin remodeling, and (6) signaling. In addition, CH genes can be categorized into (1) gain- and loss-of-function mutations and (2) M-CH and L-CH (Figure 2). The most commonly found mutations in DNMT3A, TET2, and ASXL1 responsible for M-CH are epigenetic regulators and are loss-of-function mutations. SF3B1, SRSF2, and U2AF1 are splicing factors [55], while TP53 and PPM1D regulate the DNA damage response [56]. Besides these insights, the precise molecular mechanisms regulating clonal expansion, evolution, and disease progression remain ill defined [11]. L-CH and its associated mutations have been reviewed recently by von Beck et al. [9]. For the remainder of this review, we will thus focus on understanding M-CH-related mutations and specifically focus on our mechanistic understanding of DNMT3A-, TET2-, and ASXL1-mutant CH.
Classically, HSC fates are thought to be regulated by cell-intrinsic and -extrinsic mechanisms. Intrinsic mechanisms are the inherent properties of the cell, such as the cell cycle, transcription factors, and epigenetic modification, while extrinsic mechanisms refer to signals from the microenvironment, including growth factors and cell–cell and cell–matrix interactions [57,58]. Although mutant HSCs rely on the same cell-intrinsic and -extrinsic processes as normal HSCs, some of these processes are deregulated, causing mutant HSCs to behave and respond differently to external cues. As these cell-intrinsic and -extrinsic processes are interrelated, it has been difficult to identify the key pathways and the precise order of events, as well as the cause and consequences of clonal expansion. It is thus maybe not surprising that clonal outgrowth appears to be highly context-dependent. Factors driving the expansion of one mutant clone might not affect or even inhibit the relative fitness of another [54]. Environmental factors including infection, inflammation, and chemotherapy have been shown to exert context-dependent effects and clonal outgrowth by increasing HSC fitness through changing survival, proliferation, self-renewal, or differentiation in response to different stressors—factors and mechanisms we will discuss in detail below.
6. Inflammation and Clonal Hematopoiesis
A central role of the hematopoietic system is to fight infections by viruses, bacteria, and other insults that are detected by mature blood cells. When mature blood cells detect a threat, they secrete inflammatory cytokines to alarm neighboring cells to coordinate and amplify the immune response. This response typically involves changes in cellular activity, migration, cell divisions, and differentiation and also affects HSCs [59,60]. Normal HSCs, which are mostly quiescent and do not cycle, become activated, divide, and return to a quiescent state when the infection has passed and inflammatory cytokine levels drop [61]. Chronic inflammation, i.e., a situation when inflammatory cytokine levels do not return to baseline, impairs normal HSC function by inducing proliferative stress and premature exhaustion [62]. As inflammatory cytokine levels increase during aging [63], and CH correlates with age [64], identifying the drivers of inflammation and understanding how HSPCs and more mature blood cells respond to inflammatory cytokines have been of great interest and attracted a lot of attention in recent years.
6.1. Mutant Mature Myeloid Cells Produce Aberrant Levels of Inflammatory Cytokines
Pro-inflammatory cytokines influence normal and mutant HSCs and are thought to promote the selective expansion of clones over time [53]. Increased levels of Interleukin-6 (IL-6), IL-1β, IL-12p70, IFN-γ, IL-4, IL-5, IL-10, TNF-α, and C-reactive protein, a marker for systemic inflammation, were found in the blood of individuals with CH [65,66]. As part of the immune system, terminally differentiated blood cells are a major source of inflammatory cytokines, suggesting that somatic mutations driving selective clonal expansion can also affect the behavior and function of mature cell types [67]. Indeed, macrophages, a class of myeloid cells that possess the ability to promote and restrain inflammation [68], secrete increased levels of inflammatory cytokines and promote the development of cardiovascular disease in Tet2-mutant cells in atherosclerosis-prone, low-density lipoprotein receptor-deficient (Ldlr−/−) mice. In line with this observation, Tet2-mutant macrophages are hypersensitive to low-density lipoprotein (LDL) or bacterial lipopolysaccharide (LPS) and IFN-γ and secrete more IL-1β and IL-6, a process mediated by the NLRP3 inflammasome [69]. The specific deletion of Tet2 in myeloid cells, but not other lineages, in mice further suggests that the altered response of Tet+/− or Tet−/− macrophages to high levels of LDL (i.e., a high-cholesterol diet) is sufficient to increase aortic lesions and to induce atherosclerosis in Ldlr knock-out animals [32]. These insights thus provide a mechanistic link between Tet2-mutant CH and the increased risk of myocardial infarction [32]. Besides Tet2, other CH-associated mutations can also lead to elevated levels of pro-inflammatory cytokines. For instance, Ppm1d-mutant mouse macrophages produce more IL-1β and IL-18 after LPS stimulation in vitro [70], and macrophages in asxl1-, tp53-, and dnmt8 (ortholog of DNMT3A)-mutant zebrafish produce more IL-1β and TNF-α [71]. While these findings suggest that mutant macrophages might be commonly deregulated in CH, evidence for increased inflammatory cytokine production was also found in asxl1- [71], Tet2- [72], and Runx1-mutant neutrophils [73], and Dnmt3a-deficient mast cells [74], which produce more IL-6, TNF-α, and IL-13 [74]. While only a few studies have looked into the role of mutant mast cells in CH, mutations in DNMT3A, TET2, or ASXL1 were shown to affect the prognosis and reduce the overall survival of patients with mastocytosis, a hematological neoplasm with aberrant accumulation of mast cells in multiple organs [75,76]. Together, these studies suggest that elevated levels of pro-inflammatory cytokines found in individuals with CH are produced by the aberrant behavior and function of mature cell types, such as macrophages, neutrophils, and mast cells. Higher levels of pro-inflammatory cytokines produced by mutant mature cells are thus likely create a positive feedback loop to reinforce and/or stabilize an inflammatory environment that allows mutant hematopoietic cells to thrive.
Beyond simply secreting inflammatory cytokines, the cells of the innate immune system can also be in direct contact with HSCs. For instance, during trogocytosis, a quality assurance mechanism, macrophages ‘groom’ HSCs and remove cytoplasmic material from HSCs [77]. Macrophage ‘dooming’ leads to the engulfment and death of damaged HSCs with high levels of reactive oxygen species. The role of trogocytosis in CH is, however, unclear.
6.2. Changes in the Adaptive Immune System
The aberrant behavior of mature blood cells with CH mutations is not restricted to myeloid cells as both the T and B cell function is altered by the loss of Dnmt3a or Tet2 [78]. Studies of Dnmt3a-deficient T cells in mice, for instance, showed that DNMT3A is required to regulate cytokine production. In the absence of DNMT3A, T cells are unable to silence the expression of Il2, Il4, and Infg and produce elevated cytokine levels [79,80,81,82]. Conditional and B cell-specific deletion of Dnmt3a causes chronic lymphocytic leukemia [83] and the immunization of B cells deficient for Dnmt3a and Dnmt3b resulted in the antigen-specific expansion of B cells in germinal centers, plasma cell accumulation, and elevated levels of serum antibodies [84]. Tet2 loss impairs the differentiation of B-2 cells into plasma cells [85] and increases B-1 cell numbers and IgM production in the bone marrow and spleen [86]. Besides these changes in differentiation, the disruption of either Dnmt3a [87] or Tet2 [88] was found to promote the therapeutic efficacy of CAR T cells, suggesting that these mutant CAR T cells do not become exhausted and maintain their capacity to proliferate. This prolonged anti-tumor activity might be leveraged to improve future CAR T cell therapies.
6.3. An Anti-Inflammatory Program Protects Mutant HSPCs
Contrary to mutant mature cells, which upregulate pro-inflammatory programs, recent observations suggest that DNMT3A-, TET2-, and ASXL1-mutant HSCs upregulate anti-inflammatory programs to counteract the effects of inflammation. Although systematic studies on the effects of pro-inflammatory cytokines are lacking, and some of the reported effects of inflammation on mutant HSCs are not consistent, overall, the following model emerges. Inflammation depletes and/or impairs the self-renewal capacity of normal HSCs [62,89], while mutant HSCs evade and/or suppress inflammation to improve their fitness in conditions detrimental to normal HSCs (Figure 4).
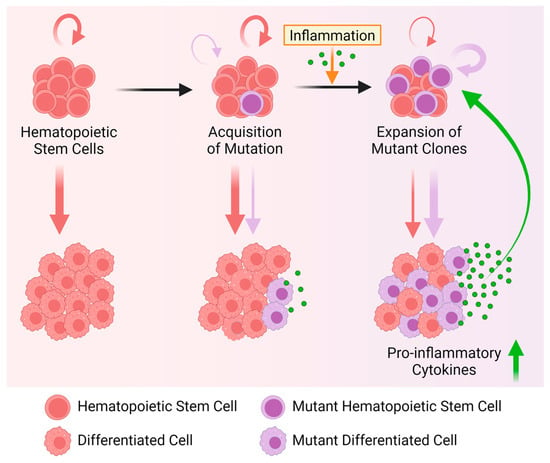
Figure 4.
Inflammation drives clonal expansion of mutant HSCs. Normal HSCs (round red cells) self-renew to maintain the stem cell pool and differentiate to produce mature blood cells (left). Eventually, HSCs acquire mutations. While most mutations remain inconsequential, others provide a fitness advantage (round blue cells) under certain circumstances. In the absence of stress, these mutant HSCs self-renew and differentiate similarly to normal HSCs (middle). When exposed to inflammatory factors, mutant HSCs can outcompete normal HSCs through different cytokine-mediated and context-dependent mechanisms. The following clonal outgrowth of mature mutant blood cells leads to aberrant production of pro-inflammatory cytokines, reinforcing inflammation. As mutant HSCs are more stress-resilient in this pro-inflammatory environment, this selective advantage further promotes clonal expansion and perpetuates the cycle. Created with https://biorender.com.
In a zebrafish model of asxl1 CH, the fitness advantage of mutant HSPCs depends on the upregulation of the negative regulators of inflammation socs3a and nr4a1 [71], as the homozygous deletion of nr4a1 alone was able to prevent clonal outgrowth (Figure 5A). This demonstrates that nr4a1 is required for the competitive advantage of asxl1-mutant HSCs [71]. The upregulation of Socs3 and Nr4a1 was also found in Dnmt3afl-R878H/+ [90] and in VavCre;Tet2fl/fl [91] mutant HSCs in mice, suggesting that the outgrowth of different mutant clones might share mechanisms to improve their fitness, possibly by increasing their resilience to inflammatory stress (Figure 6C and Figure 7C). However, in a mouse model of chronic infection using M. avium, IFN-γ signaling led to the downregulation of Socs3 and Nr4a1 in Dnmt3a−/− HSPCs [92]. At this point, it thus remains unclear whether these discrepancies are caused by cytokine-mediated context-specific effects or if HSCs adapt and execute different programs to boost resilience in response to chronic inflammation mediated by a ‘real’ infection (Figure 6D). While these discrepancies and presumably context-specific effects require more studies, it is well established that normal HSCs need to suppress inflammation and proliferative stress to prevent exhaustion. The ability to suppress inflammation might be limited in normal HSCs while mutant HSCs can suppress inflammation more efficiently and/or for longer. In line with this hypothesis, the loss of negative regulators of inflammation, such as the Activating Transcription Factor 3 (ATF3), leads to HSC exhaustion after 5-fluorouracil (5-FU) treatment of VavCre;Atf3fl/fl mice [93]. The nuclear orphan receptor NR4A1 restricts HSC proliferation and DNA damage and represses inflammation via C/EBPα [94], while the loss of SOCS3, a negative regulator of cytokine signaling, increases inflammation [94]. Importantly, NR4A2 and other anti-inflammatory genes are also upregulated in HSCs isolated from individuals with DNMT3A and TET2 CH [95]. It thus seems plausible that upregulation of ATF3, SOCS3, NR4A1, and other anti-inflammatory genes could be beneficial and protect mutant HSCs. If correct, how precisely do these anti-inflammatory programs protect mutant HSCs? Recent studies, mostly using genetic models of CH in mice, started to shed light on this question and suggest that complex context-dependent but interrelated mechanisms affect HSC survival, proliferation, differentiation, and self-renewal to improve fitness in Asxl1 (Figure 5), Dnmt3a (Figure 6), and Tet2 mutants (Figure 7) [45]. However, how individual inflammatory cytokines affect mutant HSC fates remains poorly understood.
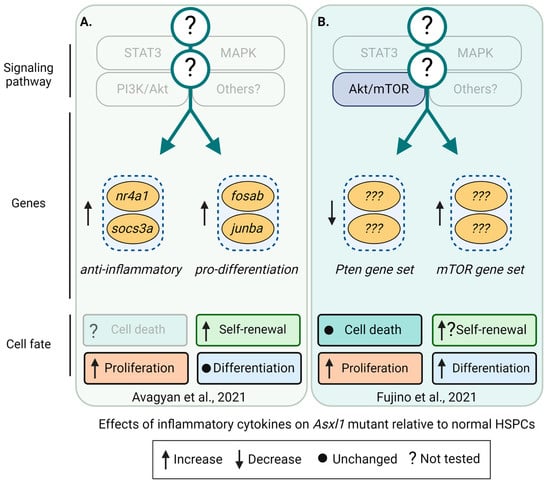
Figure 5.
Differentially regulated pathways, genes, and cell fates in Asxl1-mutant HSPCs. Differentially regulated signaling pathways, genes, and their effects on cell fates, including cell death, self-renewal, proliferation, and differentiation of Asxl1 mutant relative to normal HSPCs. Cell-extrinsic factors driving the clonal expansion of Asxl1 mutants are currently unknown. Transparent boxes and annotations indicate unknown signaling pathways, genes, and cell fates. (A) In a zebrafish model, asxl1-mutant HSPCs exhibit increased proliferation and self-renewal. These cells express higher levels of anti-inflammatory nr4a1 and socs3a and the pro-differentiation genes junba and fosab. Deletion of nr4a1 showed that the fitness advantage of asxl1-mutant clones depends on nr4a1 [71]. (B) Using a mutant Asxl1 knock-in mouse model, clonal outgrowth and increased proliferation of Asxl1-mutant HSPCs were mediated by Akt/mTOR activation. However, transplants impaired Asxl1-mutant HSPC function [96]. Created with https://biorender.com.
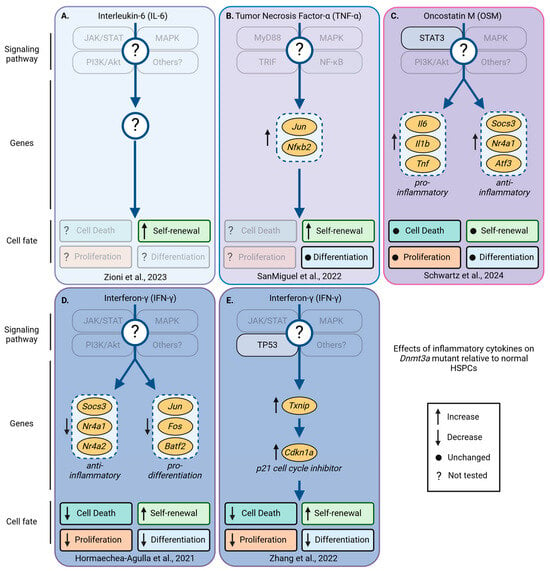
Figure 6.
Pro-inflammatory cytokine-induced changes in signaling, gene expression, and cell fates that occur in Dnmt3a-mutant, but not in wild-type HSPCs. Dnmt3a-mutant and normal HSPCs respond differently to inflammatory cytokines. Depicted are the currently known differentially regulated signaling pathways, genes, and their effects on survival, proliferation, self-renewal, and differentiation of Dnmt3a-mutant relative to normal HSPCs in response to inflammatory cytokines. Several genes, including Socs3, Nr4a1, Fos, Jun, and JunB, are deregulated in Dnmt3a-mutant HSPCs; these genes are sometimes up- or downregulated, suggesting context-dependent effects of pro-inflammatory cytokines. Transparent boxes and labels indicate unknown pathways, genes, and/or HSPC fates not investigated. (A) In fatty bone marrow, elevated levels of IL-6 promote Dnmt3afl-R882H/+ HSPC self-renewal [97]. (B) Knock-out of TNF-α receptor 1 (TNF-αR1), but not TNF-αR2 increases HSPC self-renewal, without changing myeloid vs. lymphoid differentiation. TNF-αR1 signaling induces Jun, Nfkb2, and CD69 more strongly in Dnmt3aR878H/+ than in normal HSPCs [98]. (C) In young Dnmt3aR878H/+ HSPCs, OSM activates STAT3, induces the expression of pro- and anti-inflammatory genes, and inhibits JunB expression. Despite these changes, OSM did not change HSPC fates, including cell death, proliferation, self-renewal, and differentiation [90]. (D,E) In response to IFN-γ, Dnmt3a−/− HSPCs die less and proliferate slower than wild-type HSPCs. In addition, Dnmt3a−/− HSPC self-renewal increased and differentiation decreased compared to wild-type HSPCs exposed to IFN-γ. (D) M. avium infection-mediated IFN-γ induces pro-inflammatory genes and inhibits pro-differentiation genes at the same time [92]. (E) IFN-γ induces TP53 signaling in Dnmt3a−/− HSPCs and inhibits cell cycle progression via Txnip-p21 [99]. Created with https://biorender.com.
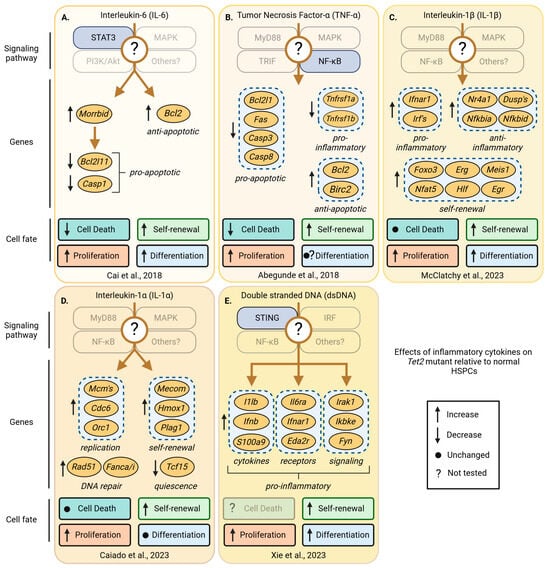
Figure 7.
Pro-inflammatory cytokine-induced changes in signaling, gene expression, and cell fates that occur in Tet2-mutant, but not in wild-type HSPCs. The effects of inflammatory cytokines on Tet2-mutant HSPCs across multiple studies show that changes in signaling, gene expression, and HSPC fates vary between cytokines and relative to normal HSPCs. Shown are known differentially regulated signaling pathways, changes in gene expression, and their effects on Tet2-mutant relative to wild-type HSPC fates including cell death, self-renewal, proliferation, and differentiation after exposure to inflammatory cytokines. Tet2-mutant HSPCs have increased self-renewal and proliferate more compared to wild-type HSPCs in response to inflammatory signals. Transparent boxes and labels indicate unknown effects on signaling pathways, genes, and HSPC fates. (A) IL-6 mediated downregulation of pro- and upregulation of anti-apoptotic genes reduces cell death of Tet+/− and Tet2−/− HSPCs [100]. (B) TNF-α-induced NF-κB signaling reduces cell death in Tet2−/− relative to wild-type HSPCs and correlates with the up-and downregulation of anti- and pro-apoptotic genes, respectively [101]. (C) IL-1β induces the expression of pro- as well as anti-inflammatory genes in Vav-Cre Tet2fl/fl HSPCs [91]. (D) The increase in self-renewal, proliferation, DNA replication, and DNA repair gene expression programs of Tet2+/− HSPCs depends on IL-1α receptor 1-mediated signaling [102]. (E) Studies using Tet2fl/fl;Sting−/− mice show that the cGAS-STING pathway is required for the competitive advantage of Tet2−/− HSPCs. Genes encoding pro-inflammatory cytokines, receptors, and signaling factors depend on STING activation in Tet2fl/fl HSPCs [103]. Created with https://biorender.com.
7. Changes in Normal and Mutant HSC Fate Decisions
While inflammation is advantageous to mutant HSCs (Figure 4), the precise role of HSC fate decisions contributing to clonal expansion remains poorly understood. Given that mutant HSCs are typically present for decades without noticeable outgrowth and/or consequences, the initial changes in HSCs are likely subtle. However, even slight differences in mutant vs. normal HSCs’ survival in response to stress can amplify over many decades and cause large differences in the relative size of clones. Although this process starts slowly, it progressively accelerates over time, very much like earning interest on savings in a bank account. As the yearly interest earned is based on the cumulative earnings of prior years, the largest ‘gains’ are typically obtained many years later in life. Although this model is a helpful analogy to understand how small changes in HSC numbers can lead to large clonal outgrowth many years later, it is important to remember that clonal competition is more complex. The rates of clonal expansion (i.e., the ‘interest rate’) can change over time as the cells adapt to new stresses and can also involve changes in HSC proliferation, self-renewal, and differentiation. In line with these considerations, recent evidence from longitudinal studies shows that clone numbers can shrink and/or remain stable in size for years (Figure 3). But how do mutant clones change these fate decisions to increase their fitness, and can we use these insights to create new therapies?
7.1. Improved Survival of Mutant Clones
The most intuitive way to improve cellular fitness might be increased cell survival. A reduction in cell death can, either under steady-state conditions or stress, lead to a shift in the relative frequencies of clones as the surviving cells can divide again to expand further. Increased cell survival as a mechanism of clonal expansion has been described for PPM1D-mutant clones [104]. In this study, Hsu et al. treated PPM1D-mutant clones in vitro and in vivo with cytotoxic agents and discovered that among others, cisplatin treatment led to a relative expansion of PPM1D-mutant cells. As PPM1D suppresses P53, it is not surprising that PPM1D-mutant clones partially phenocopy p53-mutant HSCs, which outcompete their normal counterparts [105]. Whether changes in survival affect mutant HSCs with other mutations that are not directly linked to p53 is, however, less clear. In a tamoxifen-inducible Tet2+/− and VavCre;Tet2fl/fl model of Tet2 CH, cell death between Tet2-mutant and wild-type HSCs, before and after treatment with IL-1α [102] and IL-1β [91], respectively, was comparable (Figure 7C,D). However, other studies reported lineage-negative Tet2−/− cells died less after TNF-α [101] and IL-6 stimulation [100] (Figure 7A,B). In the latter study, increased levels of IL-6 induced hyperactive SHP2-STAT3 signaling, which resulted in the upregulation of the anti-apoptotic long non-coding RNA morrbid in Tet2-KO myeloid cells and progenitors. In general, these studies suggest that the improved survival of Tet2-mutant HSPCs in response to IL-6 and/or TNF-α is mediated by the increased expression of anti-apoptotic Bcl2 and Birc2, while the pro-apoptotic genes including Bcl2l11, Casp1, Bcl2l1, Fas, Casp3, and Casp8 are downregulated [100]. However, as both studies used Tet2-mutant myeloid cells and their progenitors, whether this mechanism also extends to highly purified HSCs remains unclear. Similar to Tet2, also, Dnmt3a−/−- and Dnmt3aR878/+-mutant HSCs show increased survival in response to inflammatory cytokines. However, contrary to Tet2 mutants, Dnmt3a-mutant HSPC survival is unaltered and comparable to that of normal HSPCs exposed to IL-6, TNF-α, and IFN-α [99]. However, Dnmt3a-mutant HSPCs were more resilient to IFN-γ-mediated stress as suggested by reduced Caspase 3/7 luminescence levels in vitro (Figure 6D,E). Although the survival of Dnmt3a-mutant HSCs in vivo was unaltered, similar to Tet2, Dnmt3a-mutant HSPCs had elevated levels of Bcl2. However, as the Bcl2 levels were already increased in Dnmt3a-mutant cells before inflammatory cytokine stimulation, it is surprising that Bcl2 did not protect against the deleterious effects of other inflammatory cytokines. The interpretation of this these data is not straightforward, and further, more systematic studies using more sensitive tools with single-cell resolution are needed to clarify the role of cell death in response to individual cytokines in mutant HSPC survival.
7.2. Changes in Proliferation of Mutant Clones
Besides changes in survival, mutant HSC clones can also expand by increasing proliferation. This can either be accomplished by shortening the overall cell cycle duration between divisions and/or by increasing the number of divisions before HSCs return to quiescence after entering the cell cycle. Overall, few studies have looked into how the proliferation and/or cell cycle kinetics of highly purified mutant HSCs change [71], and the context-specific changes in response to different cytokines are poorly understood. In line with previous studies, using a Tet2+/− mouse model, Caiado et al. observed the expansion of myeloid-biased multipotent progenitors with a simultaneous reduction in the numbers of downstream Lin−cKIT+ progenitors 14 days post IL-1α injection (Figure 7D). Although fewer Tet+/− HSPCs remained in G0 after the IL-1α injections, the overall frequencies of the HSPCs did not change compared to the wild-type animals treated with IL-1α [102]. Tet2−/− HSPCs isolated from VavCre;Tet2fl/fl mice after IL-1β injection show similar trends, with no to minor changes in HSPCs in G0 and a slight increase in HSPCs in S/G2, suggesting Tet2-mutant HSCs proliferate more [91]. While these studies suggest Tet2-mutant HSPCs proliferate slightly more than their wild-type counterparts, how these seemingly small changes in proliferation account for the observed expansion of immunophenotypic HSCs remains obscure. While increased proliferation might be expected, it is not required for clonal outgrowth. As shown recently by the Challen group, where at least in the context of IFN-γ, highly purified Dnmt3a-mutant HSCs seem to cycle less. Although key insights are still missing, it appears that inflammatory cytokines tend to induce the proliferation of Asxl1- (Figure 5) and Tet2-mutant HSPCs (Figure 7), while the proliferation of Dnmt3a mutants remains unaltered or slows down (Figure 6C–E).
7.3. Self-Renewal and Differentiation
Although a reduction in the proliferation of Dnmt3a-mutant HSCs might seem counterintuitive to accomplish clonal expansion, excessive proliferation is not required when self-renewal increases or differentiation decreases (Figure 8). At first glance, increased self-renewal and decreased differentiation seem to be the same as both lead to clonal expansion. However, the identification of self-renewal and differentiation gene signatures suggests that there are different programs and that HSCs can expand by either increasing self-renewal and/or by reducing differentiation [106]. Evidence from multiple independent groups has firmly established that both Dnmt3a- [107] and Tet2 [108]-mutant mouse HSCs have increased self-renewal and outcompete wild-type HSCs in competitive transplants in vivo. The self-renewal capacity of Dnmt3a-mutant HSCs thereby seems to outcompete HSCs with mutations in Tet2 [109]. The Challen lab illustrated this by showing that Dnmt3aKO HSCs can be serially transplanted at least 12 times without exhaustion [110]. Tet2-deficient HSCs, on the other hand, have been suggested to lose their self-renewal ability and become exhausted after tertiary transplant [109]. Although a detailed mechanistic understanding of how these mutations lead to increased self-renewal and/or differentiation is lacking, a molecular analysis of Dnmt3a- and Tet2-mutant HSCs showed progressive and localized reductions in DNA methylation at crucial regulatory sites linked with self-renewal genes [110]. Compared to Dnmt3a and Tet2, how Asxl1 mutations affect self-renewal and differentiation is less clear. Similar to individuals with CH, Asxl1-mutant mouse HSCs expand with age [96]. However, contrary to Dnmt3a and Tet2 mutants, Asxl1-mutant HSCs do not outcompete wild-type HSCs in competitive transplants [96]. This suggests that transplant data, which are often referred to as the ‘gold standard’ to assess HSC function, should be interpreted carefully, as transplantation might expose mutants and/or wild-type HSCs to stress they would not experience under normal circumstances.
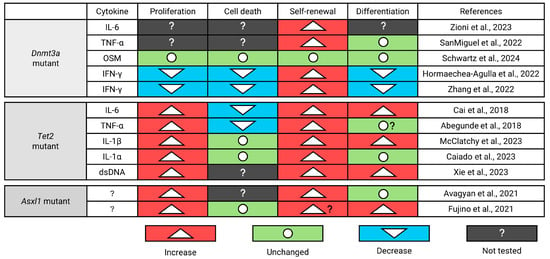
Figure 8.
Summary of known differentially regulated cell fates of Dnmt3a-, Tet2-, and Asxl1-mutant mouse HSPCs in response to pro-inflammatory cytokines [71,90,91,92,96,97,98,99,100,101,102,103,111]. IL-6: Interleukin-6; TNF-α: Tumor Necrosis Factor-α; OSM: Oncostatin M; IFN-γ: Interferon-γ; IL-1β: Interleukin-1β; dsDNA: Double stranded DNA.
Taken together, understanding the mechanisms regulating the survival, proliferation, self-renewal, and differentiation of normal and mutant HSPCs is critical to developing novel therapies, as altering HSPC decisions can slow down and/or prevent clonal expansion and disease progression. As mutant HSPCs seem to be more resilient to inflammatory cytokines than normal HSPCs, reducing inflammation therapeutically should reduce the risk of developing hematological and non-hematological diseases associated with CH (Figure 1). If successful, these therapies might lower the risk of many diseases, including cardiovascular diseases, one of the leading causes of death.
8. DNA Methylation
The DNA methylation of cytosine–guanine dinucleotides, also referred to as “CpG sites”, is crucial for regulating gene expression, chromatin accessibility, and nuclear architecture [112] and is mediated by DNMT1, DNMT3A, and DNMT3B [113]. Changes in DNA methylation are a hallmark of cancer [114] and were found in HSCs harboring CH mutations, including mouse models of Dnmt3a−/− [107], Dnmt3aR878H/+ [115], Tet2−/− [116], Tet2+/− [117], and Asxl1 [118]. These studies showed that DNA methylation is largely maintained throughout the genome in mutant HSCs. However, specific genomic regions are differentially methylated. Based on their overall size and ‘shape’ in the genomic landscape, these regions have been termed CpG islands [119], shores [120], shelves [121], canyons [122], and the more recently described mesas [123]. CpG islands are smaller regions approximately 1000 bp long with little or no DNA methylation and are typically located at promoters, gene bodies, and enhancer regions and make up approximately 1–2% of the mammalian genome [124,125]. CpG shores span less than 2 kb and flank CpG Islands, and CpG shelves span less than 2 kb and flank outwards from CpG shores [122,126]. Approximately 1000 DNA methylation canyons in the genome represent regions with low average methylation levels that are larger than 3.5kb [113], while mesas or promotor exon 1—intron 1 (PrExI) regions are suggested to act as demethylation firing centers [123].
Paradoxically, DNMT3A and TET2, the first and second most frequently mutated genes in CH, respectively, have opposing roles but lead both to increased HSC self-renewal and promoted clonal outgrowth. DNMT3A adds methyl groups, and TET2 demethylates CpGs by adding a hydroxyl group and converting methylcytosine into 5-hydroxymethylcytosine [127]. The detailed mechanism of how site-specific changes in DNA methylation regulate self-renewal remains to be elucidated, but several observations provide clues. For instance, in Dnmt3aKO (Dnmt3a−/−) mouse HSCs, DNA methylation at enhancers and canyons is reduced [128]. As these canyons are enriched for genes associated with HSC self-renewal, including HoxA9, Meis1, and Evi1, reduced DNA methylation and increased expression of self-renewal genes in Dnmt3a−/− HSCs could be linked to changes in HSC function. Contrary to WT HSCs, which cease to engraft after 3–4 rounds of serial transplantation, Dnmt3a−/− HSCs can be transplanted for at least 12 consecutive rounds [110]. Based on these observations, Challen and Goodell [52] proposed that when instructed to differentiate, DNMT3A silences the HSC self-renewal program to allow differentiation. In the absence of DNMT3A or in CH, where DNMT3A function is greatly reduced, HSCs cannot efficiently differentiate. While this model is appealing, many questions remain. How DNMT3A localizes and targets promoters and enhancers associated with HSC-specific genes for DNA methylation and silencing is poorly understood. It also appears that one would expect a more severe phenotype if differentiation was less efficient than proposed. However, even complete Dnmt3aKO HSCs are still able to differentiate and produce mature blood cells. Also, if the lack of DNMT3A increases self-renewal because self-renewal genes cannot be shut down, is the increased fitness of Tet2-mutant HSCs mediated because genes required for effective differentiation cannot be turned on? While the answer to this question is unclear, changes in DNA methylation might provide additional insights [129,130]. Studies using algorithms to analyze changes in the DNA methylation of blood cells during aging have shown that the biological age of individuals can be predicted with high accuracy [131,132]. These algorithms, including Horvath [133], Hannum [134], PhenoAge [135], and others, have been termed DNA methylation clocks and have recently been used on blood samples from individuals with CH and hematological malignancies [136,137]. A study of 154 Danish twins using the Hannum and GrimAge clocks found that the biological age of individuals with CH is accelerated when compared to individuals without mutant clones [138]. Accelerated aging was also found in individuals with mutations in DNMT3A and TET2 by Nachun et al., who studied 5522 individuals [136]. While clinical studies using DNA methylation clocks to assess the biological age of individuals relied on the analysis of bulk blood cells isolated from the peripheral blood, Mick Milsom’s group recently showed for the first time that DNA methylation clocks are also accelerated in highly purified HSCs in a mouse model of chronic inflammation [62]. Similar studies of mutant HSCs might help to decipher how changes in DNA methylation regulate genes required for HSC self-renewal and differentiation.
9. Conclusions
Ten years after the discovery of genetic variants in the peripheral blood cells of healthy elderly individuals, the phenomenon of clonal hematopoiesis, in essence, the expansion of mutant HSC clones, has taken the stem cell field by storm [23,24,29]. The heightened interest is fueled by the opportunity and desire to discover and understand the very origins of hematological diseases, and the hope that these efforts will lead to the development of novel early therapeutic interventions. These research efforts promise nothing less than the possibility to detect and fight potential mutant HSC clones before they progress to a more difficult-to-treat malignant state. After the discovery that CH affects both hematological and non-hematological diseases, including cardiovascular disease and diabetes [139], which, together, claim millions of lives worldwide every year [140], the interest in CH exploded as the eradication of mutant blood clones might help to treat and/or prevent a plethora of diseases (Figure 1). Although we have seen tremendous progress in the last decade, we are still in the early stages of understanding the ramifications of this phenomenon and many mechanisms remain to be discovered.
Despite many open questions, inflammation is emerging as a common theme and driver of clonal expansion. Recent evidence suggests that mutant HSCs are more resistant to the effects of inflammation than normal HSCs [71,92] and that mutant mature cells, including macrophages, neutrophils, mast cells, and others, fuel disease progression by producing more inflammatory cytokines than normal mature cells [141]. Elevated levels of the classical pro-inflammatory factors of IFN-γ, IL-1β, IL-6, and TNF-α have been repeatedly reported in both mouse models [91] and individuals with CH [142], but additional less well-studied factors such as Oncostatin M are involved, too [90]. However, systematic studies that map how mutant HSCs and their offspring differ from normal cells in their response to cytokines are lacking. Why mutant myeloid cells produce more cytokines is incompletely understood, but a cell-intrinsic pro-inflammatory profile and a hyper-responsive phenotype to environmental cues have been proposed [53]. These studies highlight that external factors, including infections [143], smoking [144], and prior exposure to chemotherapy [104], promote CH. The biological consequences of these cues are likely highly context-dependent and will vary depending on the specific mutation and genetic predispositions, as recently described for an inherited polymorphism in the Tcl1a promotor [145]. Despite these insights, whether CH is induced by an existing pro-inflammatory state or creates the pro-inflammatory environment required for clonal expansion remains unclear. As inflammatory cytokines such as IFN-α were suggested to induce asymmetric cell division (ACD) in normal HSCs [146], changes in cell polarity and ACD might play a role in regulating the self-renewal and differentiation of mutant HSCs. ACD is an important and evolutionarily conserved process that regulates metabolic activation and the return to quiescence after HSC activation through the asymmetric mitotic segregation of lysosomes and other organelles [147,148,149,150]. How inflammation affects the cell polarity and division of mutant HSCs remains poorly understood and/or has not been studied, but might provide important clues to understand how mutant HSCs expand. So far, sequencing technologies have been at the forefront of discovery and novel and more sensitive approaches such as error-corrected targeted sequencing [151], deep-targeted sequencing [152], and ultra-sensitive sequencing [34] are now available to detect even small clones with a VAF below 0.01%. Also, computational tools developed to analyze clonal dynamics and the sequential acquisition of mutations over the human lifespan continue to provide new insights [78,79]. While sequencing will continue to shape our understanding of CH and novel rare mutations will be identified in the years to come, the discovery of new mechanistic insights as a basis for novel targeted therapies will require that the field moves beyond ‘simple’ sequencing studies. In the not-so-distant future, hundreds of millions of genomes will have been sequenced. Sequencing a few more genomes will then seem insignificant as we quickly approach a world where sequencing blood samples will be part of annual routine health checks. Emerging single-cell technologies, such as multi-omics approaches that integrate gene expression, epigenetics, and mutations, will continue to shape our understanding of clonal evolution [153]. However, lineage tracing tools to reconstruct clonal dynamics over human lifespans [4,111], and the direct observation of clonal evolution over many weeks and months using novel long-term live cell imaging [148,149], will be crucial moving forward and will likely be the new frontier.
Author Contributions
S.P.: writing—original draft, writing—review and editing. D.L.: conceptualization, funding acquisition, supervision, writing—original draft, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Alex’s Lemonade Stand Foundation and RUNX1 Research Program (Grant # 22-27080) (D.L.) and St. Jude Comprehensive Cancer Center Developmental Funds Grant (#PROJ-0003066 720202211) (D.L.).
Acknowledgments
The authors thank all current members of the Loeffler lab and the Department of Hematology at St. Jude for the helpful discussions and feedback.
Conflicts of Interest
The authors declare no conflicts of interest, financial or otherwise.
References
- Jagannathan-Bogdan, M.; Zon, L.I. Hematopoiesis. Development 2013, 140, 2463–2467. [Google Scholar] [CrossRef] [PubMed]
- de Haan, G.; Lazare, S.S. Aging of hematopoietic stem cells. Blood 2018, 131, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Mitchell, E.; Chapman, M.S.; Williams, N.; Dawson, K.J.; Mende, N.; Calderbank, E.F.; Jung, H.; Mitchell, T.; Coorens, T.H.H.; Spencer, D.H.; et al. Clonal dynamics of haematopoiesis across the human lifespan. Nature 2022, 606, 343–350. [Google Scholar]
- Steensma, D.P.; Bejar, R.; Jaiswal, S.; Lindsley, R.C.; Sekeres, M.A.; Hasserjian, R.P.; Ebert, B.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015, 126, 9–16. [Google Scholar] [PubMed]
- Shlush, L.I. Age-related clonal hematopoiesis. Blood 2018, 131, 496–504. [Google Scholar] [PubMed]
- Trowbridge, J.J.; Starczynowski, D.T. Innate immune pathways and inflammation in hematopoietic aging, clonal hematopoiesis, and MDS. J. Exp. Med. 2021, 218, e20201544. [Google Scholar] [CrossRef]
- Warren, J.T.; Link, D.C. Clonal hematopoiesis and risk for hematologic malignancy. Blood 2020, 136, 1599–1605. [Google Scholar]
- von Beck, K.; von Beck, T.; Ferrell, P.B.; Bick, A.G.; Kishtagari, A. Lymphoid clonal hematopoiesis: Implications for malignancy, immunity, and treatment. Blood Cancer J. 2023, 13, 5. [Google Scholar] [CrossRef]
- Vlasschaert, C.; Lanktree, M.B.; Rauh, M.J.; Kelly, T.N.; Natarajan, P. Clonal haematopoiesis, ageing and kidney disease. Nat. Rev. Nephrol. 2024, 20, 161–174. [Google Scholar] [CrossRef]
- Mendez, L.M.; Patnaik, M.M. Clonal Hematopoiesis: Origins and determinants of evolution. Leuk. Res. 2023, 129, 107076. [Google Scholar] [CrossRef] [PubMed]
- Fabre, M.A.; de Almeida, J.G.; Fiorillo, E.; Mitchell, E.; Damaskou, A.; Rak, J.; Orru, V.; Marongiu, M.; Chapman, M.S.; Vijayabaskar, M.S.; et al. The longitudinal dynamics and natural history of clonal haematopoiesis. Nature 2022, 606, 335–342. [Google Scholar]
- Bolton, K.L.; Koh, Y.; Foote, M.B.; Im, H.; Jee, J.; Sun, C.H.; Safonov, A.; Ptashkin, R.; Moon, J.H.; Lee, J.Y.; et al. Clonal hematopoiesis is associated with risk of severe COVID-19. Nat. Commun. 2021, 12, 5975. [Google Scholar]
- Lyon, M.F. Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 1962, 14, 135–148. [Google Scholar]
- Beutler, E.; Yeh, M.; Fairbanks, V.F. The normal human female as a mosaic of X-chromosome activity: Studies using the gene for C-6-PD-deficiency as a marker. Proc. Natl. Acad. Sci. USA 1962, 48, 9–16. [Google Scholar]
- Linder, D.; Gartler, S.M. Glucose-6-phosphate dehydrogenase mosaicism: Utilization as a cell marker in the study of leiomyomas. Science 1965, 150, 67–69. [Google Scholar]
- Fialkow, P.J.; Gartler, S.M.; Yoshida, A. Clonal origin of chronic myelocytic leukemia in man. Proc. Natl. Acad. Sci. USA 1967, 58, 1468–1471. [Google Scholar]
- Allen, R.C.; Zoghbi, H.Y.; Moseley, A.B.; Rosenblatt, H.M.; Belmont, J.W. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am. J. Hum. Genet. 1992, 51, 1229–1239. [Google Scholar] [PubMed]
- Gale, R.E.; Wheadon, H.; Boulos, P.; Linch, D.C. Tissue specificity of X-chromosome inactivation patterns. Blood 1994, 83, 2899–2905. [Google Scholar] [PubMed]
- Champion, K.M.; Gilbert, J.G.; Asimakopoulos, F.A.; Hinshelwood, S.; Green, A.R. Clonal haemopoiesis in normal elderly women: Implications for the myeloproliferative disorders and myelodysplastic syndromes. Br. J. Haematol. 1997, 97, 920–926. [Google Scholar]
- Fey, M.F.; Liechti-Gallati, S.; von Rohr, A.; Borisch, B.; Theilkäs, L.; Schneider, V.; Oestreicher, M.; Nagel, S.; Ziemiecki, A.; Tobler, A. Clonality and X-inactivation patterns in hematopoietic cell populations detected by the highly informative M27 beta DNA probe. Blood 1994, 83, 931–938. [Google Scholar] [PubMed]
- Ayachi, S.; Buscarlet, M.; Busque, L. 60 Years of clonal hematopoiesis research: From X-chromosome inactivation studies to the identification of driver mutations. Exp. Hematol. 2020, 83, 2–11. [Google Scholar] [PubMed]
- Genovese, G.; Kähler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [PubMed]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [PubMed]
- Min, K.D.; Polizio, A.H.; Kour, A.; Thel, M.C.; Walsh, K. Experimental ASXL1-Mediated Clonal Hematopoiesis Promotes Inflammation and Accelerates Heart Failure. J. Am. Heart Assoc. 2022, 11, e026154. [Google Scholar]
- Jongen-Lavrencic, M.; Grob, T.; Hanekamp, D.; Kavelaars, F.G.; Al Hinai, A.; Zeilemaker, A.; Erpelinck-Verschueren, C.A.J.; Gradowska, P.L.; Meijer, R.; Cloos, J.; et al. Molecular Minimal Residual Disease in Acute Myeloid Leukemia. N. Engl. J. Med. 2018, 378, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Pløen, G.G.; Nederby, L.; Guldberg, P.; Hansen, M.; Ebbesen, L.H.; Jensen, U.B.; Hokland, P.; Aggerholm, A. Persistence of DNMT3A mutations at long-term remission in adult patients with AML. Br. J. Haematol. 2014, 167, 478–486. [Google Scholar] [PubMed]
- Niroula, A.; Sekar, A.; Murakami, M.A.; Trinder, M.; Agrawal, M.; Wong, W.J.; Bick, A.G.; Uddin, M.M.; Gibson, C.J.; Griffin, G.K.; et al. Distinction of lymphoid and myeloid clonal hematopoiesis. Nat. Med. 2021, 27, 1921–1927. [Google Scholar] [PubMed]
- Xie, M.; Lu, C.; Wang, J.; McLellan, M.D.; Johnson, K.J.; Wendl, M.C.; McMichael, J.F.; Schmidt, H.K.; Yellapantula, V.; Miller, C.A.; et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014, 20, 1472–1478. [Google Scholar] [CrossRef]
- Young, A.L.; Challen, G.A.; Birmann, B.M.; Druley, T.E. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun. 2016, 7, 12484. [Google Scholar]
- Miller, P.G.; Qiao, D.; Rojas-Quintero, J.; Honigberg, M.C.; Sperling, A.S.; Gibson, C.J.; Bick, A.G.; Niroula, A.; McConkey, M.E.; Sandoval, B.; et al. Association of clonal hematopoiesis with chronic obstructive pulmonary disease. Blood 2022, 139, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Natarajan, P.; Silver, A.J.; Gibson, C.J.; Bick, A.G.; Shvartz, E.; McConkey, M.; Gupta, N.; Gabriel, S.; Ardissino, D.; et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 2017, 377, 111–121. [Google Scholar] [PubMed]
- Acuna-Hidalgo, R.; Sengul, H.; Steehouwer, M.; van de Vorst, M.; Vermeulen, S.H.; Kiemeney, L.; Veltman, J.A.; Gilissen, C.; Hoischen, A. Ultra-sensitive Sequencing Identifies High Prevalence of Clonal Hematopoiesis-Associated Mutations throughout Adult Life. Am. J. Hum. Genet. 2017, 101, 50–64. [Google Scholar] [PubMed]
- Uddin, M.M.; Zhou, Y.; Bick, A.G.; Burugula, B.B.; Jaiswal, S.; Desai, P.; Honigberg, M.C.; Love, S.A.; Barac, A.; Hayden, K.M.; et al. Longitudinal profiling of clonal hematopoiesis provides insight into clonal dynamics. Immun. Ageing 2022, 19, 23. [Google Scholar] [PubMed]
- Robertson, N.A.; Latorre-Crespo, E.; Terradas-Terradas, M.; Lemos-Portela, J.; Purcell, A.C.; Livesey, B.J.; Hillary, R.F.; Murphy, L.; Fawkes, A.; MacGillivray, L.; et al. Longitudinal dynamics of clonal hematopoiesis identifies gene-specific fitness effects. Nat. Med. 2022, 28, 1439–1446. [Google Scholar] [PubMed]
- Andersen, M.A.; Bjerrum, O.W.; Ranjan, A.; Skov, V.; Kruse, T.A.; Thomassen, M.; Skytthe, A.; Hasselbalch, H.C.; Christensen, K. Myeloproliferative Neoplasms in Danish Twins. Acta Haematol. 2018, 139, 195–198. [Google Scholar] [PubMed]
- Kohnke, T.; Majeti, R. Clonal Hematopoiesis: From Mechanisms to Clinical Intervention. Cancer Discov. 2021, 11, 2987–2997. [Google Scholar] [PubMed]
- Lee-Six, H.; Øbro, N.F.; Shepherd, M.S.; Grossmann, S.; Dawson, K.; Belmonte, M.; Osborne, R.J.; Huntly, B.J.P.; Martincorena, I.; Anderson, E.; et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 2018, 561, 473–478. [Google Scholar]
- Calvanese, V.; Capellera-Garcia, S.; Ma, F.; Fares, I.; Liebscher, S.; Ng, E.S.; Ekstrand, S.; Aguadé-Gorgorió, J.; Vavilina, A.; Lefaudeux, D.; et al. Mapping human haematopoietic stem cells from haemogenic endothelium to birth. Nature 2022, 604, 534–540. [Google Scholar]
- Williams, N.; Lee, J.; Mitchell, E.; Moore, L.; Baxter, E.J.; Hewinson, J.; Dawson, K.J.; Menzies, A.; Godfrey, A.L.; Green, A.R.; et al. Life histories of myeloproliferative neoplasms inferred from phylogenies. Nature 2022, 602, 162–168. [Google Scholar]
- Fabre, M.A.; McKerrell, T.; Zwiebel, M.; Vijayabaskar, M.S.; Park, N.; Wells, P.M.; Rad, R.; Deloukas, P.; Small, K.; Steves, C.J.; et al. Concordance for clonal hematopoiesis is limited in elderly twins. Blood 2020, 135, 269–273. [Google Scholar]
- Behjati, S.; Huch, M.; van Boxtel, R.; Karthaus, W.; Wedge, D.C.; Tamuri, A.U.; Martincorena, I.; Petljak, M.; Alexandrov, L.B.; Gundem, G.; et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature 2014, 513, 422–425. [Google Scholar] [PubMed]
- Welch, J.S.; Ley, T.J.; Link, D.C.; Miller, C.A.; Larson, D.E.; Koboldt, D.C.; Wartman, L.D.; Lamprecht, T.L.; Liu, F.; Xia, J.; et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012, 150, 264–278. [Google Scholar] [PubMed]
- Tovy, A.; Reyes, J.M.; Gundry, M.C.; Brunetti, L.; Lee-Six, H.; Petljak, M.; Park, H.J.; Guzman, A.G.; Rosas, C.; Jeffries, A.R.; et al. Tissue-Biased Expansion of DNMT3A-Mutant Clones in a Mosaic Individual Is Associated with Conserved Epigenetic Erosion. Cell Stem Cell 2020, 27, 326–335.e4. [Google Scholar] [PubMed]
- Waldvogel, S.M.; Posey, J.E.; Goodell, M.A. Human embryonic genetic mosaicism and its effects on development and disease. Nat. Rev. Genet. 2024. ahead of print. [Google Scholar]
- Dykstra, B.; Kent, D.; Bowie, M.; McCaffrey, L.; Hamilton, M.; Lyons, K.; Lee, S.J.; Brinkman, R.; Eaves, C. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell 2007, 1, 218–229. [Google Scholar]
- Sieburg, H.B.; Cho, R.H.; Dykstra, B.; Uchida, N.; Eaves, C.J.; Muller-Sieburg, C.E. The hematopoietic stem compartment consists of a limited number of discrete stem cell subsets. Blood 2006, 107, 2311–2316. [Google Scholar]
- Jurecic, R. Hematopoietic Stem Cell Heterogeneity. Adv. Exp. Med. Biol. 2019, 1169, 195–211. [Google Scholar]
- Challen, G.A.; Boles, N.C.; Chambers, S.M.; Goodell, M.A. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell 2010, 6, 265–278. [Google Scholar]
- Zink, F.; Stacey, S.N.; Norddahl, G.L.; Frigge, M.L.; Magnusson, O.T.; Jonsdottir, I.; Thorgeirsson, T.E.; Sigurdsson, A.; Gudjonsson, S.A.; Gudmundsson, J.; et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 2017, 130, 742–752. [Google Scholar]
- Bowman, R.L.; Busque, L.; Levine, R.L. Clonal Hematopoiesis and Evolution to Hematopoietic Malignancies. Cell Stem Cell 2018, 22, 157–170. [Google Scholar] [PubMed]
- Challen, G.A.; Goodell, M.A. Clonal hematopoiesis: Mechanisms driving dominance of stem cell clones. Blood 2020, 136, 1590–1598. [Google Scholar] [PubMed]
- Avagyan, S.; Zon, L.I. Clonal hematopoiesis and inflammation—The perpetual cycle. Trends Cell Biol. 2023, 33, 695–707. [Google Scholar] [PubMed]
- Florez, M.A.; Tran, B.T.; Wathan, T.K.; DeGregori, J.; Pietras, E.M.; King, K.Y. Clonal hematopoiesis: Mutation-specific adaptation to environmental change. Cell Stem Cell 2022, 29, 882–904. [Google Scholar] [PubMed]
- Chen, S.; Benbarche, S.; Abdel-Wahab, O. Splicing factor mutations in hematologic malignancies. Blood 2021, 138, 599–612. [Google Scholar]
- Chen, C.W.; Zhang, L.; Dutta, R.; Niroula, A.; Miller, P.G.; Gibson, C.J.; Bick, A.G.; Reyes, J.M.; Lee, Y.T.; Tovy, A.; et al. SRCAP mutations drive clonal hematopoiesis through epigenetic and DNA repair dysregulation. Cell Stem Cell 2023, 30, 1503–1519.e8. [Google Scholar] [PubMed]
- Lin, S.; Zhao, R.; Xiao, Y.; Li, P. Mechanisms determining the fate of hematopoietic stem cells. Stem Cell Investig. 2015, 2, 10. [Google Scholar] [PubMed]
- Lee, Y.; Decker, M.; Lee, H.; Ding, L. Extrinsic regulation of hematopoietic stem cells in development, homeostasis and diseases. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e279. [Google Scholar] [CrossRef]
- Baldridge, M.T.; King, K.Y.; Boles, N.C.; Weksberg, D.C.; Goodell, M.A. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature 2010, 465, 793–797. [Google Scholar]
- Essers, M.A.; Offner, S.; Blanco-Bose, W.E.; Waibler, Z.; Kalinke, U.; Duchosal, M.A.; Trumpp, A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 2009, 458, 904–908. [Google Scholar]
- Wilson, A.; Laurenti, E.; Oser, G.; van der Wath, R.C.; Blanco-Bose, W.; Jaworski, M.; Offner, S.; Dunant, C.F.; Eshkind, L.; Bockamp, E.; et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 2008, 135, 1118–1129. [Google Scholar] [CrossRef]
- Bogeska, R.; Mikecin, A.M.; Kaschutnig, P.; Fawaz, M.; Büchler-Schäff, M.; Le, D.; Ganuza, M.; Vollmer, A.; Paffenholz, S.V.; Asada, N.; et al. Inflammatory exposure drives long-lived impairment of hematopoietic stem cell self-renewal activity and accelerated aging. Cell Stem Cell 2022, 29, 1273–1284.e8. [Google Scholar] [PubMed]
- Michaud, M.; Balardy, L.; Moulis, G.; Gaudin, C.; Peyrot, C.; Vellas, B.; Cesari, M.; Nourhashemi, F. Proinflammatory cytokines, aging, and age-related diseases. J. Am. Med. Dir. Assoc. 2013, 14, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, W.; Yi, L.; Zhao, M.; Li, P.-Y.; Fu, M.-H.; Lin, R.; Zhu, Y.-M.; Li, J.-F.; Yang, W.-P.; et al. The impact of age and number of mutations on the size of clonal hematopoiesis. Proc. Natl. Acad. Sci. USA 2024, 121, e2319364121. [Google Scholar]
- Bick, A.G.; Weinstock, J.S.; Nandakumar, S.K.; Fulco, C.P.; Bao, E.L.; Zekavat, S.M.; Szeto, M.D.; Liao, X.; Leventhal, M.J.; Nasser, J.; et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 2020, 586, 763–768. [Google Scholar]
- Busque, L.; Sun, M.; Buscarlet, M.; Ayachi, S.; Zada, Y.F.; Provost, S.; Bourgoin, V.; Mollica, L.; Meisel, M.; Hinterleitner, R.; et al. High-sensitivity C-reactive protein is associated with clonal hematopoiesis of indeterminate potential. Blood Adv. 2020, 4, 2430–2438. [Google Scholar] [PubMed]
- Weeks, L.D.; Ebert, B.L. Causes and consequences of clonal hematopoiesis. Blood 2023, 142, 2235–2246. [Google Scholar] [PubMed]
- Seyfried, A.N.; Maloney, J.M.; MacNamara, K.C. Macrophages Orchestrate Hematopoietic Programs and Regulate HSC Function during Inflammatory Stress. Front. Immunol. 2020, 11, 1499. [Google Scholar]
- Fuster, J.J.; MacLauchlan, S.; Zuriaga, M.A.; Polackal, M.N.; Ostriker, A.C.; Chakraborty, R.; Wu, C.L.; Sano, S.; Muralidharan, S.; Rius, C.; et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 2017, 355, 842–847. [Google Scholar] [CrossRef]
- Yura, Y.; Miura-Yura, E.; Katanasaka, Y.; Min, K.D.; Chavkin, N.; Polizio, A.H.; Ogawa, H.; Horitani, K.; Doviak, H.; Evans, M.A.; et al. The Cancer Therapy-Related Clonal Hematopoiesis Driver Gene Ppm1d Promotes Inflammation and Non-Ischemic Heart Failure in Mice. Circ. Res. 2021, 129, 684–698. [Google Scholar] [CrossRef]
- Avagyan, S.; Henninger, J.E.; Mannherz, W.P.; Mistry, M.; Yoon, J.; Yang, S.; Weber, M.C.; Moore, J.L.; Zon, L.I. Resistance to inflammation underlies enhanced fitness in clonal hematopoiesis. Science 2021, 374, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Quin, C.; DeJong, E.N.; Cook, E.K.; Luo, Y.Z.; Vlasschaert, C.; Sadh, S.; McNaughton, A.J.; Buttigieg, M.M.; Breznik, J.A.; Kennedy, A.E.; et al. Neutrophil-mediated innate immune resistance to bacterial pneumonia is dependent on Tet2 function. J. Clin. Investig. 2024, 134, e171002. [Google Scholar] [CrossRef]
- Bellissimo, D.C.; Chen, C.H.; Zhu, Q.; Bagga, S.; Lee, C.T.; He, B.; Wertheim, G.B.; Jordan, M.; Tan, K.; Worthen, G.S.; et al. Runx1 negatively regulates inflammatory cytokine production by neutrophils in response to Toll-like receptor signaling. Blood Adv. 2020, 4, 1145–1158. [Google Scholar]
- Leoni, C.; Montagner, S.; Rinaldi, A.; Bertoni, F.; Polletti, S.; Balestrieri, C.; Monticelli, S. Dnmt3a restrains mast cell inflammatory responses. Proc. Natl. Acad. Sci. USA 2017, 114, E1490–E1499. [Google Scholar] [CrossRef]
- Reszka, E.; Jabłońska, E.; Wieczorek, E.; Valent, P.; Arock, M.; Nilsson, G.; Nedoszytko, B.; Niedoszytko, M. Epigenetic Changes in Neoplastic Mast Cells and Potential Impact in Mastocytosis. Int. J. Mol. Sci. 2021, 22, 2964. [Google Scholar] [CrossRef] [PubMed]
- Traina, F.; Visconte, V.; Jankowska, A.M.; Makishima, H.; O’Keefe, C.L.; Elson, P.; Han, Y.; Hsieh, F.H.; Sekeres, M.A.; Mali, R.S.; et al. Single nucleotide polymorphism array lesions, TET2, DNMT3A, ASXL1 and CBL mutations are present in systemic mastocytosis. PLoS ONE 2012, 7, e43090. [Google Scholar] [CrossRef]
- Wattrus, S.J.; Smith, M.L.; Rodrigues, C.P.; Hagedorn, E.J.; Kim, J.W.; Budnik, B.; Zon, L.I. Quality assurance of hematopoietic stem cells by macrophages determines stem cell clonality. Science 2022, 377, 1413–1419. [Google Scholar] [CrossRef]
- Asada, S.; Kitamura, T. Clonal hematopoiesis and associated diseases: A review of recent findings. Cancer Sci. 2021, 112, 3962–3971. [Google Scholar] [CrossRef] [PubMed]
- Kersh, E.N.; Fitzpatrick, D.R.; Murali-Krishna, K.; Shires, J.; Speck, S.H.; Boss, J.M.; Ahmed, R. Rapid demethylation of the IFN-gamma gene occurs in memory but not naive CD8 T cells. J. Immunol. 2006, 176, 4083–4093. [Google Scholar] [CrossRef]
- Makar, K.W.; Pérez-Melgosa, M.; Shnyreva, M.; Weaver, W.M.; Fitzpatrick, D.R.; Wilson, C.B. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat. Immunol. 2003, 4, 1183–1190. [Google Scholar]
- Bruniquel, D.; Schwartz, R.H. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat. Immunol. 2003, 4, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Leoni, C.; Vincenzetti, L.; Emming, S.; Monticelli, S. Epigenetics of T lymphocytes in health and disease. Swiss Med. Wkly. 2015, 145, w14191. [Google Scholar] [CrossRef] [PubMed]
- Biran, A.; Yin, S.; Kretzmer, H.; Hacken, E.T.; Parvin, S.; Lucas, F.; Uduman, M.; Gutierrez, C.; Dangle, N.; Billington, L.; et al. Activation of Notch and Myc Signaling via B-cell-Restricted Depletion of Dnmt3a Generates a Consistent Murine Model of Chronic Lymphocytic Leukemia. Cancer Res. 2021, 81, 6117–6130. [Google Scholar] [PubMed]
- Barwick, B.G.; Scharer, C.D.; Martinez, R.J.; Price, M.J.; Wein, A.N.; Haines, R.R.; Bally, A.P.R.; Kohlmeier, J.E.; Boss, J.M. B cell activation and plasma cell differentiation are inhibited by de novo DNA methylation. Nat. Commun. 2018, 9, 1900. [Google Scholar] [PubMed]
- Dominguez, P.M.; Ghamlouch, H.; Rosikiewicz, W.; Kumar, P.; Béguelin, W.; Fontán, L.; Rivas, M.A.; Pawlikowska, P.; Armand, M.; Mouly, E.; et al. TET2 Deficiency Causes Germinal Center Hyperplasia, Impairs Plasma Cell Differentiation, and Promotes B-cell Lymphomagenesis. Cancer Discov. 2018, 8, 1632–1653. [Google Scholar] [PubMed]
- Dennis, E.; Murach, M.; Blackburn, C.M.R.; Marshall, M.; Root, K.; Pattarabanjird, T.; Deroissart, J.; Erickson, L.D.; Binder, C.J.; Bekiranov, S.; et al. Loss of TET2 increases B-1 cell number and IgM production while limiting CDR3 diversity. Front. Immunol. 2024, 15, 1380641. [Google Scholar] [CrossRef] [PubMed]
- Prinzing, B.; Zebley, C.C.; Petersen, C.T.; Fan, Y.; Anido, A.A.; Yi, Z.; Nguyen, P.; Houke, H.; Bell, M.; Haydar, D.; et al. Deleting DNMT3A in CAR T cells prevents exhaustion and enhances antitumor activity. Sci. Transl. Med. 2021, 13, eabh0272. [Google Scholar] [PubMed]
- Fraietta, J.A.; Nobles, C.L.; Sammons, M.A.; Lundh, S.; Carty, S.A.; Reich, T.J.; Cogdill, A.P.; Morrissette, J.J.D.; DeNizio, J.E.; Reddy, S.; et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 2018, 558, 307–312. [Google Scholar]
- Matatall, K.A.; Jeong, M.; Chen, S.; Sun, D.; Chen, F.; Mo, Q.; Kimmel, M.; King, K.Y. Chronic Infection Depletes Hematopoietic Stem Cells through Stress-Induced Terminal Differentiation. Cell Rep. 2016, 17, 2584–2595. [Google Scholar]
- Schwartz, L.S.; Young, K.A.; Stearns, T.M.; Boyer, N.; Mujica, K.D.; Trowbridge, J.J. Transcriptional and functional consequences of Oncostatin M signaling on young Dnmt3a-mutant hematopoietic stem cells. Exp. Hematol. 2024, 130, 104131. [Google Scholar] [CrossRef]
- McClatchy, J.; Strogantsev, R.; Wolfe, E.; Lin, H.Y.; Mohammadhosseini, M.; Davis, B.A.; Eden, C.; Goldman, D.; Fleming, W.H.; Conley, P.; et al. Clonal hematopoiesis related TET2 loss-of-function impedes IL1β-mediated epigenetic reprogramming in hematopoietic stem and progenitor cells. Nat. Commun. 2023, 14, 8102. [Google Scholar] [PubMed]
- Hormaechea-Agulla, D.; Matatall, K.A.; Le, D.T.; Kain, B.; Long, X.; Kus, P.; Jaksik, R.; Challen, G.A.; Kimmel, M.; King, K.Y. Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNγ signaling. Cell Stem Cell 2021, 28, 1428–1442.e6. [Google Scholar] [PubMed]
- Liu, Y.; Chen, Y.; Deng, X.; Zhou, J. ATF3 Prevents Stress-Induced Hematopoietic Stem Cell Exhaustion. Front. Cell Dev. Biol. 2020, 8, 585771. [Google Scholar] [CrossRef] [PubMed]
- Ushiki, T.; Huntington, N.D.; Glaser, S.P.; Kiu, H.; Georgiou, A.; Zhang, J.G.; Metcalf, D.; Nicola, N.A.; Roberts, A.W.; Alexander, W.S. Rapid Inflammation in Mice Lacking Both SOCS1 and SOCS3 in Hematopoietic Cells. PLoS ONE 2016, 11, e0162111. [Google Scholar]
- Jakobsen, N.A.; Turkalj, S.; Zeng, A.G.X.; Stoilova, B.; Metzner, M.; Rahmig, S.; Nagree, M.S.; Shah, S.; Moore, R.; Usukhbayar, B.; et al. Selective advantage of mutant stem cells in human clonal hematopoiesis is associated with attenuated response to inflammation and aging. Cell Stem Cell 2024. ahead of print. [Google Scholar]
- Fujino, T.; Goyama, S.; Sugiura, Y.; Inoue, D.; Asada, S.; Yamasaki, S.; Matsumoto, A.; Yamaguchi, K.; Isobe, Y.; Tsuchiya, A.; et al. Mutant ASXL1 induces age-related expansion of phenotypic hematopoietic stem cells through activation of Akt/mTOR pathway. Nat. Commun. 2021, 12, 1826. [Google Scholar] [PubMed]
- Zioni, N.; Bercovich, A.A.; Chapal-Ilani, N.; Bacharach, T.; Rappoport, N.; Solomon, A.; Avraham, R.; Kopitman, E.; Porat, Z.; Sacma, M.; et al. Inflammatory signals from fatty bone marrow support DNMT3A driven clonal hematopoiesis. Nat. Commun. 2023, 14, 2070. [Google Scholar] [CrossRef]
- San Miguel, J.M.; Eudy, E.; Loberg, M.A.; Young, K.A.; Mistry, J.J.; Mujica, K.D.; Schwartz, L.S.; Stearns, T.M.; Challen, G.A.; Trowbridge, J.J. Distinct Tumor Necrosis Factor Alpha Receptors Dictate Stem Cell Fitness versus Lineage Output in Dnmt3a-Mutant Clonal Hematopoiesis. Cancer Discov. 2022, 12, 2763–2773. [Google Scholar]
- Zhang, C.R.; Ostrander, E.L.; Kukhar, O.; Mallaney, C.; Sun, J.; Haussler, E.; Celik, H.; Koh, W.K.; King, K.Y.; Gontarz, P.; et al. Txnip Enhances Fitness of Dnmt3a-Mutant Hematopoietic Stem Cells via p21. Blood Cancer Discov. 2022, 3, 220–239. [Google Scholar] [CrossRef]
- Cai, Z.; Kotzin, J.J.; Ramdas, B.; Chen, S.; Nelanuthala, S.; Palam, L.R.; Pandey, R.; Mali, R.S.; Liu, Y.; Kelley, M.R.; et al. Inhibition of Inflammatory Signaling in Tet2 Mutant Preleukemic Cells Mitigates Stress-Induced Abnormalities and Clonal Hematopoiesis. Cell Stem Cell 2018, 23, 833–849.e5. [Google Scholar]
- Abegunde, S.O.; Buckstein, R.; Wells, R.A.; Rauh, M.J. An inflammatory environment containing TNFalpha favors Tet2-mutant clonal hematopoiesis. Exp. Hematol. 2018, 59, 60–65. [Google Scholar]
- Caiado, F.; Kovtonyuk, L.V.; Gonullu, N.G.; Fullin, J.; Boettcher, S.; Manz, M.G. Aging drives Tet2+/− clonal hematopoiesis via IL-1 signaling. Blood 2023, 141, 886–903. [Google Scholar] [PubMed]
- Xie, J.; Sheng, M.; Rong, S.; Zhou, D.; Wang, C.; Wu, W.; Huang, J.; Sun, Y.; Wang, Y.; Chen, P.; et al. STING activation in TET2-mutated hematopoietic stem/progenitor cells contributes to the increased self-renewal and neoplastic transformation. Leukemia 2023, 37, 2457–2467. [Google Scholar]
- Hsu, J.I.; Dayaram, T.; Tovy, A.; De Braekeleer, E.; Jeong, M.; Wang, F.; Zhang, J.; Heffernan, T.P.; Gera, S.; Kovacs, J.J.; et al. PPM1D Mutations Drive Clonal Hematopoiesis in Response to Cytotoxic Chemotherapy. Cell Stem Cell 2018, 23, 700–713.e6. [Google Scholar] [CrossRef]
- Bondar, T.; Medzhitov, R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell 2010, 6, 309–322. [Google Scholar] [CrossRef]
- Kowalczyk, M.S.; Tirosh, I.; Heckl, D.; Rao, T.N.; Dixit, A.; Haas, B.J.; Schneider, R.K.; Wagers, A.J.; Ebert, B.L.; Regev, A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015, 25, 1860–1872. [Google Scholar]
- Challen, G.A.; Sun, D.; Jeong, M.; Luo, M.; Jelinek, J.; Berg, J.S.; Bock, C.; Vasanthakumar, A.; Gu, H.; Xi, Y.; et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2011, 44, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Moran-Crusio, K.; Reavie, L.; Shih, A.; Abdel-Wahab, O.; Ndiaye-Lobry, D.; Lobry, C.; Figueroa, M.E.; Vasanthakumar, A.; Patel, J.; Zhao, X.; et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011, 20, 11–24. [Google Scholar] [PubMed]
- Ostrander, E.L.; Kramer, A.C.; Mallaney, C.; Celik, H.; Koh, W.K.; Fairchild, J.; Haussler, E.; Zhang, C.R.C.; Challen, G.A. Divergent Effects of Dnmt3a and Tet2 Mutations on Hematopoietic Progenitor Cell Fitness. Stem Cell Rep. 2020, 14, 551–560. [Google Scholar]
- Jeong, M.; Park, H.J.; Celik, H.; Ostrander, E.L.; Reyes, J.M.; Guzman, A.; Rodriguez, B.; Lei, Y.; Lee, Y.; Ding, L.; et al. Loss of Dnmt3a Immortalizes Hematopoietic Stem Cells In Vivo. Cell Rep. 2018, 23, 1–10. [Google Scholar]
- Ludwig, L.S.; Lareau, C.A.; Ulirsch, J.C.; Christian, E.; Muus, C.; Li, L.H.; Pelka, K.; Ge, W.; Oren, Y.; Brack, A.; et al. Lineage Tracing in Humans Enabled by Mitochondrial Mutations and Single-Cell Genomics. Cell 2019, 176, 1325–1339.e22. [Google Scholar] [PubMed]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [PubMed]
- Wiehle, L.; Raddatz, G.; Musch, T.; Dawlaty, M.M.; Jaenisch, R.; Lyko, F.; Breiling, A. Tet1 and Tet2 Protect DNA Methylation Canyons against Hypermethylation. Mol. Cell Biol. 2016, 36, 452–461. [Google Scholar] [PubMed]
- Momparler, R.L.; Bovenzi, V. DNA methylation and cancer. J. Cell Physiol. 2000, 183, 145–154. [Google Scholar] [PubMed]
- Liao, M.; Chen, R.; Yang, Y.; He, H.; Xu, L.; Jiang, Y.; Guo, Z.; He, W.; Jiang, H.; Wang, J. Aging-elevated inflammation promotes DNMT3A R878H-driven clonal hematopoiesis. Acta Pharm. Sin. B 2022, 12, 678–691. [Google Scholar]
- Wang, Y.; Sano, S.; Yura, Y.; Ke, Z.; Sano, M.; Oshima, K.; Ogawa, H.; Horitani, K.; Min, K.D.; Miura-Yura, E.; et al. Tet2-mediated clonal hematopoiesis in nonconditioned mice accelerates age-associated cardiac dysfunction. JCI Insight 2020, 5, e135204. [Google Scholar] [PubMed]
- Li, Z.; Cai, X.; Cai, C.L.; Wang, J.; Zhang, W.; Petersen, B.E.; Yang, F.C.; Xu, M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 2011, 118, 4509–4518. [Google Scholar] [PubMed]
- Nagase, R.; Inoue, D.; Pastore, A.; Fujino, T.; Hou, H.A.; Yamasaki, N.; Goyama, S.; Saika, M.; Kanai, A.; Sera, Y.; et al. Expression of mutant Asxl1 perturbs hematopoiesis and promotes susceptibility to leukemic transformation. J. Exp. Med. 2018, 215, 1729–1747. [Google Scholar]
- Stadler, M.B.; Murr, R.; Burger, L.; Ivanek, R.; Lienert, F.; Schöler, A.; van Nimwegen, E.; Wirbelauer, C.; Oakeley, E.J.; Gaidatzis, D.; et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011, 480, 490–495. [Google Scholar] [CrossRef]
- Mitsumori, R.; Sakaguchi, K.; Shigemizu, D.; Mori, T.; Akiyama, S.; Ozaki, K.; Niida, S.; Shimoda, N. Lower DNA methylation levels in CpG island shores of CR1, CLU, and PICALM in the blood of Japanese Alzheimer’s disease patients. PLoS ONE 2020, 15, e0239196. [Google Scholar] [CrossRef]
- Edgar, R.; Tan, P.P.; Portales-Casamar, E.; Pavlidis, P. Meta-analysis of human methylomes reveals stably methylated sequences surrounding CpG islands associated with high gene expression. Epigenetics Chromatin 2014, 7, 28. [Google Scholar] [PubMed]
- Jeong, M.; Sun, D.; Luo, M.; Huang, Y.; Challen, G.A.; Rodriguez, B.; Zhang, X.; Chavez, L.; Wang, H.; Hannah, R.; et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat. Genet. 2014, 46, 17–23. [Google Scholar]
- Liu, Y.V.; Bassal, M.A.; Jayasinghe, M.K.; Lin, Q.X.X.; Wu, C.-S.; Tang, J.P.; Kwon, J.; Zhou, Q.; Tan, H.K.; Ebralidze, A.K.; et al. Targeted intragenic demethylation initiates chromatin rewiring for gene activation. bioRxiv 2024. bioRxiv:2020.07.16.205922. [Google Scholar]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes. Dev. 2011, 25, 1010–1022. [Google Scholar] [PubMed]
- Strichman-Almashanu, L.Z.; Lee, R.S.; Onyango, P.O.; Perlman, E.; Flam, F.; Frieman, M.B.; Feinberg, A.P. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 2002, 12, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Trino, S.; Zoppoli, P.; Carella, A.M.; Laurenzana, I.; Weisz, A.; Memoli, D.; Calice, G.; La Rocca, F.; Simeon, V.; Savino, L.; et al. DNA methylation dynamic of bone marrow hematopoietic stem cells after allogeneic transplantation. Stem Cell Res. Ther. 2019, 10, 138. [Google Scholar] [PubMed]
- Cypris, O.; Božić, T.; Wagner, W. Chicken or Egg: Is Clonal Hematopoiesis Primarily Caused by Genetic or Epigenetic Aberrations? Front. Genet. 2019, 10, 785. [Google Scholar]
- Zhang, X.; Su, J.; Jeong, M.; Ko, M.; Huang, Y.; Park, H.J.; Guzman, A.; Lei, Y.; Huang, Y.H.; Rao, A.; et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat. Genet. 2016, 48, 1014–1023. [Google Scholar]
- Bergstedt, J.; Azzou, S.A.K.; Tsuo, K.; Jaquaniello, A.; Urrutia, A.; Rotival, M.; Lin, D.T.S.; MacIsaac, J.L.; Kobor, M.S.; Albert, M.L.; et al. The immune factors driving DNA methylation variation in human blood. Nat. Commun. 2022, 13, 5895. [Google Scholar]
- Refn, M.R.; Andersen, M.M.; Kampmann, M.-L.; Tfelt-Hansen, J.; Sørensen, E.; Larsen, M.H.; Morling, N.; Børsting, C.; Pereira, V. Longitudinal changes and variation in human DNA methylation analysed with the Illumina MethylationEPIC BeadChip assay and their implications on forensic age prediction. Sci. Rep. 2023, 13, 21658. [Google Scholar]
- Aanes, H.; Bleka, Ø.; Dahlberg, P.S.; Carm, K.T.; Lehtimäki, T.; Raitakari, O.; Kähönen, M.; Hurme, M.; Rolseth, V. A new blood based epigenetic age predictor for adolescents and young adults. Sci. Rep. 2023, 13, 2303. [Google Scholar] [PubMed]
- Salameh, Y.; Bejaoui, Y.; El Hajj, N. DNA Methylation Biomarkers in Aging and Age-Related Diseases. Front. Genet. 2020, 11, 171. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.H.; Marioni, R.E.; Colicino, E.; Peters, M.J.; Ward-Caviness, C.K.; Tsai, P.C.; Roetker, N.S.; Just, A.C.; Demerath, E.W.; Guan, W.; et al. DNA methylation-based measures of biological age: Meta-analysis predicting time to death. Aging 2016, 8, 1844–1865. [Google Scholar] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Bergsma, T.; Rogaeva, E. DNA Methylation Clocks and Their Predictive Capacity for Aging Phenotypes and Healthspan. Neurosci. Insights 2020, 15, 2633105520942221. [Google Scholar] [PubMed]
- Nachun, D.; Lu, A.T.; Bick, A.G.; Natarajan, P.; Weinstock, J.; Szeto, M.D.; Kathiresan, S.; Abecasis, G.; Taylor, K.D.; Guo, X.; et al. Clonal hematopoiesis associated with epigenetic aging and clinical outcomes. Aging Cell 2021, 20, e13366. [Google Scholar] [CrossRef] [PubMed]
- Feldkamp, J.D.; Vetter, V.M.; Arends, C.M.; Lang, T.J.L.; Bullinger, L.; Damm, F.; Demuth, I.; Frick, M. Clonal hematopoiesis of indeterminate potential-related epigenetic age acceleration correlates with clonal hematopoiesis of indeterminate potential clone size in patients with high morbidity. Haematologica 2022, 107, 1703–1708. [Google Scholar] [CrossRef] [PubMed]
- Soerensen, M.; Tulstrup, M.; Hansen, J.W.; Weischenfeldt, J.; Grønbæk, K.; Christensen, K. Clonal Hematopoiesis and Epigenetic Age Acceleration in Elderly Danish Twins. HemaSphere 2022, 6, e768. [Google Scholar] [PubMed]
- Lin, A.E.; Rauch, P.J.; Jaiswal, S.; Ebert, B.L. Clonal Hematopoiesis: Confluence of Malignant and Nonmalignant Diseases. Annu. Rev. Cancer Biol. 2022, 6, 187–200. [Google Scholar] [CrossRef]
- Di Cesare, M.; Perel, P.; Taylor, S.; Kabudula, C.; Bixby, H.; Gaziano, T.A.; McGhie, D.V.; Mwangi, J.; Pervan, B.; Narula, J.; et al. The Heart of the World. Glob. Heart 2024, 19, 11. [Google Scholar]
- Deng, J.; Fleming, J.B. Inflammation and Myeloid Cells in Cancer Progression and Metastasis. Front. Cell Dev. Biol. 2021, 9, 759691. [Google Scholar]
- Zhang, C.R.C.; Nix, D.; Gregory, M.; Ciorba, M.A.; Ostrander, E.L.; Newberry, R.D.; Spencer, D.H.; Challen, G.A. Inflammatory cytokines promote clonal hematopoiesis with specific mutations in ulcerative colitis patients. Exp. Hematol. 2019, 80, 36–41.e3. [Google Scholar]
- Shen, Q.; Zhang, Q.; Shi, Y.; Shi, Q.; Jiang, Y.; Gu, Y.; Li, Z.; Li, X.; Zhao, K.; Wang, C.; et al. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature 2018, 554, 123–127. [Google Scholar]
- Levin, M.G.; Nakao, T.; Zekavat, S.M.; Koyama, S.; Bick, A.G.; Niroula, A.; Ebert, B.; Damrauer, S.M.; Natarajan, P. Genetics of smoking and risk of clonal hematopoiesis. Sci. Rep. 2022, 12, 7248. [Google Scholar]
- Weinstock, J.S.; Gopakumar, J.; Burugula, B.B.; Uddin, M.M.; Jahn, N.; Belk, J.A.; Bouzid, H.; Daniel, B.; Miao, Z.; Ly, N.; et al. Aberrant activation of TCL1A promotes stem cell expansion in clonal haematopoiesis. Nature 2023, 616, 755–763. [Google Scholar] [CrossRef]
- Girotra, M.; Trachsel, V.; Roch, A.; Lutolf, M.P. In Vivo Pre-Instructed HSCs Robustly Execute Asymmetric Cell Divisions In Vitro. Int. J. Mol. Sci. 2020, 21, 8225. [Google Scholar] [CrossRef]
- Wehling, A.; Loeffler, D.; Zhang, Y.; Kull, T.; Donato, C.; Szczerba, B.; Ortega, G.C.; Lee, M.; Moor, A.; Göttgens, B.; et al. Combining single-cell tracking and omics improves blood stem cell fate regulator identification. Blood 2022, 140, 1482–1495. [Google Scholar]
- Loeffler, D.; Schneiter, F.; Wang, W.; Wehling, A.; Kull, T.; Lengerke, C.; Manz, M.G.; Schroeder, T. Asymmetric organelle inheritance predicts human blood stem cell fate. Blood 2022, 139, 2011–2023. [Google Scholar] [CrossRef]
- Loeffler, D.; Wehling, A.; Schneiter, F.; Zhang, Y.; Müller-Bötticher, N.; Hoppe, P.S.; Hilsenbeck, O.; Kokkaliaris, K.D.; Endele, M.; Schroeder, T. Asymmetric lysosome inheritance predicts activation of haematopoietic stem cells. Nature 2019, 573, 426–429. [Google Scholar]
- Nunes, J.; Loeffler, D. Asymmetric cell division of hematopoietic stem cells: Recent advances, emerging concepts, and future perspectives. Front. Hematol. 2024, 3, 1373554. [Google Scholar] [CrossRef]
- Marchetti, F.; Cardoso, R.; Chen, C.L.; Douglas, G.R.; Elloway, J.; Escobar, P.A.; Harper, T.; Heflich, R.H.; Kidd, D.; Lynch, A.M.; et al. Error-corrected next generation sequencing—Promises and challenges for genotoxicity and cancer risk assessment. Mutat. Res. Rev. Mutat. Res. 2023, 792, 108466. [Google Scholar] [PubMed]
- Chan, I.C.C.; Panchot, A.; Schmidt, E.; McNulty, S.; Wiley, B.J.; Liu, J.; Turner, K.; Moukarzel, L.; Wong, W.S.W.; Tran, D.; et al. ArCH: Improving the performance of clonal hematopoiesis variant calling and interpretation. Bioinformatics 2024, 40, btae121. [Google Scholar]
- Chen, C.; Wang, J.; Pan, D.; Wang, X.; Xu, Y.; Yan, J.; Wang, L.; Yang, X.; Yang, M.; Liu, G.P. Applications of multi-omics analysis in human diseases. MedComm 2023, 4, e315. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).