Extracellular Vesicles, Circadian Rhythms, and Cancer: A Comprehensive Review with Emphasis on Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Exosome Biogenesis
3. The Role of Exosomes/ECVs in Oncogenesis
3.1. Hepatic-Cell-Derived ECVs: Cargo and Functions
3.2. Exosomes in HCC Initiation, Progression, Metastasis, and Angiogenesis
3.3. ECVs and Tumor Microenvironment
3.4. Diagnostic Biomarkers in HCC
3.5. ECVs in Immunotherapy and Therapy Resistance
4. The Molecular Basis of Circadian Rhythms in Mammals
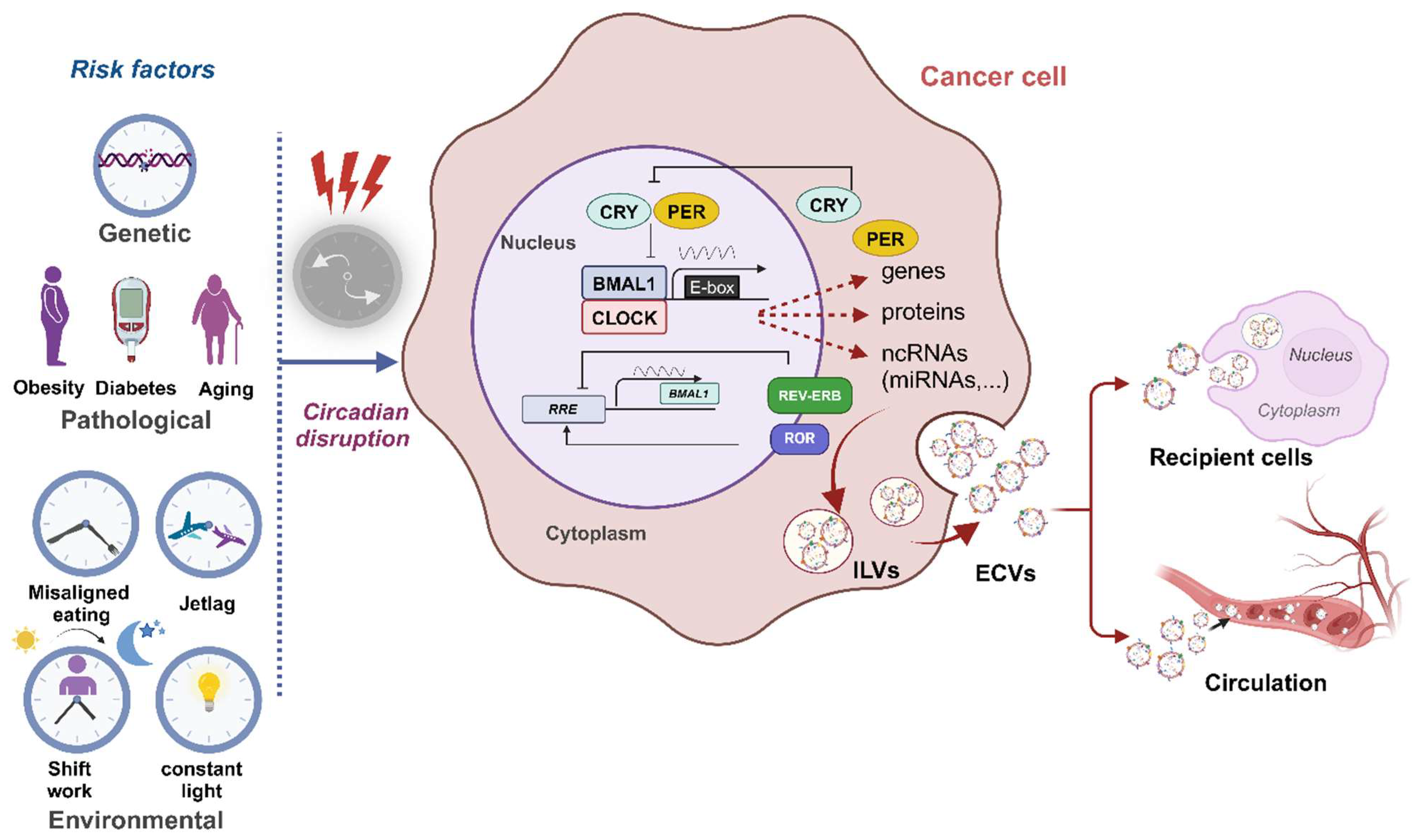
4.1. Circadian Regulation of ECVs
4.2. Circadian Clock and HCC
4.3. The Impact of the Circadian Clock on Cancer Progression via ECVs
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.E.; Lehtiö, J.; EL Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef]
- Dhar, R.; Gorai, S.; Devi, A.; Muthusamy, R.; Alexiou, A.; Papadakis, M. Decoding of exosome heterogeneity for cancer theranostics. Clin. Transl. Med. 2023, 13, e1288. [Google Scholar] [CrossRef]
- Almeria, C.; Kreß, S.; Weber, V.; Egger, D.; Kasper, C. Heterogeneity of mesenchymal stem cell-derived extracellular vesicles is highly impacted by the tissue/cell source and culture conditions. Cell Biosci. 2022, 12, 51. [Google Scholar] [CrossRef]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef]
- Zhu, L.; Sun, H.-T.; Wang, S.; Huang, S.-L.; Zheng, Y.; Wang, C.-Q.; Hu, B.-Y.; Qin, W.; Zou, T.-T.; Fu, Y.; et al. Isolation and characterization of exosomes for cancer research. J. Hematol. Oncol. 2020, 13, 152. [Google Scholar] [CrossRef]
- Huda, M.N.; Nafiujjaman, M.; Deaguero, I.G.; Okonkwo, J.; Hill, M.L.; Kim, T.; Nurunnabi, M. Potential Use of Exosomes as Diagnostic Biomarkers and in Targeted Drug Delivery: Progress in Clinical and Preclinical Applications. ACS Biomater. Sci. Eng. 2021, 7, 2106–2149. [Google Scholar] [CrossRef]
- Kojima, S.; Shingle, D.L.; Green, C.B. Post-transcriptional control of circadian rhythms. J. Cell Sci. 2011, 124 Pt 3, 311–320. [Google Scholar] [CrossRef]
- Guo, J.; Cheng, P.; Yuan, H.; Liu, Y. The Exosome Regulates Circadian Gene Expression in a Posttranscriptional Negative Feedback Loop. Cell 2009, 138, 1236–1246. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Y.; Xu, X.; Gu, J.; Chen, H.; Gui, Y. Exosomes rich in Wnt5 improved circadian rhythm dysfunction via enhanced PPARγ activity in the 6-hydroxydopamine model of Parkinson’s disease. Neurosci. Lett. 2023, 802, 137139. [Google Scholar] [CrossRef] [PubMed]
- Hahn, K.; Sundar, I.K. Current Perspective on the Role of the Circadian Clock and Extracellular Matrix in Chronic Lung Diseases. Int. J. Environ. Res. Public Health 2023, 20, 2455. [Google Scholar] [CrossRef]
- Chen, W.-H.; Huang, Q.-Y.; Wang, Z.-Y.; Zhuang, X.-X.; Lin, S.; Shi, Q.-Y. Therapeutic potential of exosomes/miRNAs in polycystic ovary syndrome induced by the alteration of circadian rhythms. Front. Endocrinol. 2022, 13, 918805. [Google Scholar] [CrossRef]
- Sulli, G.; Lam, M.T.Y.; Panda, S. Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment. Trends Cancer 2019, 5, 475–494. [Google Scholar] [CrossRef]
- Altman, B.J.; Hsieh, A.L.; Sengupta, A.; Krishnanaiah, S.Y.; Stine, Z.E.; Walton, Z.E.; Gouw, A.M.; Venkataraman, A.; Li, B.; Goraksha-Hicks, P.; et al. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metab. 2015, 22, 1009–1019. [Google Scholar] [CrossRef]
- Fekry, B.; Eckel-Mahan, K. The circadian clock and cancer: Links between circadian disruption and disease Pathology. J. Biochem. 2022, 171, 477–486. [Google Scholar] [CrossRef]
- Verlande, A.; Masri, S. Circadian Clocks and Cancer: Timekeeping Governs Cellular Metabolism. Trends Endocrinol. Metab. 2019, 30, 445–458. [Google Scholar] [CrossRef]
- Fekry, B.; Ribas-Latre, A.; Baumgartner, C.; Mohamed, A.M.T.; Kolonin, M.G.; Sladek, F.M.; Younes, M.; Eckel-Mahan, K.L. HNF4α-Deficient Fatty Liver Provides a Permissive Environment for Sex-Independent Hepatocellular Carcinoma. Cancer Res. 2019, 79, 5860–5873. [Google Scholar] [CrossRef]
- Dong, P.; Wang, Y.; Liu, Y.; Zhu, C.; Lin, J.; Qian, R.; Hua, L.; Lu, C. BMAL1 induces colorectal cancer metastasis by stimulating exosome secretion. Mol. Biol. Rep. 2022, 49, 373–384. [Google Scholar] [CrossRef]
- Ortega-Campos, S.M.; Verdugo-Sivianes, E.M.; Amiama-Roig, A.; Blanco, J.R.; Carnero, A. Interactions of circadian clock genes with the hallmarks of cancer. Biochim. Biophys. Acta BBA Rev. Cancer 2023, 1878, 188900. [Google Scholar] [CrossRef]
- Duffy, M.J. Tumor markers in clinical practice: A review focusing on common solid cancers. Med. Princ. Pr. 2012, 22, 4–11. [Google Scholar] [CrossRef]
- Locker, G.Y.; Hamilton, S.; Harris, J.; Jessup, J.M.; Kemeny, N.; Macdonald, J.S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol. 2006, 24, 5313–5327. [Google Scholar] [CrossRef]
- He, C.-Z.; Zhang, K.-H.; Li, Q.; Liu, X.-H.; Hong, Y.; Lv, N.-H. Combined use of AFP, CEA, CA125 and CAl9-9 improves the sensitivity for the diagnosis of gastric cancer. BMC Gastroenterol. 2013, 13, 87. [Google Scholar] [CrossRef]
- Gupta, D.; Lis, C.G. Role of CA125 in predicting ovarian cancer survival—A review of the epidemiological literature. J. Ovarian Res. 2009, 2, 13. [Google Scholar] [CrossRef]
- Wong, C.H.; Chen, Y.C. Clinical significance of exosomes as potential biomarkers in cancer. World J. Clin. Cases 2019, 7, 171–190. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Krishnan, A.; Bhattacharya, B.; Mandal, D.; Dhar, R.; Muthu, S. Salivary exosomes: A theranostics secret of oral cancer—Correspondence. Int. J. Surg. 2022, 108, 106990. [Google Scholar] [CrossRef]
- Wang, X.; Tian, L.; Lu, J.; Ng, I.O. Exosomes and cancer—Diagnostic and prognostic biomarkers and therapeutic vehicle. Oncogenesis 2022, 11, 54. [Google Scholar] [CrossRef]
- Makler, A.; Asghar, W. Exosomal biomarkers for cancer diagnosis and patient monitoring. Expert. Rev. Mol. Diagn. 2020, 20, 387–400. [Google Scholar] [CrossRef]
- Hanjani, N.A.; Esmaelizad, N.; Zanganeh, S.; Gharavi, A.T.; Heidarizadeh, P.; Radfar, M.; Omidi, F.; MacLoughlin, R.; Doroudian, M. Emerging role of exosomes as biomarkers in cancer treatment and diagnosis. Crit. Rev. Oncol. Hematol. 2022, 169, 103565. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Anwanwan, D.; Singh, S.K.; Singh, S.; Saikam, V.; Singh, R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta BBA Rev. Cancer 2020, 1873, 188314. [Google Scholar] [CrossRef]
- National Comprehensive Cancer Nwtwork. NCCN Clinical Practice Guidelines in Oncology: Hepatobiliary Cancers; Version 1.2023; National Comprehensive Cancer Network: Plymouth Meeting, PA, USA, 2023; Available online: https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1438.2023 (accessed on 10 June 2024).
- Singal, A.G.; Llovet, J.M.; Yarchoan, M.; Mehta, N.; Heimbach, J.K.; Dawson, L.A.; Jou, J.H.; Kulik, L.M.; Agopian, V.G.; Marrero, J.A.; et al. AASLD Practice Guidance on prevention, diagnosis, and treatment of hepatocellular carcinoma. Hepatology 2023, 78, 1922–1965. [Google Scholar] [CrossRef]
- Choi, D.T.; Kum, H.C.; Park, S.; Ohsfeldt, R.L.; Shen, Y.; Parikh, N.D.; Singal, A.G. Hepatocellular Carcinoma Screening Is Associated with Increased Survival of Patients with Cirrhosis. Clin. Gastroenterol. Hepatol. 2019, 17, 976–987.e4. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- Lok, A.S.; Sterling, R.K.; Everhart, J.E.; Wright, E.C.; Hoefs, J.C.; Di Bisceglie, A.M.; Morgan, T.R.; Kim, H.; Lee, W.M.; Bonkovsky, H.L.; et al. Des-γ-carboxy prothrombin and α-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology 2010, 138, 493–502. [Google Scholar] [CrossRef]
- Pinyol, R.; Montal, R.; Bassaganyas, L.; Sia, D.; Takayama, T.; Chau, G.-Y.; Mazzaferro, V.; Roayaie, S.; Lee, H.C.; Kokudo, N.; et al. Molecular predictors of prevention of recurrence in HCC with sorafenib as adjuvant treatment and prognostic factors in the phase 3 STORM trial. Gut 2019, 68, 1065–1075. [Google Scholar] [CrossRef]
- Parikh, N.D.; Mehta, A.S.; Singal, A.G.; Block, T.; Marrero, J.A.; Lok, A.S. Biomarkers for the Early Detection of Hepatocellular Carcinoma. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2495–2503. [Google Scholar] [CrossRef]
- Chen, W.; Mao, Y.; Liu, C.; Wu, H.; Chen, S. Exosome in Hepatocellular Carcinoma: An update. J. Cancer 2021, 12, 2526–2536. [Google Scholar] [CrossRef]
- Szabo, G.; Momen-Heravi, F. Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 455–466. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Edwards, K.A.; Leete, J.J.; Smith, E.G.; Quick, A.; Modica, C.M.; Wassermann, E.M.; Polejaeva, E.; Dell, K.C.; LoPresti, M.; Walker, P.; et al. Elevations in Tumor Necrosis Factor Alpha and Interleukin 6 from Neuronal-Derived Extracellular Vesicles in Repeated Low-Level Blast Exposed Personnel. Front. Neurol. 2022, 13, 723923. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Sheta, M.; Taha, E.A.; Lu, Y.; Eguchi, T. Extracellular Vesicles: New Classification and Tumor Immunosuppression. Biology 2023, 12, 110. [Google Scholar] [CrossRef]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Paskeh, M.D.A.; Entezari, M.; Mirzaei, S.; Zabolian, A.; Saleki, H.; Naghdi, M.J.; Sabet, S.; Khoshbakht, M.A.; Hashemi, M.; Hushmandi, K.; et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J. Hematol. Oncol. 2022, 15, 83. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Wang, X.; Huang, J.; Chen, W.; Li, G.; Li, Z.; Lei, J. The updated role of exosomal proteins in the diagnosis, prognosis, and treatment of cancer. Exp. Mol. Med. 2022, 54, 1390–1400. [Google Scholar] [CrossRef]
- Smolarz, M.; Pietrowska, M.; Matysiak, N.; Mielańczyk, Ł.; Widłak, P. Proteome profiling of exosomes purified from a small amount of human serum: The problem of co-purified serum components. Proteomes 2019, 7, 18. [Google Scholar] [CrossRef]
- Liang, Y.; Lehrich, B.M.; Zheng, S.; Lu, M. Emerging methods in biomarker identification for extracellular vesicle-based liquid biopsy. J. Extracell. Vesicles 2021, 10, e12090. [Google Scholar] [CrossRef]
- Wang, N.; Song, X.; Liu, L.; Niu, L.; Wang, X.; Song, X.; Xie, L. Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. 2018, 109, 1701–1709. [Google Scholar] [CrossRef]
- Mazurov, D.; Barbashova, L.; Filatov, A. Tetraspanin protein CD 9 interacts with metalloprotease CD 10 and enhances its release via exosomes. FEBS J. 2013, 280, 1200–1213. [Google Scholar] [CrossRef]
- Malla, R.R.; Pandrangi, S.; Kumari, S.; Gavara, M.M.; Badana, A.K. Exosomal tetraspanins as regulators of cancer progression and metastasis and novel diagnostic markers. Asia Pac. J. Clin. Oncol. 2018, 14, 383–391. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- Nolte-’t Hoen, E.N.; Buermans, H.P.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.; ’t Hoen, P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Ge, L.; Zhang, N.; Li, D.; Wu, Y.; Wang, H.; Wang, J. Circulating exosomal small RNAs are promising non-invasive diagnostic biomarkers for gastric cancer. J. Cell Mol. Med. 2020, 24, 14502–14513. [Google Scholar] [CrossRef]
- Li, X.; Chen, R.; Kemper, S.; Brigstock, D.R. Dynamic changes in function and proteomic composition of extracellular vesicles from hepatic stellate cells during cellular activation. Cells 2020, 9, 290. [Google Scholar] [CrossRef]
- Malhi, H. Emerging role of extracellular vesicles in liver diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G739–G749. [Google Scholar] [CrossRef]
- Kostallari, E.; Hirsova, P.; Prasnicka, A.; Verma, V.K.; Yaqoob, U.; Wongjarupong, N.; Roberts, L.R.; Shah, V.H. Hepatic stellate cell–derived platelet-derived growth factor receptor-alpha-enriched extracellular vesicles promote liver fibrosis in mice through SHP2. Hepatology 2018, 68, 333–348. [Google Scholar] [CrossRef]
- Dasgupta, D.; Nakao, Y.; Mauer, A.S.; Thompson, J.M.; Sehrawat, T.S.; Liao, C.-Y.; Krishnan, A.; Lucien, F.; Guo, Q.; Liu, M.; et al. IRE1A stimulates hepatocyte-derived extracellular vesicles that promote inflammation in mice with steatohepatitis. Gastroenterology 2020, 159, 1487–1503.e17. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef]
- Osaki, M.; Okada, F. Exosomes and their role in cancer progression. Yonago Acta Medica 2019, 62, 182–190. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, H.; Li, P.; Chen, Y.; Zhang, M.; Yuan, Z.; Zhang, Y.; Xu, Z.; Luo, G.; Fang, Y.; et al. Exosomes: A New Pathway for Cancer Drug Resistance. Front. Oncol. 2021, 11, 743556. [Google Scholar] [CrossRef]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, D.; Guan, S.; Dong, M. Diagnostic role of circulating MiR-21 in colorectal cancer: A update meta-analysis. Ann. Med. 2021, 53, 87–102. [Google Scholar] [CrossRef]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef]
- Thind, A.; Wilson, C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 2016, 5, 31292. [Google Scholar] [CrossRef] [PubMed]
- Eichelser, C.; Stückrath, I.; Müller, V.; Milde-Langosch, K.; Wikman, H.; Pantel, K.; Schwarzenbach, H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014, 5, 9650–9663. [Google Scholar] [CrossRef] [PubMed]
- Que, R.; Ding, G.; Chen, J.; Cao, L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J. Surg. Oncol. 2013, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.; Chang, P.; LeBlanc, A.; Li, D.; Abbruzzesse, J.L.; Frazier, M.L.; Killary, A.M.; Sen, S. MicroRNAs in Plasma of Pancreatic Ductal Adenocarcinoma Patients as Novel Blood-Based Biomarkers of Disease. Cancer Prev. Res. 2009, 2, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.-S.; Liu, X.-B.; Wong, B.Y.-H.; Ng, R.W.-M.; Yuen, A.P.-W.; Wei, W.I. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin. Cancer Res. 2008, 14, 2588–2592. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kosaka, N.; Tanaka, M.; Koizumi, F.; Kanai, Y.; Mizutani, T.; Murakami, Y.; Kuroda, M.; Miyajima, A.; Kato, T.; et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers 2009, 14, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Dong, W.; Zhang, H.; Zhao, H.; Zeng, Z.; Zhang, F.; Li, Q.; Duan, X.; Hu, Y.; Xiao, W. Exosomal microRNA panel as a diagnostic biomarker in patients with hepatocellular carcinoma. Front. Cell Dev. Biol. 2022, 10, 927251. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Wu, J.; Wu, J.; Ji, A.; Qiang, G.; Jiang, Y.; Jiang, C.; Ding, Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget 2017, 8, 80666. [Google Scholar] [CrossRef]
- Sohn, W.; Kim, J.; Kang, S.H.; Yang, S.R.; Cho, J.-Y.; Cho, H.C.; Shim, S.G.; Paik, Y.-H. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp. Mol. Med. 2015, 47, e184. [Google Scholar] [CrossRef]
- Tomimaru, Y.; Eguchi, H.; Nagano, H.; Wada, H.; Kobayashi, S.; Marubashi, S.; Tanemura, M.; Tomokuni, A.; Takemasa, I.; Umeshita, K.; et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J. Hepatol. 2012, 56, 167–175. [Google Scholar] [CrossRef]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, Characterization, and Clinical Utility for Oral Cancer Detection. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.-F.; Gao, C.; Huang, X.-Y.; Lu, J.-C.; Guo, X.-J.; Shi, G.-M.; Cai, J.-B.; Ke, A.-W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 2020, 19, 110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.-F.; Wei, C.-Y.; Huang, X.-Y.; Peng, R.; Yang, X.; Lu, J.-C.; Zhang, C.; Gao, C.; Cai, J.-B.; Gao, P.-T.; et al. Circular RNA circTRIM33–12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol. Cancer 2019, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.-Y.; Huang, Z.-L.; Huang, J.; Xu, B.; Huang, X.-Y.; Xu, Y.-H.; Zhou, J.; Tang, Z.-Y. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 20. [Google Scholar] [CrossRef]
- Chen, W.; Quan, Y.; Fan, S.; Wang, H.; Liang, J.; Huang, L.; Chen, L.; Liu, Q.; He, P.; Ye, Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020, 475, 119–128. [Google Scholar] [CrossRef]
- Xian, J.; Su, W.; Liu, L.; Rao, B.; Lin, M.; Feng, Y.; Qiu, F.; Chen, J.; Zhou, Q.; Zhao, Z.; et al. Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of non–small-cell lung cancer in the Chinese population. J. Mol. Diagn. 2020, 22, 1096–1108. [Google Scholar] [CrossRef]
- Kang, Y.; You, J.; Gan, Y.; Chen, Q.; Huang, C.; Chen, F.; Xu, X.; Chen, L. Serum and serum exosomal CircRNAs hsa_circ_0001492, hsa_circ_0001439, and hsa_circ_0000896 as diagnostic biomarkers for lung adenocarcinoma. Front. Oncol. 2022, 12, 912246. [Google Scholar] [CrossRef]
- Luo, Y.; Ma, J.; Liu, F.; Guo, J.; Gui, R. Diagnostic value of exosomal circMYC in radioresistant nasopharyngeal carcinoma. Head. Neck 2020, 42, 3702–3711. [Google Scholar] [CrossRef]
- Xia, D.; Gu, X. Plasmatic exosome-derived circRNAs panel act as fingerprint for glioblastoma. Aging 2021, 13, 19575–19586. [Google Scholar] [CrossRef]
- He, Y.-D.; Tao, W.; He, T.; Wang, B.-Y.; Tang, X.-M.; Zhang, L.-M.; Wu, Z.-Q.; Deng, W.-M.; Zhang, L.-X.; Shao, C.-K.; et al. A urine extracellular vesicle circRNA classifier for detection of high-grade prostate cancer in patients with prostate-specific antigen 2–10 ng/mL at initial biopsy. Mol. Cancer 2021, 20, 96. [Google Scholar] [CrossRef] [PubMed]
- Royo, F.; Moreno, L.; Mleczko, J.; Palomo, L.; Gonzalez, E.; Cabrera, D.; Cogolludo, A.; Vizcaino, F.P.; Van-Liempd, S.; Falcon-Perez, J.M. Hepatocyte-secreted extracellular vesicles modify blood metabolome and endothelial function by an arginase-dependent mechanism. Sci. Rep. 2017, 7, 42798. [Google Scholar] [CrossRef] [PubMed]
- Nojima, H.; Freeman, C.M.; Schuster, R.M.; Japtok, L.; Kleuser, B.; Edwards, M.J.; Gulbins, E.; Lentsch, A.B. Hepatocyte exosomes mediate liver repair and regeneration via sphingosine-1-phosphate. J. Hepatol. 2016, 64, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Ding, Q.; Yaqoob, U.; de Assuncao, T.M.; Verma, V.K.; Hirsova, P.; Cao, S.; Mukhopadhyay, D.; Huebert, R.C.; Shah, V.H. Exosome adherence and internalization by hepatic stellate cells triggers sphingosine 1-phosphate-dependent migration. J. Biol. Chem. 2015, 290, 30684–30696. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Charrier, A.; Zhou, Y.; Chen, R.; Yu, B.; Agarwal, K.; Tsukamoto, H.; Lee, L.J.; Paulaitis, M.E.; Brigstock, D.R. Epigenetic regulation of connective tissue growth factor by MicroRNA-214 delivery in exosomes from mouse or human hepatic stellate cells. Hepatology 2014, 59, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, R.; Velazquez, V.M.; Brigstock, D.R. Fibrogenic signaling is suppressed in hepatic stellate cells through targeting of connective tissue growth factor (CCN2) by cellular or exosomal microRNA-199a-5p. Am. J. Pathol. 2016, 186, 2921–2933. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, F.; Huang, P.; Wang, X.; Zhou, K.; Zhou, C.; Yu, L.; Peng, Y.; Fan, J.; Zhou, J.; et al. Exosome-depleted MiR-148a-3p derived from Hepatic Stellate Cells Promotes Tumor Progression via ITGA5/PI3K/Akt Axis in Hepatocellular Carcinoma. Int. J. Biol. Sci. 2022, 18, 2249–2260. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, R.; Huang, Z.; Gurley, E.C.; Wang, X.; Wang, J.; He, H.; Yang, H.; Lai, G.; Zhang, L.; et al. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology 2018, 68, 599–615. [Google Scholar] [CrossRef] [PubMed]
- Witek, R.P.; Yang, L.; Liu, R.; Jung, Y.; Omenetti, A.; Syn, W.K.; Choi, S.S.; Cheong, Y.; Fearing, C.M.; Agboola, K.M.; et al. Liver cell–derived microparticles activate hedgehog signaling and alter gene expression in hepatic endothelial cells. Gastroenterology 2009, 136, 320–330.e2. [Google Scholar] [CrossRef] [PubMed]
- Newman, L.A.; Muller, K.; Rowland, A. Circulating cell-specific extracellular vesicles as biomarkers for the diagnosis and monitoring of chronic liver diseases. Cell Mol. Life Sci. 2022, 79, 232. [Google Scholar] [CrossRef]
- Sung, S.; Kim, J.; Jung, Y. Liver-derived exosomes and their implications in liver pathobiology. Int. J. Mol. Sci. 2018, 19, 3715. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Rao, H.; Liu, F.; Wei, L.; Li, H.; Wu, C. Recent Advances in Adipose Tissue Dysfunction and Its Role in the Pathogenesis of Non-Alcoholic Fatty Liver Disease. Cells 2021, 10, 3300. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zhang, Y. Hepatocyte nuclear factor 4α in the pathogenesis of non-alcoholic fatty liver disease. Chin. Med. J. 2022, 135, 1172–1181. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, A.; Yan, R.; Pan, S.Q.; Wu, R.; Kim, J.; Chen, Y.; Ansong, C.; Smith, R.D.; Tempaku, M.; Ohno-Machado, L.; et al. Comprehensive characterization of hepatocyte-derived extracellular vesicles identifies direct miRNA-based regulation of hepatic stellate cells and DAMP-based hepatic macrophage IL-1β and IL-17 upregulation in alcoholic hepatitis mice. J. Mol. Med. 2020, 98, 1021–1034. [Google Scholar] [CrossRef] [PubMed]
- Keinicke, H.; Sun, G.; Mentzel, C.M.J.; Fredholm, M.; John, L.M.; Andersen, B.; Raun, K.; Kjaergaard, M. FGF21 regulates hepatic metabolic pathways to improve steatosis and inflammation. Endocr. Connect. 2020, 9, 755–768. [Google Scholar] [CrossRef]
- Julich-Haertel, H.; Urban, S.K.; Krawczyk, M.; Willms, A.; Jankowski, K.; Patkowski, W.; Kruk, B.; Krasnodębski, M.; Ligocka, J.; Schwab, R.; et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J. Hepatol. 2017, 67, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Arbelaiz, A.; Azkargorta, M.; Krawczyk, M.; Santos-Laso, A.; Lapitz, A.; Perugorria, M.J.; Erice, O.; Gonzalez, E.; Jimenez-Agüero, R.; La Casta, A.; et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017, 66, 1125–1143. [Google Scholar] [CrossRef]
- Chen, L.; Guo, P.; He, Y.; Chen, Z.; Chen, L.; Luo, Y.; Qi, L.; Liu, Y.; Wu, Q.; Cui, Y.; et al. HCC-derived exosomes elicit HCC progression and recurrence by epithelial-mesenchymal transition through MAPK/ERK signalling pathway. Cell Death Dis. 2018, 9, 513. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.-q.; Yang, X.-w.; Chen, Y.-b.; Zhang, D.-w.; Jiang, X.-F.; Xue, P. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol. Cancer 2019, 18, 1–14. [Google Scholar] [CrossRef]
- Xue, X.; Wang, X.; Zhao, Y.; Hu, R.; Qin, L. Exosomal miR-93 promotes proliferation and invasion in hepatocellular carcinoma by directly inhibiting TIMP2/TP53INP1/CDKN1A. Biochem. Biophys. Res. Commun. 2018, 502, 515–521. [Google Scholar] [CrossRef]
- Verma, V.K.; Li, H.; Wang, R.; Hirsova, P.; Mushref, M.; Liu, Y.; Cao, S.; Contreras, P.C.; Malhi, H.; Kamath, P.S.; et al. Alcohol stimulates macrophage activation through caspase-dependent hepatocyte derived release of CD40L containing extracellular vesicles. J. Hepatol. 2016, 64, 651–660. [Google Scholar] [CrossRef]
- Liu, D.; Kang, H.; Gao, M.; Jin, L.; Zhang, F.; Chen, D.; Li, M.; Xiao, L. Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol. Oncol. 2020, 14, 1365–1380. [Google Scholar] [CrossRef]
- Zhu, C.; Su, Y.; Liu, L.; Wang, S.; Liu, Y.; Wu, J. Circular RNA hsa_circ_0004277 Stimulates Malignant Phenotype of Hepatocellular Carcinoma and Epithelial-Mesenchymal Transition of Peripheral Cells. Front. Cell Dev. Biol. 2020, 8, 585565. [Google Scholar] [CrossRef]
- Chen, C.; Luo, F.; Liu, X.; Lu, L.; Xu, H.; Yang, Q.; Xue, J.; Shi, L.; Li, J.; Zhang, A.; et al. NF-kB-regulated exosomal miR-155 promotes the inflammation associated with arsenite carcinogenesis. Cancer Lett. 2017, 388, 21–33. [Google Scholar] [CrossRef]
- Wu, Q.; Zhou, L.; Lv, D.; Zhu, X.; Tang, H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J. Hematol. Oncol. 2019, 12, 53. [Google Scholar] [CrossRef]
- Gai, X.; Tang, B.; Liu, F.; Wu, Y.; Wang, F.; Jing, Y.; Huang, F.; Jin, D.; Wang, L.; Zhang, H. mTOR/miR-145-regulated exosomal GOLM1 promotes hepatocellular carcinoma through augmented GSK-3β/MMPs. J. Genet. Genom. 2019, 46, 235–245. [Google Scholar] [CrossRef]
- Leonardi, G.C.; Candido, S.; Cervello, M.; Nicolosi, D.; Raiti, F.; Travali, S.; Spandidos, D.A.; Libra, M. The tumor microenvironment in hepatocellular carcinoma. Int. J. Oncol. 2012, 40, 1733–1747. [Google Scholar]
- Tian, Y.; Li, S.; Song, J.; Ji, T.; Zhu, M.; Anderson, G.J.; Wei, J.; Nie, G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef]
- Conigliaro, A.; Costa, V.; Lo Dico, A.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M.; et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 155. [Google Scholar] [CrossRef]
- Huang, A.; Dong, J.; Li, S.; Wang, C.; Ding, H.; Li, H.; Su, X.; Ge, X.; Sun, L.; Bai, C.; et al. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int. J. Biol. Sci. 2015, 11, 961–969. [Google Scholar] [CrossRef]
- Moh-Moh-Aung, A.; Fujisawa, M.; Ito, S.; Katayama, H.; Ohara, T.; Ota, Y.; Yoshimura, T.; Matsukawa, A. Decreased miR-200b-3p in cancer cells leads to angiogenesis in HCC by enhancing endothelial ERG expression. Sci. Rep. 2020, 10, 10418. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhang, Q.; Cheng, Y.; Chen, X.; Wang, G.; Shi, M.; Zhang, T.; Cao, Y.; Pan, H.; Zhang, L.; et al. Tumor-derived exosomal HMGB1 fosters hepatocellular carcinoma immune evasion by promoting TIM-1(+) regulatory B cell expansion. J. Immunother. Cancer 2018, 6, 145. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Lee, Y.-T.; Zhang, R.Y.; Kao, R.; Teng, P.-C.; Yang, Y.; Yang, P.; Wang, J.J.; Smalley, M.; Chen, P.-J.; et al. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat. Commun. 2020, 11, 4489. [Google Scholar] [CrossRef]
- Kooijmans, S.A.; Vader, P.; van Dommelen, S.M.; van Solinge, W.W.; Schiffelers, R.M. Exosome mimetics: A novel class of drug delivery systems. Int. J. Nanomed. 2012, 7, 1525–1541. [Google Scholar]
- Khamisipour, G.; Jadidi-Niaragh, F.; Jahromi, A.S.; Zandi, K.; Hojjat-Farsangi, M. Mechanisms of tumor cell resistance to the current targeted-therapy agents. Tumor Biol. 2016, 37, 10021–10039. [Google Scholar] [CrossRef]
- Xue, D.; Han, J.; Liu, Y.; Tuo, H.; Peng, Y. Current perspectives on exosomes in the diagnosis and treatment of hepatocellular carcinoma (review). Cancer Biol. Ther. 2021, 22, 279–290. [Google Scholar] [CrossRef]
- Liu, G.; Ouyang, X.; Sun, Y.; Xiao, Y.; You, B.; Gao, Y.; Yeh, S.; Li, Y.; Chang, C. The miR-92a-2-5p in exosomes from macrophages increases liver cancer cells invasion via altering the AR/PHLPP/p-AKT/β-catenin signaling. Cell Death Differ. 2020, 27, 3258–3272. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, B.; Xiao, S.; Li, Y.; Chen, Q. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J Cell Biochem 2019, 120, 3046–3055. [Google Scholar] [CrossRef]
- Wang, G.; Luo, G.; Zhao, M.; Miao, H. Significance of exosomes in hepatocellular carcinoma. Front. Oncol. 2022, 12, 1056379. [Google Scholar] [CrossRef]
- Tian, X.-P.; Wang, C.-Y.; Jin, X.-H.; Li, M.; Wang, F.-W.; Huang, W.-J.; Yun, J.-P.; Xu, R.-H.; Cai, Q.-Q.; Xie, D. Acidic microenvironment up-regulates exosomal miR-21 and miR-10b in early-stage hepatocellular carcinoma to promote cancer cell proliferation and metastasis. Theranostics 2019, 9, 1965–1979. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Schmidberger, H.; Mayer, A. The Warburg effect: Essential part of metabolic reprogramming and central contributor to cancer progression. Int. J. Radiat. Biol. 2019, 95, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Qian, D.; Xie, Y.; Huang, M.; Gu, J. Tumor-derived exosomes in hypoxic microenvironment: Release mechanism, biological function and clinical application. J. Cancer 2022, 13, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Lv, H.; Lv, G.; Li, T.; Wang, C.; Han, Q.; Yu, L.; Su, B.; Guo, L.; Huang, S.; et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018, 9, 191. [Google Scholar] [CrossRef]
- Yugawa, K.; Yoshizumi, T.; Mano, Y.; Itoh, S.; Harada, N.; Ikegami, T.; Kohashi, K.; Oda, Y.; Mori, M. Cancer-associated fibroblasts promote hepatocellular carcinoma progression through downregulation of exosomal miR-150-3p. Eur. J. Surg. Oncol. 2021, 47, 384–393. [Google Scholar] [CrossRef]
- Li, S.; Qi, Y.; Huang, Y.; Guo, Y.; Huang, T.; Jia, L. Exosome-derived SNHG16 sponging miR-4500 activates HUVEC angiogenesis by targeting GALNT1 via PI3K/Akt/mTOR pathway in hepatocellular carcinoma. J. Physiol. Biochem. 2021, 77, 667–682. [Google Scholar] [CrossRef]
- Dai, W.; Wang, Y.; Yang, T.; Wang, J.; Wu, W.; Gu, J. Downregulation of exosomal CLEC3B in hepatocellular carcinoma promotes metastasis and angiogenesis via AMPK and VEGF signals. Cell Commun. Signal. 2019, 17, 1–17. [Google Scholar] [CrossRef]
- Wang, W.; Wei, C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020, 7, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Manea, I.; Iacob, R.; Iacob, S.; Cerban, R.; Dima, S.; Oniscu, G.; Popescu, I.; Gheorghe, L. Liquid biopsy for early detection of hepatocellular carcinoma. Front. Med. 2023, 10, 1218705. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Zhang, P.; Guo, G.; Jiang, T.; Zhao, X.; Jiang, J.; Huang, X.; Tong, H.; Tian, Y. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018, 7, 1670–1679. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, Y.; Wu, X.; Zhang, C.; Li, G. Diagnostic Value of lncRNAs as Biomarker in Hepatocellular Carcinoma: An Updated Meta-Analysis. Can. J. Gastroenterol. Hepatol. 2018, 2018, 8410195. [Google Scholar] [CrossRef]
- Xu, H.; Chen, Y.; Dong, X.; Wang, X. Serum Exosomal Long Noncoding RNAs ENSG00000258332.1 and LINC00635 for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Cancer Epidemiol. Biomark. Prev. 2018, 27, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Ferracin, M.; Trerè, D.; Milazzo, M.; Marinelli, S.; Galassi, M.; Venerandi, L.; Pollutri, D.; Patrizi, C.; Borghi, A.; et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, Identify Cirrhotic Patients with HCC. PLoS ONE 2015, 10, e0141448. [Google Scholar] [CrossRef]
- Yu, D.; Li, Y.; Wang, M.; Gu, J.; Xu, W.; Cai, H.; Fang, X.; Zhang, X. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer 2022, 21, 56. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Kato, T.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles 2013, 2, 20424. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, J.-Q.; Liu, J.-L.; Tian, L. Exosomes in tumor microenvironment: Novel transporters and biomarkers. J. Transl. Med. 2016, 14, 1–9. [Google Scholar] [CrossRef]
- Larssen, P.; Wik, L.; Czarnewski, P.; Eldh, M.; Löf, L.; Ronquist, K.G.; Dubois, L.; Freyhult, E.; Gallant, C.J.; Oelrich, J.; et al. Tracing cellular origin of human exosomes using multiplex proximity extension assays. Mol. Cell. Proteom. 2017, 16, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, L.; Darbinian, N.; Goetzl, E.J. Novel window on early human neurodevelopment via fetal exosomes in maternal blood. Ann. Clin. Transl. Neurol. 2016, 3, 381–385. [Google Scholar] [CrossRef]
- Goetzl, L.; Darbinian, N.; Merabova, N. Noninvasive assessment of fetal central nervous system insult: Potential application to prenatal diagnosis. Prenat. Diagn. 2019, 39, 609–615. [Google Scholar] [CrossRef]
- Goetzl, L.; Merabova, N.; Darbinian, N.; Martirosyan, D.; Poletto, E.; Fugarolas, K.; Menkiti, O. Diagnostic Potential of Neural Exosome Cargo as Biomarkers for Acute Brain Injury. Ann. Clin. Transl. Neurol. 2018, 5, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, E.J.; Boxer, A.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Miller, B.L.; Kapogiannis, D. Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology 2015, 85, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Newman, L.A.; Useckaite, Z.; Johnson, J.; Sorich, M.J.; Hopkins, A.M.; Rowland, A. Selective Isolation of Liver-Derived Extracellular Vesicles Redefines Performance of miRNA Biomarkers for Non-Alcoholic Fatty Liver Disease. Biomedicines 2022, 10, 195. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Hu, S.; Luo, Y.; He, K. Exosome Cargos as Biomarkers for Diagnosis and Prognosis of Hepatocellular Carcinoma. Pharmaceutics 2023, 15, 2365. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xu, M.; Li, X.; Su, X.; Xiao, X.; Keating, A.; Zhao, R.C. Exosomes released by hepatocarcinoma cells endow adipocytes with tumor-promoting properties. J. Hematol. Oncol. 2018, 11, 82. [Google Scholar] [CrossRef]
- Liu, W.-h.; Ren, L.-n.; Wang, X.; Wang, T.; Zhang, N.; Gao, Y.; Luo, H.; Navarro-Alvarez, N.; Tang, L.-J. Combination of exosomes and circulating microRNAs may serve as a promising tumor marker complementary to alpha-fetoprotein for early-stage hepatocellular carcinoma diagnosis in rats. J. Cancer Res. Clin. Oncol. 2015, 141, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, C.; Zhang, L.; Wu, M.; Cao, K.; Jiang, F.; Chen, D.; Li, N.; Li, W. The significance of exosomes in the development and treatment of hepatocellular carcinoma. Mol. Cancer 2020, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Chapuy-Regaud, S.; Dubois, M.; Plisson-Chastang, C.; Bonnefois, T.; Lhomme, S.; Bertrand-Michel, J.; You, B.; Simoneau, S.; Gleizes, P.-E.; Flan, B.; et al. Characterization of the lipid envelope of exosome encapsulated HEV particles protected from the immune response. Biochimie 2017, 141, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Haraszti, R.A.; Didiot, M.-C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; DiFiglia, M.; Kiebish, M.A.; et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 2016, 5, 32570. [Google Scholar] [CrossRef]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta 2013, 1831, 1302–1309. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.I.; Jiao, J.; Kwan, S.Y.; Veillon, L.; Warmoes, M.O.; Tan, L.; Odewole, M.; Rich, N.E.; Wei, P.; Lorenzi, P.L.; et al. Lipidomic Profiles of Plasma Exosomes Identify Candidate Biomarkers for Early Detection of Hepatocellular Carcinoma in Patients with Cirrhosis. Cancer Prev. Res. 2021, 14, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; d’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.X.; Fan, X.; Drummond, C.A.; Majumder, R.; Xie, Y.; Chen, T.; Liu, L.; Haller, S.T.; Brewster, P.S.; Dworkin, L.D.; et al. MicroRNA profiling in kidney disease: Plasma versus plasma-derived exosomes. Gene 2017, 627, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, H.; Liu, Y.; Li, J.; Wu, C.; Tang, H. Exosomal Non-Coding RNAs: New Insights into the Biology of Hepatocellular Carcinoma. Curr. Oncol. 2022, 29, 5383–5406. [Google Scholar] [CrossRef]
- Cho, H.J.; Eun, J.W.; Baek, G.O.; Seo, C.W.; Ahn, H.R.; Kim, S.S.; Cho, S.W.; Cheong, J.Y. Serum exosomal microRNA, miR-10b-5p, as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. J. Clin. Med. 2020, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Baek, G.O.; Seo, C.W.; Ahn, H.R.; Sung, S.; Son, J.A.; Kim, S.S.; Cho, S.W.; Jang, J.W.; Nam, S.W.; et al. Exosomal microRNA-4661-5p–based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 2020, 9, 5459–5472. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Jiang, Y.; Yang, L.; Yan, S.; Wang, Y.G.; Lu, X.J. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J. Cell. Biochem. 2018, 119, 4711–4716. [Google Scholar] [CrossRef]
- Li, W.; Ding, X.; Wang, S.; Xu, L.; Yin, T.; Han, S.; Geng, J.; Sun, W. Downregulation of serum exosomal miR-320d predicts poor prognosis in hepatocellular carcinoma. J. Clin. Lab. Anal. 2020, 34, e23239. [Google Scholar] [CrossRef]
- Yan, L.; Chen, Y.; Zhou, J.; Zhao, H.; Zhang, H.; Wang, G. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int. J. Infect. Dis. 2018, 67, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.W.; Han, C.L.; Dong, Z.R.; Tan, S.Y.; Wang, D.X.; Li, T. Role of Exosomes in Immunotherapy of Hepatocellular Carcinoma. Cancers 2022, 14, 4036. [Google Scholar] [CrossRef] [PubMed]
- Poggio, M.; Hu, T.; Pai, C.-C.; Chu, B.; Belair, C.D.; Chang, A.; Montabana, E.; Lang, U.E.; Fu, Q.; Fong, L.; et al. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell 2019, 177, 414–427.e13. [Google Scholar] [CrossRef] [PubMed]
- Kar, R.; Dhar, R.; Mukherjee, S.; Nag, S.; Gorai, S.; Mukerjee, N.; Mukherjee, D.; Vatsa, R.; Jadhav, M.C.; Ghosh, A.; et al. Exosome-Based Smart Drug Delivery Tool for Cancer Theranostics. ACS Biomater. Sci. Eng. 2023, 9, 577–594. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Hadla, M.; Palazzolo, S.; Corona, G.; Caligiuri, I.; Canzonieri, V.; Toffoli, G.; Rizzolio, F. Exosomes increase the therapeutic index of doxorubicin in breast and ovarian cancer mouse models. Nanomedicine 2016, 11, 2431–2441. [Google Scholar] [CrossRef]
- Wei, H.; Chen, J.; Wang, S.; Fu, F.; Zhu, X.; Wu, C.; Liu, Z.; Zhong, G.; Lin, J. A nanodrug consisting of doxorubicin and exosome derived from mesenchymal stem cells for osteosarcoma treatment in vitro. Int. J. Nanomed. 2019, 14, 8603–8610. [Google Scholar] [CrossRef]
- Najafi, S.; Majidpoor, J.; Mortezaee, K. Extracellular vesicle-based drug delivery in cancer immunotherapy. Drug Deliv. Transl. Res. 2023, 13, 2790–2806. [Google Scholar] [CrossRef]
- Morad, G.; Helmink, B.A.; Sharma, P.; Wargo, J.A. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 2021, 184, 5309–5337. [Google Scholar] [CrossRef]
- Seo, N.; Akiyoshi, K.; Shiku, H. Exosome-mediated regulation of tumor immunology. Cancer Sci. 2018, 109, 2998–3004. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Liu, H.; Yan, H.; Che, R.; Jin, Y.; Yang, X.; Zhou, X.; Yang, H.; Ge, K.; Liang, X.-J.; et al. A CAR T-inspiring platform based on antibody-engineered exosomes from antigen-feeding dendritic cells for precise solid tumor therapy. Biomaterials 2022, 282, 121424. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wei, Q.; Lu, L.; Cui, S.; Ma, K.; Zhang, W.; Ma, F.; Li, H.; Fu, X.; Zhang, C. Immunomodulatory potential of mesenchymal stem cell-derived extracellular vesicles: Targeting immune cells. Front. Immunol. 2023, 14, 1094685. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Lei, C.; Liu, S.; Cui, Y.; Wang, C.; Qian, K.; Li, T.; Shen, Y.; Fan, X.; Lin, F.; et al. CAR exosomes derived from effector CAR-T cells have potent antitumour effects and low toxicity. Nat. Commun. 2019, 10, 4355. [Google Scholar] [CrossRef] [PubMed]
- Taghikhani, A.; Farzaneh, F.; Sharifzad, F.; Mardpour, S.; Ebrahimi, M.; Hassan, Z.M. Engineered Tumor-Derived Extracellular Vesicles: Potentials in Cancer Immunotherapy. Front. Immunol. 2020, 11, 221. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Chen, H.; Selaru, F.M. Extracellular Vesicles in Hepatocellular Carcinoma: Progress and Challenges in the Translation from the Laboratory to Clinic. Medicina 2023, 59, 1599. [Google Scholar] [CrossRef] [PubMed]
- Jesus, S.; Soares, E.; Cruz, M.T.; Borges, O. Exosomes as adjuvants for the recombinant hepatitis B antigen: First report. Eur. J. Pharm. Biopharm. 2018, 133, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.C.; Economides, K.D.; Moniz, R.J.; Sia, C.L.; Lewis, N.; McCoy, C.; Zi, T.; Zhang, K.; Harrison, R.A.; Lim, J.; et al. ExoSTING, an extracellular vesicle loaded with STING agonists, promotes tumor immune surveillance. Commun. Biol. 2021, 4, 497. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ji, L.; Liang, Y.; Wan, Z.; Zheng, W.; Song, X.; Gorshkov, K.; Sun, Q.; Lin, H.; Zheng, X.; et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct. Target. Ther. 2020, 5, 298. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.C.; Zhang, P.F.; Huang, X.Y.; Guo, X.J.; Gao, C.; Zeng, H.Y.; Zheng, Y.M.; Wang, S.W.; Cai, J.B.; Sun, Q.M.; et al. Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J. Hematol. Oncol. 2021, 14, 200. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Y.; Lu, J.; Wang, X. The role of exosomes in therapeutic resistance of hepatocellular carcinoma. Hepatoma Res. 2023, 9, 46. [Google Scholar] [CrossRef]
- He, C.; Dong, X.; Zhai, B.; Jiang, X.; Dong, D.; Li, B.; Jiang, H.; Xu, S.; Sun, X. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget 2015, 6, 28867–28881. [Google Scholar] [CrossRef] [PubMed]
- Lou, G.; Song, X.; Yang, F.; Wu, S.; Wang, J.; Chen, Z.; Liu, Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J. Hematol. Oncol. 2015, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yin, Z.; Qi, Y.; Peng, H.; Ma, W.; Wang, R.; Li, W. Golgi phosphoprotein 3 promotes angiogenesis and sorafenib resistance in hepatocellular carcinoma via upregulating exosomal miR-494-3p. Cancer Cell Int. 2022, 22, 35. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B.; Dorval, T.; Chaput, N.; André, F.; Caby, M.-P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: Results of thefirst phase I clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Besse, B.; Charrier, M.; Lapierre, V.; Dansin, E.; Lantz, O.; Planchard, D.; Le Chevalier, T.; Livartoski, A.; Barlesi, F.; Laplanche, A.; et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology 2016, 5, e1071008. [Google Scholar] [CrossRef] [PubMed]
- Gehrmann, U.; Näslund, T.I.; Hiltbrunner, S.; Larssen, P.; Gabrielsson, S. (Eds.) Harnessing the exosome-induced immune response for cancer immunotherapy. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Katzenstein, H.M.; Langham, M.R.; Malogolowkin, M.H.; Krailo, M.D.; Towbin, A.J.; McCarville, M.B.; Finegold, M.J.; Ranganathan, S.; Dunn, S.; McGahren, E.D.; et al. Minimal adjuvant chemotherapy for children with hepatoblastoma resected at diagnosis (AHEP0731): A Children’s Oncology Group, multicentre, phase 3 trial. Lancet Oncol. 2019, 20, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Yeung, C.C.; Dondelinger, F.; Schoof, E.M.; Georg, B.; Lu, Y.; Zheng, Z.; Zhang, J.; Hannibal, J.; Fahrenkrug, J.; Kjaer, M. Circadian regulation of protein cargo in extracellular vesicles. Sci. Adv. 2022, 8, eabc9061. [Google Scholar] [CrossRef] [PubMed]
- Partch, C.L.; Green, C.B.; Takahashi, J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014, 24, 90–99. [Google Scholar] [CrossRef]
- Patke, A.; Young, M.W.; Axelrod, S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84. [Google Scholar] [CrossRef]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Shafi, A.A.; Knudsen, K.E. Cancer and the circadian clock. Cancer Res. 2019, 79, 3806–3814. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A.; Van Gelder, R.N. Clocks, cancer, and chronochemotherapy. Science 2021, 371, eabb0738. [Google Scholar] [CrossRef] [PubMed]
- Buhr, E.D.; Takahashi, J.S. Molecular components of the Mammalian circadian clock. Handb. Exp. Pharmacol. 2013, 217, 3–27. [Google Scholar]
- Panda, S.; Hogenesch, J.B.; Kay, S.A. Circadian rhythms from flies to human. Nature 2002, 417, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Bass, J.; Takahashi, J.S. Circadian integration of metabolism and energetics. Science 2010, 330, 1349–1354. [Google Scholar] [CrossRef]
- Battaglin, F.; Chan, P.; Pan, Y.; Soni, S.; Qu, M.; Spiller, E.R.; Castanon, S.; Torres, E.T.R.; Mumenthaler, S.M.; Kay, S.A.; et al. Clocking cancer: The circadian clock as a target in cancer therapy. Oncogene 2021, 40, 3187–3200. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhao, J.; Tang, Q.; Li, H.; Zhang, C.; Yu, R.; Zhao, Y.; Huo, Y.; Wu, C. PFKFB3 control of cancer growth by responding to circadian clock outputs. Sci. Rep. 2016, 6, 24324. [Google Scholar] [CrossRef]
- Tang, Q.; Cheng, B.; Xie, M.; Chen, Y.; Zhao, J.; Zhou, X.; Chen, L. Circadian clock gene Bmal1 inhibits tumorigenesis and increases paclitaxel sensitivity in tongue squamous cell carcinoma. Cancer Res. 2017, 77, 532–544. [Google Scholar] [CrossRef]
- Lucassen, E.A.; Coomans, C.P.; van Putten, M.; de Kreij, S.R.; van Genugten, J.H.; Sutorius, R.P.; de Rooij, K.E.; van der Velde, M.; Verhoeve, S.L.; Smit, J.W.; et al. Environmental 24-hr cycles are essential for health. Curr. Biol. 2016, 26, 1843–1853. [Google Scholar] [CrossRef]
- Pan, A.; Schernhammer, E.S.; Sun, Q.; Hu, F.B. Rotating night shift work and risk of type 2 diabetes: Two prospective cohort studies in women. PLoS Med. 2011, 8, e1001141. [Google Scholar] [CrossRef] [PubMed]
- Logan, R.W.; McClung, C.A. Rhythms of life: Circadian disruption and brain disorders across the lifespan. Nat. Rev. Neurosci. 2019, 20, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Dooner, M.S.; Stewart, C.; Deng, Y.; Papa, E.; Pereira, M.; Del Tatto, M.; Johnson, S.; Wen, S.; Amaral, A.; Aliotta, J.; et al. Daily rhythms influence the ability of lung-derived extracellular vesicles to modulate bone marrow cell phenotype. PLoS ONE 2018, 13, e0207444. [Google Scholar] [CrossRef] [PubMed]
- Khalyfa, A.; Gaddameedhi, S.; Crooks, E.; Zhang, C.; Li, Y.; Qiao, Z.; Trzepizur, W.; Kay, S.A.; Andrade, J.; Satterfield, B.C.; et al. Circulating Exosomal miRNAs Signal Circadian Misalignment to Peripheral Metabolic Tissues. Int. J. Mol. Sci. 2020, 21, 6396. [Google Scholar] [CrossRef] [PubMed]
- Danielson, K.M.; Estanislau, J.; Tigges, J.; Toxavidis, V.; Camacho, V.; Felton, E.J.; Khoory, J.; Kreimer, S.; Ivanov, A.R.; Mantel, Y.; et al. Diurnal Variations of Circulating Extracellular Vesicles Measured by Nano Flow Cytometry. PLoS ONE 2016, 11, e0144678. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sundaram, K.; Mu, J.; Dryden, G.W.; Sriwastva, M.K.; Lei, C.; Zhang, L.; Qiu, X.; Xu, F.; Yan, J.; et al. High-fat diet-induced upregulation of exosomal phosphatidylcholine contributes to insulin resistance. Nat. Commun. 2021, 12, 213. [Google Scholar] [CrossRef]
- Noh, J. The Effect of Circadian and Sleep Disruptions on Obesity Risk. J. Obes. Metab. Syndr. 2018, 27, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Chaput, J.-P.; McHill, A.W.; Cox, R.C.; Broussard, J.L.; Dutil, C.; da Costa, B.G.G.; Sampasa-Kanyinga, H.; Wright, K.P. The role of insufficient sleep and circadian misalignment in obesity. Nat. Rev. Endocrinol. 2023, 19, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Koritzinsky, E.H.; Street, J.M.; Chari, R.R.; Glispie, D.M.; Bellomo, T.R.; Aponte, A.M.; Star, R.A.; Yuen, P.S.T. Circadian variation in the release of small extracellular vesicles can be normalized by vesicle number or TSG101. Am. J. Physiol. Physiol. 2019, 317, F1098–F1110. [Google Scholar] [CrossRef]
- Eckel-Mahan, K.L.; Patel, V.R.; De Mateo, S.; Orozco-Solis, R.; Ceglia, N.J.; Sahar, S.; Dilag-Penilla, S.A.; Dyar, K.A.; Baldi, P.; Sassone-Corsi, P. Reprogramming of the circadian clock by nutritional challenge. Cell 2013, 155, 1464–1478. [Google Scholar] [CrossRef]
- Vollmers, C.; Gill, S.; DiTacchio, L.; Pulivarthy, S.R.; Le, H.D.; Panda, S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 21453–21458. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Tuccinardi, D.; Ernesti, I.; Basciani, S.; Mariani, S.; Genco, A.; Manfrini, S.; Lubrano, C.; Gnessi, L. Scientific evidence underlying contraindications to the ketogenic diet: An update. Obes. Rev. 2020, 21, e13053. [Google Scholar] [CrossRef] [PubMed]
- Tognini, P.; Murakami, M.; Liu, Y.; Eckel-Mahan, K.L.; Newman, J.C.; Verdin, E.; Baldi, P.; Sassone-Corsi, P. Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab. 2017, 26, 523–538.e5. [Google Scholar] [CrossRef] [PubMed]
- Ribas-Latre, A.; Fekry, B.; Kwok, C.; Baumgartner, C.; Shivshankar, S.; Sun, K.; Chen, Z.; Eckel-Mahan, K. Rosiglitazone reverses high fat diet-induced changes in BMAL1 function in muscle, fat, and liver tissue in mice. Int. J. Obes. 2019, 43, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Lamia, K.A.; Storch, K.-F.; Weitz, C.J. Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. USA 2008, 105, 15172–15177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.E.; Liu, Y.; Dentin, R.; Pongsawakul, P.Y.; Liu, A.C.; Hirota, T.; Nusinow, D.A.; Sun, X.; Landais, S.; Kodama, Y.; et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 2010, 16, 1152–1156. [Google Scholar] [CrossRef]
- Erren, T.C.; Falaturi, P.; Morfeld, P.; Knauth, P.; Reiter, R.J.; Piekarski, C. Shift work and cancer: The evidence and the challenge. Dtsch. Arztebl. Int. 2010, 107, 657–662. [Google Scholar] [PubMed]
- Erren, T.C.; Morfeld, P.; Groß, J.V.; Wild, U.; Lewis, P. “Night shift work” is probably carcinogenic: What about disturbed chronobiology in all walks of life? J. Occup. Med. Toxicol. 2019, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Kinouchi, K.; Sassone-Corsi, P. Metabolic rivalry: Circadian homeostasis and tumorigenesis. Nat. Rev. Cancer 2020, 20, 645–661. [Google Scholar] [CrossRef]
- Koritala, B.S.; Porter, K.I.; Arshad, O.A.; Gajula, R.P.; Mitchell, H.D.; Arman, T.; Manjanatha, M.G.; Teeguarden, J.; Van Dongen, H.P.A.; McDermott, J.E.; et al. Night shift schedule causes circadian dysregulation of DNA repair genes and elevated DNA damage in humans. J. Pineal Res. 2021, 70, e12726. [Google Scholar] [CrossRef]
- Chen, W.-D.; Wen, M.-S.; Shie, S.-S.; Lo, Y.-L.; Wo, H.-T.; Wang, C.-C.; Hsieh, I.C.; Lee, T.H.; Wang, C.Y. The circadian rhythm controls telomeres and telomerase activity. Biochem. Biophys. Res. Commun. 2014, 451, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Yuan, X.; Eltzschig, H.K. Circadian rhythm as a therapeutic target. Nat. Rev. Drug Discov. 2021, 20, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhang, C.; Cao, Y.; Li, J.; Bi, F. Major roles of the circadian clock in cancer. Cancer Biol. Med. 2023, 20, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Kettner, N.M.; Voicu, H.; Finegold, M.J.; Coarfa, C.; Sreekumar, A.; Putluri, N.; Katchy, C.A.; Lee, C.; Moore, D.D.; Fu, L. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 2016, 30, 909–924. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Xiang, Y.; Ozguc, F.M.; Kim, Y.; Liu, C.-J.; Park, P.K.; Hu, Q.; Diao, L.; Lou, Y.; Lin, C.; et al. The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cell Syst. 2018, 6, 314–328.e2. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Zhang, G.; Qu, M.; Gimple, R.C.; Wu, Q.; Qiu, Z.; Prager, B.C.; Wang, X.; Kim, L.J.; Morton, A.R.; et al. Targeting Glioblastoma Stem Cells through Disruption of the Circadian Clock. Cancer Discov. 2019, 9, 1556–1573. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Hsu, W.-H.; Chang, A.; Tan, Z.; Lan, Z.; Zhou, A.; Spring, D.J.; Lang, F.F.; Wang, Y.A.; DePinho, R.A. Circadian regulator CLOCK recruits immune-suppressive microglia into the GBM tumor microenvironment. Cancer Discov. 2020, 10, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Papagiannakopoulos, T.; Bauer, M.R.; Davidson, S.M.; Heimann, M.; Subbaraj, L.; Bhutkar, A.; Bartlebaugh, J.; Heiden, M.G.V.; Jacks, T. Circadian rhythm disruption promotes lung tumorigenesis. Cell Metab. 2016, 24, 324–331. [Google Scholar] [CrossRef]
- Qu, M.; Zhang, G.; Qu, H.; Vu, A.; Wu, R.; Tsukamoto, H.; Jia, Z.; Huang, W.; Lenz, H.-J.; Rich, J.N.; et al. Circadian regulator BMAL1::CLOCK promotes cell proliferation in hepatocellular carcinoma by controlling apoptosis and cell cycle. Proc. Natl. Acad. Sci. USA 2023, 120, e2214829120. [Google Scholar] [CrossRef]
- Fekry, B.; Ribas-Latre, A.; Drunen, R.V.; Santos, R.B.; Shivshankar, S.; Dai, Y.; Zhao, Z.; Yoo, S.; Chen, Z.; Sun, K.; et al. Hepatic circadian and differentiation factors control liver susceptibility for fatty liver disease and tumorigenesis. Faseb J. 2022, 36, e22482. [Google Scholar] [CrossRef]
- Crespo, M.; Leiva, M.; Sabio, G. Circadian Clock and Liver Cancer. Cancers 2021, 13, 3631. [Google Scholar] [CrossRef] [PubMed]
- Fekry, B.; Ribas-Latre, A.; Baumgartner, C.; Deans, J.R.; Kwok, C.; Patel, P.; Fu, L.; Berdeaux, R.; Sun, K.; Kolonin, M.G.; et al. Incompatibility of the circadian protein BMAL1 and HNF4α in hepatocellular carcinoma. Nat. Commun. 2018, 9, 4349. [Google Scholar] [CrossRef] [PubMed]
- Zisapel, N. New perspectives on the role of melatonin in human sleep, circadian rhythms and their regulation. Br. J. Pharmacol. 2018, 175, 3190–3199. [Google Scholar] [CrossRef] [PubMed]
- Talib, W.H.; Alsayed, A.R.; Abuawad, A.; Daoud, S.; Mahmod, A.I. Melatonin in Cancer Treatment: Current Knowledge and Future Opportunities. Molecules 2021, 26, 2506. [Google Scholar] [CrossRef]
- Cheng, L.; Liu, J.; Liu, Q.; Liu, Y.; Fan, L.; Wang, F.; Yu, H.; Li, Y.; Bu, L.; Li, X.; et al. Exosomes from melatonin treated hepatocellularcarcinoma cells alter the immunosupression status through STAT3 pathway in macrophages. Int. J. Biol. Sci. 2017, 13, 723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Pant, S.M.; Ritch, C.C.; Tang, H.-Y.; Shao, H.; Dweep, H.; Gong, Y.-Y.; Brooks, R.; Brafford, P.; Wolpaw, A.J.; et al. Cell state dependent effects of Bmal1 on melanoma immunity and tumorigenicity. Nat. Commun. 2024, 15, 633. [Google Scholar] [CrossRef] [PubMed]
- Schibler, U.; Sassone-Corsi, P. A web of circadian pacemakers. Cell 2002, 111, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Valekunja, U.K.; Stangherlin, A.; Howell, S.A.; Snijders, A.P.; Damodaran, G.; Reddy, A.B. Circadian rhythms in the absence of the clock gene Bmal1. Science 2020, 367, 800–806. [Google Scholar] [CrossRef]
- Khalyfa, A.; Poroyko, V.A.; Qiao, Z.; Gileles-Hillel, A.; Khalyfa, A.A.; Akbarpour, M.; Farré, R.; Gozal, D. Exosomes and metabolic function in mice exposed to alternating dark-light cycles mimicking night shift work schedules. Front. Physiol. 2017, 8, 311175. [Google Scholar] [CrossRef]
- Chang, H.C.; Guarente, L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 2013, 153, 1448–1460. [Google Scholar] [CrossRef]
- Asher, G.; Gatfield, D.; Stratmann, M.; Reinke, H.; Dibner, C.; Kreppel, F.; Mostoslavsky, R.; Alt, F.W.; Schibler, U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 2008, 134, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Forterre, A.; Jalabert, A.; Chikh, K.; Pesenti, S.; Euthine, V.; Granjon, A.; Errazuriz, E.; Lefai, E.; Vidal, H.; Rome, S. Myotube-derived exosomal miRNAs downregulate Sirtuin1 in myoblasts during muscle cell differentiation. Cell Cycle 2014, 13, 78–89. [Google Scholar] [CrossRef]
- Shi, X.-F.; Wang, H.; Kong, F.-X.; Xu, Q.-Q.; Xiao, F.-J.; Yang, Y.-F.; Ge, R.-L.; Wang, L.-S. Exosomal miR-486 regulates hypoxia-induced erythroid differentiation of erythroleukemia cells through targeting Sirt1. Exp. Cell Res. 2017, 351, 74–81. [Google Scholar] [CrossRef]
- Ding, L.; Li, Z.-l.; Zhou, Y.; Liu, N.-c.; Liu, S.-s.; Zhang, X.-j.; Liu, C.-C.; Zhang, D.-J.; Wang, G.-H.; Ma, R.-X. Loss of Sirt1 promotes exosome secretion from podocytes by inhibiting lysosomal acidification in diabetic nephropathy. Mol. Cell. Endocrinol. 2023, 568–569, 111913. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Guo, W.; Yi, F.; Zhou, T.; Li, X.; Feng, Y.; Guo, Q.; Xu, H.; Song, X.; Cao, L. The Regulatory Effect of SIRT1 on Extracellular Microenvironment Remodeling. Int. J. Biol. Sci. 2021, 17, 89–96. [Google Scholar] [CrossRef] [PubMed]
- McAndrews, K.M.; LeBleu, V.S.; Kalluri, R. SIRT1 Regulates Lysosome Function and Exosome Secretion. Dev. Cell 2019, 49, 302–303. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Das, A.; Vernekar, M.; Mandal, S.; Chatterjee, N. Understanding the Correlation between Metabolic Regulator SIRT1 and Exosomes with CA-125 in Ovarian Cancer: A Clinicopathological Study. BioMed Res. Int. 2022, 2022, 5346091. [Google Scholar] [CrossRef]
- Liu, L.; Jin, X.; Hu, C.-F.; Li, R.; Zhou, Z.; Shen, C.-X. Exosomes derived from mesenchymal stem cells rescue myocardial ischaemia/reperfusion injury by inducing cardiomyocyte autophagy via AMPK and Akt pathways. Cell. Physiol. Biochem. 2017, 43, 52–68. [Google Scholar] [CrossRef]
- Liu, F.; Bu, Z.; Zhao, F.; Xiao, D. Increased T-helper 17 cell differentiation mediated by exosome-mediated micro RNA-451 redistribution in gastric cancer infiltrated T cells. Cancer Sci. 2018, 109, 65–73. [Google Scholar] [CrossRef]
- Arslan, F.; Lai, R.C.; Smeets, M.B.; Akeroyd, L.; Choo, A.; Aguor, E.N.; Timmers, L.; Van Rijen, H.V.; Doevendans, P.A.; Pasterkamp, G.; et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013, 10, 301–312. [Google Scholar] [CrossRef]
- Mukherji, A.; Bailey, S.M.; Staels, B.; Baumert, T.F. The circadian clock and liver function in health and disease. J. Hepatol. 2019, 71, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guan, J.; Jiang, Z.; Cheng, S.; Hou, W.; Yao, J.; Wang, Z. Microglial Exosome miR-7239-3p Promotes Glioma Progression by Regulating Circadian Genes. Neurosci. Bull. 2021, 37, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Qu, H.; Jia, Z.; Kay, S.A. HNF4A defines tissue-specific circadian rhythms by beaconing BMAL1::CLOCK chromatin binding and shaping the rhythmic chromatin landscape. Nat. Commun. 2021, 12, 6350. [Google Scholar] [CrossRef] [PubMed]
- Deans, J.R.; Deol, P.; Titova, N.; Radi, S.H.; Vuong, L.M.; Evans, J.R.; Pan, S.; Fahrmann, J.; Yang, J.; Hammock, B.D.; et al. HNF4α isoforms regulate the circadian balance between carbohydrate and lipid metabolism in the liver. Front. Endocrinol. 2023, 14, 1266527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, X.; Wang, S.; Zheng, L.; Guo, H.; Ren, Y.; Qiao, B.; Wu, J.; Zhao, D.; Xu, L.; et al. Adipocyte-derived exosomal miR-22-3p modulated by circadian rhythm disruption regulates insulin sensitivity in skeletal muscle cells. J. Biol. Chem. 2023, 299, 105476. [Google Scholar] [CrossRef] [PubMed]
- Pusic, A.D.; Kraig, R.P. Youth and environmental enrichment generate serum exosomes containing miR-219 that promote CNS myelination. Glia 2014, 62, 284–299. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.C.; Guo, S.C. Extracellular Vesicles: Potential Participants in Circadian Rhythm Synchronization. Int. J. Biol. Sci. 2018, 14, 1610–1620. [Google Scholar] [CrossRef] [PubMed]
- Shende, V.R.; Goldrick, M.M.; Ramani, S.; Earnest, D.J. Expression and rhythmic modulation of circulating microRNAs targeting the clock gene Bmal1 in mice. PLoS ONE 2011, 6, e22586. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38 (Suppl. S2), W214–W220. [Google Scholar] [CrossRef]
- Liu, W.; Yu, M.; Xie, D.; Wang, L.; Ye, C.; Zhu, Q.; Liu, F.; Yang, L. Melatonin-stimulated MSC-derived exosomes improve diabetic wound healing through regulating macrophage M1 and M2 polarization by targeting the PTEN/AKT pathway. Stem Cell Res. Ther. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.M.; Lee, J.H.; Song, K.H.; Noh, H.; Lee, S.H. Melatonin-stimulated exosomes enhance the regenerative potential of chronic kidney disease-derived mesenchymal stem/stromal cells via cellular prion proteins. J. Pineal Res. 2020, 68, e12632. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, R.; Wang, J.; Hao, Y.; Xie, Q.; Wang, L.; Wang, X. Melatonin pretreatment on exosomes: Heterogeneity, therapeutic effects, and usage. Front. Immunol. 2022, 13, 933736. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.X.; Lv, L.H.; Wan, Y.L.; Cao, Y.; Li, G.L.; Lin, H.M.; Zhou, R.; Shang, C.Z.; Cao, J.; He, H.; et al. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology 2015, 61, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Li, P.; Zhang, Z.; Wu, M. Insights into Exosomal Non-Coding RNAs Sorting Mechanism and Clinical Application. Front. Oncol. 2021, 11, 664904. [Google Scholar]
- Lee, Y.J.; Shin, K.J.; Chae, Y.C. Regulation of cargo selection in exosome biogenesis and its biomedical applications in cancer. Exp. Mol. Med. 2024, 56, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martin, R.; Brandao, B.B.; Thomou, T.; Altindis, E.; Kahn, C.R. Tissue differences in the exosomal/small extracellular vesicle proteome and their potential as indicators of altered tissue metabolism. Cell Rep. 2022, 38, 110277. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-Z.; Ma, Z.-J.; Kang, X.-W. Current status and outlook of advances in exosome isolation. Anal. Bioanal. Chem. 2022, 414, 7123–7141. [Google Scholar] [CrossRef]
- Gao, J.; Li, A.; Hu, J.; Feng, L.; Liu, L.; Shen, Z. Recent developments in isolating methods for exosomes. Front. Bioeng. Biotechnol. 2022, 10, 1100892. [Google Scholar] [CrossRef]
- Jin, X.; Zhang, J.; Zhang, Y.; He, J.; Wang, M.; Hei, Y.; Guo, S.; Xu, X.; Liu, Y. Different origin-derived exosomes and their clinical advantages in cancer therapy. Front. Immunol. 2024, 15, 1401852. [Google Scholar] [CrossRef]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T.S. Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Innominato, P.F.; Roche, V.P.; Palesh, O.G.; Ulusakarya, A.; Spiegel, D.; Lévi, F.A. The circadian timing system in clinical oncology. Ann. Med. 2014, 46, 191–207. [Google Scholar] [CrossRef] [PubMed]



| Exosomal ncRNA | Type of Disease | Source of Exosome | Expression | References |
|---|---|---|---|---|
| miRNA-21, miRNA-1224, miRNA-1229, miRNA-1246, miRNA-150, miRNA-21, miRNA-223, miRNA-23a | Colorectal cancer | Blood | ↑, ↑, ↑, ↑, ↑, ↑, ↑, ↑ | [70,71] |
| miRNA-21, miRNA-105, miRNA-372 | Breast cancer | Blood | ↑, ↑, ↑ | [72,73] |
| miRNA-373 | Triple-negative breast cancer | Blood | ↑ | [74] |
| miRNA-21, miRNA-141, miRNA-200a, miRNA-200c, miRNA-200b, miRNA-203, miRNA-205, miRNA-214 | Ovarian cancer | Blood | ↑, ↑, ↑, ↑, ↑, ↑, ↑, ↑ | |
| miRNA-17-5p, miRNA-21, miRNA-106a, miRNA-106b | Pancreatic cancer | Blood | ↑, ↑, ↑, ↑ | [75,76] |
| miRNA-184 | Tongue cancer | Blood | ↑ | [77] |
| miRNA-21, miRNA-500, mi-RNA-26a, mi-RNA-26c, miRNA-224, miRNA-665, miRNA-10b-5p, miRNA-18a-5p, miRNA-215-5p, miRNA-940mi-RNA-199a, miRNA-18a, miRNA-221, miRNA-222 | Hepatocellular carcinoma | Blood | ↑, ↓, ↓, ↓, ↑, ↑, ↑, ↑, ↑, ↑, ↑, ↑, ↑ | [78,79,80,81,82] |
| miRNA-125a, miRNA-200a | Oral squamous cell carcinoma | Blood | ↓ | [83] |
| circ-DB, circCUHRF1, circTMEM4A, circ-100338, circ-0051443 | Hepatocelluar carcinoma | Blood | ↑, ↑, ↑, ↑, ↓, ↓ | [84,85,86,87] |
| circ_0047921, circ_0056285, circ_0007761 | Non-small cell lung cancer | Blood | ↓, ↓, ↑ | [88] |
| circ_0001439, circ_0001492, circ_0000896 | Lung adenocarcinoma | Blood | ↑, ↑, ↑ | [89] |
| circMYC | Nasopharyngeal carcinoma | Blood | ↑ | [90] |
| circ_0047921, circ_0056285, circ_0007761 | Non-small cell lung cancer | Blood | ↓, ↓, ↑ | [88] |
| hsa_circ_0055202, hsa_circ_0074920 hsa_circ_0043722 | Glioblastoma | Blood | ↑, ↑, ↑ | [91] |
| circPDLIM5 | Prostate cancer | Urine | ↑ | [92] |
| Exosomal Component | Effect | Mechanism of Impact | References |
|---|---|---|---|
| miRNA-93 | Tumor proliferation | Enhances the growth of HCC cells | [111,112] |
| PDGFRα, Hedgehog ligands | Fibrogenesis, angiogenesis | Promotes tissue remodeling and blood vessel formation | [64] |
| miRNA-1247-3p | Lung metastasis | Converts cells to cancer-associated fibroblasts and increases pro-inflammatory cytokines (IL-6, IL-8) | [18] |
| Circ-0004277, miRNA-136-5p | EMT, tissue invasion | Induces epithelial-to-mesenchymal transition in surrounding cells | [113,114] |
| miRNA-21, miRNA-10b | Tumor proliferation, metastasis | Regulation of the tumor suppressor gene PTEN through modulation of Tet methylcytosine dioxygenase expression. Increases HIF-1α mRNA and promotes survival under hypoxia | [110] |
| miRNA-155 | Inflammation | Intensifies IL-6 and IL-8 levels and enhances inflammatory response | [115,116] |
| GOLM1 | Tumor occurrence, metastasis | Activates GSK-3β/MMP signaling, a potential biomarker | [38,117] |
| miRNA-320a, miRNA-451a | Anti-tumorigenic effects | Inhibits proliferation, migration, angiogenesis | [118,119] |
| LncRNA H19 | Enhanced angiogenesis | Increases VEGF secretion and VEGF-R1 production | [120] |
| miRNA-32 | Angiogenesis | Suppresses PTEN and activates PI3K/Akt pathway | [110] |
| VASN | Tumor development, angiogenesis | Stimulates endothelial cell proliferation and neovascularization | [121] |
| miRNA-200b-3p | Anti-angiogenic | Inhibits erythroblast transformation-related genes | [122] |
| HMGB1 | Immune evasion | Expands regulatory B cells, facilitating tumor survival | [123] |
| Exosomal SMAD, Caveolin, MET, caveolins, S100, ITGαvβ5, OXL4, SDF-1α, IL-6, IL-8, AFP, and GGT | HCC progression | Cell adhesion, motility, invasive abilities, metastasis, angiogenesis, and cell proliferation | [124,125,126,127,128] |
| miRNA-92a-2-5p | Promotes metastasis | Enhances cancer cell invasion via AR/PHLPP/p-AKT/β-catenin | [129] |
| miRNA-125a/b | Impedes tumor-associated macrophages | Targets CD90 in tumor-associated macrophages | [130] |
| Function | Example | Description | Context and Patient Use | Reference |
|---|---|---|---|---|
| Immunomodulation | PD-L1-enriched exosomes | Exosomes carrying PD-L1 inhibited T-cell activity, modulating the immune environment to favor tumor growth and impacting responses to immunotherapy | Oncology immunotherapy research—research phase | [174] |
| Drug Resistance | Exosomes carrying miRNA-1247-3p | Enhanced resistance to sorafenib in HCC by altering gene expression related to drug metabolism and cellular survival pathways | Chemoresistance mechanisms study—research phase | [135] |
| Chemotherapy Delivery | Exosomes for Doxorubicin delivery | Facilitated targeted delivery of Doxorubicin, enhancing drug specificity and minimizing off-target effects | Targeted chemotherapy research—early clinical trials | [178,179] |
| Gene Therapy | miRNA-220a/220b/429 mimics | Delivered miRNAs that regulate oncogenic pathways, providing a method for precision gene therapy | Gene therapy innovation—research phase | [174] |
| Chemoresistance Mechanisms | Exosomes carrying circRNA-SORE and miRNAs in chemoresistance | Studied circRNAs and miRNAs that enhance cellular mechanisms of resistance to chemotherapy agents | Molecular oncology exploration—research phase | [84,190,191,192,193,194,195] |
| Enhanced Immune Surveillance | Exosomes carrying HMGB1 | Examined the role of HMGB1-bearing exosomes in modulating immune surveillance in HCC | Immune surveillance enhancement—research phase | [176] |
| Prognostic Biomarkers | Circulating exosomal PD-L1 levels | Developed and validated exosomal PD-L1 as a biomarker for assessing responses to immunotherapy | Biomarker development—early clinical trials | [174] |
| Therapeutic Drug Delivery | Exosomes in targeted drug delivery | Employed engineered exosomes for specific drug conveyance to tumor sites, reducing systemic toxicity | Drug delivery system development—early clinical trials | [175,176,177,178,179] |
| Circadian Influence on Therapy | Variability in exosome release by circadian rhythms | Circadian rhythms affect the secretion and composition of exosomes, influencing the response | Chronotherapy research—research phase | [8,200] |
| Example | Finding | Reference |
|---|---|---|
| Circadian Variation in ECV Quantity | ECV quantity extracted from peripheral blood, bone marrow, and lungs of mice exhibited time-dependent changes | [215] |
| Circadian Control of Exosomal Cargo | Flot1 regulated circadian control over MMP 14 in tendon fibroblast small ECVs | [200] |
| Impact of Night Shift Work on ECV Cargo | Night shift work disrupted circadian rhythms and exosomal cargo, influencing metabolic health | [216] |
| Circadian Variations in Plasma ECV Characteristics | Plasma ECVs were larger at 10:00 compared to 22:00 in HIV patients | [217] |
| Influence of Exosomal Cargo on Insulin Resistance | Exosomes from obese mice or patients with type II diabetes could induce insulin resistance in lean mice | [218,219,220] |
| Circadian Normalization Factor for Small ECV Biomarkers | Biomarker TSG101 levels in urine showed a circadian correlation with small ECV excretion in healthy rats | [221] |
| Impact of Circadian Clock | Description | Associated Conditions/Models | Reference |
|---|---|---|---|
| Circadian Component and Cancer Role | |||
| BMAL1 Function | BMAL1 promotes metastasis in colorectal cancer through increased exosome secretion | Colorectal cancer The link between circadian control and tumor progression | [17] |
| SIRT1 Mechanisms | SIRT1 interacts with CLOCK-BMAL1 to modulate PER2 stability, affecting exosome secretion and tumor environment interactions | Facilitates tumor microenvironment degradation Breast cancer | [252,253,254,255] |
| SIRT1 Mechanisms | SIRT1 loss leads to enhanced exosome secretion, impacting breast cancer and diabetic nephropathy | Diabetic nephropathy | [256,257,258] |
| SIRT1 in Cancer | Shift-work-related miRNA-22-3p uptake by nurses is linked to increased insulin resistance, highlighting its potential as a biomarker for diabetes prevention | Ovarian cancer | [259] |
| Shift Work and its Effects | |||
| Shift-Work-Induced Changes | Chronic shift work in mice alters intestinal flora and increases colonic permeability, affecting circadian gene expression via changes in plasma ECV components | Mouse model of chronic shift work | [251] |
| Night Shift Exosomal Impact | Night shift conditions lead to reduced insulin sensitivity in adipocytes through alterations in exosome content, affecting core clock genes | Simulated shift work study Affects core clock genes and metabolic functions | [216] |
| Shift Work and Diabetes Risk | Shift-work-related miRNA-22-3p uptake by nurses is linked to increased insulin resistance, highlighting its potential as a biomarker for diabetes prevention | Shift nurses | [267] |
| Exosomal miRNAs and Circadian Genes | miRNA-3614-5p as a messenger of circadian misalignment in night shift workers | Contributes to metabolic dysfunction | [216] |
| Circadian Genes and miRNA | Circulating miRNAs like miRNA-219, miRNA-152, miRNA-494, and miRNA-142-3p regulate clock genes | Affects BMAL1 and PER1 in circadian regulation | [268,269,270,271,272] |
| Circadian Genes and miRNA | Exosomal miRNAs modulate peripheral circadian oscillators in various disease models | Has impact on glioma progression and Parkinson’s disease | [9,264] |
| Melatonin’s Therapeutic Role | |||
| Melatonin-Enhanced Exosomes | Melatonin pre-treatment enhances anticancer and anti-inflammatory properties of exosomes, improving their therapeutic effectiveness in various clinical settings | Cancer therapies, various models | [273,274,275] |
| Anti-Inflammatory Effects | Melatonin-treated exosomes significantly reduce inflammatory markers and PD-L1 expression in macrophages, aiding in cancer therapy and reducing immune evasion | Hepatocellular carcinoma | [247] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fekry, B.; Ugartemendia, L.; Esnaola, N.F.; Goetzl, L. Extracellular Vesicles, Circadian Rhythms, and Cancer: A Comprehensive Review with Emphasis on Hepatocellular Carcinoma. Cancers 2024, 16, 2552. https://doi.org/10.3390/cancers16142552
Fekry B, Ugartemendia L, Esnaola NF, Goetzl L. Extracellular Vesicles, Circadian Rhythms, and Cancer: A Comprehensive Review with Emphasis on Hepatocellular Carcinoma. Cancers. 2024; 16(14):2552. https://doi.org/10.3390/cancers16142552
Chicago/Turabian StyleFekry, Baharan, Lierni Ugartemendia, Nestor F. Esnaola, and Laura Goetzl. 2024. "Extracellular Vesicles, Circadian Rhythms, and Cancer: A Comprehensive Review with Emphasis on Hepatocellular Carcinoma" Cancers 16, no. 14: 2552. https://doi.org/10.3390/cancers16142552
APA StyleFekry, B., Ugartemendia, L., Esnaola, N. F., & Goetzl, L. (2024). Extracellular Vesicles, Circadian Rhythms, and Cancer: A Comprehensive Review with Emphasis on Hepatocellular Carcinoma. Cancers, 16(14), 2552. https://doi.org/10.3390/cancers16142552






