Transcriptome-Wide Association Study Reveals New Molecular Interactions Associated with Melanoma Pathogenesis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. GWAS Summary Statistics Dataset for Melanoma
2.2. Gene Expression Dataset for Melanoma
2.3. Transcriptome-Wide Association Study (TWAS)
2.4. Statistical Test
2.5. Expansion of TWAS-Associated Genes with Partner Proteins
2.6. Gene Ontology (GO) and Enrichment Analysis
2.7. Corresponding MicroRNA and Disease Enrichment Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bush, W.S.; Moore, J.H. Chapter 11: Genome-Wide Association Studies. PLoS Comput. Biol. 2012, 8, e1002822. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Xing, L.; He, H.; Zhang, X. A short review on Genome-Wide Association Studies. Bioinformation 2020, 16, 393–397. [Google Scholar]
- Wang, M.H.; Cordell, H.J.; Van Steen, K. Statistical methods for genome-wide association studies. Semin. Cancer Biol. 2019, 55, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Zhao, H. Statistical Methods in Genome-Wide Association Studies. Annu. Rev. Biomed. Data Sci. 2020, 3, 265–288. [Google Scholar] [CrossRef]
- Schaid, D.J.; Chen, W.; Larson, N.B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018, 19, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, M.; Sinnott-Armstrong, N.; Mancuso, N.; Barbeira, A.N.; Knowles, D.A.; Golan, D.; Ermel, R.; Ruusalepp, A.; Quertermous, T.; Hao, K.; et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019, 51, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.J.; Mihm, M.C. Melanoma. N. Engl. J. Med. 2006, 355, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Schadendorf, D.; Fisher, D.E.; Garbe, C.; Gershenwald, J.E.; Grob, J.-J.; Halpern, A.; Herlyn, M.; Marchetti, M.A.; McArthur, G.; Ribas, A.; et al. Melanoma. Nat. Rev. Dis. Primers 2015, 1, 15003. [Google Scholar] [CrossRef]
- Xia, Y.; Xu, F.; Xiong, M.; Yang, H.; Lin, W.; Xie, Y.; Xi, H.; Xue, Q.; Ye, T.; Yu, L. Repurposing of antipsychotic trifluoperazine for treating brain metastasis, lung metastasis and bone metastasis of melanoma by disrupting autophagy flux. Pharmacol. Res. 2020, 163, 105295. [Google Scholar] [CrossRef]
- Landi, M.T.; Bishop, D.T.; MacGregor, S.; Machiela, M.J.; Stratigos, A.J.; Ghiorzo, P.; Brossard, M.; Calista, D.; Choi, J.; Fargnoli, M.C.; et al. Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility. Nat. Genet. 2020, 52, 494–504. [Google Scholar] [CrossRef]
- Zhang, T.; Choi, J.; Kovacs, M.A.; Shi, J.; Xu, M.; Goldstein, A.M.; Trower, A.J.; Bishop, D.T.; Iles, M.M.; Duffy, D.L.; et al. Cell-type–specific eQTL of primary melanocytes facilitates identification of melanoma susceptibility genes. Genome Res. 2018, 28, 1621–1635. [Google Scholar] [CrossRef]
- Fidalgo, F.; Torrezan, G.T.; de Sá, B.C.S.; Barros, B.D.d.F.; Moredo, L.F.; Valieris, R.; de Souza, S.J.; Duprat, J.P.; Krepischi, A.C.V.; Carraro, D.M. Family-based whole-exome sequencing identifies rare variants potentially related to cutaneous melanoma predisposition in Brazilian melanoma-prone families. PLoS ONE 2022, 17, e0262419. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Chen, J.; Zhao, Y.; Gu, L.; Shao, X.; Li, J.; Xu, Y.; Liu, Z.; Xu, Q. Key candidate genes of STAT1 and CXCL10 in melanoma identified by integrated bioinformatical analysis. IUBMB Life 2019, 71, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Song, Z.; Xu, Z.; Tao, Y.; Wu, Y.; Wan, X. Screening of gene markers related to the prognosis of metastatic skin cutaneous melanoma based on Logit regression and survival analysis. BMC Med. Genom. 2021, 14, 96. [Google Scholar] [CrossRef] [PubMed]
- Arnoff, T.E.; El-Deiry, W.S. MDM2/MDM4 amplification and CDKN2A deletion in metastatic melanoma and glioblastoma multiforme may have implications for targeted therapeutics and immunotherapy. Am. J. Cancer Res. 2022, 12, 2102–2117. [Google Scholar] [PubMed]
- Calbet-Llopart, N.; Combalia, M.; Kiroglu, A.; Potrony, M.; Tell-Martí, G.; Combalia, A.; Brugues, A.; Podlipnik, S.; Carrera, C.; Puig, S.; et al. Common genetic variants associated with melanoma risk or naevus count in patients with wildtype MC1R melanoma. Br. J. Dermatol. 2022, 187, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Walbrecq, G.; Lecha, O.; Gaigneaux, A.; Fougeras, M.R.; Philippidou, D.; Margue, C.; Nomigni, M.T.; Bernardin, F.; Dittmar, G.; Behrmann, I.; et al. Hypoxia-Induced Adaptations of miRNomes and Proteomes in Melanoma Cells and Their Secreted Extracellular Vesicles. Cancers 2020, 12, 692. [Google Scholar] [CrossRef] [PubMed]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- Canela-Xandri, O.; Rawlik, K.; Tenesa, A. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018, 50, 1593–1599. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M.; The Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.W.J.H.; Jansen, R.; de Geus, E.J.C.; I Boomsma, D.; Wright, F.A.; et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016, 48, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Gamazon, E.R.; Wheeler, H.E.; Shah, K.P.; Mozaffari, S.V.; Aquino-Michaels, K.; Carroll, R.J.; Eyler, A.E.; Denny, J.C.; Nicolae, D.L.; et al.; GTEx Consortium A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015, 47, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to accessing data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef] [PubMed]
- Boughton, A.P.; Welch, R.P.; Flickinger, M.; VandeHaar, P.; Taliun, D.; Abecasis, G.R.; Boehnke, M. LocusZoom.js: Interactive and embeddable visualization of genetic association study results. Bioinformatics 2021, 37, 3017–3018. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef]
- Li, C.; Tang, Z.; Zhang, W.; Ye, Z.; Liu, F. GEPIA2021: Integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021, 49, W242–W246. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer Nature: Singapore, 2022; pp. 27–56. [Google Scholar]
- Nazarieh, M.; Hamed, M.; Spaniol, C.; Will, T.; Helms, V. TFmiR2: Constructing and analyzing disease-, tissue- and process-specific transcription factor and microRNA co-regulatory networks. Bioinformatics 2019, 36, 2300–2302. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, X.; Wan, Y.; Zhang, S.; Zhao, Y.; Fan, R.; Cui, Q.; Zhou, Y. TAM 2.0: Tool for MicroRNA set analysis. Nucleic Acids Res. 2018, 46, W180–W185. [Google Scholar] [CrossRef] [PubMed]
- Urtatiz, O.; Haage, A.; Tanentzapf, G.; Van Raamsdonk, C.D. Crosstalk with keratinocytes causes GNAQ oncogene specificity in melanoma. eLife 2021, 10, 71825. [Google Scholar] [CrossRef] [PubMed]
- Kushima, I.; Aleksic, B.; Nakatochi, M.; Shimamura, T.; Okada, T.; Uno, Y.; Morikawa, M.; Ishizuka, K.; Shiino, T.; Kimura, H.; et al. Comparative Analyses of Copy-Number Variation in Autism Spectrum Disorder and Schizophrenia Reveal Etiological Overlap and Biological Insights. Cell Rep. 2018, 24, 2838–2856. [Google Scholar] [CrossRef] [PubMed]
- Bracalente, C.; Ibañez, I.L.; Berenstein, A.; Notcovich, C.; Cerda, M.B.; Klamt, F.; Chernomoretz, A.; Durán, H. Reprogramming human A375 amelanotic melanoma cells by catalase overexpression: Upregulation of antioxidant genes correlates with regression of melanoma malignancy and with malignant progression when downregulated. Oncotarget 2016, 7, 41154–41171. [Google Scholar] [CrossRef] [PubMed]
- Gembarska, A.; Luciani, F.; Fedele, C.; Russell, E.A.; Dewaele, M.; Villar, S.; Zwolinska, A.; Haupt, S.; de Lange, J.; Yip, D.; et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat. Med. 2012, 18, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- AbuHammad, S.; Cullinane, C.; Martin, C.; Bacolas, Z.; Ward, T.; Chen, H.; Slater, A.; Ardley, K.; Kirby, L.; Chan, K.T.; et al. Regulation of PRMT5–MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc. Natl. Acad. Sci. USA 2019, 116, 17990–18000. [Google Scholar] [CrossRef]
- Alatawi, A.; Kho, S.; Markey, M.P. MDM4 Isoform Expression in Melanoma Supports an Oncogenic Role for MDM4-A. J. Ski. Cancer 2021, 2021, 3087579. [Google Scholar] [CrossRef]
- Tusup, M.; Cheng, P.F.; Picardi, E.; Raziunaite, A.; Dummer, R.; Levesque, M.P.; French, L.E.; Guenova, E.; Kundig, T.M.; Pascolo, S. Evaluation of the Interplay between the ADAR Editome and Immunotherapy in Melanoma. Non-Coding RNA 2021, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Nykamp, K.; Anderson, M.; Powers, M.; Garcia, J.; Herrera, B.; Ho, Y.-Y.; Kobayashi, Y.; Patil, N.; Thusberg, J.; Westbrook, M.; et al. Sherloc: A comprehensive refinement of the ACMG–AMP variant classification criteria. Anesth. Analg. 2017, 19, 1105–1117. [Google Scholar] [CrossRef] [PubMed]
- Watkin, L.B.; Jessen, B.; Wiszniewski, W.; Vece, T.J.; Jan, M.; Sha, Y.; Thamsen, M.; Santos-Cortez, R.L.P.; Lee, K.; Gambin, T.; et al. COPA mutations impair ER-Golgi transport and cause hereditary autoimmune-mediated lung disease and arthritis. Nat. Genet. 2015, 47, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Ohbayashi, N.; Fukuda, M. Recent advances in understanding the molecular basis of melanogenesis in melanocytes. F1000Research 2020, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- McDonald, S.L.; Edington, H.D.; Kirkwood, J.M.; Becker, D. Expression Analysis of Genes identified by Molecular Profiling of VGP Melanomas and MGP Melanoma-Positive Lymph Nodes. Cancer Biol. Ther. 2004, 3, 110–120. [Google Scholar] [CrossRef]
- Lazar, I.; Fabre, B.; Feng, Y.; Khateb, A.; Turko, P.; Gomez, J.M.M.; Frederick, D.T.; Levesque, M.P.; Feld, L.; Zhang, G.; et al. SPANX Control of Lamin A/C Modulates Nuclear Architecture and Promotes Melanoma Growth. Mol. Cancer Res. 2020, 18, 1560–1573. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.Y.; Lee, J.H.; Gide, T.N.; Menzies, A.M.; Guminski, A.; Carlino, M.S.; Breen, E.J.; Yang, J.Y.H.; Ghazanfar, S.; Kefford, R.F.; et al. Circulating Cytokines Predict Immune-Related Toxicity in Melanoma Patients Receiving Anti-PD-1–Based Immunotherapy. Clin. Cancer Res. 2019, 25, 1557–1563. [Google Scholar] [CrossRef] [PubMed]
- Jandova, J.; Wondrak, G.T. Genomic GLO1 deletion modulates TXNIP expression, glucose metabolism, and redox homeostasis while accelerating human A375 malignant melanoma tumor growth. Redox Biol. 2020, 39, 101838. [Google Scholar] [CrossRef] [PubMed]
- Kholmanskikh, O.; van Baren, N.; Brasseur, F.; Ottaviani, S.; Vanacker, J.; Arts, N.; van der Bruggen, P.; Coulie, P.; De Plaen, E. Interleukins 1α and 1β secreted by some melanoma cell lines strongly reduce expression of MITF-M and melanocyte differentiation antigens. Int. J. Cancer 2010, 127, 1625–1636. [Google Scholar] [CrossRef]
- Young, H.L.; Rowling, E.J.; Bugatti, M.; Giurisato, E.; Luheshi, N.; Arozarena, I.; Acosta, J.-C.; Kamarashev, J.; Frederick, D.T.; Cooper, Z.A.; et al. An adaptive signaling network in melanoma inflammatory niches confers tolerance to MAPK signaling inhibition. J. Exp. Med. 2017, 214, 1691–1710. [Google Scholar] [CrossRef]
- Rovera, C.; Berestjuk, I.; Lecacheur, M.; Tavernier, C.; Diazzi, S.; Pisano, S.; Irondelle, M.; Mallavialle, A.; Albrengues, J.; Gaggioli, C.; et al. Secretion of IL1 by Dedifferentiated Melanoma Cells Inhibits JAK1-STAT3–Driven Actomyosin Contractility of Lymph Node Fibroblastic Reticular Cells. Cancer Res. 2022, 82, 1774–1788. [Google Scholar] [CrossRef] [PubMed]
- Pich, C.; Meylan, P.; Mastelic-Gavillet, B.; Nguyen, T.N.; Loyon, R.; Trang, B.K.; Moser, H.; Moret, C.; Goepfert, C.; Hafner, J.; et al. Induction of Paracrine Signaling in Metastatic Melanoma Cells by PPARγ Agonist Rosiglitazone Activates Stromal Cells and Enhances Tumor Growth. Cancer Res. 2018, 78, 6447–6461. [Google Scholar] [CrossRef] [PubMed]
- Õunap, K.; Ilus, T.; Laidre, P.; Uibo, O.; Tammur, P.; Bartsch, O. A new case of 2q duplication supports either a locus for orofacial clefting between markers D2S1897 and D2S2023 or a locus for cleft palate only on chromosome 2q13-q21. Am. J. Med. Genet. Part A 2005, 137, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Wenger, S.L.; Bleigh, O.C.; Hummel, M. Cleft Palate in a Newborn with Duplication 2(q13q23). Cleft Palate-Craniofacial J. 2004, 41, 568–570. [Google Scholar] [CrossRef]
- Kazenwadel, J.; Secker, G.A.; Liu, Y.J.; Rosenfeld, J.A.; Wildin, R.S.; Cuellar-Rodriguez, J.; Hsu, A.P.; Dyack, S.; Fernandez, C.V.; Chong, C.-E.; et al. Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood 2012, 119, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Neira, J.; Pehlivan, D.; Santiago-Sim, T.; Song, X.; Rosenfeld, J.; Posey, J.E.; Patel, V.; Jin, W.; Adam, M.P.; et al. Clinical exome sequencing reveals locus heterogeneity and phenotypic variability of cohesinopathies. Anesth. Analg. 2019, 21, 663–675. [Google Scholar] [CrossRef] [PubMed]
- McCannel, T.A.; Burgess, B.L.; Nelson, S.F.; Eskin, A.; Straatsma, B.R. Genomic Identification of Significant Targets in Ciliochoroidal Melanoma. Investig. Opthalmol. Vis. Sci. 2011, 52, 3018–3022. [Google Scholar] [CrossRef]
- Beesley, C.E.; Meaney, C.A.; Greenland, G.; Adams, V.; Vellodi, A.; Young, E.P.; Winchester, B.G. Mutational analysis of 85 mucopolysaccharidosis type I families: Frequency of known mutations, identification of 17 novel mutations and in vitro expression of missense mutations. Hum. Genet. 2001, 109, 503–511. [Google Scholar]
- Wang, X.; Zhang, W.; Shi, H.; Qiu, Z.; Meng, Y.; Yao, F.; Wei, M. Mucopolysaccharidosis I mutations in Chinese patients: Identification of 27 novel mutations and 6 cases involving prenatal diagnosis. Clin. Genet. 2011, 81, 443–452. [Google Scholar] [CrossRef]
- Breen, C.; Mercer, J.; Jones, S.A.; Jahic, A.; Heptinstall, L.; Tylee, K.; Newman, W.G.; Beetz, C. Maternal mosaicism for IDUA deletion clarifies recurrence risk in MPS I. Hum. Genome Var. 2016, 3, 16031. [Google Scholar] [CrossRef]
- Lindberg, J.; Mills, I.G.; Klevebring, D.; Liu, W.; Neiman, M.; Xu, J.; Wikström, P.; Wiklund, P.; Wiklund, F.; Egevad, L.; et al. The Mitochondrial and Autosomal Mutation Landscapes of Prostate Cancer. Eur. Urol. 2013, 63, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Preiksaitiene, E.; Kasnauskiene, J.; Ciuladaite, Z.; Tumiene, B.; Patsalis, P.C.; Kučinskas, V. Clinical and molecular characterization of a second case of 7p22.1 microduplication. Am. J. Med. Genet. Part A 2012, 158, 1200–1203. [Google Scholar] [CrossRef] [PubMed]
- Caselli, R.; Ballarati, L.; Vignoli, A.; Peron, A.; Recalcati, M.P.; Catusi, I.; Larizza, L.; Giardino, D. 7p22.1 microduplication syndrome: Clinical and molecular characterization of an adult case and review of the literature. Eur. J. Med. Genet. 2015, 58, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Ronzoni, L.; Grassi, F.S.; Pezzani, L.; Tucci, A.; Baccarin, M.; Esposito, S.; Milani, D. 7p22.1 microduplication syndrome: Refinement of the critical region. Eur. J. Med. Genet. 2017, 60, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhao, H.; Wang, X.; Sun, J.; Su, J. Analysis of long noncoding RNAs highlights region-specific altered expression patterns and diagnostic roles in Alzheimer’s disease. Brief. Bioinform. 2018, 20, 598–608. [Google Scholar] [CrossRef]
- Lancaster, J.M.; Zgheib, N.B.; Xiong, Y.; Marchion, D.C.; Bicaku, E.; Chon, H.S.; Stickles, X.B.; Al Sawah, E.; Judson, P.L.; Hakam, A.; et al. The O-glycan pathway is associated with in vitro sensitivity to gemcitabine and overall survival from ovarian cancer. Int. J. Oncol. 2012, 41, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wu, N.; Gao, F.; Han, J.; Yang, Y.; Zhou, C.; Sun, W.; Xian, L.; Cheng, Y.; Li, B.; et al. MTERFD1 functions as an oncogene. Oncotarget 2016, 5, 11140. [Google Scholar] [CrossRef][Green Version]
- Garrisi, V.M.; Strippoli, S.; De Summa, S.; Pinto, R.; Perrone, A.; Guida, G.; Azzariti, A.; Guida, M.; Stefania, T. Proteomic Profile and In Silico Analysis in Metastatic Melanoma with and without BRAF Mutation. PLoS ONE 2014, 9, e112025. [Google Scholar] [CrossRef]
- Zhao, X.; Lei, Y.; Li, G.; Cheng, Y.; Yang, H.; Xie, L.; Long, H.; Jiang, R. Integrative analysis of cancer driver genes in prostate adenocarcinoma. Mol. Med. Rep. 2019, 19, 2707–2715. [Google Scholar] [CrossRef]
- Larsson, M.; Duffy, D.L.; Zhu, G.; Liu, J.Z.; Macgregor, S.; McRae, A.F.; Wright, M.J.; Sturm, R.A.; Mackey, D.A.; Montgomery, G.W.; et al. GWAS Findings for Human Iris Patterns: Associations with Variants in Genes that Influence Normal Neuronal Pattern Development. Am. J. Hum. Genet. 2011, 89, 334–343. [Google Scholar] [CrossRef]
- Kasak, L.; Rull, K.; Vaas, P.; Teesalu, P.; Laan, M. Extensive load of somatic CNVs in the human placenta. Sci. Rep. 2015, 5, 8342. [Google Scholar] [CrossRef]
- Haymaker, C.L.; Wu, R.C.; Ritthipichai, K.; Bernatchez, C.; Forget, M.-A.; Chen, J.Q.; Liu, H.; Wang, E.; Marincola, F.; Hwu, P.; et al. BTLA marks a less-differentiated tumor-infiltrating lymphocyte subset in melanoma with enhanced survival properties. OncoImmunology 2015, 4, e1014246. [Google Scholar] [CrossRef]
- Orgaz, J.L.; Benguria, A.; Sanchez-Martinez, C.; Ladhani, O.; Volpert, O.V.; Jimenez, B. Changes in the gene expression profile of A375 human melanoma cells induced by overexpression of multifunctional pigment epithelium-derived factor. Melanoma Res. 2011, 21, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Turro, E.; Astle, W.J.; Megy, K.; Gräf, S.; Greene, D.; Shamardina, O.; Allen, H.L.; Sanchis-Juan, A.; Frontini, M.; Thys, C.; et al. Whole-genome sequencing of patients with rare diseases in a national health system. Nature 2020, 583, 96–102. [Google Scholar] [CrossRef]
- Miao, Y.; Zhang, W.; Liu, S.; Leng, X.; Hu, C.; Sun, H. HOXC10 promotes growth and migration of melanoma by regulating Slug to activate the YAP/TAZ signaling pathway. Discov. Oncol. 2021, 12, 12. [Google Scholar] [CrossRef]
- Cillo, C.; Cantile, M.; Mortarini, R.; Barba, P.; Parmiani, G.; Anichini, A. Differential patterns of HOX gene expression are associated with specific integrin and ICAM profiles in clonal populations isolated from a single human melanoma metastasis. Int. J. Cancer 1996, 66, 692–697. [Google Scholar] [CrossRef]
- Bhattacharya, C.; Wang, X.; Becker, D. The DEAD/DEAH box helicase, DDX11, is essential for the survival of advanced melanomas. Mol. Cancer 2012, 11, 82. [Google Scholar] [CrossRef] [PubMed]
- Capo-Chichi, J.-M.; Bharti, S.K.; Sommers, J.A.; Yammine, T.; Chouery, E.; Patry, L.; Rouleau, G.A.; Samuels, M.E.; Hamdan, F.F.; Michaud, J.L.; et al. Identification and Biochemical Characterization of a Novel Mutation in DDX11 Causing Warsaw Breakage Syndrome. Hum. Mutat. 2012, 34, 103–107. [Google Scholar] [CrossRef]
- Sun, X.; Chen, H.; Deng, Z.; Hu, B.; Luo, H.; Zeng, X.; Han, L.; Cai, G.; Ma, L. The Warsaw breakage syndrome-related protein DDX11 is required for ribosomal RNA synthesis and embryonic development. Hum. Mol. Genet. 2015, 24, 4901–4915. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Pazhoor, S.; Bao, M.; Zhang, Z.; Hanabuchi, S.; Facchinetti, V.; Bover, L.; Plumas, J.; Chaperot, L.; Qin, J.; et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA 2010, 107, 15181–15186. [Google Scholar] [CrossRef]
- Rabin, R.; Hirsch, Y.; Johansson, M.M.; Ekstein, J.; Zeevi, D.A.; Keena, B.; Zackai, E.H.; Pappas, J. Study of carrier frequency of Warsaw breakage syndrome in the Ashkenazi Jewish population and presentation of two cases. Am. J. Med. Genet. Part A 2019, 179, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Van Schie, J.J.M.; Faramarz, A.; Balk, J.A.; Stewart, G.S.; Cantelli, E.; Oostra, A.B.; Rooimans, M.A.; Parish, J.L.; de Almeida Estéves, C.; Dumic, K.; et al. Warsaw breakage syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion. Nat. Commun. 2020, 11, 4287. [Google Scholar] [CrossRef] [PubMed]
- van der Lelij, P.; Chrzanowska, K.H.; Godthelp, B.C.; Rooimans, M.A.; Oostra, A.B.; Stumm, M.; Zdzienicka, M.Z.; Joenje, H.; de Winter, J.P. Warsaw Breakage Syndrome, a Cohesinopathy Associated with Mutations in the XPD Helicase Family Member DDX11/ChlR1. Am. J. Hum. Genet. 2010, 86, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Alkhunaizi, E.; Shaheen, R.; Bharti, S.K.; Joseph-George, A.M.; Chong, K.; Abdel-Salam, G.M.H.; Alowain, M.; Blaser, S.I.; Papsin, B.C.; Butt, M.; et al. Warsaw breakage syndrome: Further clinical and genetic delineation. Am. J. Med. Genet. Part A 2018, 176, 2404–2418. [Google Scholar] [CrossRef] [PubMed]
- Megy, K.; Downes, K.; Morel-Kopp, M.; Bastida, J.M.; Brooks, S.; Bury, L.; Leinoe, E.; Gomez, K.; Morgan, N.V.; Othman, M.; et al. GoldVariants, a resource for sharing rare genetic variants detected in bleeding, thrombotic, and platelet disorders: Communication from the ISTH SSC Subcommittee on Genomics in Thrombosis and Hemostasis. J. Thromb. Haemost. 2021, 19, 2612–2617. [Google Scholar] [CrossRef] [PubMed]
- Souri, M.; Koseki-Kuno, S.; Iwata, H.; Kemkes-Matthes, B.; Ichinose, A. A naturally occurring E30Q mutation in the Gla domain of protein Z causes its impaired secretion and subsequent deficiency. Blood 2005, 105, 3149–3154. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Kokubu, A.; Miyamoto, M.; Sasajima, Y.; Yamazaki, N. Mutant IDH1 Confers an in Vivo Growth in a Melanoma Cell Line with BRAF Mutation. Am. J. Pathol. 2011, 178, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Richard, E.M.; Bakhtiari, S.; Marsh, A.P.L.; Kaiyrzhanov, R.; Wagner, M.; Shetty, S.; Pagnozzi, A.; Nordlie, S.M.; Guida, B.S.; Cornejo, P.; et al. Bi-allelic variants in SPATA5L1 lead to intellectual disability, spastic-dystonic cerebral palsy, epilepsy, and hearing loss. Am. J. Hum. Genet. 2021, 108, 2006–2016. [Google Scholar] [CrossRef] [PubMed]
- Feichtinger, J.; Larcombe, L.; McFarlane, R.J. Meta-analysis of expression of l(3)mbt tumor-associated germline genes supports the model that a soma-to-germline transition is a hallmark of human cancers. Int. J. Cancer 2013, 134, 2359–2365. [Google Scholar] [CrossRef]
- Shiohama, Y.; Ohtake, J.; Ohkuri, T.; Noguchi, D.; Togashi, Y.; Kitamura, H.; Nishimura, T. Identification of a meiosis-specific protein, MEIOB, as a novel cancer/testis antigen and its augmented expression in demethylated cancer cells. Immunol. Lett. 2014, 158, 175–182. [Google Scholar] [CrossRef]
- Jay, A.; Reitz, D.; Namekawa, S.H.; Heyer, W.-D. Cancer testis antigens and genomic instability: More than immunology. DNA Repair 2021, 108, 103214. [Google Scholar] [CrossRef] [PubMed]
- Gershoni, M.; Hauser, R.; Yogev, L.; Lehavi, O.; Azem, F.; Yavetz, H.; Pietrokovski, S.; Kleiman, S.E. A familial study of azoospermic men identifies three novel causative mutations in three new human azoospermia genes. Anesth. Analg. 2017, 19, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Rendtorff, N.D.; Bjerregaard, B.; Frödin, M.; Kjaergaard, S.; Hove, H.; Skovby, F.; Brøndum-Nielsen, K.; Schwartz, M.; Danish Tuberous Sclerosis Group. Analysis of 65 tuberous sclerosis complex (TSC) patients by TSC2 DGGE, TSC1/TSC2 MLPA, and TSC1 long-range PCR sequencing, and report of 28 novel mutations. Hum. Mutat. 2005, 26, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, P.; Roberts, P.; Dabora, S.; Franz, D.; Bissler, J.; Northrup, H.; Au, K.S.; Lazarus, R.; Domanska-Pakiela, D.; Kotulska, K.; et al. Identification of 54 large deletions/duplications in TSC1 and TSC2 using MLPA, and genotype-phenotype correlations. Hum. Genet. 2007, 121, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Crino, P.B.; Aronica, E.; Baltuch, G.; Nathanson, K.L. Biallelic TSC gene inactivation in tuberous sclerosis complex. Neurology 2010, 74, 1716–1723. [Google Scholar] [CrossRef] [PubMed]
- Glushkova, M.; Bojinova, V.; Koleva, M.; Dimova, P.; Bojidarova, M.; Litvinenko, I.; Todorov, T.; Iluca, E.; Calusaru, C.; Neagu, E.; et al. Molecular genetic diagnostics of tuberous sclerosis complex in Bulgaria: Six novel mutations in the TSC1 and TSC2 genes. J. Genet. 2018, 97, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Griffin, N.G.; Cronin, K.D.; Walley, N.M.; Hulette, C.M.; Grant, G.A.; Mikati, M.A.; LaBreche, H.G.; Rehder, C.W.; Allen, A.S.; Crino, P.B.; et al. Somatic uniparental disomy of Chromosome 16p in hemimegalencephaly. Mol. Case Stud. 2017, 3, a001735. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wan, Q.; Tang, J.; Lu, J.; Jin, L.; Su, Y.; Wang, S.; Cheng, Y.; Liu, Y.; Li, C.; Wang, Z. Six-gene-based prognostic model predicts overall survival in patients with uveal melanoma. Cancer Biomark. 2020, 27, 343–356. [Google Scholar] [CrossRef]
- Yin, J.; Liu, H.; Liu, Z.; Wang, L.-E.; Chen, W.V.; Zhu, D.; Amos, C.I.; Fang, S.; Lee, J.E.; Wei, Q. Genetic Variants in Fanconi Anemia Pathway Genes BRCA2 and FANCA Predict Melanoma Survival. J. Investig. Dermatol. 2015, 135, 542–550. [Google Scholar] [CrossRef]
- Bourseguin, J.; Bonet, C.; Renaud, E.; Pandiani, C.; Boncompagni, M.; Giuliano, S.; Pawlikowska, P.; Karmous-Benailly, H.; Ballotti, R.; Rosselli, F.; et al. FANCD2 functions as a critical factor downstream of MiTF to maintain the proliferation and survival of melanoma cells. Sci. Rep. 2016, 6, 36539. [Google Scholar] [CrossRef]
- Yu, Y.; Hu, H.; Chen, J.-S.; Hu, F.; Fowler, J.; Scheet, P.; Zhao, H.; Huff, C.D. Integrated case-control and somatic-germline interaction analyses of melanoma susceptibility genes. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2018, 1864, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Qureshi, A.A.; Guo, Q.; Han, J. Genetic variation in DNA repair pathway genes and melanoma risk. DNA Repair 2011, 10, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Leong, S.P.L.; Singer, M.I.; Parrett, B.M.; Moretto, J.; Minor, D.R.; Vosoughi, E.; Millis, S.Z.; Ross, J.S.; Kashani-Sabet, M. Frequency of genetic homologous recombination (HR) alterations in metastatic cutaneous melanoma. J. Clin. Oncol. 2017, 35, e21033. [Google Scholar] [CrossRef]
- Kim, K.B.; Soroceanu, L.; de Semir, D.; Millis, S.Z.; Ross, J.; Vosoughi, E.; Dar, A.A.; Nosrati, M.; Desprez, P.-Y.; Ice, R.; et al. Prevalence of Homologous Recombination Pathway Gene Mutations in Melanoma: Rationale for a New Targeted Therapeutic Approach. J. Investig. Dermatol. 2021, 141, 2028–2036.e2. [Google Scholar] [CrossRef] [PubMed]
- Kimble, D.C.; Lach, F.P.; Gregg, S.Q.; Donovan, F.X.; Flynn, E.K.; Kamat, A.; Young, A.; Vemulapalli, M.; Thomas, J.W.; Mullikin, J.C.; et al. A comprehensive approach to identification of pathogenic FANCA variants in Fanconi anemia patients and their families. Hum. Mutat. 2017, 39, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Moghrabi, N.N.; Johnson, M.A.; Yoshitomi, M.J.; Zhu, X.; Al-Dhalimy, M.J.; Olson, S.B.; Grompe, M.; Richards, C.S. Validation of Fanconi anemia complementation Group A assignment using molecular analysis. Anesth. Analg. 2009, 11, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Fransson, S.; Siaw, J.T.; Treis, D.; Van den Eynden, J.; Chand, D.; Umapathy, G.; Ruuth, K.; Svenberg, P.; Wessman, S.; et al. Clinical response of the novel activating ALK-I1171T mutation in neuroblastoma to the ALK inhibitor ceritinib. Mol. Case Stud. 2018, 4, a002550. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Riker, A.; Shevde-Samant, L.; Samant, R.; Morris, C.; Gavin, E.; Fodstad, O.; Ju, J. Global Comparative Gene Expression Analysis of Melanoma Patient Samples, Derived Cell Lines and Corresponding Tumor Xenografts. Cancer Genom. Proteom. 2008, 5, 1–35. [Google Scholar]
- Kang, J.; Lee, J.; Lee, A.; Lee, Y.S. Prediction of BRAF V600E variant from cancer gene expression data. Transl. Cancer Res. 2022, 11, 4051–4056. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Zhang, J.; Xu, G. Prognostic Implication and Oncogenic Role of PNPO in Pan-Cancer. Front. Cell Dev. Biol. 2022, 9, 763674. [Google Scholar] [CrossRef]
- Chen, X.; Yang, M.; Hao, W.; Han, J.; Ma, J.; Wang, C.; Sun, S.; Zheng, Q. Differentiation-inducing and anti-proliferative activities of isoliquiritigenin and all-trans-retinoic acid on B16F0 melanoma cells: Mechanisms profiling by RNA-seq. Gene 2016, 592, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Um, H.N.; Han, J.M.; Hwang, J.; Hong, S.I.; Vaudry, H.; Seong, J.Y. Molecular coevolution of kisspeptins and their receptors from fish to mammals. Ann. N. Y. Acad. Sci. 2010, 1200, 67–74. [Google Scholar] [CrossRef] [PubMed]
- De Francesco, E.M.; Bonuccelli, G.; Maggiolini, M.; Sotgia, F.; Lisanti, M.P. Vitamin C and Doxycycline: A synthetic lethal combination therapy targeting metabolic flexibility in cancer stem cells (CSCs). Oncotarget 2017, 8, 67269–67286. [Google Scholar] [CrossRef]
- Ghosh, S.; Spagnoli, G.C.; Martin, I.; Ploegert, S.; Demougin, P.; Heberer, M.; Reschner, A. Three-dimensional culture of melanoma cells profoundly affects gene expression profile: A high density oligonucleotide array study. J. Cell. Physiol. 2005, 204, 522–531. [Google Scholar] [CrossRef]
- Munteanu, C.V.; Chirițoiu, G.N.; Chirițoiu, M.; Ghenea, S.; Petrescu, A.-J.; Petrescu, Ș.M. Affinity Proteomics and Deglycoproteomics Uncover Novel EDEM2 Endogenous Substrates and an Integrative ERAD Network. Mol. Cell. Proteom. 2021, 20, 100125. [Google Scholar] [CrossRef]
- Munteanu, C.V.A.; Chiriţoiu, G.N.; Petrescu, A.-J.; Petrescu, Ș.M. Profiling Optimal Conditions for Capturing EDEM Proteins Complexes in Melanoma Using Mass Spectrometry. In Advancements of Mass Spectrometry in Biomedical Research; Woods, A.G., Darie, C.C., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 155–167. [Google Scholar]
- Jangi, S.-M.; Ruiz-Larrea, M.B.; Nicolau-Galmés, F.; Andollo, N.; Arroyo-Berdugo, Y.; Ortega-Martínez, I.; Díaz-Pérez, J.L.; Boyano, M.D. Terfenadine-induced apoptosis in human melanoma cells is mediated through Ca2+ homeostasis modulation and tyrosine kinase activity, independently of H1 histamine receptors. Carcinogenesis 2008, 29, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Jangi, S.-M.; Díaz-Pérez, J.L.; Ochoa-Lizarralde, B.; Martín-Ruiz, I.; Asumendi, A.; Pérez-Yarza, G.; Gardeazabal, J.; Díaz-Ramón, J.L.; Boyano, M.D. H1 histamine receptor antagonists induce genotoxic and caspase-2-dependent apoptosis in human melanoma cells. Carcinog. 2006, 27, 1787–1796. [Google Scholar] [CrossRef]
- Pradhan, D.; Jour, G.; Milton, D.; Vasudevaraja, V.; Tetzlaff, M.T.; Nagarajan, P.; Curry, J.L.; Ivan, D.; Long, L.; Ding, Y.; et al. Aberrant DNA Methylation Predicts Melanoma-Specific Survival in Patients with Acral Melanoma. Cancers 2019, 11, 2031. [Google Scholar] [CrossRef] [PubMed]
- Warnatz, K.; Salzer, U.; Rizzi, M.; Fischer, B.; Gutenberger, S.; Böhm, J.; Kienzler, A.-K.; Pan-Hammarström, Q.; Hammarström, L.; Rakhmanov, M.; et al. B-cell activating factor receptor deficiency is associated with an adult-onset antibody deficiency syndrome in humans. Proc. Natl. Acad. Sci. USA 2009, 106, 13945–13950. [Google Scholar] [CrossRef]
- Marchione, R.; Laurin, D.; Liguori, L.; Leibovitch, M.P.; Leibovitch, S.A.; Lenormand, J.-L. MD11-mediated delivery of recombinant eIF3f induces melanoma and colorectal carcinoma cell death. Mol. Ther. Methods Clin. Dev. 2015, 2, 14056. [Google Scholar] [CrossRef]
- Guo, S.; Guo, W.; Li, S.; Dai, W.; Zhang, N.; Zhao, T.; Wang, H.; Ma, J.; Yi, X.; Ge, R.; et al. Serum miR-16: A Potential Biomarker for Predicting Melanoma Prognosis. J. Investig. Dermatol. 2016, 136, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Alderman, C.; Sehlaoui, A.; Xiao, Z.; Yang, Y. MicroRNA-15a inhibits the growth and invasiveness of malignant melanoma and directly targets on CDCA4 gene. Tumor Biol. 2016, 37, 13941–13950. [Google Scholar] [CrossRef] [PubMed]
- Gajos-Michniewicz, A.; Czyz, M. Role of miRNAs in Melanoma Metastasis. Cancers 2019, 11, 326. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.U.; Ullah, Z.; Shaukat, H.; Unab, S.; Jannat, S.; Ali, W.; Ali, A.; Irfan, M.; Khan, M.F.; Cervantes-Villagrana, R.D. TP53 and its Regulatory Genes as Prognosis of Cutaneous Melanoma. Cancer Inform. 2023, 22, 11769351231177267. [Google Scholar] [CrossRef] [PubMed]
- Potrony, M.; Badenas, C.; Aguilera, P.; Puig-Butille, J.A.; Carrera, C.; Malvehy, J.; Puig, S. Update in genetic susceptibility in melanoma. Ann. Transl. Med. 2015, 3, 210. [Google Scholar] [CrossRef] [PubMed]
- Güllülü, Ö.; Hehlgans, S.; Rödel, C.; Fokas, E.; Rödel, F. Tumor Suppressor Protein p53 and Inhibitor of Apoptosis Proteins in Colorectal Cancer—A Promising Signaling Network for Therapeutic Interventions. Cancers 2021, 13, 624. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guo, M.; Wei, H.; Chen, Y. Targeting p53 pathways: Mechanisms, structures, and advances in therapy. Signal Transduct. Target. Ther. 2023, 8, 92. [Google Scholar] [CrossRef]
- Patel, N.; Garikapati, K.R.; Pandita, R.K.; Singh, D.K.; Pandita, T.K.; Bhadra, U.; Bhadra, M.P. miR-15a/miR-16 down-regulates BMI1, impacting Ub-H2A mediated DNA repair and breast cancer cell sensitivity to doxorubicin. Sci. Rep. 2017, 7, 4263. [Google Scholar] [CrossRef] [PubMed]
- Varrone, F.; Caputo, E. The miRNAs Role in Melanoma and in Its Resistance to Therapy. Int. J. Mol. Sci. 2020, 21, 878. [Google Scholar] [CrossRef]
- Dong, L.; Tian, X.; Zhao, Y.; Tu, H.; Wong, A.; Yang, Y. The Roles of MiRNAs (MicroRNAs) in Melanoma Immunotherapy. Int. J. Mol. Sci. 2022, 23, 14775. [Google Scholar] [CrossRef]
- Poniewierska-Baran, A.; Słuczanowska-Głąbowska, S.; Małkowska, P.; Sierawska, O.; Zadroga, Ł.; Pawlik, A.; Niedźwiedzka-Rystwej, P. Role of miRNA in Melanoma Development and Progression. Int. J. Mol. Sci. 2022, 24, 201. [Google Scholar] [CrossRef] [PubMed]
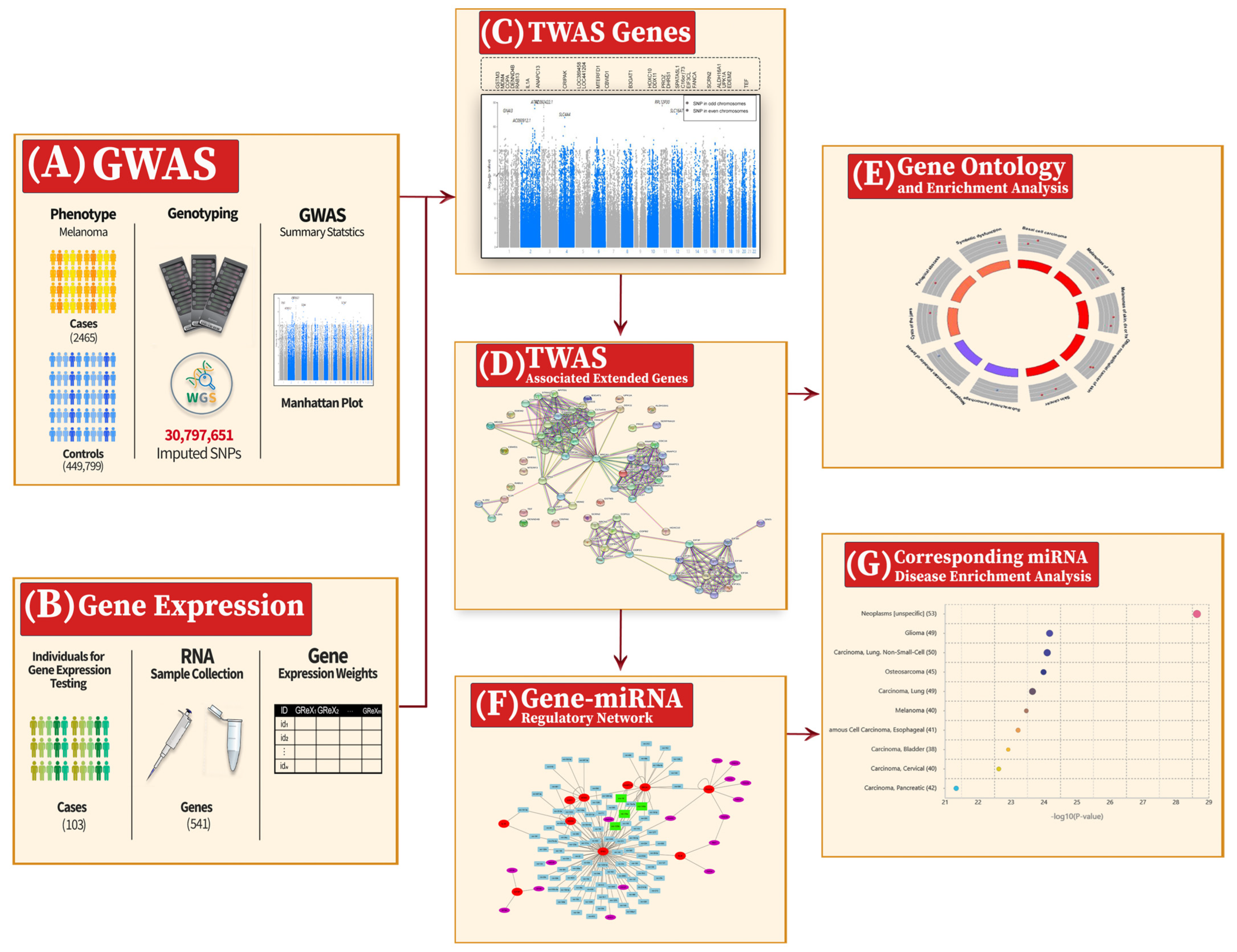

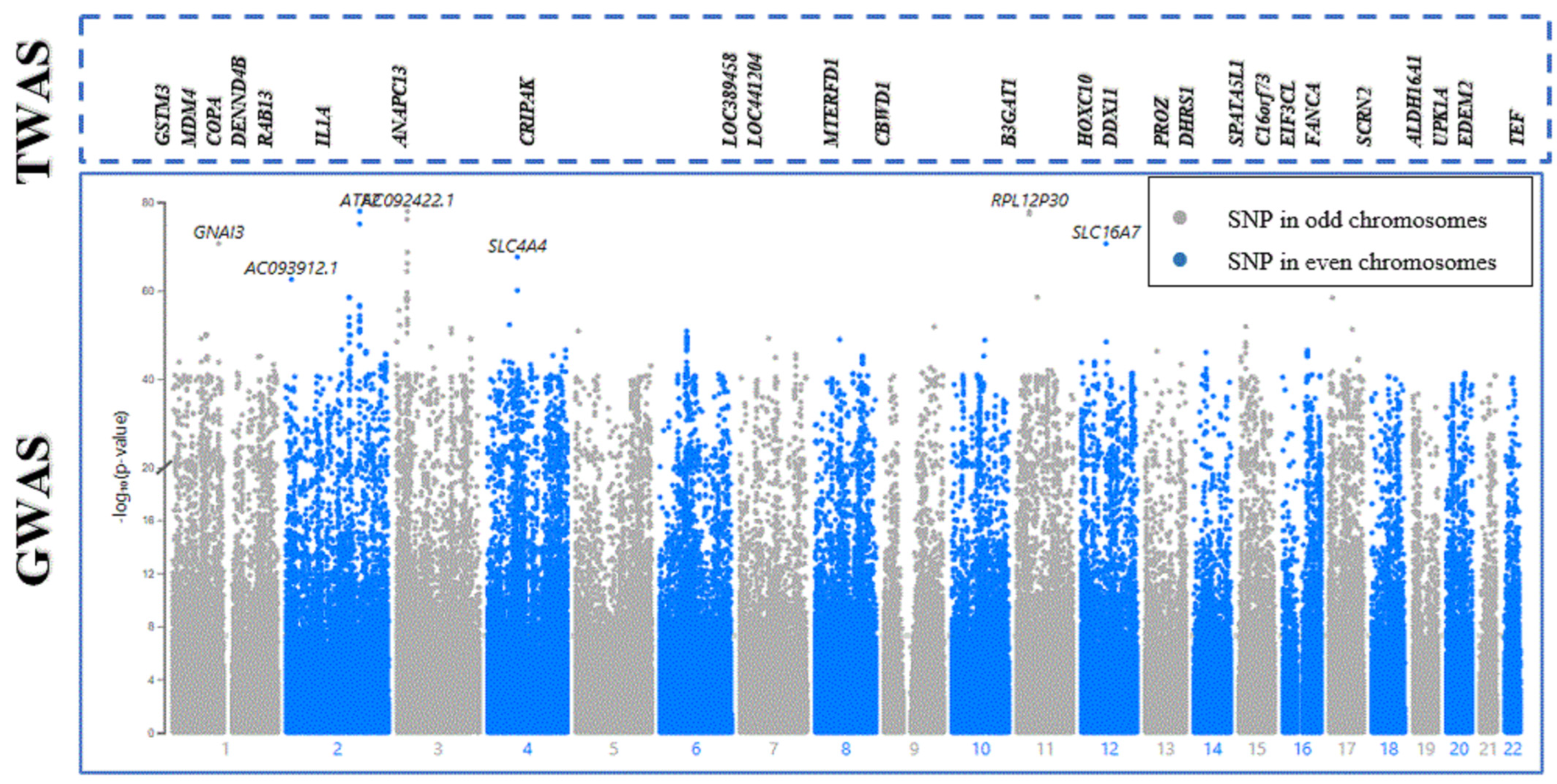
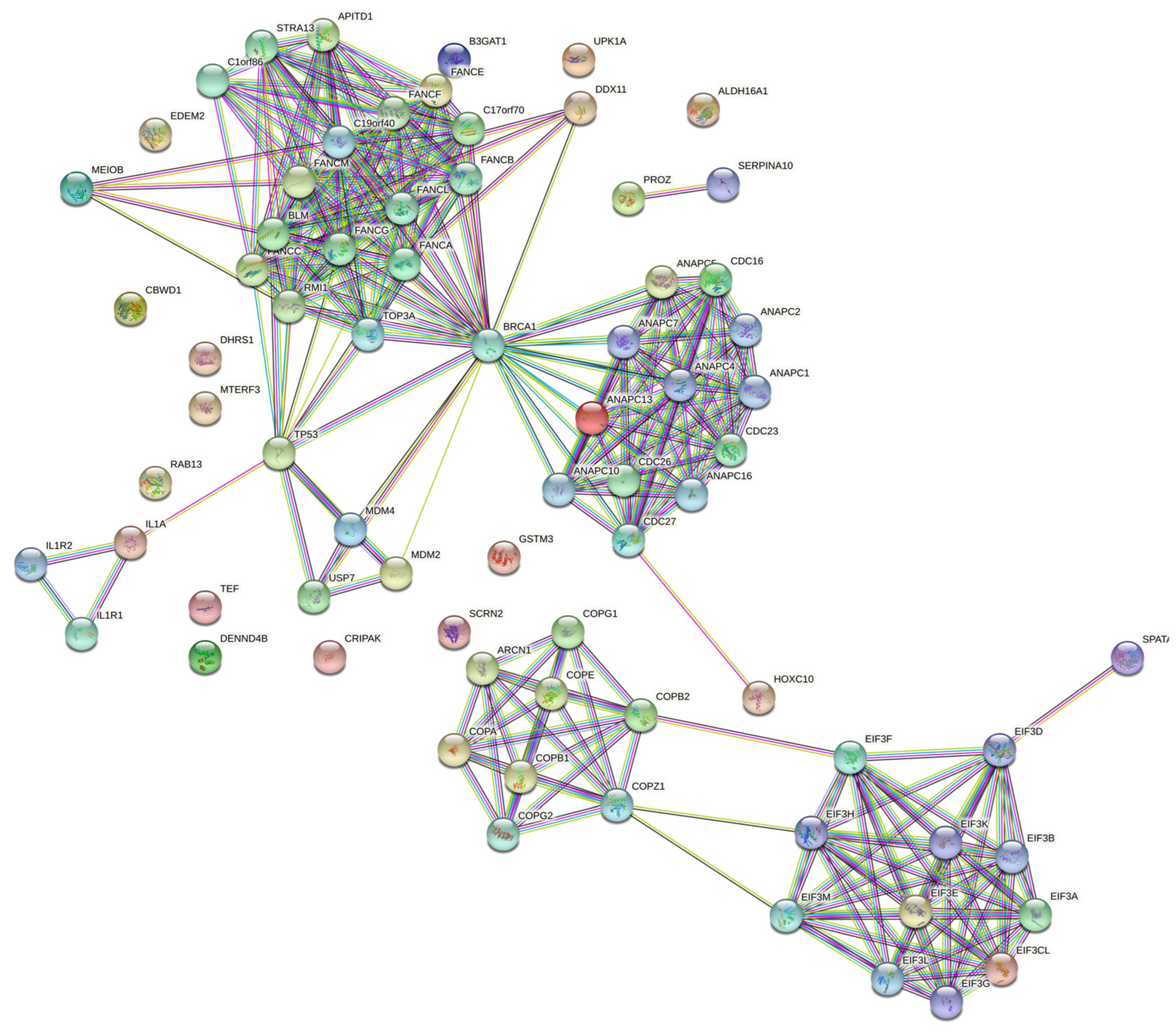
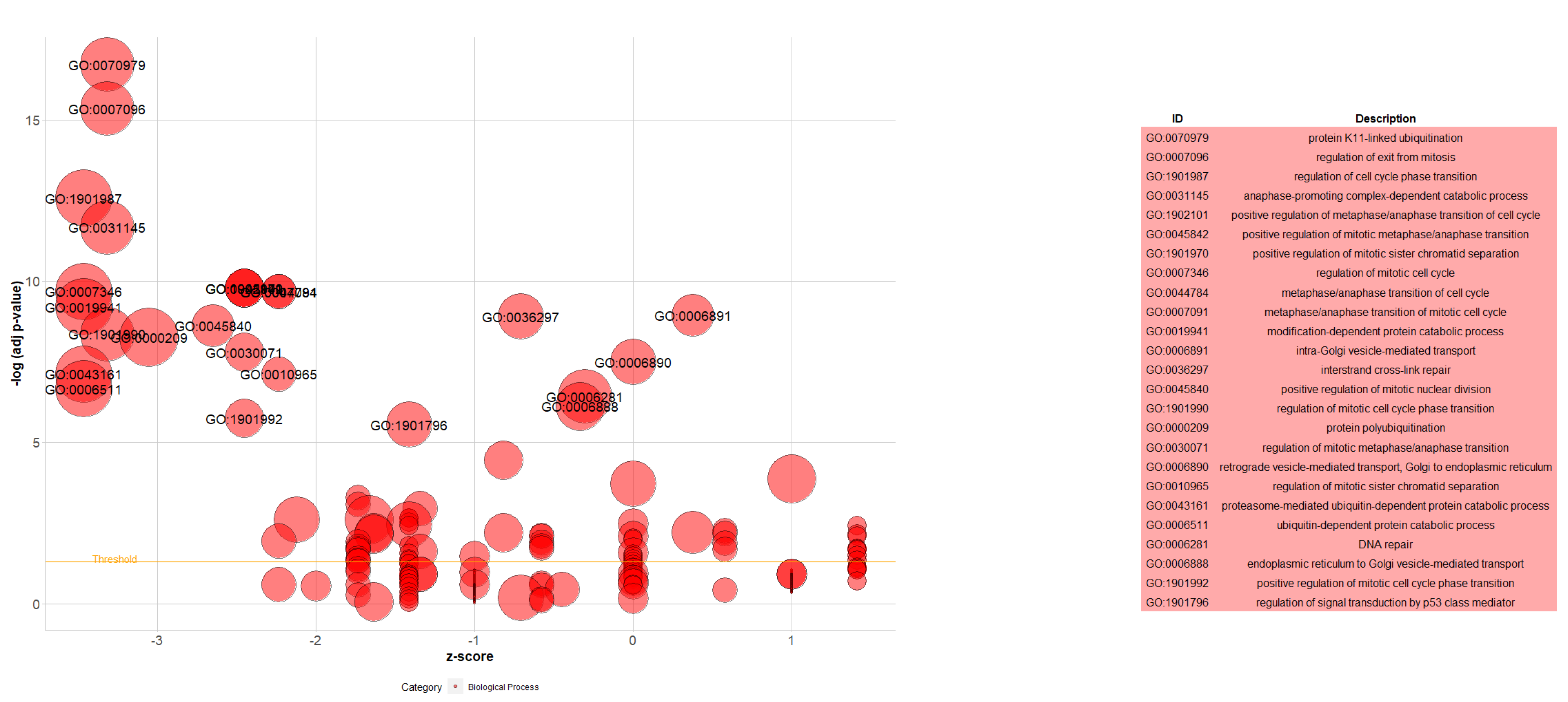
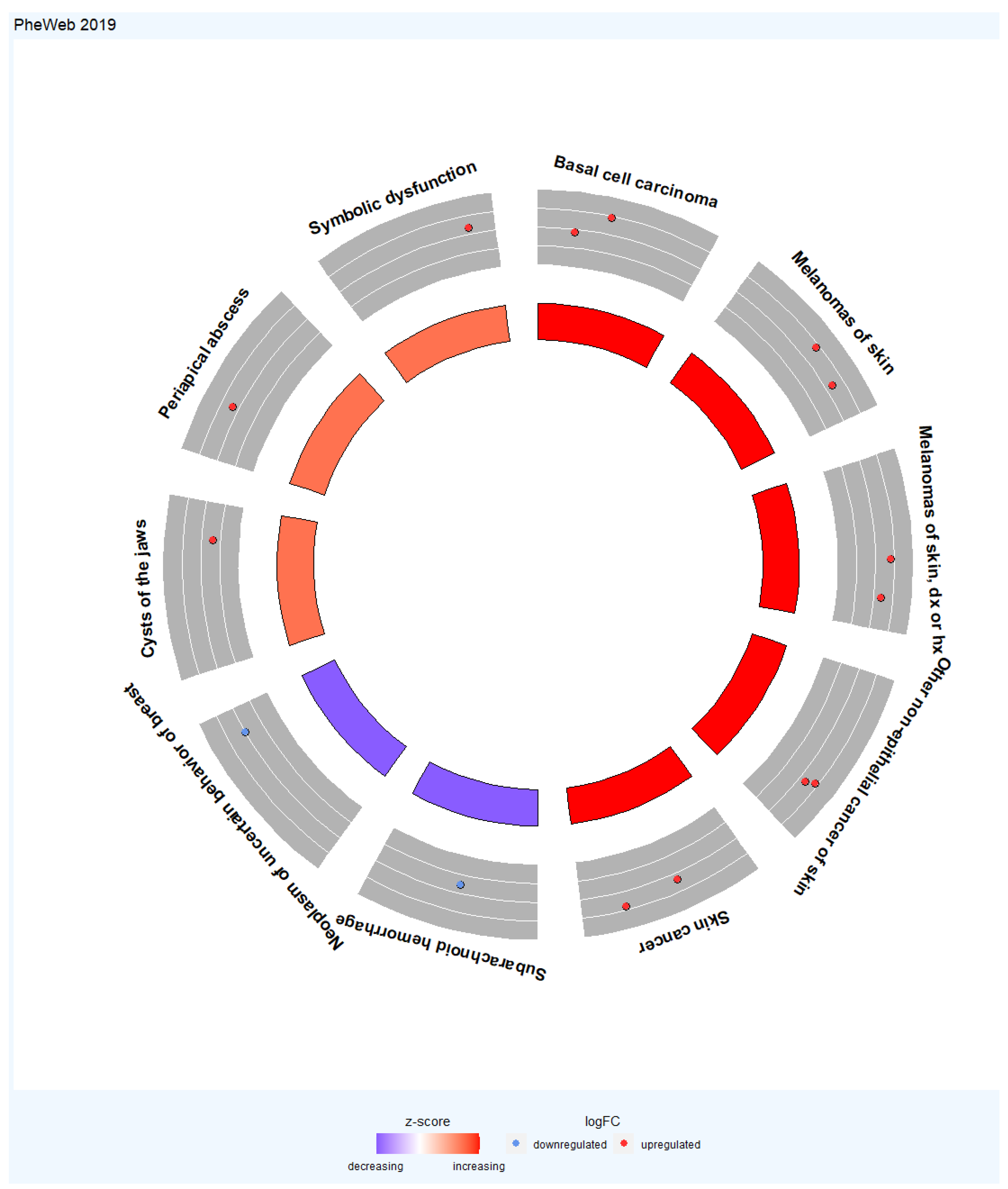
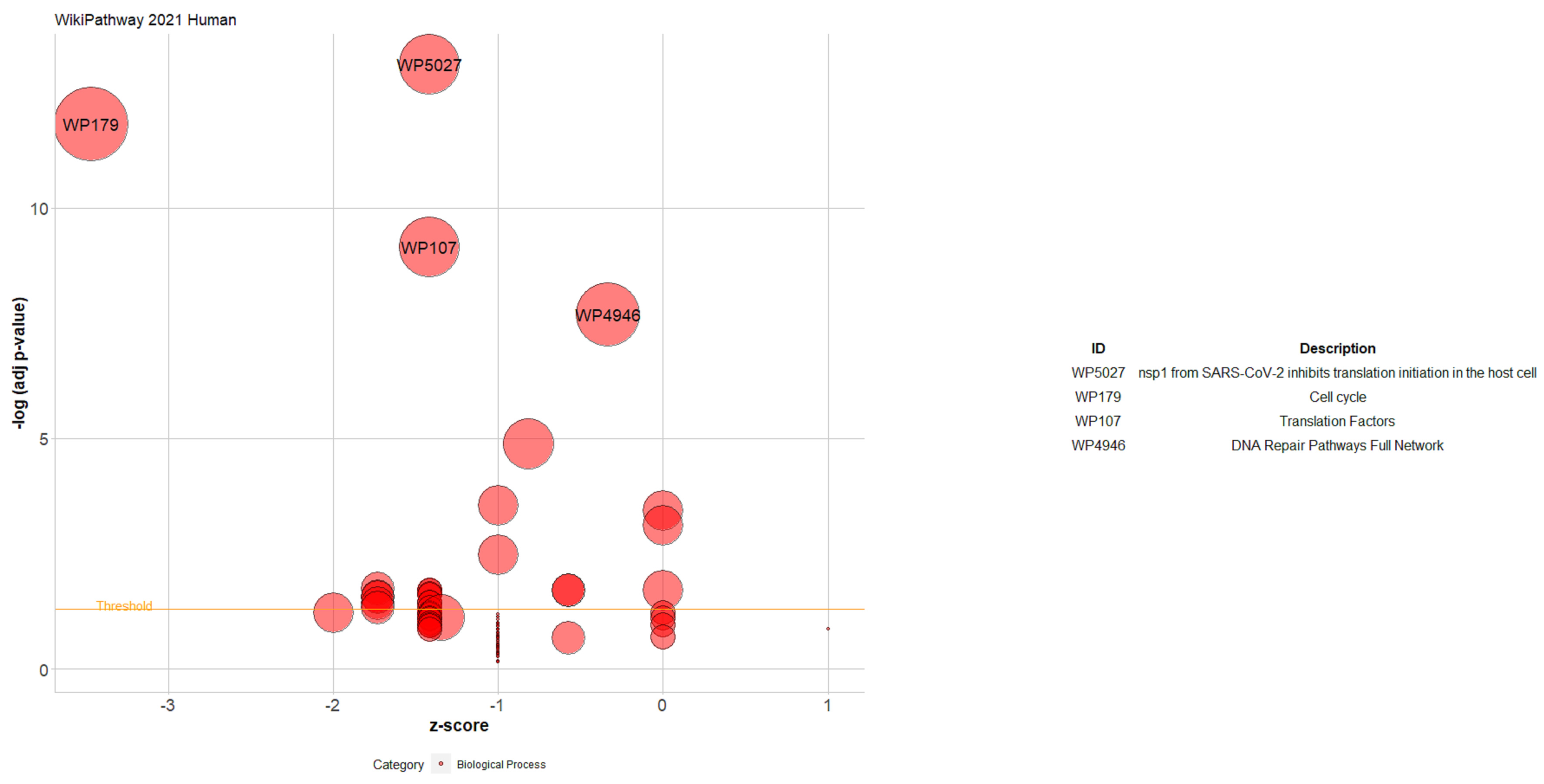
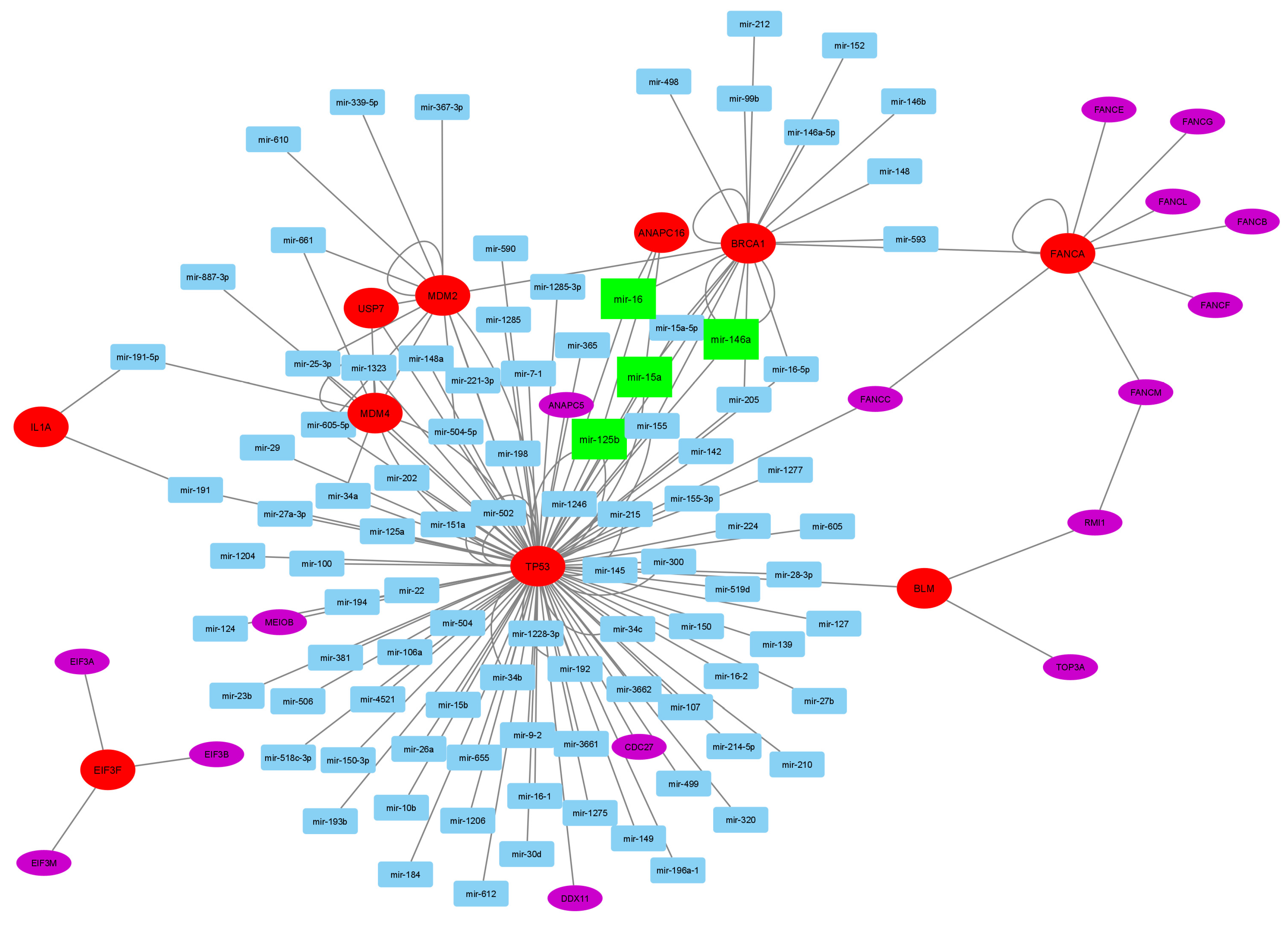
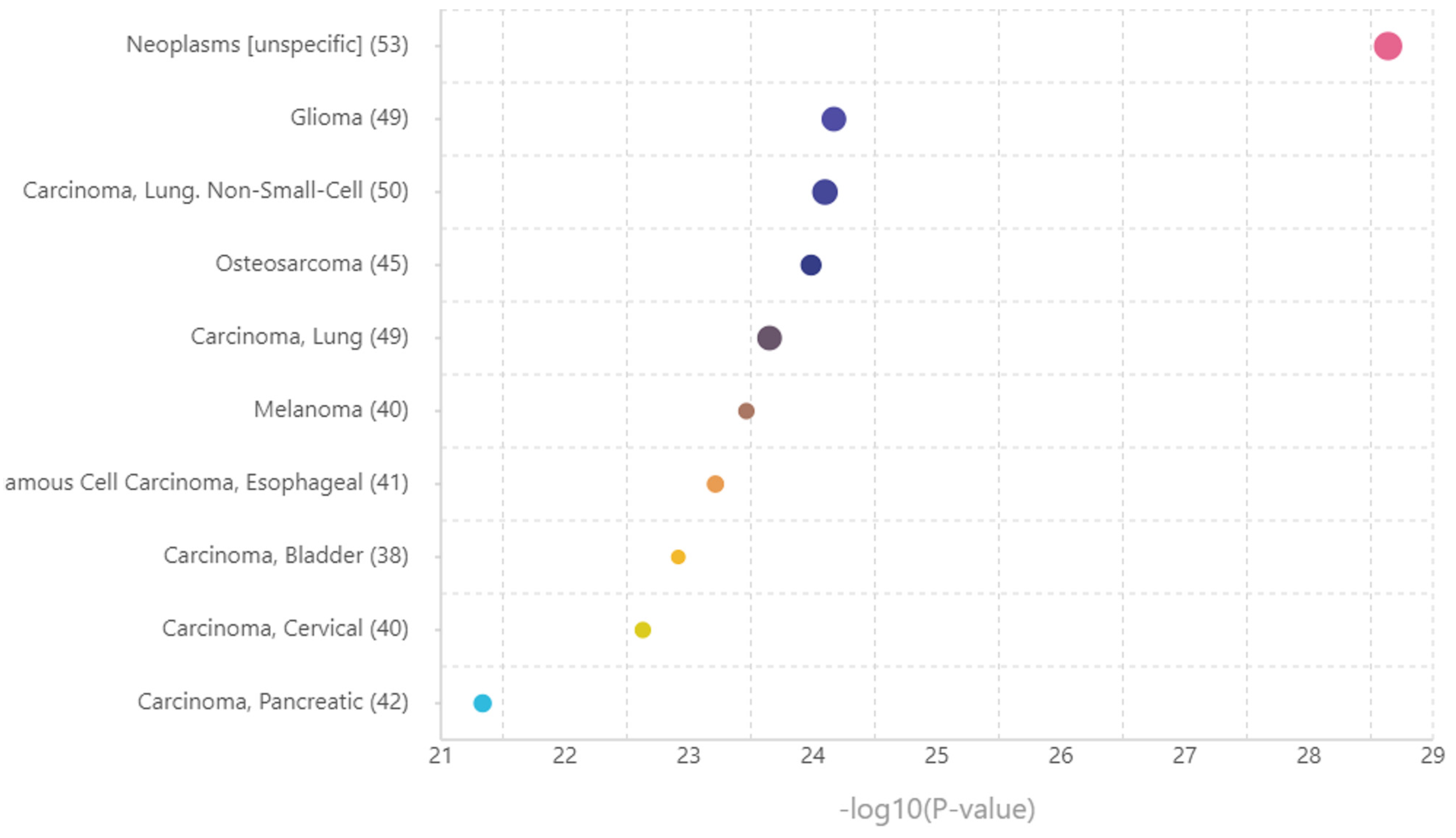
| Gene | CHR | Gene Start | Gene End | BEST.GWAS.ID | NSN | TWAS.p | Fold Change | Previously Studied in | Other Diseases Affected by the Gene |
|---|---|---|---|---|---|---|---|---|---|
| AMIGO1 | 1 | 110049447 | 110052336 | rs12135954 | 46 | 0.00593 | 0.77 | [36] | Autism [37] |
| GSTM3 | 1 | 110276554 | 110283660 | rs3754446 | 51 | 0.01209 | 0.83 | [38] | Autism [37] |
| MDM4 | 1 | 204485511 | 204677661 | rs10458588 | 134 | 0.01635 | 0.33 | [15,39,40,41] | Autism [37] |
| COPA | 1 | 160258378 | 160313354 | rs12140856 | 52 | 0.01683 | 2.42 | [42] | Autoimmune interstitial lung disease-arthritis syndrome [43], acute respiratory failure [44] |
| DENND4B | 1 | 153901977 | 153919154 | rs11581644 | 37 | 0.04715 | 0.97 | [45] | - |
| RAB13 | 1 | 153954128 | 153958806 | rs12048137 | 30 | 0.04715 | 1.97 | [17,46] | - |
| IL1A | 2 | 113531493 | 113542971 | rs4848300 | 51 | 0.0493 | 0.1 | [16,47,48,49,50,51,52,53] | 2q13q22.3 microduplication syndrome [54,55] |
| ANAPC13 | 3 | 134196547 | 134204865 | rs9790169 | 38 | 0.0261 | 1.35 | - | GATA2 deficiency with susceptibility to MDS/AMLDeafness-lymphedema-leukemia syndrome [56], mental retardation, autosomal dominant 47 [57] |
| CRIPAK | 4 | 1385340 | 1389782 | rs4974619 | 29 | 0.047 | 0.17 | [58] | Mucopolysaccharidosis type 1 [59,60,61] |
| LOC389458 | 7 | 5023302 | 5112853 | rs10234867 | 63 | 0.000236 | 1.2 | - | Malignant tumor of prostate [62], Baraitser–Winter syndrome 1 [63,64,65] |
| LOC441204 | 7 | 26438339 | 26538594 | rs1229677 | 138 | 0.015916 | - | - | Alzheimer’s disease [66], gemcitabine chemoresistance [67] |
| MTERFD1 | 8 | 97251645 | 97273796 | rs10092898 | 42 | 0.0183 | 2.35 | [68,69] | Prostate adenocarcinoma [70], Furrow contractions [71] |
| CBWD1 | 9 | 121039 | 179075 | rs636922 | 30 | 0.00317 | 0.6 | [10,11] | Normal pregnancy [72] |
| B3GAT1 | 11 | 134248400 | 134281812 | rs7123380 | 75 | 0.031 | 1 | [73,74] | Schizophrenia [37], Paris–Trousseau thrombocytopenia [75] |
| HOXC10 | 12 | 54378946 | 54384060 | rs4759316 | 56 | 0.00643 | 0.46 | [76,77] | - |
| DDX11 | 12 | 31226779 | 31257725 | rs2287465 | 26 | 0.04 | 0.78 | [78] | Warsaw breakage syndrome [79,80,81,82,83,84,85] |
| PROZ | 13 | 113812968 | 113826694 | rs473598 | 38 | 0.0399 | 0.5 | - | Factor X deficiency, factor VII deficiency [86], protein Z deficiency [87] |
| DHRS1 | 14 | 24759806 | 24769039 | rs2180196 | 77 | 0.0111 | 0.32 | [88] | - |
| SPATA5L1 | 15 | 45694519 | 45713614 | rs8025019 | 33 | 0.039 | 0.54 | - | Neurodevelopmental disorder with hearing loss and spasticity, deafness, autosomal recessive 119 [89] |
| C16orf73 | 16 | 1883984 | 1934295 | rs12149777 | 91 | 0.000395 | 0.11 | [90,91,92] | Spermatogenic failure 22 [93], tuberous sclerosis 2 [94,95,96,97], hemimegalencephaly [98] |
| EIF3CL | 16 | 28699879 | 28747052 | rs12448482 | 18 | 0.002754 | 2.12 | [99] | Schizophrenia [37], hemimegalencephaly [98] |
| FANCA | 16 | 89803959 | 89883065 | rs258322 | 82 | 0.04941 | 2.66 | [12,100,101,102,103,104,105] | Fanconi anemia [106], Fanconi anemia complementation group A [107], neuroblastoma [108] |
| SCRN2 | 17 | 45915049 | 45918699 | rs17856536 | 53 | 0.033 | 0.81 | [109,110,111] | - |
| ALDH16A1 | 19 | 49956473 | 49974304 | rs11669675 | 45 | 0.0234 | 0.85 | [112,113,114] | - |
| UPK1A | 19 | 36157715 | 36169365 | rs3761093 | 41 | 0.033 | 0.04 | [13,115] | Dystonic disorder [75] |
| EDEM2 | 20 | 33703160 | 33865928 | rs2425025 | 102 | 0.0421 | 2.27 | [116,117] | Long QT syndrome [43] |
| TEF | 22 | 41763392 | 41795328 | rs2234059 | 36 | 0.00331 | 0.33 | [118,119,120] | Immunodeficiency, common variable, 4 [121] |
| Extended Genes | Aliases | Ensembl ID | |
|---|---|---|---|
| 1 | ALDH16A1 | ENSG00000161618.9 | |
| 2 | ANAPC1 | ENSG00000153107.11 | |
| 3 | ANAPC10 | ENSG00000164162.12 | |
| 4 | ANAPC13 | ENSG00000129055.12 | |
| 5 | ANAPC16 | ENSG00000166295.8 | |
| 6 | ANAPC2 | ENSG00000176248.8 | |
| 7 | ANAPC4 | ENSG00000053900.10 | |
| 8 | ANAPC5 | ENSG00000089053.12 | |
| 9 | ANAPC7 | ENSG00000196510.12 | |
| 10 | APITD1 | CENPS | ENSG00000175279.21 |
| 11 | ARCN1 | ENSG00000095139.13 | |
| 12 | B3GAT1 | ENSG00000109956.12 | |
| 13 | BLM | ENSG00000197299.10 | |
| 14 | BRCA1 | ENSG00000012048.19 | |
| 15 | C16orf73 | MEIOB | ENSG00000162039.14 |
| 16 | C17orf70 | FAAP100 | ENSG00000185504.16 |
| 17 | C19orf40 | FAAP24 | ENSG00000131944.9 |
| 18 | C1orf86 | FAAP20 | ENSG00000162585.16 |
| 19 | CBWD1 | ENSG00000172785.18 | |
| 20 | CDC16 | ENSG00000130177.14 | |
| 21 | CDC23 | ENSG00000094880.10 | |
| 22 | CDC26 | ENSG00000176386.8 | |
| 23 | CDC27 | ENSG00000004897.11 | |
| 24 | COPA | ENSG00000122218.14 | |
| 25 | COPB1 | ENSG00000129083.12 | |
| 26 | COPB2 | ENSG00000184432.9 | |
| 27 | COPE | ENSG00000105669.12 | |
| 28 | COPG1 | ENSG00000181789.14 | |
| 29 | COPG2 | ENSG00000158623.14 | |
| 30 | COPZ1 | ENSG00000111481.9 | |
| 31 | CRIPAK | ENSG00000179979.8 | |
| 32 | DDX11 | CHL1 | ENSG00000013573.16 |
| 33 | DENND4B | ENSG00000198837.9 | |
| 34 | DHRS1 | ENSG00000157379.13 | |
| 35 | EDEM2 | ENSG00000088298.12 | |
| 36 | EIF3A | ENSG00000107581.12 | |
| 37 | EIF3B | ENSG00000106263.17 | |
| 38 | EIF3CL | ENSG00000205609.12 | |
| 39 | EIF3D | ENSG00000100353.17 | |
| 40 | EIF3E | ENSG00000104408.9 | |
| 41 | EIF3F | ENSG00000175390.12 | |
| 42 | EIF3G | ENSG00000130811.10 | |
| 43 | EIF3H | ENSG00000147677.10 | |
| 44 | EIF3K | ENSG00000178982.9 | |
| 45 | EIF3L | ENSG00000100129.17 | |
| 46 | EIF3M | ENSG00000149100.12 | |
| 47 | FANCA | ENSG00000187741.14 | |
| 48 | FANCB | ENSG00000181544.13 | |
| 49 | FANCC | ENSG00000158169.11 | |
| 50 | FANCE | ENSG00000112039.3 | |
| 51 | FANCF | ENSG00000183161.4 | |
| 52 | FANCG | ENSG00000221829.9 | |
| 53 | FANCL | ENSG00000115392.11 | |
| 54 | FANCM | ENSG00000187790.10 | |
| 55 | GSTM3 | ENSG00000134202.10 | |
| 56 | HOXC10 | ENSG00000180818.4 | |
| 57 | IL1A | ENSG00000115008.5 | |
| 58 | IL1R1 | ENSG00000115594.11 | |
| 59 | IL1R2 | ENSG00000115590.13 | |
| 60 | LOC389458 | RBAK | ENSG00000146587.17 |
| 61 | LOC441204 | RPL36AP26 | ENSG00000235828.5 |
| 62 | MDM2 | ENSG00000135679.21 | |
| 63 | MDM4 | ENSG00000198625.12 | |
| 64 | MTERFD1 | MTERF3 | ENSG00000156469.8 |
| 65 | PROZ | ENSG00000126231.13 | |
| 66 | RAB13 | ENSG00000143545.8 | |
| 67 | RMI1 | ENSG00000178966.15 | |
| 68 | SCRN2 | ENSG00000141295.13 | |
| 69 | SERPINA10 | ENSG00000140093.9 | |
| 70 | SPATA5L1 | ENSG00000171763.17 | |
| 71 | STRA13 | BHLHE40 or CENPX | ENSG00000169689.14 |
| 72 | TEF | ENSG00000167074.14 | |
| 73 | TOP3A | ENSG00000177302.14 | |
| 74 | TP53 | ENSG00000141510.15 | |
| 75 | UPK1A | ENSG00000105668.7 | |
| 76 | USP7 | ENSG00000187555.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saad, M.N.; Hamed, M. Transcriptome-Wide Association Study Reveals New Molecular Interactions Associated with Melanoma Pathogenesis. Cancers 2024, 16, 2517. https://doi.org/10.3390/cancers16142517
Saad MN, Hamed M. Transcriptome-Wide Association Study Reveals New Molecular Interactions Associated with Melanoma Pathogenesis. Cancers. 2024; 16(14):2517. https://doi.org/10.3390/cancers16142517
Chicago/Turabian StyleSaad, Mohamed N., and Mohamed Hamed. 2024. "Transcriptome-Wide Association Study Reveals New Molecular Interactions Associated with Melanoma Pathogenesis" Cancers 16, no. 14: 2517. https://doi.org/10.3390/cancers16142517
APA StyleSaad, M. N., & Hamed, M. (2024). Transcriptome-Wide Association Study Reveals New Molecular Interactions Associated with Melanoma Pathogenesis. Cancers, 16(14), 2517. https://doi.org/10.3390/cancers16142517







