Simple Summary
STING is a key element in the cGAS/STING cytosolic sensing pathway and several STING agonists are currently being evaluated as anticancer drugs in the field of cancer immunotherapy. This study provides a unique catalog of STING expressions in tumor cells as well as its clinical relevance and association with PD-L1 expression in tumor and inflammatory cells in more than 130 different tumor entities.
Abstract
Stimulator of interferon genes protein (STING) activates the immune response in inflammatory cells. STING expression in cancer cells is less well characterized, but STING agonists are currently being evaluated as anticancer drugs. A tissue microarray containing 18,001 samples from 139 different tumor types was analyzed for STING by immunohistochemistry. STING-positive tumor cells were found in 130 (93.5%) of 139 tumor entities. The highest STING positivity rates occurred in squamous cell carcinomas (up to 96%); malignant mesothelioma (88.5%–95.7%); adenocarcinoma of the pancreas (94.9%), lung (90.3%), cervix (90.0%), colorectum (75.2%), and gallbladder (68.8%); and serous high-grade ovarian cancer (86.0%). High STING expression was linked to adverse phenotypes in breast cancer, clear cell renal cell carcinoma, colorectal adenocarcinoma, hepatocellular carcinoma, and papillary carcinoma of the thyroid (p < 0.05). In pTa urothelial carcinomas, STING expression was associated with low-grade carcinoma (p = 0.0002). Across all tumors, STING expression paralleled PD-L1 positivity of tumor and inflammatory cells (p < 0.0001 each) but was unrelated to the density of CD8+ lymphocytes. STING expression is variable across tumor types and may be related to aggressive tumor phenotype and PD-L1 positivity. The lack of relationship with tumor-infiltrating CD8+ lymphocytes argues against a significant IFN production by STING positive tumor cells.
1. Introduction
The recognition of pathogens such as viruses, bacteria, and fungi occurs through the perception of pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) through a set of specialized receptors. Exogenous DNA derived from pathogens or self-DNA in the cytosol represent highly efficient PAMPs/DAMPs, which can induce strong innate immune responses through the stimulation of downstream signaling cascades including the production of proinflammatory mediators and type I interferons (IFNs). Stimulator of interferon genes (STING) is a key component in this pathogen response system (summarized in [1]). STING “activation” is triggered by cytosolic nucleic acids derived from DNA viruses and bacteria as well as damaged self-DNA in the cyclic GMP-AMP (cGAS)/STING cytosolic DNA-sensing pathway (summarized in [2]). Activated STING acts as an adapter protein to enable, for example, phosphorylation of interferon regulatory factor 3 (IRF3) by serine/threonine protein kinase (TBK1), which leads to type I IFN activation (summarized in [1]). Due to the capability of STING to stimulate immune response, numerous STING agonists are currently being evaluated for their suitability as anticancer drugs in the field of cancer immunotherapy (summarized in [3,4]), and recent evidence indicates that the combination of STING agonists with STAT3 inhibitors can further enhance antitumor immunity [5].
Most studies evaluating the function of STING have focused on inflammatory cells, but STING can also be expressed in cancer cells, in which the role of STING is less well characterized (summarized in [6]). The available data suggest a high complexity of the STING pathway in cancer cells. For example, in vitro STING knockout and STING agonist experiments have revealed reduced, unchanged, or increased cell proliferation and survival in studies employing cell lines from different cancer types (summarized in [6]). Several recent studies have suggested that the STING pathway in cancer cells can be important for regulating the antitumor immune response. For example, programmed cell death 1 ligand 1 (PD-L1) expression in tumor cells could be stimulated by STING agonist in a cancer murine model [7], by elevated STING expression in DNA damage-response-deficient breast cancer cells [8] and by radiation-therapy-induced STING expression in human and mouse liver cancer cells [9]. Furthermore, activation of the cGAS/STING pathway led to natural killer cell (NK) infiltration [10,11], whereas an absence of STING in cancer cells was found to be associated with low numbers of NKs and CD8-positive lymphocytes [12,13]. While these findings could be explained by IFN production in STING positive tumor cells, other studies have questioned the STING-induced secretion of biologically relevant quantities of IFN by tumor cells (summarized in [6]).
Most available data on STING expression in cancer are RNA-based and, therefore, cannot distinguish the specific role of STING expression in cancer cells. Studies evaluating STING expression of tumor cells by immunohistochemistry (IHC) are so far limited in number and the obtained results suggest a rather complex and variable role of STING expression in cancer. For example, STING downregulation as compared to adjacent normal tissue has been reported in colorectal [14] and gastric carcinomas [15], while STING upregulation, as compared to adjacent normal tissue, was described in squamous cell carcinomas (SQCC) of the tongue [16]. Associations of high STING levels with unfavorable tumor features were found in carcinomas of the cervix uteri [17], ovary [18], stomach [19], colon [20], lung [21], larynx [22] tongue [16], and kidney [23], while low STING levels were related to unfavorable tumor features in carcinomas of the breast [24], head and neck [25], the urinary tract [26], lung [27,28], and stomach [15], as well as in T-cell lymphoma [29].
Considering the potential clinical and therapeutic relevance of STING expression in cancer, we aimed at a comprehensive characterization of tumor cell STING expression in human neoplasms. For this purpose, a pre-existing set of tissue microarrays (TMAs) containing more than 18,000 tumor samples from 139 different tumor types and subtypes as well as 76 non-neoplastic tissue categories were analyzed for STING expression by IHC.
2. Materials and Methods
2.1. Tissue Microarrays (TMAs)
Two sets of TMAs made from formalin-fixed, paraffin-embedded tissue samples were employed in our study. These included a normal TMA with a total of 608 samples that was built from 8 samples (from 8 different donors) from each of 76 different normal tissue types, as well as cancer TMAs. The cancer TMAs included a total of 18,001 primary tumors from 139 different tumor types and subtypes. The TMAs contained one 0.6 mm spot per tumor and were manufactured as described earlier [30,31]. A histopathological and molecular database was attached to the TMAs containing data of cancers of the breast (n = 1208), kidney (n = 1534), liver (n = 231), bladder (n = 1073), thyroid (n = 518), colon (n = 2351), pancreas (n = 598), stomach (n = 327), endometrium (n = 182), and ovary (n = 369). Clinical follow-up data were available from 877 patients with invasive breast carcinomas of no special type with a median follow-up time of 43 months. From an earlier study, data on PD-L1 expression were available for 11,938 cancers and on the density of CD8-positive lymphocytes for 6,897 cancers [32]. Details on the normal tissues and cancers represented in the TMAs are given in Section 3. All samples were obtained from the routine archives of the Institute of Pathology, University Hospital of Hamburg, Germany; the Institute of Pathology, Clinical Center Osnabrueck, Germany; and the Department of Pathology, Academic Hospital Fuerth, Germany. The use of archived remnants of diagnostic tissues for TMA manufacturing, their analysis for research purposes, and the use of patient data complied with local laws (HmbKHG, §12) and analysis had been approved by the local ethics committee (Ethics commission Hamburg, WF-049/09). All work has been carried out in compliance with the Declaration of Helsinki.
2.2. Immunohistochemistry (IHC)
All experiments were carried out on the same day and with the same batch of reagents. For best immunostaining results, all TMA blocks were freshly cut one day in advance of immunostaining. Slides were deparaffinized with xylol, rehydrated through a graded alcohol series, and exposed to heat-induced antigen retrieval for 5 minutes in an autoclave at 121 °C in pH 7.8 Tris-EDTA-Citrate (TEC) buffer. Endogenous peroxidase activity was blocked with Dako REAL Peroxidase-Blocking Solution (Agilent Technologies, Santa Clara, CA, USA; #S2023) for 10 minutes. Primary antibody specific for STING (mouse monoclonal, MSVA-515M, MS Validated Antibodies, Hamburg, Germany; #5473-515M) was applied at 37 °C for 60 minutes at a dilution of 1:150. For the purpose of antibody validation, the normal tissue TMA was also analyzed by the rabbit recombinant monoclonal STING antibody clone D2P2F (Cell Signaling Technologies®, Danvers, MA, USA; #13647) at a dilution of 1:600 and an otherwise identical protocol. Bound antibody was visualized using the Dako REAL EnVision Detection System Peroxidase/DAB+ Rabbit/Mouse kit (Agilent Technologies, Santa Clara, CA, USA; #K5007), according to the manufacturer’s directions. The sections were counterstained with hemalaun. Scoring of tumor samples was performed as described before [33]. In brief, the percentage of positive neoplastic cells was estimated, and the staining intensity was semi-quantitatively recorded (0, 1+, 2+, or 3+). For statistical analyses, the staining results were categorized into four groups (see Supplementary Table S1). Tumors without any staining were considered negative. Tumors with 1+ staining intensity in ≤70% of tumor cells and 2+ intensity in ≤30% of tumor cells were considered weakly positive. Tumors with 1+ staining intensity in >70% of tumor cells, 2+ intensity in 31–70%, or 3+ intensity in ≤30% of tumor cells were considered moderately positive. Tumors with 2+ intensity in >70% or 3+ intensity in >30% of tumor cells were considered strongly positive. We used this scoring system in many earlier TMA studies and found it suitable for the identification of numerous known and novel prognostic molecular features in various tumor types [33].
2.3. Statistics
The JMP17® software package (SAS®, Cary, NC, USA) was used for statistical analyses including the chi2-test to search for associations between STING immunostaining and tumor phenotype and PD-L1 immunostaining in tumor and immune cells, analysis of variance (ANOVA) to search for associations between STING immunostaining and the density of CD8-positive lymphocytes, and the Log-rank test along with Kaplan–Meier plots for survival analysis.
3. Results
3.1. Technical Issues
In the tumor TMAs, a total of 15,345 (85.2%) of 18,001 tumor samples were interpretable in our analysis. In the normal tissue TMA, at least four samples were evaluable of each normal tissue type. Non-interpretable samples demonstrated an absence of specific cell types, absence of unequivocal tumor cells, or a complete lack of individual tissue spots.
3.2. STING in Normal Tissues
STING immunostaining of cells was always cytoplasmic. Particularly strong STING staining was observed in endothelial cells of vessels of all sizes; macrophages/dendritic cells; subsets of lymphocytes and of bone marrow cells, respiratory, fallopian tube, and endocervical epithelium; as well as in basal cells of the prostate. The few cell types that were always negative in our screening of up to eight samples per tissue type included heart and skeletal muscle cells; syncytiotrophoblast and cytotrophoblast cells; amnion and chorion cells of the placenta; epithelial cells of the parathyroid, thyroid, and adrenal glands; acinar cells of the pancreas and of the prostate; hepatocytes; principal cells of the caput epididymis; and Sertoli cells and germ cells of the testis. Representative images of these tissues are shown in Figure 1. In many other cell types, the STING staining pattern varied between samples. For example, STING staining ranged from negative to strong in the gastrointestinal epithelium, gallbladder, collecting ducts of the kidney, urothelium, epithelial cells of the cauda epididymis and the seminal vesicle, and epithelial and stromal cells of the endometrium. STING staining varied from negative to moderate intensity in the salivary glands, Brunner glands, breast epithelium, and in subsets of cells in the adeno- and neurohypophysis. In squamous epithelium, STING staining was usually limited to the basal cell layer, although STING staining was more intense in tonsil crypts. All these normal tissue findings were obtained by using the mouse monoclonal antibody MSVA-515M and the rabbit recombinant monoclonal antibody D2P2F and were, therefore, considered to be specific. Representative images of tissue staining are given in Supplementary Figure S1.
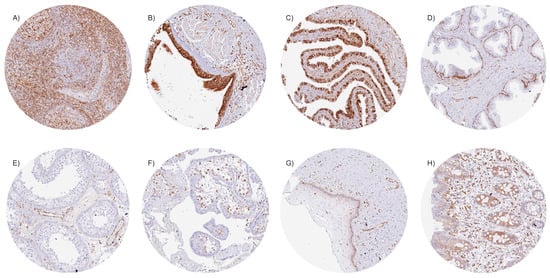
Figure 1.
STING immunostaining of normal tissues. (A) Strong cytoplasmic staining of many different cell types in the lymph node, (B) respiratory epithelial cells of the bronchus, (C) a large subset of epithelial cells of the fallopian tube, (D) basal cells of the prostate, (E) endothelial cells of the testis, and (F) stroma cells in the first trimester placenta. (G) STING staining is less intense and limited to the basal cell layer in the squamous epithelium of the ectocervix. (H) Focal staining in epithelial cells of the rectum.
3.3. STING Expression in Cancer
In tumor samples, STING immunostaining was seen in macrophages, lymphocytic cells, endothelial cells, other stroma cells, and (often) also in tumor cells. STING positivity of tumor cells was detectable in 8908 (58.1%) of the 15,345 analyzable tumors, including 4169 (27.2%) with weak, 2005 (13.1%) with moderate, and 2734 (17.8%) with strong immunostaining. Overall, 130 of 139 tumor categories showed detectable STING staining, while 96 tumor categories included at least one case with strong positivity (Table 1). Representative images of STING-positive tumors are shown in Figure 2. Particularly high rates of STING positivity occurred in SQCCs of different sites of origin (up to 96%), malignant mesothelioma (88.5–95.7%), ductal adenocarcinoma of the pancreas (94.9%), pulmonary adenocarcinoma (90.3%), cervical adenocarcinoma (90.0%), serous high-grade ovarian cancer (86.0%), anaplastic thyroid carcinoma (82.9%) colorectal adenocarcinoma (75.2%), adenocarcinoma of the gallbladder (68.8%), and in breast carcinoma (up to 66%). The prevalence of STING positivity was intermediate for urothelial neoplasms (51.3–79.6%), adenocarcinoma of the esophagus (57.4%), and several important sarcoma categories such as liposarcoma (50.9%) and osteosarcoma (48.3%). Particularly low rates of STING positivity were observed for prostatic adenocarcinomas (up to 11%), different subtypes of renal cell carcinomas (RCC; 18.8–24.5%), neuroendocrine tumors (NETs) of various sites (17.0–28.6%), neuroendocrine carcinomas (NECs; 12.5–38.5%), and Merkel cell carcinoma of the skin (5%) and small cell neuroendocrine carcinomas of the bladder (31.6%) and the prostate (17.6%). A graphical representation of a ranking order of STING-positive and strongly positive cancers is given in Supplementary Figure S2.

Table 1.
STING immunostaining in human tumors.
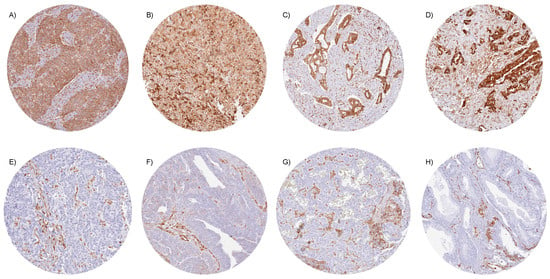
Figure 2.
STING immunostaining in cancer. In all tumor samples, cytoplasmic STING staining was seen in stroma cells, including endothelial and inflammatory cells. STING expression was more variable in tumor cells. Strong, cytoplasmic STING positivity for all tumor cells in (A) squamous cell carcinoma of the penis, (B) epithelioid malignant mesothelioma, (C) ductal adenocarcinoma of the pancreas, and (D) adenocarcinoma of the lung. STING staining was absent in tumor cells of (E) invasive urothelial carcinoma of the urinary bladder, (F) serous high-grade carcinoma of the ovary, (G) neuroendocrine tumor of the pancreas, and (H) adenocarcinoma of the prostate.
3.4. STING Expression, Tumor Phenotype, and Prognosis
The relationship between STING expression in tumor cells and clinically important histopathological and molecular tumor features in carcinomas from different sites is shown in Table 2. In clear cell renal cell carcinoma (ccRCC), high STING expression was linked to a poor histologic grade (p < 0.005), high pT category (p < 0.0001), and high UICC stage (p = 0.0060). In colorectal adenocarcinoma, high STING expression was linked to right-side tumor location (p = 0.0008), microsatellite instability (p = 0.0016), and RAS mutations (p < 0.0001). High STING expression was also related to nodal metastases in hepatocellular carcinoma (HCC; p = 0.0435) and in papillary carcinoma of the thyroid (p = 0.0074). In invasive breast cancer of no special type, high STING expression was linked to estrogen receptor (ER) and progesterone receptor (PR) expression (p < 0.0001 each), non-triple-negative status (p = 0.0028), and poor overall survival (p = 0.0196; Figure 3). However, in non-invasive urothelial carcinoma of the urinary bladder, high STING expression was associated with a low grade (p = 0.0002). A combined analysis of 480 SQCC from nine different sites showed a significant link between STING positivity and HPV infection (p = 0.0212; Supplementary Table S2). This statistical relationship was also retained in a subgroup of 50 pharyngeal SQCC (p = 0.0390). STING expression was unrelated to tumor phenotype in gastric adenocarcinoma, high-grade serous and endometrioid ovarian cancer, endometrioid endometrial carcinoma, pancreatic adenocarcinoma, and papillary RCC (Supplementary Table S3).

Table 2.
STING immunostaining and tumor phenotype.
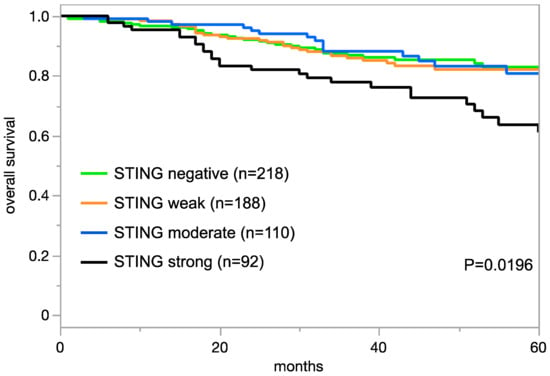
Figure 3.
STING immunostaining and prognosis in invasive breast cancer of no special type.
3.5. STING Expression, PD-L1 Status, and Tumor Microenvironment
Data on PD-L1 status were available from 10,579 and on the density of CD8 positive lymphocytes from 5880 tumors for which data on STING expression on tumor cells were collected in our project. Across all tumor entities, there was a significant relationship between high STING expression in tumor cells and PD-L1 positivity of tumor cells and tumor-associated inflammatory cells (p < 0.0001 each), while STING expression was unrelated to the density of CD8 positive cells (p = 0.4253, Table 3). Examples of PD-L1 and CD8 positive and negative immunostainings are shown in Supplementary Figure S3.

Table 3.
STING immunostaining versus PD-L1 immunostaining and density of CD8-positive lymphocytes.
4. Discussion
The main goal of our study was to provide a comprehensive overview of the prevalence of STING expression in tumor cells across a broad range of different tumor entities. Our data from 15,345 tumors from 139 tumor entities demonstrate substantial heterogeneity of tumoral STING expression between cancer types and individual patients. A total of >90% of the analyzed tumor entities had at least one STING positive case, >65% tumor entities had at least one case with strong STING positivity, >50% of all tumors were STING-positive, and 18% of all tumors were strongly STING-positive, demonstrating that STING expression is a common feature of cancer cells. It is still conspicuous that some cancer entities such as SQCCs of various sites and colorectal, pancreatic, gallbladder, pulmonary, or cervical adenocarcinomas did often show high STING expression levels while other entities such as prostatic adenocarcinomas, renal cell carcinomas, or neuroendocrine neoplasm were less frequently STING-positive. Considering that STING expression can be stimulated by the accumulation of cytosolic DAMPs, which are often found in cancer cells with a high degree of genomic instability (summarized in [34]), it could be hypothesized that cancer entities with a lesser degree of genomic instability such as prostatic adenocarcinomas [35], renal cell carcinomas [36], or neuroendocrine neoplasm [37] may be less prone to STING expression than cancers with high levels of genomic alterations such as SQCCs [38] or pulmonary [39] and pancreatic adenocarcinomas [40].
The high variability of STING expression in different samples from identical normal tissue types found in this study demonstrates that the level of STING expression fluctuates in non-neoplastic cell types. Although the reason for this variability remains elusive, it is conspicuous that many of the tissues and cell types that never showed any STING expression in our study, such as the placenta, testicular tubules, or endocrine organs, are particularly well protected from infection by viruses or bacteria as well as other causes of inflammation. The obvious inconsistency of STING expression levels in normal cell types makes it difficult to distinguish whether cancers do up- or downregulate STING, as compared to normal tissue. Most functional studies on STING in cancer have suggested that upregulation may be the prevalent mechanism (summarized in [6]). However, 42% of our tumor samples were completely STING-negative, with numerous STING-positive cells observed in the adjacent tumor stroma. This suggests that suppression of STING expression may occur in a subset of cancers.
The significant associations between high STING expression and poor overall survival in breast cancer, poor histological grade, and advanced stage in ccRCC as well as nodal metastases in HCC and papillary thyroid carcinoma suggests that STING upregulation rather than downregulation in tumor cells tends to be a feature of aggressive cancers. This observation fits well with reports of a possible tumor-promoting role of STING in cancer cells, although the molecular basis is not yet understood [41]. For example, Bakhoum et al. [42] reported that genetically instable tumors can activate STING-dependent noncanonical NF-κB signaling that facilitates metastasis. Alternatively, it is also possible that the upregulation of STING in cancer cells may represent a consequence of progressive dedifferentiation. The fact that loss of STING expression correlated with grade progression in non-invasive urothelial carcinoma shows that—depending on tumor type—the role of STING in determining the aggressiveness of tumor cells may vary. The absence of associations between STING immunostaining and parameters of cancer aggressiveness in gastric adenocarcinomas, high-grade serous and endometrioid ovarian cancer, endometrioid endometrial carcinoma, pancreatic adenocarcinoma, and papillary RCC further demonstrates that the level of STING expression in tumor cells is not likely to represent a pivotal general feature of cancer aggressiveness. Earlier studies analyzing the prognostic impact of STING expression in tumor cells have found inconsistent results. A relationship between high STING expression and unfavorable tumor phenotype or poor prognosis has been found in SQCC of the tongue [16] and—in agreement with our data—in ccRCC [23] and adenocarcinoma of the colon [20], while associations between low STING expression and poor prognosis were described in non-small cell lung carcinoma [27], gastric cancer [15], small cell lung carcinoma [28], and head and neck carcinomas [25]. Other authors could not find significant associations with patient prognosis or tumor phenotype in colorectal cancer [14], SQCC of the head and neck [43], or early gastric neoplastic lesions [44].
STING expression levels were also related to several critical molecular features of tumors. The particularly high rate of STING positivity in HPV-positive SQCC is consistent with the role of STING as a sensor for the presence of cytosolic PAMPs. In addition, data from The Cancer Genome Atlas (TCGA) also showed higher STING expression on RNA and protein levels in HPV-positive versus HPV-negative SQCC of the head and neck [45]. However, studies have shown that this correlation is maybe HPV-type specific as HPV18 E7 and HPV16 E7 lead to inhibition of the cGAS/STING pathway and reduced IFN production [46,47]. The significant link of high STING expression to MSI and RAS mutations in this study is also consistent with previous studies examining STING expression in colorectal and lung carcinomas [27,48,49], although some authors have found more RAS mutations in colorectal cancers with STING expression loss [48]. The relationship between STING positivity and ER/PR loss in breast cancer is consistent with recent studies showing elevated expression and activation of STING by DNA damage in ER/PR-negative breast cancer cell lines [50,51]. Another study found that higher STING expression in ER-positive breast cancers was associated with favorable prognosis [52].
The continuous increase in PD-L1 expression in both tumor cells and tumor-associated inflammatory cells with increasing levels of STING expression in tumor cells represents a strong confirmation of studies showing a functional relationship between STING activation and PD-L1 upregulation in cancer cells. For example, PD-L1 upregulation by STING-dependent activation of TBK1 was observed in HCC [9] and in HPV-positive cervical cancer cells [53]. Vasiyani et al. showed a STING-mediated, NF-kB-induced upregulation of PD-L1 in triple-negative breast cancer cells [50], and Grabosch et al. found PD-L1 upregulation by STING activation in an ovarian cancer mouse model [54]. Based on the assumption that STING-expressing tumor cells can produce type I IFN, a more intense population of these tumors by tumor-infiltrating lymphocytes was to be expected. The fact that we could not find a significant relationship between the level of STING expression and the density of CD8-positive, tumor-infiltrating immune cells, although data were available from more than 5500 tumors, strongly argues against functionally relevant type I IFN production in these tumors. This is consistent with data from studies exposing STING-expressing tumor cells to the STING agonist cGAMP or dsDNA and finding only low or even absent IFN-β production in mice with chronic lymphatic leukemia and colon cancer cell lines [55,56]. However, others have found that IFN-ß production is independent of cGAS-STING signaling in mismatch-repair-deficient colon cancer cells [43].
Given the high number of tumors analyzed in our study, we focused on thoroughly validating our STING IHC assay. The International Working Group for Antibody Validation (IWGAV) recommends that acceptable antibody validation for IHC on formalin-fixed tissues should involve either a comparison of results from two different independent antibodies or a comparison with expression data from another independent method [57]. Due to the high variability and cell-type specificity of STING expression, comparison with a method based on disaggregated tissue is not ideal for validating the STING antibody. Crucial evidence for the validity of our assay comes from the confirmation of all STING-positive cell types, including staining variabilities within individual tissue samples observed by MSVA-515M and by the independent second antibody D2P2F. It is noteworthy that using a broad selection of normal tissues (n = 76) for antibody validation increases the likelihood of detecting cross-reactivities, since virtually all proteins found in normal human adult cells are included in the validation process.
5. Conclusions
Our data provide an overview on the prevalence of STING expression in cancer cells across 139 different tumor types. The highest STING positivity rates occurred in squamous cell carcinomas, malignant mesothelioma, and adenocarcinomas of various origins. Comparison with the tumor phenotype demonstrates that high STING expression rather than STING deficiency tends to be linked to a more aggressive cancer phenotype. High STING expression was linked to adverse phenotypes in breast cancer, clear cell renal cell carcinoma, colorectal adenocarcinoma, hepatocellular carcinoma, and papillary carcinoma of the thyroid. Moreover, our data demonstrate that STING expression in tumor cells is tightly linked to PD-L1 expression, but it may not exert a measurable impact on the quantity of tumor-infiltrating inflammatory cells.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers16132425/s1. Supplementary Figure S1: Immunohistochemistry validation by comparison of two antibodies. The panels show a concordance of immunostaining results obtained by two independent STING antibodies with respect to both stained cell types and intensity distribution within tissues. Using MSVA-515M, cytoplasmic staining is seen in basal cells in the prostate (A); a fraction of epithelial cells of the seminal vesicle (B); basal, suprabasal, and superficial epithelial cells of the urothelium (C); basal squamous epithelial cells of the ectocervix (D); a fraction of epithelial cells of the rectal mucosa (E); endothelial cells of the testis (F); and variable subsets of cells in a normal (G) and a focally inflamed and atrophic kidney (H). Using clone D2P2F, comparable staining was seen in the prostate (I), seminal vesicle (J), urothelium (K), ectocervix (L), rectal mucosa (M), testis (N), and the kidney (O, P). The images A–H and I–P are from consecutive tissue sections. Supplementary Figure S2: Ranking order of STING immunostaining in tumors. Both the percentage of positive cases (blue dots) and the percentage of strongly positive cases (orange dots) are shown. Supplementary Figure S3: Examples of PD-L1 and CD8 immunostaining. Strong PD-L1 staining in a muscle-invasive urinary bladder cancer (a) and in a clear cell carcinoma of the ovary (b); a PD-L1-negative, muscle-invasive urinary bladder cancer (c); high intratumoral density of CD8+ lymphocytes in a squamous cell carcinoma of the cervix (d); low intratumoral density of CD8+ lymphocytes in a serous carcinoma of the ovary (e); and a CD8-negative endometrioid carcinoma of the ovary (f). Supplementary Table S1: IHC scoring criteria. Supplementary Table S2: STING immunostaining and HPV in squamous cell carcinomas. Supplementary Table S3: STING immunostaining and tumor phenotype (non-significant).
Author Contributions
A.M., J.Z., T.K., M.K., R.S. (Ronald Simon) and G.S.: contributed to conception, design, data collection, data analysis, and manuscript writing. A.M., F.V., J.Z., S.B., D.D., R.S. (Ria Schlichter), A.H., A.A.B., C.F., C.B., K.M., F.L., V.R., A.M.L., P.L., S.A.W., M.L., F.J., T.S.C., A.H.M., S.S., E.B., N.G. and S.M.: participated in pathology data analysis, data interpretation, and collection of samples. R.S. (Ria Schlichter), M.K. and C.H.-M.: data analysis. T.K., R.S. (Ronald Simon) and G.S.: study supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The usage of archived, diagnostic, left-over tissues for manufacturing TMAs, their analysis for research purposes, and patient data analysis complied by local laws (HmbKHG, §12,1) and were approved by the local ethics committee (Ethics commission Hamburg, WF-049/09). All work has been carried out in compliance with the Declaration of Helsinki.
Informed Consent Statement
Patient consent was waived due to local laws (HmbKHG, §12,1) that permit research with anonymized, diagnostic, left-over tissue samples.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Acknowledgments
We are grateful to Melanie Steurer, Laura Behm, Inge Brandt, and Sünje Seekamp for excellent technical assistance.
Conflicts of Interest
The mouse monoclonal antibody, clone MSVA-515M, was provided from MS Validated Antibodies GmbH, Hamburg, Germany (owned by a family member of G.S.).
References
- Paludan, S.R.; Bowie, A.G. Immune sensing of DNA. Immunity 2013, 38, 870–880. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, D.; Zhang, J.; Xiang, P.; Zeng, Z.; Xiong, W.; Shi, L. cGAS-STING signaling in the tumor microenvironment. Cancer Lett. 2023, 577, 216409. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Zhang, J.; Peng, Q.; Wang, X.; Xiao, X.; Shi, K. Nanomaterial-mediated modulation of the cGAS-STING signaling pathway for enhanced cancer immunotherapy. Acta Biomater. 2024, 176, 51–76. [Google Scholar] [CrossRef]
- Huang, C.; Shao, N.; Huang, Y.; Chen, J.; Wang, D.; Hu, G.; Zhang, H.; Luo, L.; Xiao, Z. Overcoming challenges in the delivery of STING agonists for cancer immunotherapy: A comprehensive review of strategies and future perspectives. Mater. Today Bio 2023, 23, 100839. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Tong, Q.; Liu, B.; Huang, W.; Tian, Y.; Fu, X. Targeting STAT3 in Cancer Immunotherapy. Mol. Cancer 2020, 19, 145. [Google Scholar] [CrossRef] [PubMed]
- Sokolowska, O.; Nowis, D. STING Signaling in Cancer Cells: Important or Not? Arch. Immunol. Ther. Exp. 2018, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Kanne, D.B.; Leong, M.; Glickman, L.H.; McWhirter, S.M.; Lemmens, E.; Mechette, K.; Leong, J.J.; Lauer, P.; Liu, W.; et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med. 2015, 7, 283ra252. [Google Scholar] [CrossRef]
- Parkes, E.E.; Walker, S.M.; Taggart, L.E.; McCabe, N.; Knight, L.A.; Wilkinson, R.; McCloskey, K.D.; Buckley, N.E.; Savage, K.I.; Salto-Tellez, M.; et al. Activation of STING-Dependent Innate Immune Signaling by S-Phase-Specific DNA Damage in Breast Cancer. J. Natl. Cancer Inst. 2017, 109, djw199. [Google Scholar] [CrossRef]
- Du, S.S.; Chen, G.W.; Yang, P.; Chen, Y.X.; Hu, Y.; Zhao, Q.Q.; Zhang, Y.; Liu, R.; Zheng, D.X.; Zhou, J.; et al. Radiation Therapy Promotes Hepatocellular Carcinoma Immune Cloaking via PD-L1 Upregulation Induced by cGAS-STING Activation. Int. J. Radiat. Oncol. Biol. Phys. 2022, 112, 1243–1255. [Google Scholar] [CrossRef]
- Yan, X.; Yao, C.; Fang, C.; Han, M.; Gong, C.; Hu, D.; Shen, W.; Wang, L.; Li, S.; Zhu, S. Rocaglamide promotes the infiltration and antitumor immunity of NK cells by activating cGAS-STING signaling in non-small cell lung cancer. Int. J. Biol. Sci. 2022, 18, 585–598. [Google Scholar] [CrossRef]
- Nicolai, C.J.; Wolf, N.; Chang, I.C.; Kirn, G.; Marcus, A.; Ndubaku, C.O.; McWhirter, S.M.; Raulet, D.H. NK cells mediate clearance of CD8(+) T cell-resistant tumors in response to STING agonists. Sci. Immunol. 2020, 5, eaaz2738. [Google Scholar] [CrossRef]
- Takashima, K.; Takeda, Y.; Oshiumi, H.; Shime, H.; Okabe, M.; Ikawa, M.; Matsumoto, M.; Seya, T. STING in tumor and host cells cooperatively work for NK cell-mediated tumor growth retardation. Biochem. Biophys. Res. Commun. 2016, 478, 1764–1771. [Google Scholar] [CrossRef]
- Woo, S.R.; Fuertes, M.B.; Corrales, L.; Spranger, S.; Furdyna, M.J.; Leung, M.Y.; Duggan, R.; Wang, Y.; Barber, G.N.; Fitzgerald, K.A.; et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 2014, 41, 830–842. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Peng, C.; Chen, Y.; Li, J.; Yang, R.; Wu, M.; Lu, P. Expression of STING and PD-L1 in colorectal cancer and their correlation with clinical prognosis. Int. J. Clin. Exp. Pathol. 2018, 11, 1256–1264. [Google Scholar]
- Song, S.; Peng, P.; Tang, Z.; Zhao, J.; Wu, W.; Li, H.; Shao, M.; Li, L.; Yang, C.; Duan, F.; et al. Decreased expression of STING predicts poor prognosis in patients with gastric cancer. Sci. Rep. 2017, 7, 39858. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zheng, Y.; Peng, X.; Li, R.; Pang, Y.; Du, Y.; Chen, Y.; Zhang, K. Low expression of PTEN and high expression of STING in human tongue squamous cell carcinoma tissues are associated with poor prognosis. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2020, 36, 1016–1020. [Google Scholar]
- Ni, H.; Zhang, H.; Li, L.; Huang, H.; Guo, H.; Zhang, L.; Li, C.; Xu, J.X.; Nie, C.P.; Li, K.; et al. T cell-intrinsic STING signaling promotes regulatory T cell induction and immunosuppression by upregulating FOXP3 transcription in cervical cancer. J. Immunother. Cancer 2022, 10, e005151. [Google Scholar] [CrossRef] [PubMed]
- Huvila, J.; Cochrane, D.R.; Ta, M.; Chow, C.; Greening, K.; Leung, S.; Karnezis, A.N.; DiFeo, A.; Huntsman, D.G. STING pathway expression in low-grade serous carcinoma of the ovary: An unexpected therapeutic opportunity? J. Pathol. Clin. Res. 2021, 7, 548–555. [Google Scholar] [CrossRef]
- Miao, L.; Qi, J.; Zhao, Q.; Wu, Q.N.; Wei, D.L.; Wei, X.L.; Liu, J.; Chen, J.; Zeng, Z.L.; Ju, H.Q.; et al. Targeting the STING pathway in tumor-associated macrophages regulates innate immune sensing of gastric cancer cells. Theranostics 2020, 10, 498–515. [Google Scholar] [CrossRef]
- Yao, H.; Wang, S.; Zhou, X.; Sun, J.; Zhou, G.; Zhou, D.; Chen, G.; Shi, X.; Chen, J.; Shi, B.; et al. STING promotes proliferation and induces drug resistance in colorectal cancer by regulating the AMPK-mTOR pathway. J. Gastrointest. Oncol. 2022, 13, 2458–2471. [Google Scholar] [CrossRef]
- Yang, B.; Rao, W.; Luo, H.; Zhang, L.; Wang, D. Relapse-related molecular signature in early-stage lung adenocarcinomas based on base excision repair, stimulator of interferon genes pathway and tumor-infiltrating lymphocytes. Cancer Sci. 2020, 111, 3493–3502. [Google Scholar] [CrossRef] [PubMed]
- Viculin, J.; Degoricija, M.; Vilovic, K.; Gabela, I.; Frankovic, L.; Vrdoljak, E.; Korac-Prlic, J. Elevated Tumor Cell-Intrinsic STING Expression in Advanced Laryngeal Cancer. Cancers 2023, 15, 3510. [Google Scholar] [CrossRef] [PubMed]
- Marletta, S.; Calio, A.; Bogina, G.; Rizzo, M.; Brunelli, M.; Pedron, S.; Marcolini, L.; Stefanizzi, L.; Gobbo, S.; Princiotta, A.; et al. STING is a prognostic factor related to tumor necrosis, sarcomatoid dedifferentiation, and distant metastasis in clear cell renal cell carcinoma. Virchows Arch. 2023, 483, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yu, S.; van der Sluis, T.; Zwager, M.C.; Schroder, C.P.; van der Vegt, B.; van Vugt, M. cGAS-STING pathway expression correlates with genomic instability and immune cell infiltration in breast cancer. NPJ Breast Cancer 2024, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Saulters, E.L.; Kennedy, P.T.; Carter, R.J.; Alsufyani, A.; Jones, T.M.; Woolley, J.F.; Dahal, L.N. Differential Regulation of the STING Pathway in Human Papillomavirus-Positive and -Negative Head and Neck Cancers. Cancer Res. Commun. 2024, 4, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.A.; Yang, B.; Rao, W.; Xiao, H.; Wang, D.; Jiang, J. The correlation of BER protein, IRF3 with CD8+ T cell and their prognostic significance in upper tract urothelial carcinoma. Onco Targets Ther. 2019, 12, 7725–7735. [Google Scholar] [CrossRef] [PubMed]
- Lohinai, Z.; Dora, D.; Caldwell, C.; Rivard, C.J.; Suda, K.; Yu, H.; Rivalland, G.; Ellison, K.; Rozeboom, L.; Dziadziuszko, R.; et al. Loss of STING expression is prognostic in non-small cell lung cancer. J. Surg. Oncol. 2022, 125, 1042–1052. [Google Scholar] [CrossRef]
- Dora, D.; Rivard, C.; Yu, H.; Pickard, S.L.; Laszlo, V.; Harko, T.; Megyesfalvi, Z.; Gerdan, C.; Dinya, E.; Hoetzenecker, K.; et al. Protein Expression of immune checkpoints STING and MHCII in small cell lung cancer. Cancer Immunol. Immunother. 2023, 72, 561–578. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi-Hara, R.; Sawada, Y.; Sugino, H.; Minokawa, Y.; Kawahara-Nanamori, H.; Itamura, M.; Tashiro, T.; Kaneoka, A.; Saito-Sasaki, N.; Yamamoto, K.; et al. STING expression is an independent prognostic factor in patients with mycosis fungoides. Sci. Rep. 2022, 12, 12739. [Google Scholar] [CrossRef]
- Kononen, J.; Bubendorf, L.; Kallioniemi, A.; Barlund, M.; Schraml, P.; Leighton, S.; Torhorst, J.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998, 4, 844–847. [Google Scholar] [CrossRef]
- Dancau, A.M.; Simon, R.; Mirlacher, M.; Sauter, G. Tissue Microarrays. Methods Mol. Biol. 2016, 1381, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Moller, K.; Knoll, M.; Bady, E.; Schmerder, M.J.; Rico, S.D.; Kluth, M.; Hube-Magg, C.; Blessin, N.C.; Mandelkow, T.; Lennartz, M.; et al. PD-L1 expression and CD8 positive lymphocytes in human neoplasms: A tissue microarray study on 11,838 tumor samples. Cancer Biomark. 2023, 36, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Mirlacher, M.; Sauter, G. Immunohistochemical analysis of tissue microarrays. Methods Mol. Biol. 2010, 664, 113–126. [Google Scholar] [CrossRef]
- Huang, J.; Xie, Y.; Sun, X.; Zeh, H.J., 3rd; Kang, R.; Lotze, M.T.; Tang, D. DAMPs, ageing, and cancer: The ‘DAMP Hypothesis’. Ageing Res. Rev. 2015, 24, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Baca, S.C.; Garraway, L.A. The genomic landscape of prostate cancer. Front. Endocrinol. 2012, 3, 69. [Google Scholar] [CrossRef] [PubMed]
- Light, A.; Ahmed, A.; Dasgupta, P.; Elhage, O. The genetic landscapes of urological cancers and their clinical implications in the era of high-throughput genome analysis. BJU Int. 2020, 126, 26–54. [Google Scholar] [CrossRef]
- van Riet, J.; van de Werken, H.J.G.; Cuppen, E.; Eskens, F.; Tesselaar, M.; van Veenendaal, L.M.; Klumpen, H.J.; Dercksen, M.W.; Valk, G.D.; Lolkema, M.P.; et al. The genomic landscape of 85 advanced neuroendocrine neoplasms reveals subtype-heterogeneity and potential therapeutic targets. Nat. Commun. 2021, 12, 4612. [Google Scholar] [CrossRef]
- Krishnamurthy, K.; Lindsey, A.M.; Estrada, C.A.; Martinez, C.C.; Cusnir, M.; Schwartz, M.; Sriganeshan, V.; Poppiti, R. Title- Genomic landscape of squamous cell carcinoma- Different genetic pathways culminating in a common phenotype. Cancer Treat. Res. Commun. 2020, 25, 100238. [Google Scholar] [CrossRef] [PubMed]
- Blakely, C.M. A New Pathway Emerges to Interpret Lung Cancer Genomic Alterations. Clin. Cancer Res. 2019, 25, 7269–7271. [Google Scholar] [CrossRef]
- Waddell, N.; Pajic, M.; Patch, A.M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef]
- Kwon, J.; Bakhoum, S.F. The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov. 2020, 10, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef]
- Lu, C.; Guan, J.; Lu, S.; Jin, Q.; Rousseau, B.; Lu, T.; Stephens, D.; Zhang, H.; Zhu, J.; Yang, M.; et al. DNA Sensing in Mismatch Repair-Deficient Tumor Cells Is Essential for Anti-tumor Immunity. Cancer Cell 2021, 39, 96–108.e6. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Chai, Y.; Chai, F.; Liu, H.; Ma, N.; Zhang, H.; Zhang, S.; Nong, L.; Li, T.; Zhang, B. Expression of SASP, DNA Damage Response, and Cell Proliferation Factors in Early Gastric Neoplastic Lesions: Correlations and Clinical Significance. Pathol. Oncol. Res. 2022, 28, 1610401. [Google Scholar] [CrossRef] [PubMed]
- Saulters, E.; Woolley, J.F.; Varadarajan, S.; Jones, T.M.; Dahal, L.N. STINGing Viral Tumors: What We Know from Head and Neck Cancers. Cancer Res. 2021, 81, 3945–3952. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, M.H.; Bortnik, V.; McMillan, N.A.; Idris, A. cGAS-STING responses are dampened in high-risk HPV type 16 positive head and neck squamous cell carcinoma cells. Microb. Pathog. 2019, 132, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Lau, L.; Gray, E.E.; Brunette, R.L.; Stetson, D.B. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 2015, 350, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Kunac, N.; Degoricija, M.; Viculin, J.; Omerovic, J.; Terzic, J.; Vilovic, K.; Korac-Prlic, J. Activation of cGAS-STING Pathway Is Associated with MSI-H Stage IV Colorectal Cancer. Cancers 2022, 15, 221. [Google Scholar] [CrossRef] [PubMed]
- Kaneta, A.; Nakajima, S.; Okayama, H.; Matsumoto, T.; Saito, K.; Kikuchi, T.; Endo, E.; Ito, M.; Mimura, K.; Kanke, Y.; et al. Role of the cGAS-STING pathway in regulating the tumor-immune microenvironment in dMMR/MSI colorectal cancer. Cancer Immunol. Immunother. 2022, 71, 2765–2776. [Google Scholar] [CrossRef] [PubMed]
- Vasiyani, H.; Mane, M.; Rana, K.; Shinde, A.; Roy, M.; Singh, J.; Gohel, D.; Currim, F.; Srivastava, R.; Singh, R. DNA damage induces STING mediated IL-6-STAT3 survival pathway in triple-negative breast cancer cells and decreased survival of breast cancer patients. Apoptosis 2022, 27, 961–978. [Google Scholar] [CrossRef]
- Vasiyani, H.; Shinde, A.; Roy, M.; Mane, M.; Singh, K.; Singh, J.; Gohel, D.; Currim, F.; Vaidya, K.; Chhabria, M.; et al. The analog of cGAMP, c-di-AMP, activates STING mediated cell death pathway in estrogen-receptor negative breast cancer cells. Apoptosis 2021, 26, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Parkes, E.E.; Humphries, M.P.; Gilmore, E.; Sidi, F.A.; Bingham, V.; Phyu, S.M.; Craig, S.; Graham, C.; Miller, J.; Griffin, D.; et al. The clinical and molecular significance associated with STING signaling in breast cancer. NPJ Breast Cancer 2021, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yan, L.; Liu, N.; Xu, M.; Cai, H. IFI16 promotes cervical cancer progression by upregulating PD-L1 in immunomicroenvironment through STING-TBK1-NF-kB pathway. Biomed. Pharmacother. 2020, 123, 109790. [Google Scholar] [CrossRef] [PubMed]
- Grabosch, S.; Bulatovic, M.; Zeng, F.; Ma, T.; Zhang, L.; Ross, M.; Brozick, J.; Fang, Y.; Tseng, G.; Kim, E.; et al. Cisplatin-induced immune modulation in ovarian cancer mouse models with distinct inflammation profiles. Oncogene 2019, 38, 2380–2393. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.H.; Zundell, J.A.; Ranatunga, S.; Lin, C.; Nefedova, Y.; Del Valle, J.R.; Hu, C.C. Agonist-Mediated Activation of STING Induces Apoptosis in Malignant B Cells. Cancer Res. 2016, 76, 2137–2152. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Konno, H.; Ahn, J.; Barber, G.N. Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates with Tumorigenesis. Cell Rep. 2016, 14, 282–297. [Google Scholar] [CrossRef]
- Uhlen, M.; Bandrowski, A.; Carr, S.; Edwards, A.; Ellenberg, J.; Lundberg, E.; Rimm, D.L.; Rodriguez, H.; Hiltke, T.; Snyder, M.; et al. A proposal for validation of antibodies. Nat. Methods 2016, 13, 823–827. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).